SUMMARY
Alternative splicing is important for the development and function of the nervous system, but little is known about the differences in alternative splicing between distinct types of neurons. Furthermore, the factors that control cell-type-specific splicing and the physiological roles of these alternative isoforms are unclear. By monitoring alternative splicing at single cell resolution in Caenorhabditis elegans, we demonstrate that splicing patterns in different neurons are often distinct and highly regulated. We identify two conserved RNA binding proteins, UNC-75/CELF and EXC-7/Hu/ELAV, which regulate overlapping networks of splicing events in GABAergic and cholinergic neurons. We use the UNC-75 exon network to discover regulators of synaptic transmission and to identify unique roles for isoforms of UNC-64/Syntaxin, a protein required for synaptic vesicle fusion. Our results indicate that combinatorial regulation of alternative splicing in distinct neurons provides a mechanism to specialize metazoan nervous systems.
INTRODUCTION
The nervous system has been recognized for over a century as a uniquely complex and diverse organ, composed of various classes of neurons forming precise synaptic connections with partners often located great distances away (Golgi, 1883; Ramón y Cajal, 1890). These anatomical and morphological observations have recently been confirmed and extended by molecular studies identifying genes and proteins that specify and/or discriminate neuronal cell types from each other (Greig et al., 2013; Hobert et al., 2010). However, much is still unknown about the molecular diversity and specification of neuronal cell types and the regulators of such diversity (Hobert et al., 2010).
Alternative pre-mRNA splicing, the process by which a single gene encodes multiple isoforms via alternative inclusion or skipping of exons in the mature mRNA, is a pervasive means of regulating gene expression that increases the diversity and complexity of the transcriptome and proteome (Braunschweig et al., 2013; Nilsen and Graveley, 2010). This increased diversity plays an important role in complex organ systems such as the nervous system (Norris and Calarco, 2012; Zheng and Black, 2013), which exhibits high levels of alternative splicing (Barbosa-Morais et al., 2012).
Several studies have identified factors responsible for differential splicing between the nervous system and other tissues (Boutz et al., 2007; Calarco et al., 2009; Gehman et al., 2011; Jensen et al., 2000), but it is not known to what extent differential splicing occurs between different neuronal cell types. While a number of individual cases have been identified (for examples see Boucard et al., 2005; Chih et al., 2006; Miura et al., 2013), it has remained difficult to study the factors that might control neuron-subtype specificity of alternative splicing. This difficulty is largely due to the technical challenge of accurately measuring splicing differences between cells of the same tissue exhibiting little spatial separation. As such, techniques for visualizing alternative splicing within individual cells of the nervous system or other tissues are highly desirable. The recent use of fluorescent reporters to monitor alternative splicing in cultured cells and in vivo has allowed an exploration of splicing regulation at cellular resolution (Kuroyanagi et al., 2006; Miura et al., 2013; Orengo et al., 2006; Zheng et al., 2013).
Investigations of alternative splicing in the nervous system and other tissues have demonstrated that individual splicing factors regulate networks of exons in sets of functionally related genes (Boutz et al., 2007; Calarco et al., 2009; Ule et al., 2005; Wang et al., 2012; Zhang et al., 2008). Complementary to these genome-wide analyses, focused studies on individual alternative splicing events demonstrate that they are often regulated by multiple splicing factors (Charlet et al., 2002; Markovtsov et al., 2000). Although these observations highlight the importance of combinatorial regulation in splicing, nothing is known about how multiple splicing factors act together to shape splicing networks at single neuron resolution.
Characterization of individual target transcripts within splicing networks have identified important splicing events that contribute to specific neuronal phenotypes (Aoto et al., 2013; Raj et al., 2011; Yano et al., 2010). However, the difficulty of systematic network interrogation in vertebrate models has made it challenging to perform functional analyses of substantial numbers of targets within splicing regulatory networks in vivo. In particular, there is little experimental evidence for how target genes and isoforms within a splicing network contribute to phenotypes associated with loss of a splicing factor.
To address these fundamental challenges in splicing research, we have exploited the nematode Caenorhabditis elegans, a well-established model system for studying neurobiology and an increasingly useful model for studying alternative splicing regulation (Barberan-Soler et al., 2011; 2008; Ohno et al., 2008; Ramani et al., 2011; Zahler, 2005). We have adopted a two color splicing reporter to provide the first visual survey of alternative splicing events at single cell resolution in the C. elegans nervous system. Remarkably, these reporters reveal that alternative splicing in specific neuronal subtypes occurs frequently and is subject to precise regulation. Characterizing one such alternative splicing event, we performed a genetic screen and identified two highly conserved RNA-binding proteins, UNC-75/CELF and EXC-7/Hu/ELAV, which act together through partially overlapping expression patterns to produce a neuron subtype-specific splicing outcome. We further demonstrate at the genome-wide level that these two factors regulate substantially overlapping target networks of alternative exons. We investigate the exon network of UNC-75 and show that phenotypes observed from the deletion of individual genes in the network are related to phenotypes observed upon loss of the splicing factor. Finally, we demonstrate that this exon network can be used to predict novel functions for previously uncharacterized genes and isoforms, including two splice variants of the conserved synaptic vesicle fusion protein UNC-64/Syntaxin.
RESULTS
Two color reporters reveal complex and diverse alternative splicing patterns within the C. elegans nervous system
To interrogate alternative splicing patterns in vivo at single neuron resolution in C. elegans, we adapted a previously described two color splicing reporter system (Kuroyanagi et al., 2010; Orengo et al., 2006) (Figure 1A). Alternative exons of interest and their neighboring introns and constitutively spliced exons were cloned upstream of two tandemly positioned open reading frames encoding either enhanced GFP (eGFP) or monomeric RFP (mRFP). The reporter is engineered such that inclusion or exclusion of the alternative exon leads to switches between the two reading frames. When the alternative exon is skipped, eGFP is read in-frame followed by a stop codon. When the exon is included, the reading-frame is shifted so that eGFP is read out of frame and without stop codons while mRFP is translated in-frame. This reporter thus enables monitoring of alternative splicing events in individual cells of live animals by fluorescence microscopy.
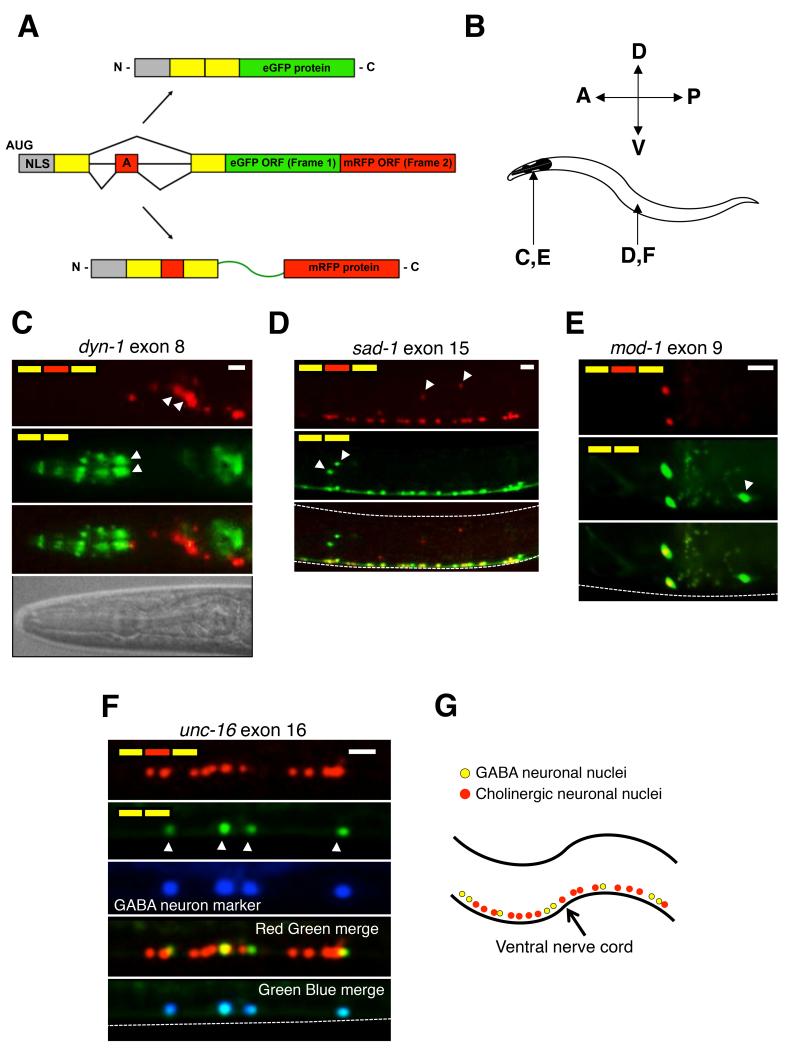
Figure 1.
