Abstract
In Drosophila oogenesis, the follicular epithelium that envelops the oocyte is patterned by a small set of inductive signals and gives rise to an elaborate three-dimensional eggshell. Several eggshell structures provide sensitive readouts of the patterning signals, but the formation of these structures is still poorly understood. In other systems, epithelial morphogenesis is guided by the spatial patterning of cell adhesion and cytoskeleton genes. As a step towards developing a comprehensive description of patterning events leading to eggshell morphogenesis, we report the expression of Drosophila cadherins, calcium dependent adhesion molecules that are repeatedly used throughout development. We found that 9/17 of Drosophila cadherins are expressed in the follicular epithelium in dynamic patterns during oogenesis. In late oogenesis, the expression patterns of cadherin genes in the main body follicle cells is summarized using a compact set of simple geometric shapes, reflecting the integration of the EGFR and DPP inductive signals. The multi-layered composite patterning of the cadherins is hypothesized to play a key role in the formation of the eggshell. Of particular note is the complex patterning of the region of the follicular epithelium that gives rise to the dorsal appendages, which are tubular structures that serve as respiratory organs for the developing embryo.
Keywords: Drosophila, cadherin, oogenesis, gene expression, morphogenesis, adhesion, pattern formation, follicle cell, epithelium
1. Results and Discussion
Epithelial morphogenesis is characterized by the sequential execution of a core set of programmed stereotypical cell movements and shape changes, which result from the differential expression of cytoskeleton and adhesion genes (Pilot and Lecuit, 2005; Ray Keller, 2003; Schock and Perrimon, 2002; Tepass, 1999). In Drosophila oogenesis, the follicular epithelium, which surrounds the nurse cells and oocyte, serves as an established model for studying epithelial patterning and morphogenesis, amenable to live imaging and sophisticated genetic perturbations (Berg, 2005; Dorman et al., 2004; Duffy, 2002; Wu et al., 2008). Particularly striking morphogenetic events occur during the later stages of oogenesis, when the main body follicle cells (MBFCs), which encapsulate the oocyte, undergo a series of cell shape changes and movements to form and secrete an elaborate three-dimensional eggshell, including tubular structures called dorsal appendages (DAs) that project out from the main eggshell body and act as respiratory tubes for the developing embryo (Hinton, 1969; Ward and Berg, 2005; Waring, 2000).
Little is known about the expression, regulation and function of effector genes required for proper eggshell morphogenesis. As one important class of effector molecules in epithelial morphogenesis, cadherins are glycoproteins that mediate Ca+2 dependent cell-cell adhesion and contain multiple, conserved cadherin domains (Halbleib and Nelson, 2006; Tepass et al., 2000). The Drosophila genome encodes 17 cadherins (Hill et al., 2001; Hynes and Zhao, 2000) with representative members in each of the known subgroups of cadherins, excluding desmosomal cadherins (Hill et al., 2001; Tepass et al., 2000) (Table 1). Drosophila cadherins are subdivided into classical cadherins, defined by the presence of a conserved catenin-binding domain (Shotgun/DE-Cad, Cadherin-N/DN-Cad, CadN2), and nonclassical cadherins, which lack any recognizable catenin-binding domain (Fung et al., 2008; Hill et al., 2001; Tepass et al., 2000). Nonclassical cadherins consist of Fat-like cadherins (Fat, Fat2, Dachsous), seven-pass transmembrane cadherins (Starry Night/Flamingo), Calsyntenin cadherins (Calsyntenin-1), protein kinase cadherins (Ret and Cad96Ca), and currently uncategorized cadherins (Cad74A, Cad86C, Cad87A, Cad88C, Cad89D, Cad96Cb, Cad99C) (Fung et al., 2008; Hill et al., 2001).
Table 1.
Cadherin classification and expression in the Drosophila genome
As a first step to providing a comprehensive analysis of the differential expression of potential effector molecules in the follicular epithelium, we have constructed a gene expression atlas of Drosophila cadherin genes. Nine expression patterns were identified in the follicle cells throughout all stages of oogenesis, five of which are newly reported: fat2, Cad86C, Cad87A, Cad88C and cals. Expression patterns for two genes, Cad74A and Cad99C were reported in the literature previously (D'Alterio et al., 2005; Schlichting et al., 2006; Zartman et al., In Press). Finally, expression of DE-Cad and DN-Cad were examined earlier, but later stages of mRNA expression were not previously shown explicitly (Becam et al., 2005; Godt and Tepass, 1998; Niewiadomska et al., 1999; Schnorr and Berg, 1996). Below we describe the expression patterns and group them into spatial and temporal categories.
1.1 Spatial and temporal expression of cadherin genes
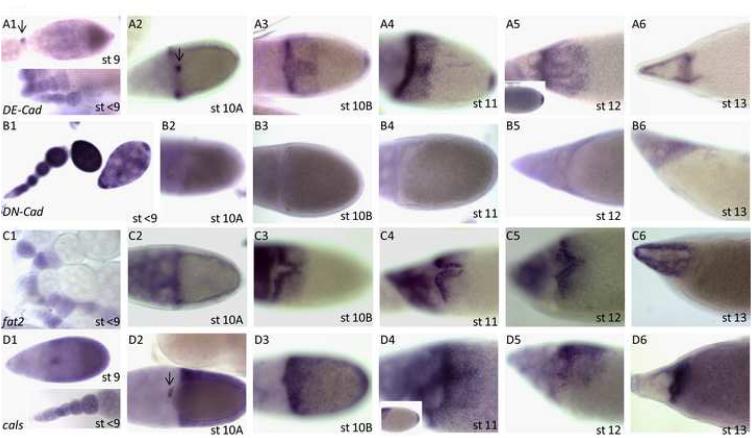
Gene expression patterns of the nine detected cadherin genes can be subdivided into two temporal stages: early oogenesis, before stage 10, as defined by (Spradling, 1993), and later stages, stages 10-14, when the diversity of patterns is greater (Fig. 1 and Table 1). During early oogenesis, seven cadherins were detected: DE-Cad (Fig. 1A1 and inset), DN-Cad (Fig. 1B1), fat2 (Fig. 1C1), cals (Fig. 1D1 and inset), Cad74A (Fig. 1E1, arrows), Cad87A (Fig. 1G1) and Cad99C (D'Alterio et al., 2005; Schlichting et al., 2006) (Fig. 1I1).
