Summary
Much of the mortality attributed to influenza virus is due to secondary bacterial pneumonia, particularly from Streptococcus pneumoniae. However, mechanisms underlying this co-infection are incompletely understood. We find that prior influenza infection enhances pneumococcal colonization of the murine nasopharynx, which in-turn promotes bacterial spread to the lungs. Influenza accelerates bacterial replication in vivo, and sialic acid, a major component of airway glycoconjugates, is identified as the host-derived metabolite that stimulates pneumococcal proliferation. Influenza infection increases sialic acid and sialylated mucin availability, and enhances desialylation of host glycoconjugates. Pneumococcal genes for sialic acid catabolism are required for influenza to promote bacterial growth. Decreasing sialic acid availability in vivo by genetic deletion of the major airway mucin Muc5ac or mucolytic treatment limits influenza-induced pneumococcal replication. Our findings suggest that higher rates of disease during co-infection could stem from influenza-provided sialic acid, which increases pneumococcal proliferation, colonization and aspiration.
Introduction
Even before the discovery of its viral cause, bacterial infections have been recognized as an important complication of influenza (McCullers, 2006). Much of the mortality attributed to this virus during both seasonal and pandemic influenza seasons is actually from secondary bacterial pneumonia, particularly due to Streptococcus pneumoniae, the pneumococcus (McCullers, 2006).
All pneumococcal disease begins with asymptomatic colonization of the nasopharynx (Bogaert et al., 2004). Though carriage is the prerequisite to invasive disease, including pneumonia, most studies of influenza-pneumococcal interaction have relied on directly inoculating bacteria into the lung, bypassing this first, conserved step in pathogenesis (McCullers, 2006; Metzger and Sun, 2013). However, there is evidence that influenza predisposes the host to acquiring pneumococcal colonization (Grijalva et al., 2014). Pneumococcal colonization in adults and children is temporally associated with viral upper respiratory tract (URT) infections, including influenza, (Kim et al., 1996; Vu et al., 2011) and experimental influenza infection leads to higher pneumococcal loads in the human nasopharynx (Wadowsky et al., 1995). Additionally, the URT is the initial site of replication for both pneumococci and influenza, (Bogaert et al., 2004; Matrosovich et al., 2004).
In addition to focusing on the lower respiratory tract, studies of the interactions between influenza and pneumococci have historically emphasized influenza-induced epithelial damage that alters lung architecture and promotes bacterial adherence (Metzger and Sun, 2013; Olitsky and Gates, 1921). More recent studies have observed in vitro that influenza neuraminidase can expose receptors for bacterial adherence by removing terminal sialic acid residues from host glycoconjugates and in vivo that neuraminidase inhibitors protect from post-influenza pneumonia (McCullers, 2004; Peltola and McCullers, 2004). However, viral strains that do not cause pathologic changes in the epithelium can still lead to secondary bacterial pneumonia in animal models, implying that tissue damage is not necessary for influenza to promote bacterial disease (Metzger and Sun, 2013).
Other work has focused on defects in anti-bacterial immunity directed by prior viral infection. Influenza influences the immune response to secondary bacterial challenge in murine models of co-infection (McCullers, 2006; Metzger and Sun, 2013). Different groups have demonstrated changes in neutrophil recruitment to the lungs, alveolar macrophage function and macrophage recruitment to the nasopharynx during post-influenza challenge (Nakamura et al., 2011; Shahangian et al., 2009; Sun and Metzger, 2008). These immune effects are general, but only a small subset of opportunistic bacterial pathogens cause the vast majority of post-influenza pneumonia, and chief among these is the pneumococcus (Klugman et al., 2009; McCullers, 2006; Metzger and Sun, 2013). The predominance of pneumococci in post-influenza disease suggests that this bacterium is particularly able to take advantage of the influenza-infected environment. We hypothesized that influenza infection predisposes the host to rapid pneumococcal growth in the nasopharynx by providing a nutrient source for replicating bacteria.
Here, we show influenza promotes pneumococcal proliferation during colonization in a mouse model of co-infection, that this rapid bacterial growth is dependent on acquisition of the host metabolite sialic acid, that sialylated airway mucins are required for this effect, and that both influenza and pneumococcal neuraminidases contribute to the release of sialic acid from host substrates in vivo.
Results
Influenza promotes pneumococcal colonization, growth and aspiration
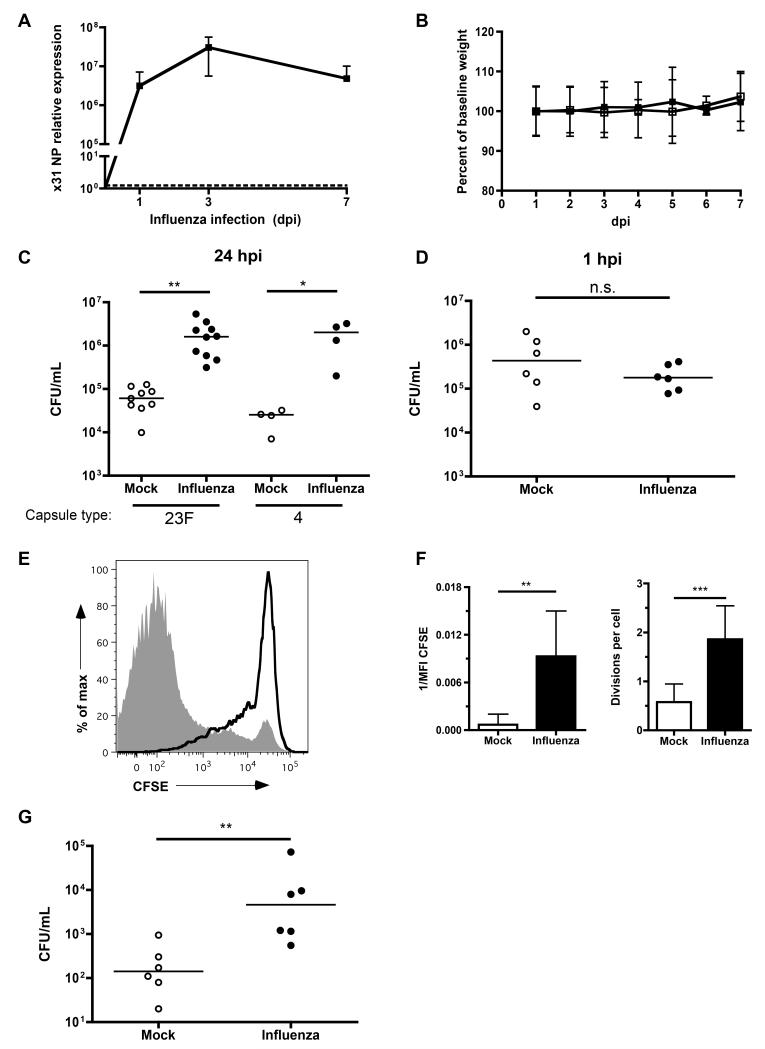
To determine whether influenza infection could promote bacterial colonization of the URT, mice were intranasally inoculated with influenza A virus or PBS (mock). This procedure was carried out using a small volume in unanesthetized mice to avoid direct introduction into the lower respiratory tract. The x31 influenza strain replicated efficiently in the URT, as measured by qRT-PCR viral titers in nasal lavages, which peaked at 3 days post-inoculation (Fig. 1A). This infection did not cause weight loss or other signs of systemic disease (Fig. 1B). Seven days after inoculation, we challenged the mice with pneumococci 24 hrs before obtaining nasal lavages to measure bacterial colonization density. Influenza infection promoted >25-fold higher density of colonizing pneumococci with clinical isolates of 2 different serotypes, 23F (strain P1121) and 4 (strain TIGR4) (Fig. 1C).
Figure 1. Influenza promotes pneumococcal colonization, growth and aspiration.
(A) Mice were infected with influenza for the indicated number of days, nasal lavages with RLT RNA lysis buffer obtained, and qRT-PCR performed to measure the relative expression of x31 viral nucleoprotein (NP). (B) Mice given PBS (mock, open symbols) or influenza (closed symbols) were weighed daily for 7 days, and the percent change in weight from baseline graphed. (C) Mice were infected with influenza or PBS, followed 7 days later by intranasal inoculation with bacteria of the indicated serotype. After 24 hrs post-bacterial inoculation (hpi), nasal lavages were obtained and plated for quantitative culture of colonizing pneumococci. (D) Mice were inoculated with influenza or PBS, followed 7 days later by challenge with strain P1121. Within 1 hr, nasal lavages were obtained and plated for quantitative culture (CFUs). (E) After 7 days of mock (black line) or influenza (grey shaded) infection, mice were inoculated with CFSE-labeled pneumococci for 8 hrs. Nasal lavages were obtained, fixed and stained for pneumococcal capsule, and flow cytometry performed to compare bacterial replication. (F) The median fluorescence intensity (MFI) of CFSE per bacterial cell was calculated for each condition, and displayed as 1/MFI, or number of divisions per cell (division index). (G) Mock or influenza-infected mice were challenged 7 days later with WT pneumococci for 24 hrs. Bronchoalveolar lavages were obtained and plated for quantitative culture. Horizontal lines indicate median values, and data in F are represented as mean +/− SD. n.s. = not significant, * = p < 0.05, ** = p < 0.01, *** = p < 0.001. All experiments were performed at least twice, with 4-11 mice per group. See also Figure S1.
