Abstract
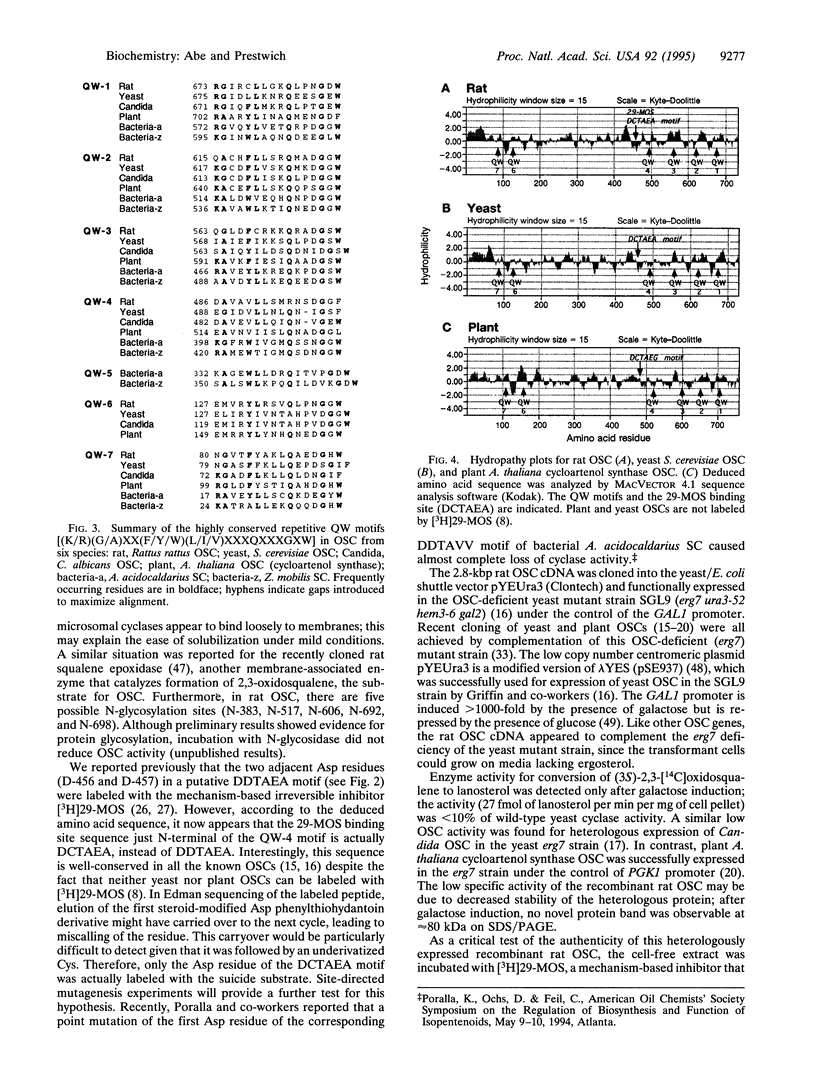
A cDNA encoding rat oxidosqualene lanosterol-cyclase [lanosterol synthase; (S)-2,3-epoxysqualene mutase (cyclizing, lanosterol-forming), EC 5.4.99.7] was cloned and sequenced by a combination of PCR amplification, using primers based on internal amino acid sequence of the purified enzyme, and cDNA library screening by oligonucleotide hybridization. An open reading frame of 2199 bp encodes a M(r) 83,321 protein with 733 amino acids. The deduced amino acid sequence of the rat enzyme showed significant homology to the known oxidosqualene cyclases (OSCs) from yeast and plant (39-44% identity) and still retained 17-26% identity to two bacterial squalene cyclases (EC 5.4.99.-). Like other cyclases, the rat enzyme is rich in aromatic amino acids and contains five so-called QW motifs, highly conserved regions with a repetitive beta-strand turn motif. The binding site sequence for the 29-methylidene-2,3-oxidosqualene (29-MOS), a mechanism-based irreversible inhibitor specific for the vertebrate cyclase, is well-conserved in all known OSCs. The hydropathy plot revealed a rather hydrophilic N-terminal region and the absence of a hydrophobic signal peptide. Unexpectedly, this microsomal membrane-associated enzyme showed no clearly delineated transmembrane domain. A full-length cDNA was constructed and subcloned into a pYEUra3 plasmid, selected in Escherichia coli cells, and used to transform the OSC-deficient uracil-auxotrophic SGL9 strain of Saccharomyces cerevisiae. The recombinant rat OSC expressed was efficiently labeled by the mechanism-based inhibitor [3H]29-MOS.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abe I., Bai M., Xiao X. Y., Prestwich G. D. Affinity labeling of vertebrate oxidosqualene cyclases with a tritiated suicide substrate. Biochem Biophys Res Commun. 1992 Aug 31;187(1):32–38. doi: 10.1016/s0006-291x(05)81454-8. [DOI] [PubMed] [Google Scholar]
- Abe I., Prestwich G. D. Active site mapping of affinity-labeled rat oxidosqualene cyclase. J Biol Chem. 1994 Jan 14;269(2):802–804. [PubMed] [Google Scholar]
- Abe I., Prestwich G. D. Identification of the active site of vertebrate oxidosqualene cyclase. Lipids. 1995 Mar;30(3):231–234. doi: 10.1007/BF02537826. [DOI] [PubMed] [Google Scholar]
- Abe I., Tomesch J. C., Wattanasin S., Prestwich G. D. Inhibitors of squalene biosynthesis and metabolism. Nat Prod Rep. 1994 Jun;11(3):279–302. doi: 10.1039/np9941100279. [DOI] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Corey E. J., Matsuda S. P., Bartel B. Isolation of an Arabidopsis thaliana gene encoding cycloartenol synthase by functional expression in a yeast mutant lacking lanosterol synthase by the use of a chromatographic screen. Proc Natl Acad Sci U S A. 1993 Dec 15;90(24):11628–11632. doi: 10.1073/pnas.90.24.11628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey E. J., Matsuda S. P., Bartel B. Molecular cloning, characterization, and overexpression of ERG7, the Saccharomyces cerevisiae gene encoding lanosterol synthase. Proc Natl Acad Sci U S A. 1994 Mar 15;91(6):2211–2215. doi: 10.1073/pnas.91.6.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duriatti A., Schuber F. Partial purification of 2,3-oxidosqualene-lanosterol cyclase from hog-liver. Evidence for a functional thiol residue. Biochem Biophys Res Commun. 1988 Mar 30;151(3):1378–1385. doi: 10.1016/s0006-291x(88)80515-1. [DOI] [PubMed] [Google Scholar]
- Gollub E. G., Liu K. P., Dayan J., Adlersberg M., Sprinson D. B. Yeast mutants deficient in heme biosynthesis and a heme mutant additionally blocked in cyclization of 2,3-oxidosqualene. J Biol Chem. 1977 May 10;252(9):2846–2854. [PubMed] [Google Scholar]