Two color splicing reporters reveal diversity of alternative splicing within the nervous system.
(A) Schematic of two color splicing reporter. An alternative exon (red box with letter A) and its neighboring intron and exon sequences (yellow boxes) are cloned upstream of EGFP and mRFP open reading frames (larger red and green boxes). Skipping of the alternative exon leads to translation of EGFP, while inclusion of the exon leads to translation of mRFP. An SV40 nuclear localization signal (grey box, NLS) is included in the reporter to focus the signal in the nucleus.
(B) Anatomical reference for panels C-F, indicating the location (head for panels C and E, or mid-body area for panels D and F)
(C-F) Fluorescence images of two color splicing reporters for neuronal alternative splicing events. White arrowheads indicate select nuclei with unique isoform usage patterns. All animals are at larval stage L4. Dashed lines delineate borders of the worms. Scale bar represents 10 μm. In panel C, a brightfield image is included to serve as a reference, highlighting that the green anterior cells (white arrowheads) are likely pharyngeal neurons. In panel F, GABAergic motor neurons are labeled with Punc-25::BFP.
(G) Simplified model of the organization of GABAergic (yellow) and cholinergic (red) motor neurons in the ventral nerve cord. See also Table S1.
We created two color splicing reporters to monitor the splicing of 14 alternative exons that have been previously confirmed by existing EST/cDNA libraries and mRNA-Seq data (Gerstein et al., 2010; Ramani et al., 2011). These exons are highly conserved in several nematode species, and are found in genes with known expression and function in the nervous system. Interestingly, when expressed under the control of a pan-neuronal promoter or endogenous promoters, 7/14 of these reporters revealed complex alternative splicing patterns within the nervous system, including differential exon inclusion between different classes of neurons (Figure 1 and Supplementary Table 1).
Monitoring alternative exon 8 in the GTPase dyn-1/Dynamin revealed that the exon-included isoform is preferentially expressed in lateral ganglion neurons posterior to the nerve ring, while the exon-excluded isoform predominates in more anteriorly positioned pharyngeal neurons (Figure 1C). The serotonin-gated chloride channel mod-1 revealed another unique pattern of differential exon usage in specific head neurons (Figure 1E). The neuronal serine/threonine protein kinase gene sad-1 encodes two isoforms that are co-expressed in most neurons in the head and ventral nerve cord, but only one isoform is expressed in a cluster of oxygen-sensing cells, while only the other isoform is expressed in a nearby cluster of lateral mechanosensory neurons (Figure 1D). A striking differential splicing pattern was observed with a reporter monitoring alternative exon 16 of unc-16/JIP3 in the ventral nerve cord, which is composed of excitatory cholinergic and inhibitory GABAergic motor neurons. The exon 16-included isoform is expressed in both cholinergic and GABAergic neurons (Figure 1F and G), while the excluded isoform is expressed only in GABAergic neurons, confirmed by co-expressing a codon-optimized blue fluorescent protein (eBFP) in GABAergic neurons (Figure 1F).
All reporters with neuron-specific splicing displayed a unique regulatory pattern. Notably, the patterns observed in each individual reporter were identical from animal to animal, indicating that splicing was being actively regulated and was not the result of purely stochastic processes. Taken together, these results demonstrate that alternative splicing patterns in C. elegans are frequently differentially regulated in specific neurons.
A pair of RNA-binding proteins, UNC-75 and EXC-7, control differential splicing in ventral cord motor neurons
To identify factors regulating motor neuron-specific alternative splicing, we conducted a microscopy-based genetic screen (Figure 2A) using the unc-16 exon 16 reporter described above. Worms carrying this reporter were subjected to EMS mutagenesis, and their F2 progeny were visually screened for disruptions in the GABAergic motor neuron-specific splicing pattern.
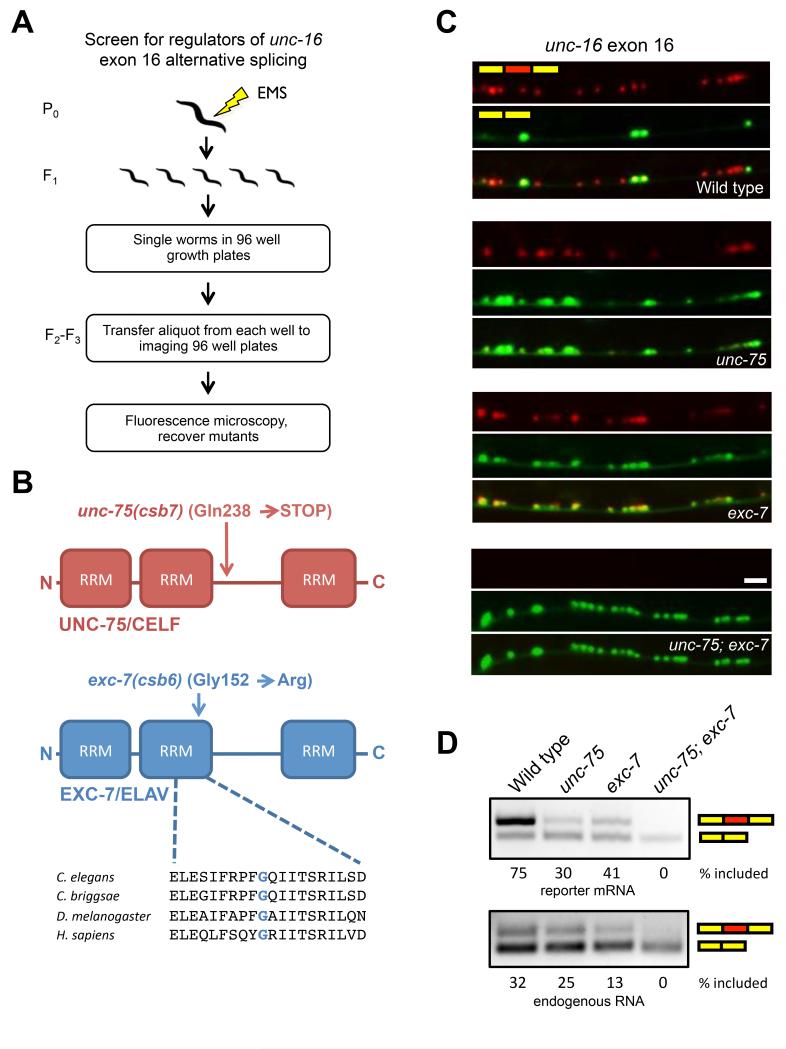
Figure 2.
Genetic screen for regulators of unc-16 alternative splicing identifies two RNA binding proteins that act combinatorially.
(A) Schematic of microscopy-based EMS mutagenesis screen
(B) Domain structure of screen hits and molecular nature of genetic lesions.
(C) unc-16 alternative splicing phenotypes in unc-75 and exc-7 mutants. Scale bar represents 10 μm.
(D) RT-PCR analysis of unc-16 two color reporter mRNA (upper panel) or native unc-16 mRNA. See also Figure S1 and S2.
From ~7000 haploid genomes screened, we recovered two viable mutants belonging to distinct complementation groups and identified the causal mutations in these animals by whole genome sequencing (Bigelow et al., 2009). These mutations were found in two genes encoding RNA-binding proteins: unc-75, a member of the CELF family, and exc-7, a member of the Hu/ELAV family (Figure 2B). Both RNA binding proteins are conserved from C. elegans to humans (Loria et al., 2003), where they are expressed in neurons, and are implicated in a number of neurological conditions (Dasgupta and Ladd, 2012). In C. elegans and mammals these factors have been shown to regulate several steps of RNA metabolism, including the regulation of splicing (Barberan-Soler et al., 2011; Darnell, 2013; Dasgupta and Ladd, 2012; Kuroyanagi et al., 2013a).
The molecular lesion in our unc-75(csb7) mutant introduces a premature termination codon in the linker region of the protein, and the lesion in our exc-7(csb6) mutant creates a Gly to Arg substitution in a conserved amino acid of the second RRM (Figure 2B). While the unc-75(csb7) allele we recovered in our screen behaved similarly to the reference unc-75(e950) null allele (Loria et al., 2003), our exc-7(csb6) allele exhibited more modest effects on alternative splicing in RT-PCR assays relative to the exc-7(rh252) null allele (Fujita et al., 2003; Supplementary Figure 2 and Figure 2D). These data suggest that our csb6 allele may be a hypomorphic allele. We therefore carried out all further experiments using the reference null alleles of unc-75 and exc-7.