To address whether this difference in colonization density was due to influenza promoting increased retention of the bacterial inoculum, we challenged mock- and influenza-infected mice for <1 hr before measuring bacterial density in nasal lavages. In contrast to observations at 24 hrs post-inoculation, there was no difference in bacterial load 1 hr post-challenge, suggesting that initial adherence to the epithelium was not responsible for the effect of prior influenza infection on colonization (Fig. 1D).
We next tested whether influenza promoted bacterial growth, rather than or in addition to inhibiting antibacterial immunity in the nasopharynx. To specifically measure bacterial replication in vivo, we developed a flow cytometric assay using carboxyfluorescein diacetate succinimidyl ester (CFSE), a dye used to track eukaryotic cell division. Each round of cell division yields bacteria with less CFSE fluorescence, which we measured in individual cells using flow cytometry (Parish, 1999). We validated the assay in vitro and in vivo to demonstrate that only dividing bacteria progressively lost CFSE fluorescence (Fig. S1). This assay was applied to influenza co-infection by inoculating CFSE-labeled pneumococci into mice that had been previously influenza- or mock-infected. Eight hrs later, we recovered the colonizing pneumococci in nasal lavages, stained the lavage with capsule type-specific antibody to distinguish pneumococci from other particles, and measured CFSE fluorescence per cell. Pneumococci colonizing mice that had been previously infected with influenza had substantially less CFSE fluorescence per cell than did those colonizing mock-treated mice (Fig. 1E). To quantify this growth effect, we measured the median fluorescence intensity (MFI) of CFSE per bacterium in each condition. As replication occurred, the MFI decreased, and 1/MFI increased (Fig. 1F). Additionally, we calculated the division index, the number of divisions per cell (Roederer, 2011). Bacteria colonizing influenza-infected mice underwent more divisions than those in mock-infected mice (Fig. 1F). Over 8 hrs, influenza promoted 3.7-fold more divisions per bacterial cell compared to PBS treatment, an effect predicted to increase bacterial numbers by 13-fold (2^3.7). This corresponded to the 12.1-fold increase in colonization density actually observed, a further validation for this assay and demonstrating the importance of bacterial growth in mediating increased colonization during co-infection.
Pneumonia generally begins with aspiration of upper airway contents into the lungs, and clinical studies have noted that pneumococcal pneumonia is associated with higher density of concurrent colonizing pneumococci in the nasopharynx (Albrich et al., 2012; Vu et al., 2011). We hypothesized that the increased bacterial growth stimulated by influenza could increase the likelihood of aspiration of pneumococci into the lungs. To test this possibility, we measured bacterial loads in the bronchoalveolar lavage fluid (BAL) of mock- and influenza-treated mice 24 hrs after establishing pneumococcal colonization of the URT. Influenza-infected mice had a higher bacterial burden in the lower respiratory tract compared to mock-treated mice (Fig. 1G). Colonization density in the URT correlated with bacterial load in the BAL (Spearman correlation coefficient, r = 0.6, P < 0.05).
Influenza increases sialic acid availability in the nasopharynx
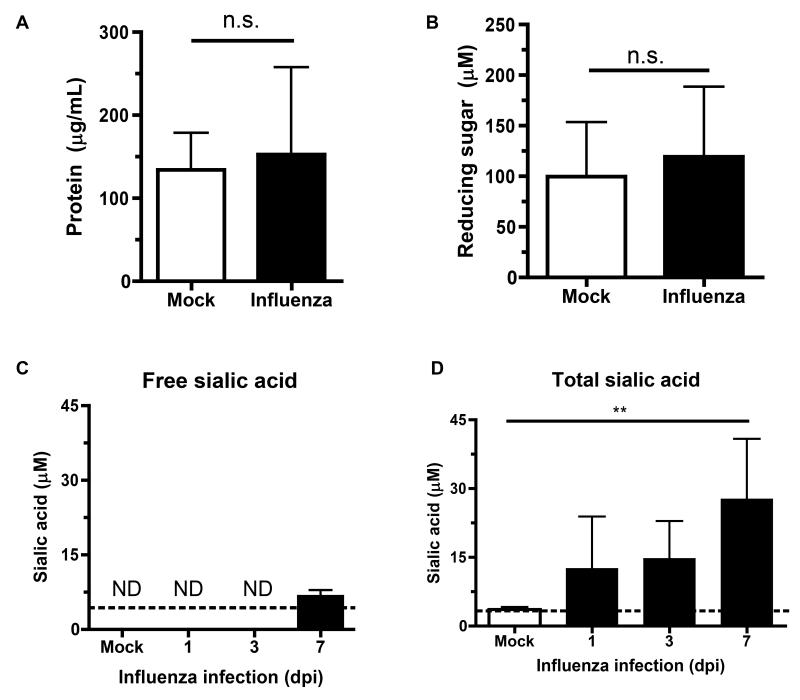
We next sought to determine the nutrient(s) influenza was providing to colonizing pneumococci. Pneumococci have complex growth requirements, and can catabolize >30 different carbohydrates as carbon sources (Buckwalter and King, 2012). To assess the availability of possible nutrient sources from inflammation and epithelial damage following influenza infection, we measured the total protein concentration of nasal lavages obtained from mice 7 days after influenza or mock infection and found no significant difference between these mice (Fig. 2A). We measured the total reducing sugar content of nasal lavages using a tetrazolium assay and found no significant difference between mock- and influenza-infected mice (Fig. 2B). The tetrazolium assay can only measure reducing sugar, however (Jue and Lipke, 1985). Sialic acid is a non-reducing sugar that is the most common terminal, and therefore accessible, modification on glycoconjugates in the airway (Angata and Varki, 2002). It is released from cells and mucus after cleavage by influenza neuraminidase during viral replication (Peltola and McCullers, 2004). Additionally, pneumococci express 2 to 3 neuraminidases per strain that liberate sialic acid (Burnaugh et al., 2008). Sialic acid can support pneumococcal growth in vitro as a sole carbon source, and pneumococci express a sialic acid transporter, SatABC, required for its utilization (Marion et al., 2011).
Figure 2. Influenza increases sialic acid availability in the nasopharynx.
(A-D) Mice were intranasally inoculated with influenza (closed bars) or mock (PBS, open bars) for 7 days, then nasal lavages obtained to measure levels of protein (A), reducing sugars (B) and sialic acid (C, D) in the lavages. Sialic acid measurements were performed without (C) or with (D) mild acid hydrolysis to measure free and total sialic acid, respectively, in lavages from mice infected with influenza for the indicated number of days (dpi). Data are represented as mean +/− SD. ND = not detected (below the limit of detection, indicated with a dotted line). n.s. = not significant, ** = p < 0.01. Experiments were performed at least twice, with 3-14 mice per group. See also Figure S2.
The amount of sialic acid freely accessible in lavage fluid was below the limit of detection in mock-infected mice but detectable in all mice infected with influenza for 7 days (Fig. 2C). To account for the total sialic acid available in lavage fluid, samples were treated with exogenous neuraminidase (data not shown) or mild acid hydrolysis to release all bound sialic acid from glycoconjugates (Nakano and Ozimek, 1999). Influenza infection was associated with significantly more total sialic acid in the nasopharynx compared to mock infection (Fig. 2D), as measured by the thiobarbituric acid assay and validated by DMB-HPLC detection (Fig. S2).