- Johnston M., Davis R. W. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Aug;4(8):1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly R., Miller S. M., Lai M. H., Kirsch D. R. Cloning and characterization of the 2,3-oxidosqualene cyclase-coding gene of Candida albicans. Gene. 1990 Mar 15;87(2):177–183. doi: 10.1016/0378-1119(90)90299-7. [DOI] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5'-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987 Oct 26;15(20):8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumpf R. A., Dougherty D. A. A mechanism for ion selectivity in potassium channels: computational studies of cation-pi interactions. Science. 1993 Sep 24;261(5129):1708–1710. doi: 10.1126/science.8378771. [DOI] [PubMed] [Google Scholar]
- Kusano M., Abe I., Sankawa U., Ebizuka Y. Purification and some properties of squalene-2,3-epoxide: lanosterol cyclase from rat liver. Chem Pharm Bull (Tokyo) 1991 Jan;39(1):239–241. doi: 10.1248/cpb.39.239. [DOI] [PubMed] [Google Scholar]
- Kusano M., Shibuya M., Sankawa U., Ebizuka Y. Molecular cloning of cDNA encoding rat 2,3-oxidosqualene: lanosterol cyclase. Biol Pharm Bull. 1995 Jan;18(1):195–197. doi: 10.1248/bpb.18.195. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Moore W. R., Schatzman G. L. Purification of 2,3-oxidosqualene cyclase from rat liver. J Biol Chem. 1992 Nov 5;267(31):22003–22006. [PubMed] [Google Scholar]
- Ochs D., Kaletta C., Entian K. D., Beck-Sickinger A., Poralla K. Cloning, expression, and sequencing of squalene-hopene cyclase, a key enzyme in triterpenoid metabolism. J Bacteriol. 1992 Jan;174(1):298–302. doi: 10.1128/jb.174.1.298-302.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poralla K., Hewelt A., Prestwich G. D., Abe I., Reipen I., Sprenger G. A specific amino acid repeat in squalene and oxidosqualene cyclases. Trends Biochem Sci. 1994 Apr;19(4):157–158. doi: 10.1016/0968-0004(94)90276-3. [DOI] [PubMed] [Google Scholar]
- Ramer S. W., Elledge S. J., Davis R. W. Dominant genetics using a yeast genomic library under the control of a strong inducible promoter. Proc Natl Acad Sci U S A. 1992 Dec 1;89(23):11589–11593. doi: 10.1073/pnas.89.23.11589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reipen I. G., Poralla K., Sahm H., Sprenger G. A. Zymomonas mobilis squalene-hopene cyclase gene (shc): cloning, DNA sequence analysis, and expression in Escherichia coli. Microbiology. 1995 Jan;141(Pt 1):155–161. doi: 10.1099/00221287-141-1-155. [DOI] [PubMed] [Google Scholar]
- Roessner C. A., Min C., Hardin S. H., Harris-Haller L. W., McCollum J. C., Scott A. I. Sequence of the Candida albicans erg7 gene. Gene. 1993 May 15;127(1):149–150. doi: 10.1016/0378-1119(93)90631-c. [DOI] [PubMed] [Google Scholar]
- Rost B., Casadio R., Fariselli P., Sander C. Transmembrane helices predicted at 95% accuracy. Protein Sci. 1995 Mar;4(3):521–533. doi: 10.1002/pro.5560040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B., Sander C. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins. 1994 May;19(1):55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- Sakakibara J., Watanabe R., Kanai Y., Ono T. Molecular cloning and expression of rat squalene epoxidase. J Biol Chem. 1995 Jan 6;270(1):17–20. doi: 10.1074/jbc.270.1.17. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl R. H., Gietz R. D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989 Dec;16(5-6):339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Shi Z., Buntel C. J., Griffin J. H. Isolation and characterization of the gene encoding 2,3-oxidosqualene-lanosterol cyclase from Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1994 Jul 19;91(15):7370–7374. doi: 10.1073/pnas.91.15.7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner P., Gärtner W. cDNA cloning of 5' terminal regions. Nucleic Acids Res. 1992 Sep 11;20(17):4669–4670. doi: 10.1093/nar/20.17.4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiner M., Gehring U. Cloning of 5' cDNA regions by inverse PCR. Biotechniques. 1994 Dec;17(6):1050-2, 1054. [PubMed] [Google Scholar]