In both unc-75 and exc-7 mutants the skipped isoform loses its restricted expression, becoming aberrantly expressed in cholinergic ventral nerve cord motor neurons (Figure 2C), indicating that UNC-75 and EXC-7 both mediate inclusion of exon 16. In unc-75; exc-7 double mutant animals, exon inclusion is lost entirely (Figure 2C). Transgenic expression of wild-type UNC-75 or EXC-7 in the respective unc-75 or exc-7 mutants rescued unc-16 splicing defects (Supplementary Figure 1). We performed RT-PCR assays with primers specific to the splicing reporter mRNA (Figure 2D, upper panel) or endogenous unc-16 mRNA (Figure 2D, lower panel), and confirmed that our reporter recapitulates the endogenous regulation of this alternative exon. The difference in overall ratios of included to skipped isoform between the splicing reporter and endogenous mRNA is likely due to unc-16 also being expressed in non-neuronal tissues, where the skipped isoform predominates (Kuroyanagi et al., 2013b), while our splicing reporter was driven by a pan-neuronal promoter.
Previous reports, which we have confirmed (data not shown), indicated that UNC-75 is expressed pan-neuronally (Loria et al., 2003), while EXC-7 is expressed in cholinergic but not in GABAergic motor neurons (Fujita et al., 1999), suggesting that their partially overlapping expression patterns could explain the differential splicing of unc-16 exon 16. If GABAergic motor neurons express only UNC-75, exon inclusion should still be observed in these neurons in exc-7 mutant animals, but not in unc-75 mutants. Indeed, in an unc-75 mutant, GABAergic motor neurons expressed only the skipped isoform, while in an exc-7 mutant, both the included and the skipped isoforms were expressed in GABAergic neurons (Supplementary Figure 2). These results indicate that in GABAergic motor neurons, UNC-75 partially stimulates exon 16 inclusion, whereas in cholinergic motor neurons, UNC-75 and EXC-7 together promote complete inclusion of exon 16.
UNC-75 and EXC-7 directly control unc-16 alternative splicing by binding to sequences in the downstream intron
To determine if UNC-75 and EXC-7 directly control unc-16 exon 16 alternative splicing by binding to its pre-mRNA, we identified consensus motifs bound in vitro by recombinant UNC-75 and EXC-7 proteins using RNAcompete (Ray et al., 2013). The UNC-75 and EXC-7 consensus sequences identified were U[G/A]UUGUG and UAAGUU, respectively (Figure 3A; Ray et al., 2013), which are in close agreement with motifs recognized by their vertebrate homologues (Ray et al., 2013). Our UNC-75 consensus motif also agrees with a recently described UNC-75-targeted U/G-rich sequence (Kuroyanagi et al., 2013b). We conducted electrophoretic mobility shift assays (EMSAs) by incubating recombinant GST-tagged UNC-75 and EXC-7 proteins with RNA oligonucleotides containing consensus motifs or sequences with nucleotide substitutions at highly constrained positions in each motif. Both factors bound RNAs containing consensus binding motifs, but displayed reduced binding to RNAs with substitutions in the motifs (Supplementary Figure 3).
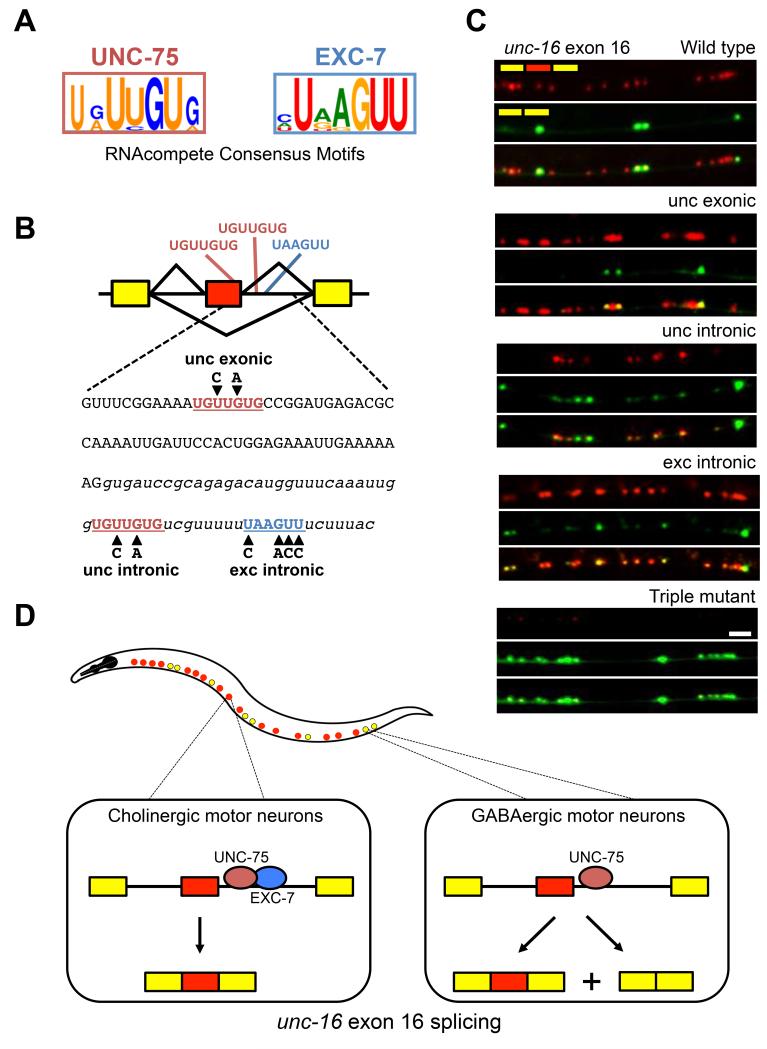
Figure 3.
UNC-75 and EXC-7 regulate unc-16 exon 16 splicing by binding to consensus motifs in its downstream intron.
(A) Consensus binding motifs as determined by RNAcompete.
(B) Presence of consensus binding motifs in unc-16 exon 16 and its downstream intron. Mutations introduced into the motifs are indicated in bold black letters.
(C) Two color splicing reporters for unc-16 exon 16 were created with the mutations shown in (B) and expressed in wild type worms. Scale bar represents 10 μm.
(D) Model for regulation of exon 16 alternative splicing. In cholinergic motor neurons (red dots in worm diagram), both UNC-75 and EXC-7 bind to cis elements in the intron downstream of unc-16 alternative exon 16, promoting full exon inclusion. In GABAergic motor neurons (yellow dots in worm diagram), only UNC-75 is present and its binding to the pre-mRNA promotes partial exon inclusion.
See also Figure S3.
The unc-16 pre-mRNA contains an EXC-7 consensus binding sequence in the intron downstream of alternative exon 16, as well as two UNC-75 consensus sequences—one in exon 16 and one in the downstream intron (Figure 3B). To test whether these sites are functional cis-elements controlling exon 16 alternative splicing in vivo, we created two color splicing reporters containing substitutions in the consensus binding motifs tested in our EMSAs (Supplementary Figure 3). While mutating the exonic UNC-75 consensus binding motif did not affect alternative splicing, mutating either the UNC-75 or EXC-7 intronic consensus binding motifs caused an increase in exon skipping (Figure 3C), consistent with what was observed in our unc-75 and exc-7 mutants (Figure 2C). When all three consensus motifs were mutated, exon inclusion was nearly undetectable, consistent with the phenotype of the unc-75; exc-7 double mutant (Figures 3C and 2C). These results strongly support a model where UNC-75 and EXC-7 combinatorially control neuron subtype-specific splicing of unc-16 exon 16 by directly binding the pre-mRNA in the downstream intron and mediating exon inclusion (Figure 3D).
UNC-75 and EXC-7 regulate extensive and overlapping exon networks
To assess whether UNC-75 and EXC-7 more generally regulate alternative splicing in a combinatorial manner, we performed high-throughput transcriptome sequencing (mRNA-Seq) analyses of wild type, unc-75, exc-7, and unc-75; exc-7 mutant whole animals. We searched for transcripts displaying statistically-significant differences of >20% in relative exon junction usage between wild type animals and each of the mutant strains (Figure 4A and B). Using these criteria, we tested 48 junction difference predictions by semi-quantitative RT-PCR, and 46/48 (96%) were validated (Figure 4B, Supplementary Figure 4A, and data not shown).