Pneumococci exploit host glycoconjugates for growth during co-infection
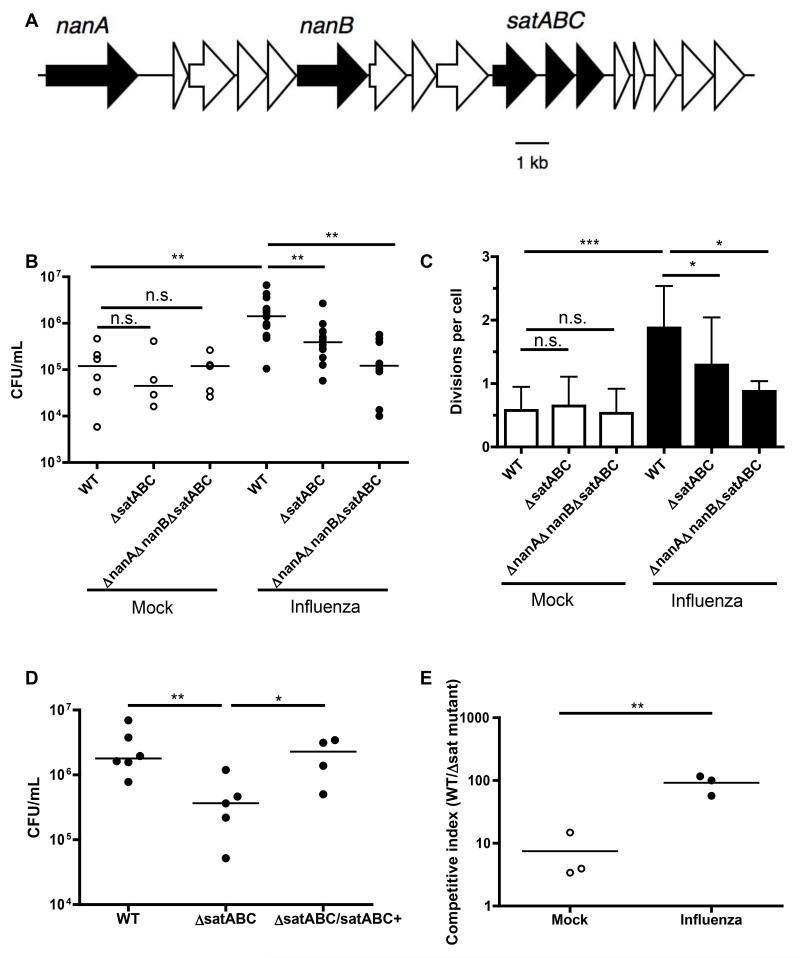
After observing that influenza infection induced a selective increase in sialic acid levels, we tested whether this increase was responsible for the increased pneumococcal growth mediated by influenza. We inoculated mock- and influenza-infected mice with pneumococci that could not catabolize sialic acid (ΔsatABC or ΔnanAΔnanBΔsatABC). The latter mutant also lacks all neuraminidases present in the parental strain, preventing release of sialic acid and thereby limiting access to additional sugars that underlie sialic acid on host glycoconjugates (Burnaugh et al., 2008). The locus containing these 5 genes is shown in Fig. 3A (Aziz et al., 2008). After 8 hrs, we measured bacterial colonization. Neither mutant strain had a defect in early colonization of mice in the absence of influenza. In contrast, pneumococci unable to metabolize sialic acid no longer exhibited increased colonization in the presence of influenza (Fig. 3B).
Figure 3. Pneumococci exploit host glycoconjugates for growth during co-infection.
(A) The genetic locus in strain P1121 containing the neuraminidase (nanA, nanB) and sialic acid transport (satABC) genes is displayed. (B) Seven days after mock (open symbols) or influenza infection (closed symbols), mice were inoculated with CFSE-labeled pneumococci of the indicated genotypes for 8 hrs. Nasal lavages were obtained and plated for quantitative culture. (C) Lavages were fixed, stained for pneumococcal capsule, and flow cytometry performed to measure bacterial replication (division index). (D) Mice were infected with influenza for 7 days, followed by 24 hrs of colonization with the indicated bacterial strains. Nasal lavages were plated for quantitative culture. (E) Seven days after mock or influenza infection, mice were given a mixed inoculum of sialic acid-catabolizing and ΔsatABC pneumococci. One day later, nasal lavages were obtained and plated for quantitative culture. Colonies from the plated inoculum and the lavages were patched onto antibiotic-containing media to determine the relative advantage in vivo of sialic acid catabolism and calculate competitive indices. CI > 1 indicates outcompetition of the streptomycin-resistant ΔsatABC pneumococci, and was defined as the ratio of WT/mutant bacteria in the output (nasal lavage), divided by the ratio of WT/mutant bacteria in the input (inoculum). Data in C are represented as mean +/− SD. Horizontal lines indicate median values. n.s. = not significant, * = p < 0.05, ** = p < 0.01, *** = p < 0.001. Experiments were performed at least twice, with 3-15 mice per group.
Next, we applied the CFSE assay to measure bacterial growth, and determined that the number of divisions per cell was lower with both sialic acid mutant strains compared to WT in the presence, but not absence, of influenza (Fig. 3C). Genetic correction of the mutation in sialic acid utilization (ΔsatABC/satABC+) restored increased bacterial load in the presence of influenza to WT levels (Fig. 3D). To test whether influenza could promote the outgrowth of sialic acid-catabolizing bacteria, mock- and influenza-infected mice were challenged with a mixed inoculum of sialic acid-catabolizing and ΔsatABC mutant pneumococci. One day later, the proportion of sialic-acid catabolizing bacteria colonizing the nasopharynx had increased 10-fold in the presence of influenza (Fig. 3E).
Sialylated airway mucins are required for influenza-induced pneumococcal growth
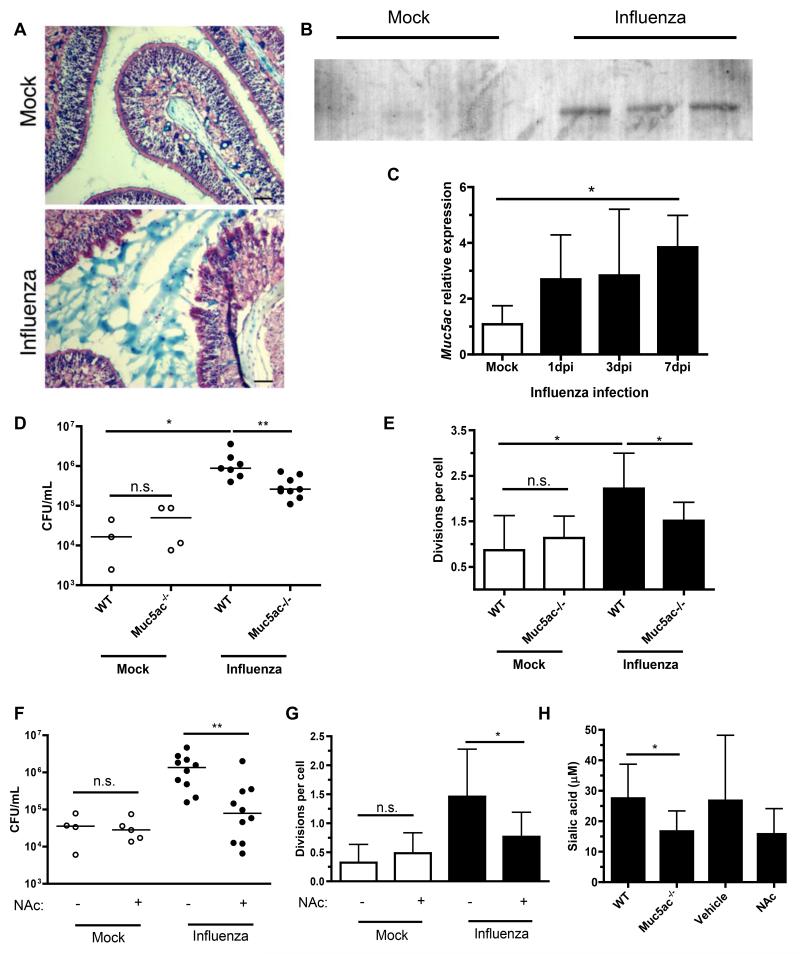
Since pneumococci can grow on mucin glycoproteins as a sole carbon source in vitro, (Yesilkaya et al., 2008) we assessed the availability of mucus, as seen by alcian blue staining for acidic mucopolysaccharides, in the URT. Influenza increased the amount of mucus present in the nasopharyngeal lumen (Fig. 4A). We next examined expression of Muc5ac, the major airway mucin secreted during inflammation and one that is highly decorated by sialic acid residues, (Koeppen et al., 2013; Rose and Voynow, 2006). Influenza induced secretion of Muc5ac onto the mucosal surface during influenza infection (Fig. 4B). This was accompanied by increased Muc5ac transcription in the URT that was maximal at 7 dpi (Fig. 4C). These results correlated with the availability of total sialic acid, which was also maximal at 7 dpi (Fig. 2D). To test whether mucins were a major source of sialic acid during influenza infection, we first examined Muc5ac−/− mice. There was no defect in pneumococcal colonization or growth in Muc5ac−/− mice in the absence of influenza, but the influenza-mediated increases in bacterial load (Fig. 4D) and replication (Fig. 4E) were attenuated in mice that lack this airway mucin. These decreases in growth and bacterial density in Muc5ac−/− mice were not due to a less robust influenza infection, as determined by viral titers (data not shown). To further assess the role of mucins in pneumococcal replication during co-infection, we intranasally treated mock- and influenza-infected WT mice with the mucolytic agent N-acetylcysteine daily until pneumococcal challenge. Mucolytic treatment reduced bacterial load (Fig. 4F) and growth (Fig. 4G), but only in influenza-infected mice. Both genetic deletion and mucolytic treatment reduced total sialic acid in the influenza-infected nasopharynx, confirming that sialylated mucins are a host-derived source of sialic acid pneumococci exploit during co-infection (Fig. 4H).
Figure 4. Sialylated airway mucins are required for influenza-induced pneumococcal growth.
(A) Mouse heads were obtained after 7 days of mock or influenza infection, fixed, decalcified and sectioned. URT sections were stained with alcian blue and nuclear fast red. Scale bars, 50 μm. (B) Nasal lavages were obtained from mice infected with influenza or PBS for 7 days, then analyzed by Western blot for the presence of Muc5ac. (C) Nasal lavages with RLT RNA lysis buffer were obtained from mice influenza or mock-infected for 7 days. qRT-PCR was performed, and relative expression of Muc5ac measured. (D) Mice of indicated genotype were mock or influenza infected for 7 days, followed by inoculation with CFSE-labeled pneumococci for 8 hrs. Nasal lavages were obtained and plated for quantitative culture. (E) Lavages were also fixed, stained for pneumococcal capsule, and flow cytometry performed to measure bacterial replication (division index). (F-G) Wildtype mice were infected with influenza or mock, followed by daily treatment for 7 days with vehicle (PBS) or 0.5 M N-acetylcysteine (NAc). CFSE-labeled pneumococci were inoculated for 8 hrs, then nasal lavages obtained and used for quantitative culture (F) and flow cytometric analysis of cell division (G). (H) Total sialic acid content was measured by thiobarbituric acid assay on samples from D-G. Data are represented as mean +/− SD. Horizontal lines indicate median values. * = p < 0.05, ** = p < 0.01. Experiments were performed at least twice, with 3-13 mice per group.
Influenza and bacterial neuraminidases desialylate host substrates
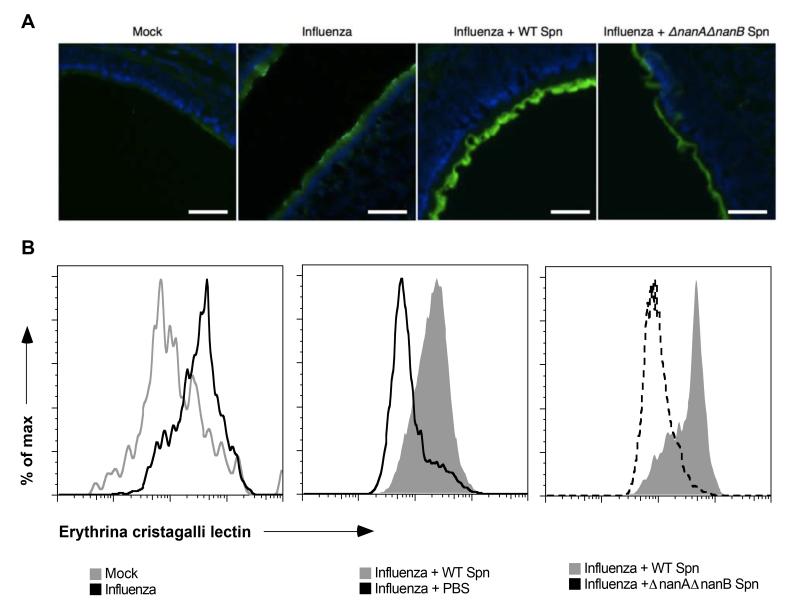
We next examined whether viral and bacterial neuraminidases released sialic acid from host sources for pneumococci to use for growth. We qualitatively measured sialic acid release from the epithelial surface by fluorescence microscopy using a fluorescently-labeled lectin from Erythrina cristagalli, which binds to galactose residues that are exposed when sialic acid is cleaved (Chang et al., 2012). After 7 days of influenza or mock infection, URT sections from mouse heads were stained with the E. cristagalli lectin. Influenza infection promoted increased desialylation of the mucosal surface in the nasopharynx. Bacterial neuraminidase desialylated host cells further, as there was additional lectin binding in samples from mice given pneumococci for 24 hrs after 7 days of influenza. This effect was specific to the pneumococcal neuraminidases, as the epithelial surface in influenza-infected mice colonized with neuraminidase-deficient (ΔnanAΔnanB) pneumococci had less lectin binding, and therefore less desialylation, than did those colonized with WT pneumococci (Fig. 5A). As a quantitative measure of desialylation, we used flow cytometry to quantify desialylation on infiltrating leukocytes (>80% neutrophils), which are abundant in the influenza-infected airway and can be easily obtained by nasal lavage (data not shown). This method confirmed the pattern of desialylation observed with fluorescence microscopy (Fig. 5B). These findings indicated that both viral and bacterial neuraminidases contribute to the desialylation in vivo of host substrates during co-infection.
Figure 5. Influenza and bacterial neuraminidases desialylate host substrates.
(A-B) Mouse heads (A) and nasal lavages (B) were obtained from separate mice after 7 days of influenza or mock infection, followed by 24 hrs of colonization with WT or ΔnanAΔnanB pneumococci, or PBS. Heads were fixed, decalcified and sectioned. URT sections were stained with DAPI and FITC-labeled lectin from E. cristagalli that binds galactose residues exposed when sialic acid is removed. All images were taken with the same optical settings. Scale bars, 50 μm. Lavages were stained with FITC-labeled E. cristagalli lectin. Flow cytometry was performed by gating on neutrophils and comparing desialylation. Experiments were performed at least twice, with 3-10 mice per group.
Sialic acid catabolism contributes to pneumococcal colonization in the absence of influenza
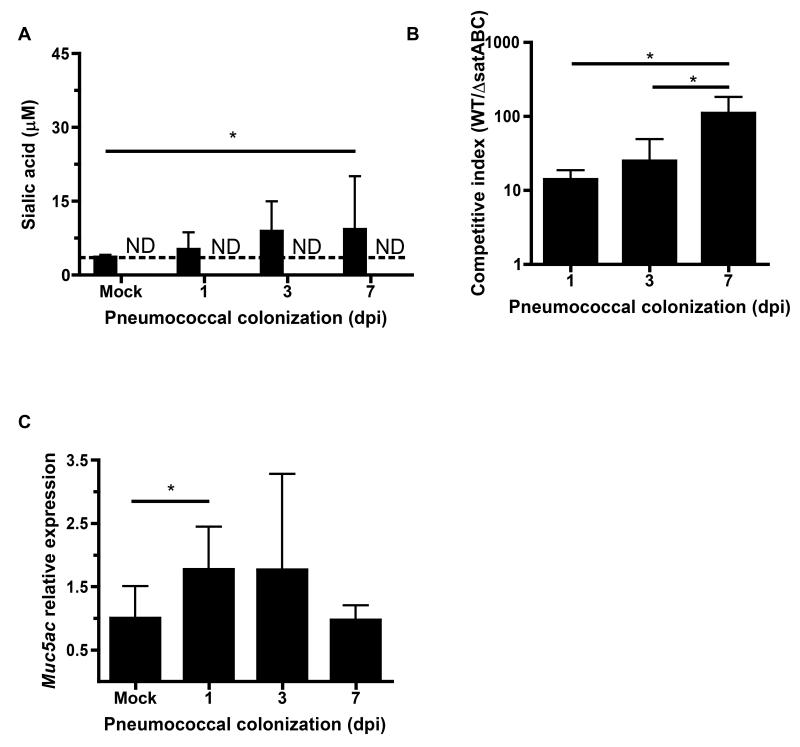
To assess the role of sialic acid catabolism during pneumococcal colonization, we measured sialic acid in nasal lavages from mice inoculated with pneumococci in the absence of influenza infection. Total sialic acid increased over the duration of colonization, though no free sialic acid was detectable (Fig. 6A). Consistent with the slow increase in sialic acid during colonization, we observed that WT pneumococci outcompeted ΔsatABC, and that this outcompetition increased over time, but was delayed compared to influenza co-infection (Fig. 6B). As with influenza infection, pneumococcal colonization induced URT transcription of the sialylated mucin Muc5ac (Fig. 6C).
Figure 6. Sialic acid catabolism contributes to pneumococcal colonization in the absence of influenza.