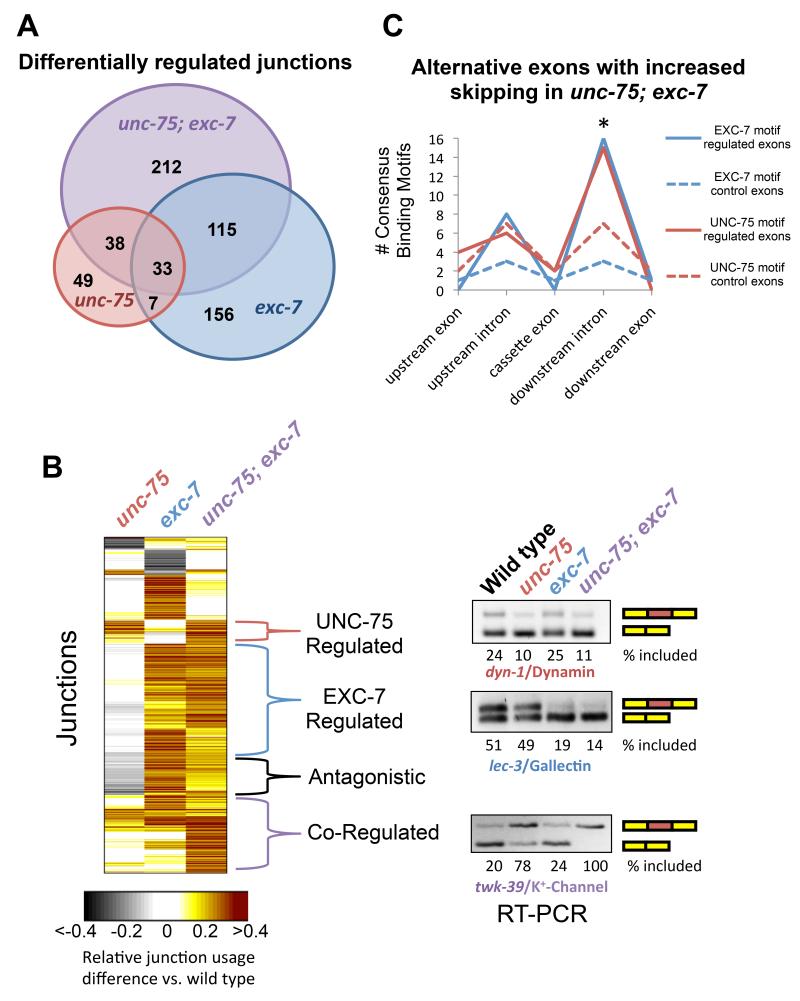
Figure 4.
UNC-75 and EXC-7 regulate partially overlapping sets of alternative junctions.
(A) Venn diagram displaying number of statistically-significant alternative junction changes between wild type and any of the three mutant conditions, as well as the overlap among the three conditions.
(B) Heat Map displaying differential junction usage (rows) between wild type and mutant conditions (columns). All junction difference values were normalized to the double mutant comparison. Right panel, representative RT-PCR validations of alternative splicing events belonging to different regulatory modes.
(C) Presence of exc-7 or unc-75 binding motifs in the vicinity of 51 cassette exons which undergo increased skipping in unc-75; exc-7 double mutants.
We identified hundreds of differentially utilized exon junctions that varied between wild type and one of the three mutant strains by >20% (Figure 4A and B, Supplementary Table 2). The most abundant classes of differential junction usage involved alternative inclusion of exons, alternative first exons and alternative 5′ splice sites, with each class representing about one quarter of the total exon junction differences observed (Supplementary Figure 4B). Among exon inclusion events that are differentially spliced between wild type and unc-75; exc-7 double mutants, 51 exons exhibit increased skipping in the double mutant, versus only 19 with increased inclusion, suggesting that UNC-75 and EXC-7 primarily mediate inclusion of exons.
Our genome-wide analysis revealed distinct modes of individual and combinatorial control of alternative junction usage by UNC-75 and/or EXC-7, and we confirmed these examples by RT-PCR analysis and two-color splicing reporters (Figure 4B and Supplementary Figure 4A and C). A substantial cluster of junctions was regulated by UNC-75 and EXC-7 (Figure 4B, ‘Co-regulated’). In these cases, unc-75 and exc-7 mutants alone have similar but modest effects on junction usage, while the double mutant has more robust changes, displaying either additive or synergistic effects on splicing regulation. The unc-16 exon 16 alternative splicing regulatory pattern belongs to this ‘Co-regulated’ category. We also identified a smaller set of junctions that were combinatorially regulated, but in these cases each single mutant displayed an opposite regulatory outcome relative to wild type and the double mutant had an intermediate effect (Figure 4B, ‘Ant gonistic’). Altogether, we found that 31% of the spliced junctions regulated by unc-75 are also regulated by exc-7 (Figure 4A and B), demonstrating overlap between UNC-75 and EXC-7 targets. Additionally, 53% of all differentially regulated junctions between wild-type and unc-75; exc-7 double mutants were not found to be differentially regulated in either of the single mutants (Figure 4A and B). These results demonstrate extensive regulatory cooperativity between these two RNA-binding proteins.
In order to extend our mechanistic studies of unc-16 exon 16 alternative splicing above, we searched full exon or intron sequences surrounding the 51 exons with increased skipping in unc-75; exc-7 double mutant animals for the presence of UNC-75 or EXC-7 binding sites identified by RNAcompete. Compared to a control set of similarly-matched exons or introns that are not regulated by either RNA binding protein, we found a significant enrichment of both unc-75 and exc-7 binding sites in the downstream intron following the cassette exon (Figure 4C, p < 0.05, Chi-squared test). Performing the same analysis on the 19 exons with increased inclusion in the double mutant yielded no enrichment of such binding-site patterns (Supplementary Figure 5). These results are consistent with our experiments with the unc-16 exon16 alternative splicing reporter (Figure 3C and D), and also agree with recent data reported (Kuroyanagi et al., 2013b) suggesting that UNC-75 generally stimulates exon inclusion by binding to downstream intronic sequences.
Gene Ontology (GO) analysis revealed that genes regulated only by UNC-75 are enriched in functions related to ion channels and transporters, while those regulated only by EXC-7 are enriched in functions related to behavior and gamete generation (Supplementary Table 3). Taken together, our genome-wide analysis indicates that UNC-75 and EXC-7 control isoform usage in partially distinct but overlapping networks of transcripts with neuronal functions.
UNC-75 and EXC-7 control cholinergic synaptic transmission through partially distinct mechanisms
Previous work has demonstrated that both unc-75 and exc-7 mutants are resistant to the acetylcholinesterase inhibitor aldicarb, implying that these mutants exhibit reduced cholinergic synaptic transmission (Loria et al., 2003). Moreover, an unc-75; exc-7 double mutant exhibits a more severe aldicarb resistance phenotype than either of the single mutants, implying that unc-75 and exc-7 have non-redundant functional roles (Loria et al., 2003).
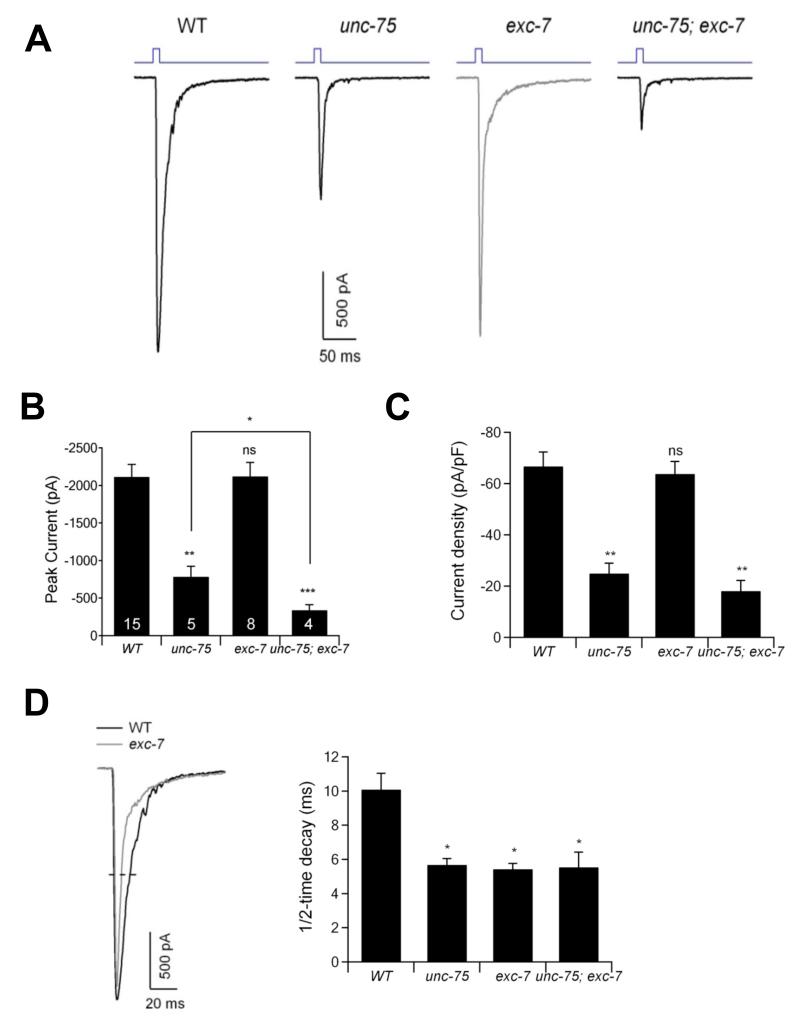
To more directly test if these mutants exhibit combinatorial synaptic transmission defects, we generated unc-75, exc-7, and double mutant animals expressing Channelrhodopsin, a light-gated ion channel which enables precise activation of neurotransmission by pulses of light (Liewald et al., 2008). We expressed Channelorhodopsin in cholinergic motor neurons and recorded spontaneous miniature post-synaptic currents (mPSCs) and evoked excitatory post-synaptic currents (EPSCs) across the neuromuscular junction in body wall muscles upon light-stimulated cholinergic motor neuron activation (Gao and Zhen, 2011; Goodman et al., 2012). Stimulation of cholinergic motor neurons in wild type animals elicited a robust EPSC (Figure 5A and B). In unc-75 mutants this evoked response was significantly diminished, whereas exc-7 mutants exhibited no change in the amplitude or density of the evoked current (Figure 5A, B and C). However, unc-75; exc-7 double mutants exhibited significantly lower evoked amplitude than unc-75 single mutants (Figure 5A and B). We also observed that the half-time decay of the evoked response is similarly shorter in exc-7, unc-75, and double mutants relative to wild-type animals (Figure 5A and D). The frequency but not the amplitude of mPSCs was significantly reduced in each of the single mutants and the double mutants (Supplementary Fig. 6). These results indicate that while both UNC-75 and EXC-7 affect cholinergic synaptic transmission, UNC-75 plays a more critical role, although UNC-75 can only partially compensate for the loss of EXC-7.
Figure 5.
unc-75 and exc-7 mutants exhibit partially overlapping cholinergic synaptic transmission defects at the neuromuscular junction.
(A) Representative traces of evoked EPSCs in wild-type (WT), unc-75(e950), exc-7(rh252) and unc-75(e950); exc-7(rh252) mutants. Muscles were holding at −60 mV. The EPSCs were induced by 10-ms blue light (top blue traces) in a zxIs6 background, expressing channelrhodopsin in cholinergic neurons.
(B-C) The amplitude (B) and density (C) of currents in wild type and mutant strains. Data represented as mean +/− SEM.
(D) The 1/2-time decay of the EPSCs in wild type and mutant animals. * P < 0.05, ** P < 0.01, *** P < 0.001, ns: not significant by Mann-Whitney U test. Numbers in the bar graph indicate the number of animals examined for each genotype.
See also Figure S6.
Analysis of the UNC-75 exon network reveals novel neuronal phenotypes for known and uncharacterized genes
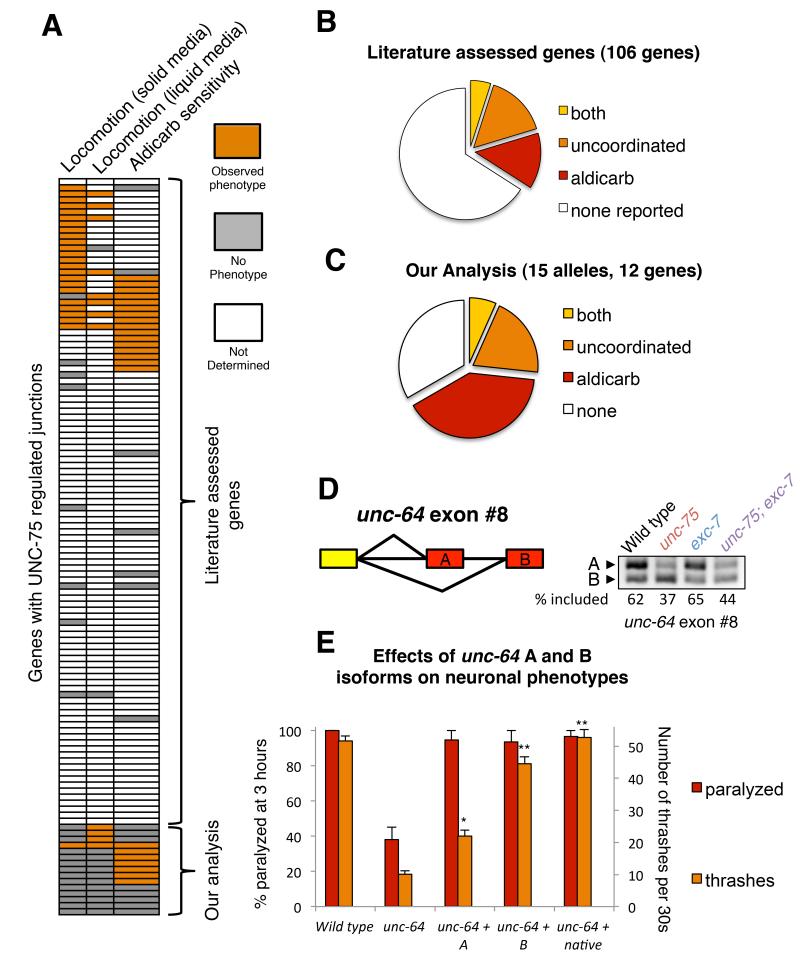
unc-75 mutants exhibit severe neuronal phenotypes, including uncoordination and altered aldicarb sensitivity. Given that UNC-75 also controls a network of exons in genes enriched for neuronal functions, we reasoned that analysis of this network might reveal phenotypic relationships among loss-of-function mutations in UNC-75 target genes. Strikingly, among 118 genes identified by our genome-wide analysis regulated by UNC-75, greater than 25% have been reported to exhibit loss-of-function uncoordinated and/or altered aldicarb sensitivity phenotypes (Figure 6A and B). This percentage is likely an underestimate of the true proportion of UNC-75-regulated genes with such phenotypes, given that most genes studied in previous reports were not assessed for changes in aldicarb sensitivity or for more subtle quantifiable effects on locomotion (Figure 6A and B).
Figure 6.
Network analysis of unc-75-regulated transcripts reveals a coherent network and predicts novel neuronal phenotypes.
(A) Heat map displaying presence or absence of uncoordination (1st and 2nd columns) or aldicarb-sensitivity phenotypes (3rd column) in the UNC-75 network of genes based on previously-reported phenotypes from the literature and our analysis of genes with previously uncharacterized phenotypes. Uncoordination phenotypes were separated on the basis of whether a phenotype could be observed on solid media (1st column), or quantitatively through thrashing assays in liquid media (2nd column).
(B-C) Proportion of target genes exhibiting one or both phenotypes in the literature search (B), or our knockout allele analysis (C).
(D) Left panel schematic showing alternative splicing at the 3′ end of unc-64 gene transcripts. Right panel: RT-PCR assay showing differential splicing patterns in unc-75, exc-7, and double mutants relative to wild type animals.
(E) Fosmid-based isoform-specific reintroduction of UNC-64/Syntaxin isoforms into an unc-64 loss-of-function background reveals isoform-specific differences among the two splice variants. Data represented as mean +/− SEM.
These results suggested that the UNC-75 exon network can be used to predict functions for genes with previously uncharacterized phenotypes. Therefore, we tested available mutant strains harboring loss of function alleles corresponding to 12 conserved genes with UNC-75-regulated exons for locomotion or aldicarb sensitivity defects. Interestingly, the majority (67%) of these alleles exhibited one or both phenotypes (Figure 6A, C and Supplementary Figure 7A and B). Several of the genes in our analysis have no known molecular function, while some have been studied in other tissues or in other organisms, which could shed light on their role in neurobiology. For example, we determined that animals carrying mutations in the gene acs-13, encoding an acyl CoA synthetase enzyme implicated in fatty acid metabolism, exhibit altered aldicarb sensitivity.
Taken together, our results indicate that interrogation of exon networks can be used as an alternative and effective approach to identify novel regulators of synaptic transmission and behavior. For instance, a previous aldicarb resistance RNAi-based screen against genes predicted to be involved in synaptic localization, cytoskeletal regulation or membrane trafficking had a positive hit rate of 8.9% (Sieburth et al., 2005), compared with our 47% positive hit rate for the aldicarb phenotype in the UNC-75 alternative splicing network analysis (Figure 6C).
Isoform-specific functions for a central gene in the UNC-75 regulatory network
In addition to using the exon network to predict gene function, we further tested whether UNC-75-regulated isoforms have distinct neuronal functions in vivo. We focused on the A and B isoforms of the gene unc-64/Syntaxin, a critical component of the SNARE complex that mediates vesicle and membrane fusion (Ogawa et al., 1998; Saifee et al., 1998). The relative usage of its mutually exclusive final exons A and B is combinatorially regulated by UNC-75 and EXC-7 (Figure 6D), and these exons encode slightly different transmembrane domains for insertion into the presynaptic membrane. unc-64 mutants, like unc-75 mutants, are uncoordinated and resistant to aldicarb (Saifee et al., 1998). We utilized fosmid recombineering (Tursun et al., 2009) to create transgenes that selectively express a single isoform and assessed the rescuing capacity of each isoform in unc-64 loss of function mutant animals. Both isoforms equivalently rescued the aldicarb resistance phenotype of mutant animals, suggesting that each isoform was functional (Figure 6E). Intriguingly however, only the B isoform efficiently rescued the uncoordination phenotype (Figure 6E and movie S1). These results indicate that unc-64 isoforms A and B have non-overlapping functions, and that the relative proportions of these isoforms are fine-tuned by UNC-75. Our data also suggest that the UNC-75 regulatory network can be used to uncover non-redundant roles for isoforms in the nervous system.