(A) Mice were colonized with pneumococci for the indicated number of days. Sialic acid concentrations in lavages were measured by thiobarbituric acid assay, with or without acid hydrolysis to measure total (solid bars) and free sialic acid (open bars), respectively. (B) WT and ΔsatABC pneumococci were competed in vivo by colonizing naive mice with mixed inocula. Nasal lavages were plated for quantitative culture and colonies from the inocula and lavages patched onto antibiotic-selective media to determine the relative advantage of sialic acid catabolism in vivo and calculate competitive indices. CI > 1 indicates WT outcompeted ΔsatABC pneumococci. (C) Nasal lavages were obtained with RLT RNA lysis buffer from mice colonized with pneumococci for the indicated number of days. qRT-PCR was performed to measure relative expression of Muc5ac. Data are represented as mean +/− SD. ND = not detected (below the limit of detection, indicated with a dotted line), * = p < 0.05. Experiments were performed at least twice, with 3-20 mice per group.
Nutrient-poor conditions on the human mucosal surface favor growth of sialic acid catabolizing pneumococci
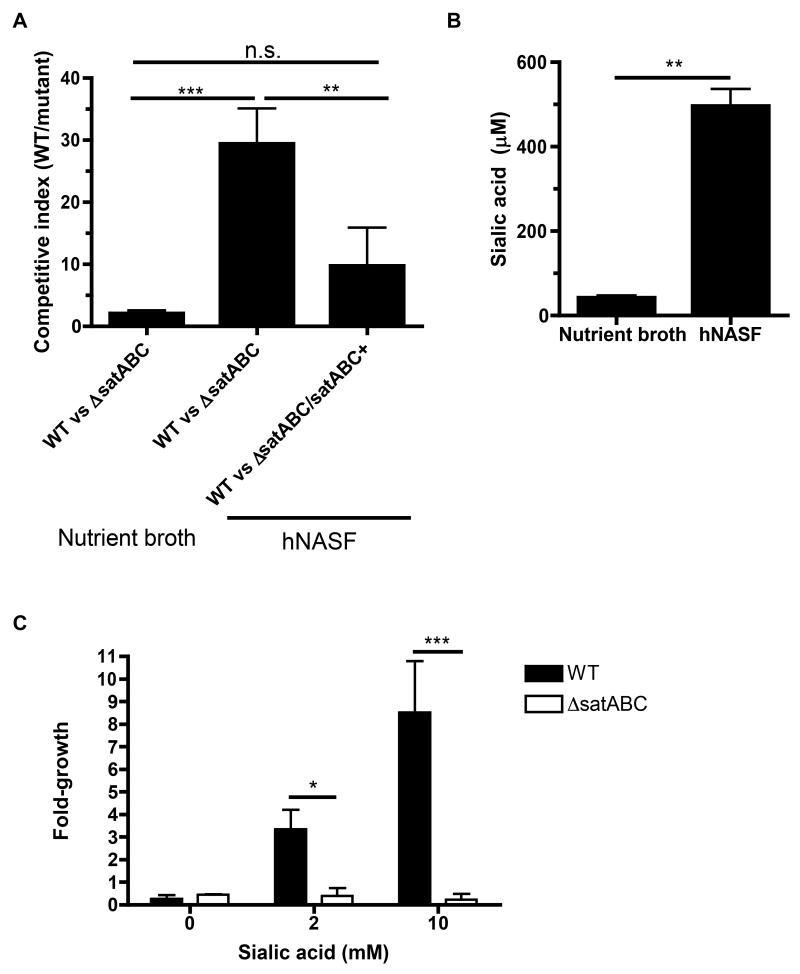
To extend our findings on sialic acid catabolism to human samples, we competed bacterial strains for growth in vitro in human nasal airway surface fluid (hNASF), the natural environment in which pneumococci replicate. There was little competition between WT and ΔsatABC pneumococci when grown in nutrient broth that contains glucose, (Fig. 7A) an in vitro preferred carbon source not found in airway secretions (Philips et al., 2003). This competitive advantage due to sialic acid catabolism increased 15-fold when pneumococci were grown in nutrient-limited hNASF. In contrast, there was no significant advantage for WT over the genetically corrected ΔsatABC/satABC+ strain in growth in hNASF (Fig. 7A). This effect correlated with the higher levels of sialic acid found in hNASF compared to nutrient broth (Fig. 7B). These sialic acid levels could not be directly compared to those found in murine nasal lavages, as the latter are dilutions of the epithelial lining fluid. The total volume of upper airway surface fluid in an adult mouse has been estimated as 3 μL (Asahi et al., 2002). Assuming the 200 μL nasal lavage contained the entire surface lining fluid, the lavages are at least a 1:67 dilution of that fluid. The ~30 μM sialic acid concentration measured, therefore, would correspond to >2 mM sialic acid on the mucosal surface during co-infection, a concentration sufficient to promote growth in vitro as a sole carbon source (Fig. 7C).
Figure 7. Nutrient-poor conditions on the human mucosal surface favor growth of sialic acid catabolizing pneumococci.
(A) Bacteria of the indicated genotypes were grown for 8 hrs in mixed inocula in tryptic soy nutrient broth, or in nutrient-limited human nasal airway surface fluid (hNASF). Aliquots were plated at the beginning and end of the growth period, and colonies patched onto antibiotic-selective media to determine the relative advantage of sialic acid catabolism in vivo and calculate competitive indices. CI > 1 indicates WT outcompeted the indicated mutant strain. (B) Total sialic acid in the growth media used in A was measured by thiobarbituric acid assay after acid hydrolysis. (C) WT (solid bars) and ΔsatABC (open bars) pneumococci were grown for 24 hrs in chemically defined medium with the indicated concentrations of sialic acid as the sole carbon source. Data show fold-growth compared to inoculum. Data are represented as mean +/− SD. n.s. = not significant, * = p < 0.05, ** = p < 0.01, *** = p < 0.001. Experiments were performed at least twice.
Discussion
Influenza has long been associated with secondary bacterial pneumonia, but how this viral infection primes the host for the rapid development of high bacterial loads has not been fully understood. Here, we demonstrated that influenza infection predisposed mice to increased pneumococcal colonization in the nasopharynx, and that faster bacterial replication contributed to this effect. This influenza-mediated growth led to increased aspiration into the lower respiratory tract, and was dependent on pneumococcal catabolism of the host sugar sialic acid and host expression of sialylated airway mucins, with both viral and bacterial neuraminidases contributing to the desialylation of host cells in vivo. Sialic acid utilization was important even during pneumococcal colonization in the absence of influenza, but to a lesser extent, consistent with the lower levels of sialic acid present without influenza infection.
In this study, we challenged mice with pneumococci after 7 days of influenza infection in the URT. At 7 days postinfection, influenza viral titers are declining from a peak reached at 3 to 5 dpi, the same time at which sialic acid levels and sialylated mucin expression in the nasopharynx are increasing. Previous animal studies have indicated this timepoint correlates with the peak of susceptibility to pneumonia when pneumococci are introduced directly into the lower respiratory tract (McCullers and Rehg, 2002; Sun and Metzger, 2008). The window of susceptibility to pneumococcal pneumonia is also 7 to 10 days following influenza infection in humans (Shrestha et al., 2013).
Correlations between URT pneumococcal colonization density and pneumonia have been observed in human patients, but it has not been clear whether higher bacterial burdens in the upper airway precede pneumonia or are an effect of lower airway disease(Albrich et al., 2012). We found that influenza, by promoting higher bacterial load in the nasopharynx, led to increased aspiration of pneumococci into the lower respiratory tract, experimentally demonstrating that higher levels of colonization can increase a key step in the early pathogenesis of pneumonia.
Previous studies of the interaction between influenza and pneumococci have focused on epithelial damage or immune dysfunction induced by the viral infection, promoting increased bacterial adherence or decreased bacterial clearance, respectively (McCullers, 2006; Metzger and Sun, 2013). We found no difference in initial adherence of the bacterial inoculum after influenza infection, and further found no increases in total protein or glycan content, suggesting this infection did not cause extensive tissue damage. Using the dilution of CFSE dye, we demonstrated a role for bacterial replication in mediating increased colonization due to influenza. Previous studies of the interaction between influenza and bacteria have measured bacterial numbers, the sum of bacterial growth and clearance. We measured both bacterial numbers and bacterial growth, allowing us to separate the contribution of bacterial growth from that of immune evasion. Our data are consistent with previous studies demonstrating the importance of viral neuraminidase for the interaction of influenza and pneumococci (Peltola and McCullers, 2004; Peltola et al., 2005). Those studies emphasized neuraminidase removing sialic acid to expose underlying receptors for pneumococci, but the same enzymatic activity would also release free sialic acid into the nasopharynx for pneumococci to utilize.