DISCUSSION
Neuron-specific alternative splicing occurs frequently and is subject to precise regulation
While cases of alternative splicing between cells in the nervous system have previously been identified by serendipity, studies have been limited by the technical challenges of observing splicing differences in individual neurons. In this study, we have developed a framework to better understand differential splicing regulation at single-neuron resolution. Using C. elegans as a model system, we found that a striking 50% of alternative exons tested with our two color reporters exhibit neuron-specific alternative splicing patterns (Figure 1). These patterns include differences between distinct functional classes of neurons. The deterministic regulation of alternative splicing in single neurons described here contrasts with recent observations of neuron-specific stochastic regulation of exon choice in Drosophila dscam1 gene transcripts (Miura et al., 2013). These observations suggest that both deterministic and stochastic variation in splicing generate cellular and molecular diversity in the nervous system. Our results highlight the importance of investigating gene regulation and alternative splicing at the level of individual cells, and complement recent transcriptome-wide analyses of splicing conducted at single cell resolution (Shalek et al., 2013).
Analysis of exon networks identifies genes with important neuronal functions
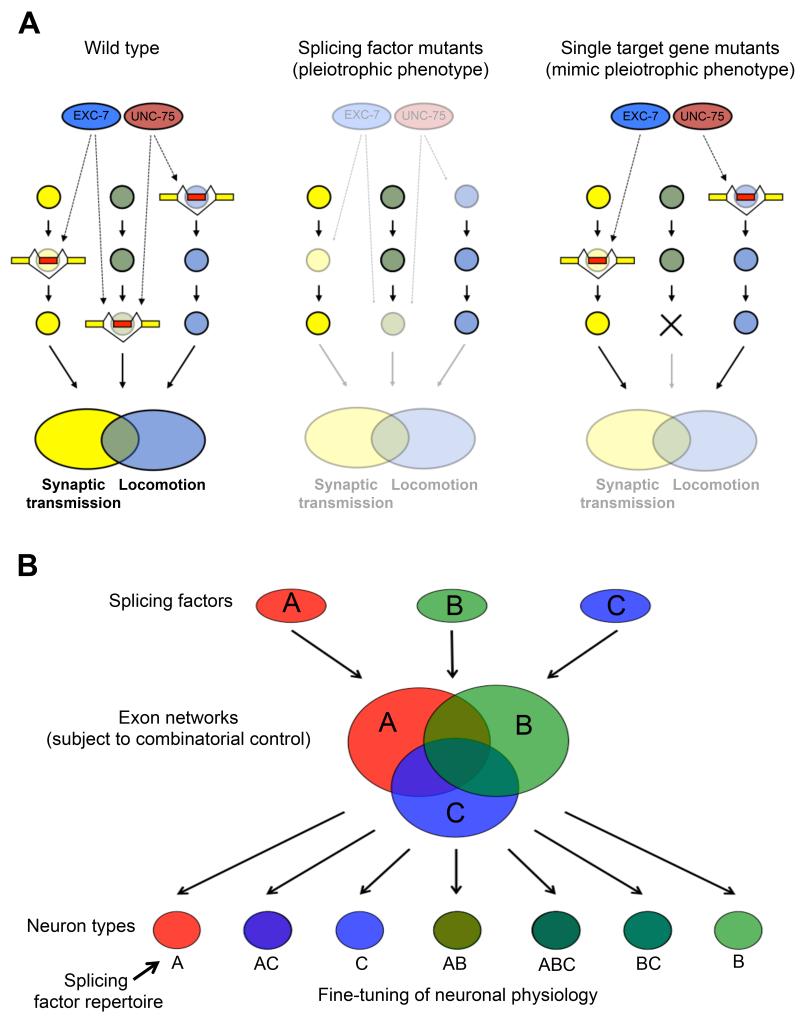
Previous studies have led to the hypothesis that splicing factor-mediated control of networks of transcripts might serve to specialize the function of specific tissues (Calarco et al., 2009; Ule et al., 2005; Wang et al., 2012; Zhang et al., 2008). Our results support and extend this paradigm at the level of individual neuronal cells. The shared phenotype between the loss of many single UNC-75 targets and loss of UNC-75 itself makes it tantalizing to hypothesize that the unc-75 phenotype may be directly caused by cumulative effects of mis-regulated splicing of target genes in the alternative splicing network (Figure 7A). Moreover in some cases, studying individual targeted alternative splicing events may also reveal neuronal phenotypes of interest mediated by alternative splicing, exemplified by our observations that unc-64 isoforms have non-overlapping functions (Figure 6E).
Figure 7.
Models for the contribution of UNC-75 and EXC-7 splicing networks to neuronal physiology.
(A) In wild-type animals (left panel), UNC-75 and EXC-7 (red and blue ovals, respectively) regulate partially distinct sets of splicing events (yellow and red boxes) in target genes (yellow, light blue, and light green circles) found in pathways controlling synaptic transmission and locomotion. When these factors are absent (middle panel), cumulative effects of mis-regulated splicing of these target gene transcripts can lead to observable locomotion and/or synaptic transmission phenotypes. However, complete loss of function of individual splicing network target genes found in critical pathways (right panel, marked by an X) are often sufficient to produce phenotypes found in the splicing factor mutants.
(B) The action of three splicing factors (Red, blue, and green ovals) with overlapping expression patterns regulate overlapping exon networks (Red, blue and green large circles). Depending on the repertoire of splicing factors in each neuronal cell (small circles at bottom), combinatorial control of the exon networks creates diverse isoform repertoires.
Such discovery-driven genetic analyses of exon networks have remained challenging to conduct in vertebrate models in vivo. For example, this study reveals a previously unknown role for the fatty acid CoA synthetase ACS-13 in maintaining proper aldicarb sensitivity. ACSL5, the human homolog of ACS-13, plays a role in fatty acid metabolism and lipid biosynthesis (Yamashita et al., 2000). While it is not immediately clear what mechanistic role acs-13 may be playing in C. elegans neuronal function, a link between the relative concentrations of long-chain fatty acids in neurons and effects on cholinergic synaptic transmission has been described in C. elegans (Lesa et al., 2003). It will be interesting to determine whether acs-13 isoforms also play a mechanistic role connecting lipid metabolism and synaptic transmission.
A role for combinatorial control of alternative exons in generating neuronal diversity
Our genetic screen has revealed how alternative splicing can be controlled in a neuron-specific manner through the overlapping expression of two splicing factors. Both UNC-75 and EXC-7 have conserved orthologs in humans (the CELF and Hu family of proteins, respectively) which also play a role in alternative splicing, are expressed in the nervous system, and have been implicated in neuronal disorders (Darnell, 2013; Dasgupta and Ladd, 2012). CELF and Hu family members have recently been shown to act together in controlling alternative splicing of exon 23a of the neuronal NF1 gene in mammalian cell culture (Fleming et al., 2012), suggesting that our model of genome-wide combinatorial control by CELF and Hu family members may apply more broadly to the mammalian nervous system as well.
Data from our mRNA-Seq experiments indicate that combinatorial control of alternative splicing by UNC-75 and EXC-7 is extensive (Figure 4). We expect that other combinations of splicing factors with overlapping expression patterns will act together to fine-tune the properties of different neuronal classes as well. This mechanism may serve to allow a relatively small number of factors with overlapping expression patterns to combinatorially create a large diversity of alternative splicing patterns within most nervous systems (Figure 7B). Future studies making use of C. elegans hold promise for yielding novel insights into the mechanisms governing alternative splicing regulation within the nervous system and the identification of functionally important genes and isoforms in distinct neuron classes.
EXPERIMENTAL PROCEDURES
Strain Maintenance and Microscopy
C. elegans were maintained by standard techniques (Brenner, 1974) on NGM agar plates seeded with OP50 E. coli. All images of two color reporter transgene-expressing animals were captured with a Zeiss Axioskop 2 fluorescence compound microscope and processed in ImageJ.