Our work has implications for the therapeutic use of neuraminidase inhibitors. Most clinical studies of neuraminidase inhibitor efficacy have focused on early treatment, (Hernán and Lipsitch, 2011; Hsu et al., 2012) but experiments in mice demonstrated survival benefits to neuraminidase inhibitor treatment during co-infection even at later timepoints when there was no effect on viral replication (McCullers, 2004). Our results may provide a mechanistic understanding of this effect, as inhibiting viral neuraminidase may limit sialic acid release even after the peak in viral replication. Additional studies on the effect of late neuraminidase inhibitor treatment on complications related to bacterial density, such as pneumonia, could be warranted. Bacterial neuraminidases, including pneumococcal NanA and NanB, are not inhibited by clinically-used neuraminidase inhibitors at concentrations reached in vivo (Nishikawa et al., 2012). Broader neuraminidase inhibitors that can prevent bacterial acquisition of sialic acid and underlying sugars may be an important, unexplored therapeutic strategy.
We observed desialylation of the mucosal surface and leukocytes during influenza infection, exacerbated by co-infection with neuraminidase-expressing pneumococci. We found that transcription of Muc5ac was upregulated in the nasopharynx during influenza infection as part of the inflammatory response, consistent with a previous report in the lungs of influenza-infected mice (Barbier et al., 2012). We also found that mucus and Muc5ac secretion increased during influenza infection, and that reducing mucins by genetic knockout of Muc5ac, or solubilizing mucus by N-acetylcysteine treatment decreased influenza-induced pneumococcal growth. Mucins are 50 to 90% glycan by mass and are heavily sialylated, particularly at the terminal, most accessible ends of sugar chains (Angata and Varki, 2002; Rose and Voynow, 2006). The epithelial surface is also heavily sialylated, and could serve as an additional source of sialic acid during influenza infection (Lewis and Lewis, 2012). The commensal flora of the nasopharynx could also provide sialic acid (Shakhnovich et al., 2002).
Sialic acid is both necessary and sufficient for pneumococcal colonization in mice (Marion et al., 2011; Trappetti et al., 2009). We noted a competitive disadvantage in vivo that increased over 7 days of colonization for pneumococci that lack the sialic acid transporter SatABC, consistent with previous work showing a defect in colonization at 5 days postinoculation (Marion et al., 2011). Increased nasal secretions, reflecting greater sialylated mucin production, have been associated with higher density of pneumococcal colonization in children (Rodrigues et al., 2013). Exogenous sialic acid, but not other amino sugars, has been shown to increase colonization density in the airway (Trappetti et al., 2009). In vitro, sialic acid can promote pneumococcal growth as a sole carbon source (Marion et al., 2011). The amount of nasopharyngeal sialic acid we measured during influenza infection is likely to be an underestimate, as it does not include sialic acid still bound to epithelial cells that would not be removed by nasal lavage. Sialic acid release by viral and bacterial neuraminidases also exposes underlying sugars, such as galactose and N-acetylglucosamine, that can be liberated by the sequential activity of pneumococcal exoglycosidases to serve as additional carbon sources (Burnaugh et al., 2008; King et al., 2006). This effect could explain why pneumococci that could not cleave or catabolize sialic acid (ΔnanAΔnanBΔsatABC) had a stronger defect in influenza-mediated growth compared to those that could not catabolize sialic acid (ΔsatABC). The viral neuraminidase could also complement mutations in the pneumococcal neuraminidases. Free sialic acid only accumulated enough to be detected after 7 days of influenza infection, even though viral titers peaked earlier. This timepoint, however, represented the peak of sialylated mucin availability. Our results suggest that the combination of increased mucin availability and neuraminidase activity is required for providing sufficient amounts of sialic acid.
Obtaining a source of carbon is necessary for bacterial growth in any environment, but how this occurs during colonization of the nutrient-poor mucosal surface has been unclear (Buckwalter and King, 2012). Free sugar is particularly limited in the nasopharynx, the pneumococcal niche, unlike in the intestines or oral cavity (Philips et al., 2003). In the gut, sialic acid released by the sialidase of one bacterial species can promote sialic acid catabolism-dependent growth of another (Ng et al., 2013). Carbon acquisition is especially important for the pneumococcus, which devotes more of its genome to sugar transporters than any other sequenced bacterium (Tettelin et al., 2001). This diversity implies that pneumococci may encounter different host sugars in abundance in distinct microenvironments; infection with neuraminidase-expressing viruses such as influenza could provide one such microenvironment. Pneumococci are not the only bacteria that can use sialic acid. Interestingly, the other prominent pathogens in secondary bacterial pneumonia following influenza, Staphylococcus aureus and Haemophilus influenzae, can also catabolize this host sugar (Olson et al., 2013; Vimr et al., 2000). Sialic acid utilization could also contribute to other common bacterial complications following influenza infection, such as acute otitis media (McCullers, 2006).
Experimental Procedures
Mice
6-8 week old C57Bl/6 mice were obtained from Jackson Laboratory. Muc5ac−/− mice on a C57Bl/6 background were previously described (Hasnain et al., 2011). Procedures were carried out according to an animal protocol approved by the University of Pennsylvania IACUC.
Influenza infection
Mice were intranasally inoculated with 2 × 104 TCID50 of influenza A virus, strain HKx31 (H3N2 from A/Hong Kong/1/1968 with the backbone of PR8 virus), diluted into 10 μL PBS.
Bacterial strains and colonization
Pneumococcal strains included TIGR4 (Tettelin et al., 2001) and P1121, (McCool et al., 2002) clinical isolates of capsule types 4 and 23F, respectively. Mutants of P1121 in the sialic acid transporter (satABC) were previously made using the Janus cassette to create unmarked and correct mutations (Marion et al., 2011). Mutants in the neuraminidase genes nanA and nanB were constructed as described previously by introducing antibiotic resistance cassettes (King et al., 2004). Pneumococci were grown in tryptic soy (TS) nutrient broth at 37° C until mid-log phase. For mouse colonization, 107 CFU bacteria were inoculated intranasally in a volume of 10 μL. Mice were sacrificed, and nasal lavages obtained with 200 μL PBS. Lavages were plated on TS agar supplemented with catalase (5,000 U/plate) (Worthington Biochemical) and 5 μg/mL neomycin for quantitative culture in 5% CO2. TS agar was supplemented with 200 μg/mL streptomycin to distinguish ΔsatABC pneumococci from WT. For bronchoalveolar lavage, 1 mL PBS was instilled into the lungs intratracheally, withdrawn with a syringe, and used for quantitative culture.
CFSE staining
Bacteria were resuspended in 1 mL PBS with 1% catalase and 10 μM carboxy-fluorescein diacetate succinimidyl diester (Molecular Probes). Reactions were incubated at 37° C for 25 min, then washed 3 times in PBS. Mice were inoculated with CFSE-labeled pneumococci. After 8 hrs, mice were sacrificed and nasal lavages obtained, then prepared for flow cytometry.
Flow Cytometry
For the in vivo CFSE assay, nasal lavages were obtained from mice inoculated with CFSE-labeled pneumococci. Samples were fixed and stained with typing serum specific to the capsule-type used (Statens Serum Institut), followed by AF647-labeled secondary antibody to rabbit IgG, then analyzed by flow cytometry. For measurement of desialylation, nasal lavages were stained with FITC-labeled lectin from Erythrina cristagalli (Vector Labs), and antibodies to identify neutrophils: anti-Ly6G (clone 1A8) and anti-CD11b. Flow cytometry was conducted using FACS Calibur and FACS Canto instruments (Becton Dickinson), and analyzed using FlowJo software (Tree Star).
Protein, reducing sugar and sialic acid measurements
Protein was measured by bicinchonic acid assay (Pierce). Reducing sugar was measured by tetrazolium blue assay (Jue and Lipke, 1985). Samples were diluted 1:100 in 0.1% tetrazolium blue, 0.05 M NaOH and 0.5 M potassium sodium tartrate. Samples were boiled for 10 min, and absorbance measured at 655 nm. Sialic acid was measured by thiobarbituric acid assay (Nakano and Ozimek, 1999; Skoza and Mohos, 1976). To measure total sialic acid, samples were first incubated with 0.1 N H2SO4 for 30 min at 80° C. All samples were then incubated with 25 μM periodic acid/62.5 mM H2SO4 for 30 min at 37° C, followed by boiling in 2% sodium arsenite/0.5 M HCl and 6% thiobarbituric acid for 10 min. Absorbance was measured at 550 nm and compared to a standard curve to calculate concentrations in lavage fluid. The specificity of this assay was confirmed by using exogenous neuraminidase instead of acid hydrolysis to enzymatically release sialic acid.
qRT-PCR
RNA was harvested from the URT epithelium by lavage with RLT buffer (Qiagen) with 1:100 β-mercaptoethanol. RNA was isolated with the RNeasy kit (Qiagen), and cDNA reverse transcribed with the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems). Reactions were carried out with Sybr Green reagents (Applied Biosystems), with 10 ng cDNA and 0.5 μM primers. Comparisons were made between conditions by the ΔΔCT method. Primer sequences are in Supplemental Experimental Procedures.
Western blot
Proteins in nasal lavages were separated by SDS-PAGE on a 10% Tris gel (Bio-Rad) and transferred onto PVDF membrane. Muc5ac was detected by polyclonal antibody (Santa Cruz Biotechnology) and rabbit anti-goat secondary antibody conjugated to alkaline phosphatase (Sigma).
Microscopy
Tissue sections of the URT were obtained as previously described (Nelson et al., 2007). Staining and microscopy was performed as described, modified by blocking in 1% gelatin in PBS when using E. cristagalli lectin.
Growth in human nasal airway surface fluid
Human nasal airway surface fluid was isolated as previously described (Gould and Weiser, 2001). Pneumococci were grown to log-phase in TS nutrient broth and diluted to a concentration of 104 CFU/mL in both hNASF and TS. Reactions were supplemented 1:100 with catalase and incubated at 37° C in 5% CO2 for 8 hrs. Samples were plated for quantitative culture at the start and end of the growth period, and colonies patched onto antibiotic selective media to calculate competitive indices. Pneumococci were grown in chemically defined medium as previously described, (Kloosterman et al., 2006) with sialic acid in place of glucose.
Statistical analysis
Prism (Graphpad) was used for statistical analysis. Comparisons were made by Mann-Whitney U-test for colonization (CFU) data, and by Student’s t-test for all other data. For multiple comparisons, we used Kruskal-Wallis with Dunn’s posttest or 1-way ANOVA with Newman-Keuls posttest, respectively.
Supplementary Material
Highlights.
-
-
Prior influenza infection increases pneumococcal colonization, replication and aspiration
-
-
Pneumococcal growth during co-infection depends on host-derived sialic acid utilization
-
-
Sialylated airway mucin Muc5ac is required for influenza-mediated pneumococcal growth
-
-
Viral and bacterial neuraminidases release sialic acid from host cells in vivo
Acknowledgments
We thank Jan Erikson (Wistar Institute), Samantha King (Nationwide Children’s Hospital) and Christopher Evans (University of Colorado Denver) for providing viral and bacterial strains and Muc5ac−/− mice, respectively. Analysis was performed by the UCSD Glycotechnology Core. This work was supported by NIH grants AI038446, AI078538 and AI105168 to JNW, and HL119030 to SJS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albrich WC, Madhi SA, Adrian PV, van Niekerk N, Mareletsi T, Cutland C, Wong M, Khoosal M, Karstaedt A, Zhao P, et al. Use of a Rapid Test of Pneumococcal Colonization Density to Diagnose Pneumococcal Pneumonia. Clin. Infect. Dis. 2012;54:601–609. doi: 10.1093/cid/cir859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angata T, Varki A. Chemical diversity in the sialic acids and related alpha-keto acids: an evolutionary perspective. Chem. Rev. 2002;102:439–469. doi: 10.1021/cr000407m. [DOI] [PubMed] [Google Scholar]
- Asahi Y, Yoshikawa T, Watanabe I, Iwasaki T, Hasegawa H, Sato Y, Shimada S-I, Nanno M, Matsuoka Y, Ohwaki M, et al. Protection against influenza virus infection in polymeric Ig receptor knockout mice immunized intranasally with adjuvant-combined vaccines. J. Immunol. 2002;168:2930–2938. doi: 10.4049/jimmunol.168.6.2930. [DOI] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier D, Garcia-Verdugo I, Pothlichet J, Khazen R, Descamps D, Rousseau K, Thornton D, Si-Tahar M, Touqui L, Chignard M, et al. Influenza A induces the major secreted airway mucin MUC5AC in a protease-EGFR-ERK-Sp1 dependent pathway. Am J Respir Cell Mol Biol. 2012;47:149–157. doi: 10.1165/rcmb.2011-0405OC. [DOI] [PubMed] [Google Scholar]
- Bogaert D, De Groot R, Hermans PWM. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect Dis. 2004;4:144–154. doi: 10.1016/S1473-3099(04)00938-7. [DOI] [PubMed] [Google Scholar]
- Buckwalter CM, King SJ. Pneumococcal carbohydrate transport: food for thought. Trends Microbiol. 2012;20:517–522. doi: 10.1016/j.tim.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnaugh AM, Frantz LJ, King SJ. Growth of Streptococcus pneumoniae on human glycoconjugates is dependent upon the sequential activity of bacterial exoglycosidases. J. Bacteriol. 2008;190:221–230. doi: 10.1128/JB.01251-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y-C, Uchiyama S, Varki A, Nizet V. Leukocyte inflammatory responses provoked by pneumococcal sialidase. MBio. 2012;3 doi: 10.1128/mBio.00220-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould JM, Weiser JN. Expression of C-reactive protein in the human respiratory tract. Infect. Immun. 2001;69:1747–1754. doi: 10.1128/IAI.69.3.1747-1754.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grijalva CG, Griffin MR, Edwards KM, Williams JV, Gil AI, Verastegui H, Hartinger SM, Vidal JE, Klugman KP, Lanata CF. The role of influenza and parainfluenza infections in nasopharyngeal pneumococcal acquisition among young children. Clin. Infect. Dis. 2014 doi: 10.1093/cid/ciu148. doi: 10.1093/cid/ciu148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasnain SZ, Evans CM, Roy M, Gallagher AL, Kindrachuk KN, Barron L, Dickey BF, Wilson MS, Wynn TA, Grencis RK, et al. Muc5ac: a critical component mediating the rejection of enteric nematodes. J. Exp. Med. 2011;208:893–900. doi: 10.1084/jem.20102057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernán MA, Lipsitch M. Oseltamivir and risk of lower respiratory tract complications in patients with flu symptoms: a meta-analysis of eleven randomized clinical trials. Clin. Infect. Dis. 2011;53:277–279. doi: 10.1093/cid/cir400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J, Santesso N, Mustafa R, Brozek J, Chen YL, Hopkins JP, Cheung A, Hovhannisyan G, Ivanova L, Flottorp SA, et al. Antivirals for Treatment of Influenza: A Systematic Review and Meta-analysis of Observational Studies. Ann. Intern. Med. 2012;156:512–524. doi: 10.7326/0003-4819-156-7-201204030-00411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jue CK, Lipke PN. Determination of reducing sugars in the nanomole range with tetrazolium blue. J. Biochem. Biophys. Methods. 1985;11:109–115. doi: 10.1016/0165-022x(85)90046-6. [DOI] [PubMed] [Google Scholar]
- Kim PE, Musher DM, Glezen WP, Rodriguez-Barradas MC, Nahm WK, Wright CE. Association of invasive pneumococcal disease with season, atmospheric conditions, air pollution, and the isolation of respiratory viruses. Clin. Infect. Dis. 1996;22:100–106. doi: 10.1093/clinids/22.1.100. [DOI] [PubMed] [Google Scholar]
- King SJ, Hippe KR, Weiser JN. Deglycosylation of human glycoconjugates by the sequential activities of exoglycosidases expressed by Streptococcus pneumoniae. Mol. Microbiol. 2006;59:961–974. doi: 10.1111/j.1365-2958.2005.04984.x. [DOI] [PubMed] [Google Scholar]
- King SJ, Hippe KR, Gould JM, Bae D, Peterson S, Cline RT, Fasching C, Janoff EN, Weiser JN. Phase variable desialylation of host proteins that bind to Streptococcus pneumoniae in vivo and protect the airway. Mol. Microbiol. 2004;54:159–171. doi: 10.1111/j.1365-2958.2004.04252.x. [DOI] [PubMed] [Google Scholar]
- Kloosterman TG, Bijlsma JJE, Kok J, Kuipers OP. To have neighbour’s fare: extending the molecular toolbox for Streptococcus pneumoniae. Microbiology. 2006;152:351–359. doi: 10.1099/mic.0.28521-0. [DOI] [PubMed] [Google Scholar]
- Klugman KP, Chien Y-W, Madhi SA. Pneumococcal pneumonia and influenza: a deadly combination. Vaccine. 2009;27(Suppl 3):C9–C14. doi: 10.1016/j.vaccine.2009.06.007. [DOI] [PubMed] [Google Scholar]
- Koeppen M, McNamee EN, Brodsky KS, Aherne CM, Faigle M, Downey GP, Colgan SP, Evans CM, Schwartz DA, Eltzschig HK. Detrimental role of the airway mucin Muc5ac during ventilator-induced lung injury. Mucosal Immunol. 2013;6:762–775. doi: 10.1038/mi.2012.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis AL, Lewis WG. Host sialoglycans and bacterial sialidases: a mucosal perspective. Cell. Microbiol. 2012;14:1174–1182. doi: 10.1111/j.1462-5822.2012.01807.x. [DOI] [PubMed] [Google Scholar]