Mutagenesis and Screening
P0 worms expressing the unc-16 exon 16 alternative splicing reporter were mutagenized with ethyl methanesulfonate (EMS) at 47 mM for four hours. Single F1s were sorted into individual wells of a 96-well plate by a Union Biometrica COPAS worm sorter. After 4-7 days of growth in NGM liquid culture media at room temperature, an aliquot of each well was transferred to an optical imaging 96 well plate (Nunc). The unc-16 alternative splicing reporter pattern in the pool of F2-F3 mutant worms was analyzed fluorescently on a Zeiss Axioskop 2 microscope. Wells containing mutant animals with altered reporter patterns were isolated. To identify causative mutations, we performed whole-genome sequencing using the software MAQgene according to previously described protocols (Bigelow et al., 2009).
mRNA-Seq analysis to identify differential junction usage
Total RNA was isolated from synchronized L4 worms using Tri reagent (Sigma Aldrich), and cDNA libraries from Poly A+ enriched RNA was prepared using the TruSeq RNA kit (Illumina) according to manufacturer’s instructions. Deep sequencing was performed on an Illumina HiSeq 2000 according to standard protocols, generating 150 base paired-end reads. Reads were mapped to the C. elegans genome (version WS190) using the RNA-Seq Unified Mapper (RUM) under default settings (Grant et al., 2011). Alternative junctions were identified using a custom-developed pipeline. For more details see the supplementary information. All raw reads (in fastq format) have been deposited to the NCBI short read archive under accession number SRX514835.
Phenotypic Analyses
Aldicarb assays were performed as previously described (Loria et al., 2003) on plates of young adult worms at 15°C and 1 mM aldicarb. A total of 20-30 worms were counted in each assay for each strain, and each assay was replicated a total of three times. Worms were considered paralyzed if they did not move in response to a light nose touch. Coordination assays were performed by placing young adult worms in a drop of M9 buffer, and after one minute of equilibration, counting the thrashing rate (frequency of number of body bends per minute) of animals recorded under a stereomicroscope. A total of 15-20 worms were analyzed from each strain tested.
Electrophysiology
The dissection of the C. elegans was described previously (Richmond and Jorgensen, 1999). The recording solutions were as described in previous studies (Richmond and Jorgensen, 1999). Specifically, the pipette solution contains (in mM): K-gluconate 115; KCl 25; CaCl2 0.1; MgCl2 5; BAPTA 1; HEPES 10; Na2ATP 5; Na2GTP 0.5; cAMP 0.5; cGMP 0.5, pH7.2 with KOH, ~320 mOsm. The bath solution consists of (in mM): NaCl 150; KCl 5; CaCl2 5; MgCl2 1; glucose 10; sucrose 5; HEPES 15, pH7.3 with NaOH, ~330 mOsm. Under these conditions, the reversal potentials are ~+20 mV for acetylcholine receptors, respectively (Gao and Zhen, 2011). Light stimulation of zxIs6 was performed with an LED lamp (KSL-70; RAPP OptoElectronic) at a wavelength of 470 nm (8 mW/mm2), controlled by the Axon amplifier software. zxIs6 was cultured in the dark at 22 °C on OP50-seeded NGM plates supplemented with all-trans retinal as described previously (Gao and Zhen, 2011). All chemicals were from Sigma. Experiments were performed at room temperatures (20–22 °).
Supplementary Material
Article Highlights.
-
-
Differential splicing regulation is frequent in distinct neuron types
-
-
Combinations of RNA binding proteins contribute to neuron-specific splicing
-
-
Regulation of partially overlapping splicing networks facilitates neuronal diversity
-
-
Splicing networks can be utilized to uncover novel biological insights
ACKNOWLEDGMENTS
We would like to thank Ben Blencowe, Andrew Murray, Vlad Denic, Bodo Stern, Nicolas Chevrier, Peter Turnbaugh, Lauren O’Connell, Arneet Saltzman, Joe Calarco, and Xico Gracida for critical reading of the manuscript. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). Other strains were provided by the NBRP (Tokyo). M.Z. is supported by grants from NSERC and CIH. J.A.C. is supported by an NIH Director’s Early Independence Award (DP5OD009153) and by Harvard University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aoto J, Martinelli DC, Malenka RC, Tabuchi K, Sudhof TC. Presynaptic neurexin-3 alternative splicing trans-synaptically controls postsynaptic AMPA receptor trafficking. Cell. 2013;154:75–88. doi: 10.1016/j.cell.2013.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ. Deciphering the splicing code. Nature. 2010;465:53–59. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
- Barberan-Soler S, Medina P, Estella J, Williams J, Zahler AM. Co-regulation of alternative splicing by diverse splicing factors in Caenorhabditis elegans. Nucleic acids research. 2011;39:666–674. doi: 10.1093/nar/gkq767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Colak R, et al. The Evolutionary Landscape of Alternative Splicing in Vertebrate Species. Science. 2012;338:1587–1593. doi: 10.1126/science.1230612. [DOI] [PubMed] [Google Scholar]
- Bigelow H, Doitsidou M, Sarin S, Hobert O. MAQGene: software to facilitate C. elegans mutant genome sequence analysis. Nature methods. 2009;6:549–549. doi: 10.1038/nmeth.f.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M, Green RE, MacArthur S, Brooks AN, Brenner SE, Eisen MB, Rio DC. Genome-wide analysis of alternative pre-mRNA splicing and RNA-binding specificities of the Drosophila hnRNP A/B family members. Molecular cell. 2009;33:438–449. doi: 10.1016/j.molcel.2009.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucard AA, Chubykin AA, Comoletti D, Taylor P, Sudhof TC. A splice code for trans-synaptic cell adhesion mediated by binding of neuroligin 1 to alpha- and beta-neurexins. Neuron. 2005;48:229–236. doi: 10.1016/j.neuron.2005.08.026. [DOI] [PubMed] [Google Scholar]
- Boutz PL, Stoilov P, Li Q, Lin CH, Chawla G, Ostrow K, Shiue L, Ares M, Black DL. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Gene Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunschweig U, Gueroussov S, Plocik AM, Graveley BR, Blencowe BJ. Dynamic integration of splicing within gene regulatory pathways. Cell. 2013;152:1252–1269. doi: 10.1016/j.cell.2013.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco JA, Superina S, O’Hanlon D, Gabut M, Raj B, Pan Q, Skalska U, Clarke L, Gelinas D, van der Kooy D, et al. Regulation of Vertebrate Nervous System Alternative Splicing and Development by an SR-Related Protein. Cell. 2009;138:898–910. doi: 10.1016/j.cell.2009.06.012. [DOI] [PubMed] [Google Scholar]
- Charlet BN, Logan P, Singh G, Cooper TA. Dynamic antagonism between ETR-3 and PTB regulates cell type-specific alternative splicing. Molecular cell. 2002;9:649–658. doi: 10.1016/s1097-2765(02)00479-3. [DOI] [PubMed] [Google Scholar]
- Chih B, Gollan L, Scheiffele P. Alternative splicing controls selective trans-synaptic interactions of the neuroligin-neurexin complex. Neuron. 2006;51:171–178. doi: 10.1016/j.neuron.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Darnell RB. RNA protein interaction in neurons. Annual review of neuroscience. 2013;36:243–270. doi: 10.1146/annurev-neuro-062912-114322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta T, Ladd AN. The importance of CELF control: molecular and biological roles of the CUG-BP, Elav-like family of RNA-binding proteins. Wiley interdisciplinary reviews RNA. 2012;3:104–121. doi: 10.1002/wrna.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming VA, Geng C, Ladd AN, Lou H. Alternative splicing of the neurofibromatosis type 1 pre-mRNA is regulated by the muscleblind-like proteins and the CUG-BP and ELAV-like factors. BMC molecular biology. 2012;13:35. doi: 10.1186/1471-2199-13-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Hawkinson D, King KV, Hall DH, Sakamoto H, Buechner M. The role of the ELAV homologue EXC-7 in the development of the Caenorhabditis elegans excretory canals. Developmental biology. 2003;256:290–301. doi: 10.1016/s0012-1606(03)00040-x. [DOI] [PubMed] [Google Scholar]
- Fujita M, Kawano T, Ohta A, Sakamoto H. Neuronal expression of a Caenorhabditis elegans elav-like gene and the effect of its ectopic expression. Biochemical and biophysical research communications. 1999;260:646–652. doi: 10.1006/bbrc.1999.0957. [DOI] [PubMed] [Google Scholar]
- Gao S, Zhen M. Action potentials drive body wall muscle contractions in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:2557–2562. doi: 10.1073/pnas.1012346108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehman LT, Stoilov P, Maguire J, Damianov A, Lin CH, Shiue L, Ares M, Jr., Mody I, Black DL. The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nature genetics. 2011;43:706–711. doi: 10.1038/ng.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golgi C. Sulla fina anatomia degli organi centrali del sistema nervoso. Rivista sperimentale di Freniatria. 1883;9:1–17. [Google Scholar]
- Goodman MB, Lindsay TH, Lockery SR, Richmond JE. Electrophysiological methods for Caenorhabditis elegans neurobiology. Methods in cell biology. 2012;107:409–436. doi: 10.1016/B978-0-12-394620-1.00014-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant GR, Farkas MH, Pizarro AD, Lahens NF, Schug J, Brunk BP, Stoeckert CJ, Hogenesch JB, Pierce EA. Comparative analysis of RNA-Seq alignment algorithms and the RNA-Seq unified mapper (RUM) Bioinformatics. 2011;27:2518–2528. doi: 10.1093/bioinformatics/btr427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig LC, Woodworth MB, Galazo MJ, Padmanabhan H, Macklis JD. Molecular logic of neocortical projection neuron specification, development and diversity. Nature reviews Neuroscience. 2013;14:755–769. doi: 10.1038/nrn3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O, Carrera I, Stefanakis N. The molecular and gene regulatory signature of a neuron. Trends in neurosciences. 2010;33:435–445. doi: 10.1016/j.tins.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KB, Dredge BK, Stefani G, Zhong R, Buckanovich RJ, Okano HJ, Yang YYL, Darnell RB. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron. 2000;25:359–371. doi: 10.1016/s0896-6273(00)80900-9. [DOI] [PubMed] [Google Scholar]
- Kuroyanagi H, Kobayashi T, Mitani S, Hagiwara M. Transgenic alternative-splicing reporters reveal tissue-specific expression profiles and regulation mechanisms in vivo. Nature methods. 2006;3:909–915. doi: 10.1038/nmeth944. [DOI] [PubMed] [Google Scholar]
- Kuroyanagi H, Ohno G, Sakane H, Maruoka H, Hagiwara M. Visualization and genetic analysis of alternative splicing regulation in vivo using fluorescence reporters in transgenic Caenorhabditis elegans. Nature protocols. 2010;5:1495–1517. doi: 10.1038/nprot.2010.107. [DOI] [PubMed] [Google Scholar]
- Kuroyanagi H, Watanabe Y, Hagiwara M. CELF family RNA-binding protein UNC-75 regulates two sets of mutually exclusive exons of the unc-32 gene in neuron-specific manners in Caenorhabditis elegans. PLoS genetics. 2013a;9:e1003337. doi: 10.1371/journal.pgen.1003337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroyanagi H, Watanabe Y, Suzuki Y, Hagiwara M. Position-dependent and neuron-specific splicing regulation by the CELF family RNA-binding protein UNC-75 in Caenorhabditis elegans. Nucleic acids research. 2013b;41:4015–4025. doi: 10.1093/nar/gkt097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesa GM, Palfreyman M, Hall DH, Clandinin MT, Rudolph C, Jorgensen EM, Schiavo G. Long chain polyunsaturated fatty acids are required for efficient neurotransmission in C. elegans. Journal of cell science. 2003;116:4965–4975. doi: 10.1242/jcs.00918. [DOI] [PubMed] [Google Scholar]
- Liewald JF, Brauner M, Stephens GJ, Bouhours M, Schultheis C, Zhen M, Gottschalk A. Optogenetic analysis of synaptic function. Nature methods. 2008;5:895–902. doi: 10.1038/nmeth.1252. [DOI] [PubMed] [Google Scholar]
- Loria PM, Duke A, Rand JB, Hobert O. Two neuronal, nuclear-localized RNA binding proteins involved in synaptic transmission. Current biology. 2003;CB 13:1317–1323. doi: 10.1016/s0960-9822(03)00532-3. [DOI] [PubMed] [Google Scholar]
- Markovtsov V, Nikolic JM, Goldman JA, Turck CW, Chou MY, Black DL. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Molecular and cellular biology. 2000;20:7463–7479. doi: 10.1128/mcb.20.20.7463-7479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura SK, Martins A, Zhang KX, Graveley BR, Zipursky SL. Probabilistic splicing of dscam1 establishes identity at the level of single neurons. Cell. 2013;155:1166–1177. doi: 10.1016/j.cell.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris AD, Calarco JA. Emerging Roles of Alternative Pre-mRNA Splicing Regulation in Neuronal Development and Function. Frontiers in neuroscience. 2012;6:122. doi: 10.3389/fnins.2012.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa H, Harada S, Sassa T, Yamamoto H, Hosono R. Functional properties of the unc-64 gene encoding a Caenorhabditis elegans syntaxin. The Journal of biological chemistry. 1998;273:2192–2198. doi: 10.1074/jbc.273.4.2192. [DOI] [PubMed] [Google Scholar]
- Ohno G, Hagiwara M, Kuroyanagi H. STAR family RNA-binding protein ASD-2 regulates developmental switching of mutually exclusive alternative splicing in vivo. Genes Dev. 2008;22:360–374. doi: 10.1101/gad.1620608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orengo JP, Bundman D, Cooper TA. A bichromatic fluorescent reporter for cell-based screens of alternative splicing. Nucleic acids research. 2006;34:e148. doi: 10.1093/nar/gkl967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj B, O’Hanlon D, Vessey JP, Pan Q, Ray D, Buckley NJ, Miller FD, Blencowe BJ. Cross-regulation between an alternative splicing activator and a transcription repressor controls neurogenesis. Molecular cell. 2011;43:843–850. doi: 10.1016/j.molcel.2011.08.014. [DOI] [PubMed] [Google Scholar]
- Ramani AK, Calarco JA, Pan Q, Mavandadi S, Wang Y, Nelson AC, Lee LJ, Morris Q, Blencowe BJ, Zhen M, et al. Genome-wide analysis of alternative splicing in Caenorhabditis elegans. Genome research. 2011;21:342–348. doi: 10.1101/gr.114645.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramón y Cajal S. Investigaciones de histología comparad en los centros ópticos de los vertebrados. Consejo Superior de Investigaciones Científicas, Instituto Ramón y Cajal; Madrid: 1890. [Google Scholar]
- Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013;499:172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond JE, Jorgensen EM. One GABA and two acetylcholine receptors function at the C. elegans neuromuscular junction. Nature neuroscience. 1999;2:791–797. doi: 10.1038/12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saifee O, Wei L, Nonet ML. The Caenorhabditis elegans unc-64 locus encodes a syntaxin that interacts genetically with synaptobrevin. Molecular biology of the cell. 1998;9:1235–1252. doi: 10.1091/mbc.9.6.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalek AK, Satija R, Adiconis X, Gertner RS, Gaublomme JT, Raychowdhury R, Schwartz S, Yosef N, Malboeuf C, Lu D, et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498:236–240. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth D, Ch’ng Q, Dybbs M, Tavazoie M, Kennedy S, Wang D, Dupuy D, Rual JF, Hill DE, Vidal M, et al. Systematic analysis of genes required for synapse structure and function. Nature. 2005;436:510–517. doi: 10.1038/nature03809. [DOI] [PubMed] [Google Scholar]
- Tursun B, Cochella L, Carrera I, Hobert O. A Toolkit and Robust Pipeline for the Generation of Fosmid-Based Reporter Genes in C. elegans. Plos One. 2009;4 doi: 10.1371/journal.pone.0004625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Ule A, Spencer J, Williams A, Hu JS, Cline M, Wang H, Clark T, Fraser C, Ruggiu M, et al. Nova regulates brain-specific splicing to shape the synapse. Nature genetics. 2005;37:844–852. doi: 10.1038/ng1610. [DOI] [PubMed] [Google Scholar]
- Wang ET, Cody NA, Jog S, Biancolella M, Wang TT, Treacy DJ, Luo S, Schroth GP, Housman DE, Reddy S, et al. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell. 2012;150:710–724. doi: 10.1016/j.cell.2012.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M, Hayakawa-Yano Y, Mele A, Darnell RB. Nova2 regulates neuronal migration through an RNA switch in disabled-1 signaling. Neuron. 2010;66:848–858. doi: 10.1016/j.neuron.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler AM. Alternative splicing in C. elegans. WormBook: the online review of C elegans biology. 2005:1–13. doi: 10.1895/wormbook.1.31.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Frias MA, Mele A, Ruggiu M, Eom T, Marney CB, Wang H, Licatalosi DD, Fak JJ, Darnell RB. Integrative modeling defines the Nova splicing-regulatory network and its combinatorial controls. Science. 2010;329:439–443. doi: 10.1126/science.1191150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhang Z, Castle J, Sun S, Johnson J, Krainer AR, Zhang MQ. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev. 2008;22:2550–2563. doi: 10.1101/gad.1703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Black DL. Alternative pre-mRNA splicing in neurons: growing up and extending its reach. Trends in genetics: TIG. 2013;29:442–448. doi: 10.1016/j.tig.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Damoiseaux R, Chen L, Black DL. A broadly applicable high-throughput screening strategy identifies new regulators of Dlg4 (Psd-95) alternative splicing. Genome research. 2013;23:998–1007. doi: 10.1101/gr.147546.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.