- Marion C, Burnaugh AM, Woodiga SA, King SJ. Sialic acid transport contributes to pneumococcal colonization. Infect. Immun. 2011;79:1262–1269. doi: 10.1128/IAI.00832-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk H-D. Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc Natl Acad Sci USA. 2004;101:4620–4624. doi: 10.1073/pnas.0308001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCool TL, Cate TR, Moy G, Weiser JN. The immune response to pneumococcal proteins during experimental human carriage. J. Exp. Med. 2002;195:359–365. doi: 10.1084/jem.20011576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullers JA. Effect of antiviral treatment on the outcome of secondary bacterial pneumonia after influenza. J. Infect. Dis. 2004;190:519–526. doi: 10.1086/421525. [DOI] [PubMed] [Google Scholar]
- McCullers JA. Insights into the interaction between influenza virus and pneumococcus. Clin. Microbiol. Rev. 2006;19:571–582. doi: 10.1128/CMR.00058-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullers JA, Rehg JE. Lethal synergism between influenza virus and Streptococcus pneumoniae: characterization of a mouse model and the role of platelet-activating factor receptor. J. Infect. Dis. 2002;186:341–350. doi: 10.1086/341462. [DOI] [PubMed] [Google Scholar]
- Metzger DW, Sun K. Immune dysfunction and bacterial coinfections following influenza. J. Immunol. 2013;191:2047–2052. doi: 10.4049/jimmunol.1301152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Davis KM, Weiser JN. Synergistic stimulation of type I interferons during influenza virus coinfection promotes Streptococcus pneumoniae colonization in mice. J. Clin. Invest. 2011;121:3657–3665. doi: 10.1172/JCI57762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Ozimek L. Determination of sialic acid by the thiobarbituric acid reaction in sweet whey and its fractions. J. Agric. Food Chem. 1999;47:2613–2616. doi: 10.1021/jf981077y. [DOI] [PubMed] [Google Scholar]
- Nelson AL, Roche AM, Gould JM, Chim K, Ratner AJ, Weiser JN. Capsule enhances pneumococcal colonization by limiting mucus-mediated clearance. Infect. Immun. 2007;75:83–90. doi: 10.1128/IAI.01475-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa T, Shimizu K, Tanaka T, Kuroda K, Takayama T, Yamamoto T, Hanada N, Hamada Y. Bacterial neuraminidase rescues influenza virus replication from inhibition by a neuraminidase inhibitor. PLoS ONE. 2012;7:e45371. doi: 10.1371/journal.pone.0045371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olitsky PK, Gates FL. Experimental studies of the nasopharyngeal secretions from influenza patients: III. Studies of the concurrent infections. J. Exp. Med. 1921;33:373–383. doi: 10.1084/jem.33.3.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson ME, King JM, Yahr TL, Horswill AR. Sialic acid catabolism in Staphylococcus aureus. J. Bacteriol. 2013;195:1779–1788. doi: 10.1128/JB.02294-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parish CR. Fluorescent dyes for lymphocyte migration and proliferation studies. Immunol. Cell Biol. 1999;77:499–508. doi: 10.1046/j.1440-1711.1999.00877.x. [DOI] [PubMed] [Google Scholar]
- Peltola VT, McCullers JA. Respiratory viruses predisposing to bacterial infections: role of neuraminidase. Pediatr. Infect. Dis. J. 2004;23:S87–S97. doi: 10.1097/01.inf.0000108197.81270.35. [DOI] [PubMed] [Google Scholar]
- Peltola VT, Murti KG, McCullers JA. Influenza virus neuraminidase contributes to secondary bacterial pneumonia. J. Infect. Dis. 2005;192:249–257. doi: 10.1086/430954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips BJ, Meguer J-X, Redman J, Baker EH. Factors determining the appearance of glucose in upper and lower respiratory tract secretions. Intensive Care Med. 2003;29:2204–2210. doi: 10.1007/s00134-003-1961-2. [DOI] [PubMed] [Google Scholar]
- Rodrigues F, Foster D, Nicoli E, Trotter C, Vipond B, Muir P, Gonçalves G, Januário L, Finn A. Relationships between rhinitis symptoms, respiratory viral infections and nasopharyngeal colonization with Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus in children attending daycare. Pediatr. Infect. Dis. J. 2013;32:227–232. doi: 10.1097/INF.0b013e31827687fc. [DOI] [PubMed] [Google Scholar]
- Roederer M. Interpretation of cellular proliferation data: avoid the panglossian. Cytometry A. 2011;79:95–101. doi: 10.1002/cyto.a.21010. [DOI] [PubMed] [Google Scholar]
- Rose MC, Voynow JA. Respiratory tract mucin genes and mucin glycoproteins in health and disease. Physiol. Rev. 2006;86:245–278. doi: 10.1152/physrev.00010.2005. [DOI] [PubMed] [Google Scholar]
- Shahangian A, Chow EK, Tian X, Kang JR, Ghaffari A, Liu SY, Belperio JA, Cheng G, Deng JC. Type I IFNs mediate development of postinfluenza bacterial pneumonia in mice. J. Clin. Invest. 2009;119:1910–1920. doi: 10.1172/JCI35412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakhnovich EA, King SJ, Weiser JN. Neuraminidase expressed by Streptococcus pneumoniae desialylates the lipopolysaccharide of Neisseria meningitidis and Haemophilus influenzae: a paradigm for interbacterial competition among pathogens of the human respiratory tract. Infect. Immun. 2002;70:7161–7164. doi: 10.1128/IAI.70.12.7161-7164.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha S, Foxman B, Weinberger DM, Steiner C, Viboud C, Rohani P. Identifying the interaction between influenza and pneumococcal pneumonia using incidence data. Sci Transl Med. 2013;5:191ra84. doi: 10.1126/scitranslmed.3005982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoza L, Mohos S. Stable thiobarbituric acid chromophore with dimethyl sulphoxide. Application to sialic acid assay in analytical de-O-acetylation. Biochem. J. 1976;159:457–462. doi: 10.1042/bj1590457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K, Metzger DW. Inhibition of pulmonary antibacterial defense by interferon-gamma during recovery from influenza infection. Nat. Med. 2008;14:558–564. doi: 10.1038/nm1765. [DOI] [PubMed] [Google Scholar]
- Tettelin H, Nelson KE, Paulsen IT, Eisen JA, Read TD, Peterson S, Heidelberg J, DeBoy RT, Haft DH, Dodson RJ, et al. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science. 2001;293:498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- Trappetti C, Kadioglu A, Carter M, Hayre J, Iannelli F, Pozzi G, Andrew PW, Oggioni MR. Sialic acid: a preventable signal for pneumococcal biofilm formation, colonization, and invasion of the host. J. Infect. Dis. 2009;199:1497–1505. doi: 10.1086/598483. [DOI] [PubMed] [Google Scholar]
- Vimr E, Lichtensteiger C, Steenbergen S. Sialic acid metabolism’s dual function in Haemophilus influenzae. Mol. Microbiol. 2000;36:1113–1123. doi: 10.1046/j.1365-2958.2000.01925.x. [DOI] [PubMed] [Google Scholar]
- Vu HTT, Yoshida LM, Suzuki M, Nguyen HAT, Nguyen CDL, Nguyen ATT, Oishi K, Yamamoto T, Watanabe K, Vu TD. Association between nasopharyngeal load of Streptococcus pneumoniae, viral coinfection, and radiologically confirmed pneumonia in Vietnamese children. Pediatr. Infect. Dis. J. 2011;30:11–18. doi: 10.1097/INF.0b013e3181f111a2. [DOI] [PubMed] [Google Scholar]
- Wadowsky RM, Mietzner SM, Skoner DP, Doyle WJ, Fireman P. Effect of experimental influenza A virus infection on isolation of Streptococcus pneumoniae and other aerobic bacteria from the oropharynges of allergic and nonallergic adult subjects. Infect. Immun. 1995;63:1153–1157. doi: 10.1128/iai.63.4.1153-1157.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yesilkaya H, Manco S, Kadioglu A, Terra VS, Andrew PW. The ability to utilize mucin affects the regulation of virulence gene expression in Streptococcus pneumoniae. FEMS Microbiol. Lett. 2008;278:231–235. doi: 10.1111/j.1574-6968.2007.01003.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.