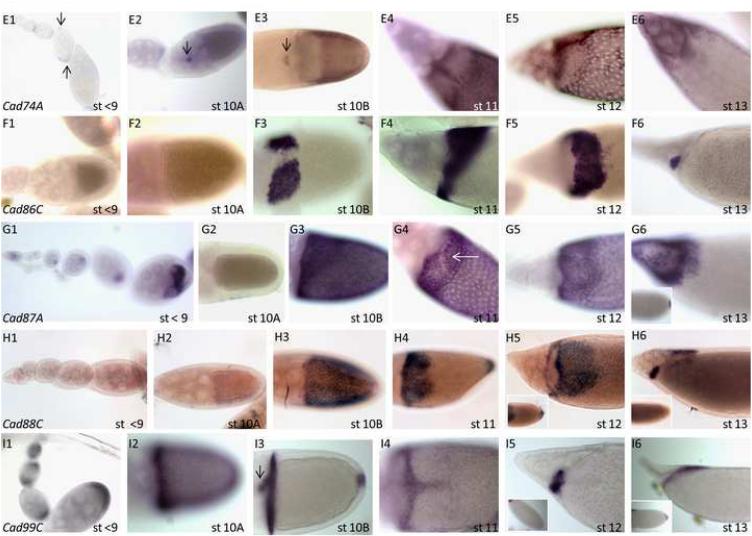
Figure 1. Expression patterns of cadherin genes in Drosophila oogenesis.
Expression of cadherin genes at stages <10 (1), Stage 10A (2), Stage 10B (3), Stages 11 (4), Stage12 (5) and Stage 13 (6) for (A) DE-Cad, (B) DN-Cad, (C) fat2, (D) calsyntenin-1 (cals), (E) Cad74A, (F) Cad86C, (G) Cad87A, (H) Cad88C, and (I) Cad99C. Inserts show expression in the earliest stages or the posterior in late stages. Dark arrows point to visible staining in the border cells. (G4): Arrow points to repression in the floor cells.
Late FC morphogenesis begins in stage 10B when the main body follicle cells (MBFCs) undergo a series of cell shape changes and movements to form the dorsal-anterior eggshell structures (Fig. 2A): the operculum, which is formed by midline cells (M), the micropyle, which is shaped by the centripetal migrating follicle cells consisting of a row of anterior follicle cells (A) and border cells (Montell et al., 1992), and dorsal appendages, which are formed by the roof (R) and floor (F) primordia, two cell populations that form the dorsal and ventral sides of each of the DAs (Dorman et al., 2004; Ward and Berg, 2005). While the patterning by EGFR and DPP signaling and the morphogenesis of the follicle cells has been studied extensively, less is known about the effectors of FC morphogenesis (Berg, 2005; Dorman et al., 2004; Horne-Badovinac and Bilder, 2005; Ward and Berg, 2005; Wu et al., 2008). Two-dimensional expression patterns are most easily categorized in stages 10-12, when the midline, floor, and roof cells have already been specified, but before significant three-dimensional shaping of the dorsal appendages occurs (Yakoby et al., under review). The dynamics of the nine expressed cadherin genes during oogenesis are discussed below.
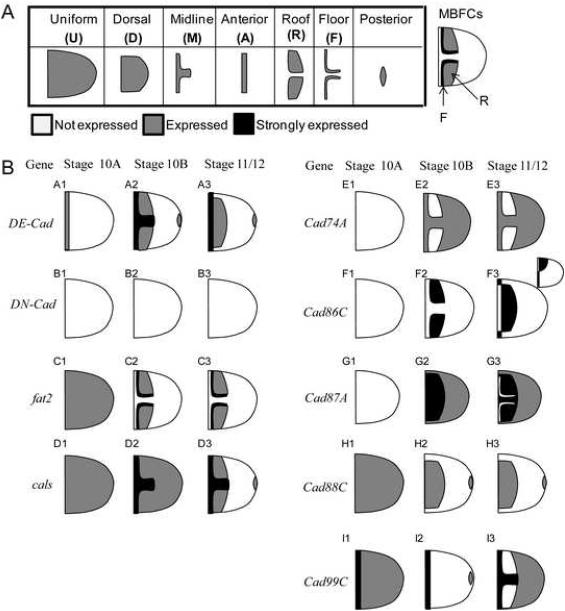
Figure 2. Pattern classification and abstracted expression.
(A) Pattern annotation scheme for the main body follicle cells (MBFCs), as proposed in (Yakoby et al., under review). Patterns have been described using three differential levels of expression: Expression not detected (no shading), basal expression (grey) and strong expression (black). (B) Abstracted expression patterns are shown as cartoons for the nine cadherin genes. Roof versus floor plus roof expression was determined by counting the number of cells between the patches (2 cells for floor plus roof, 4 for roof patterns).
1. shotgun/DE-Cad
Strong shotgun (DE-Cad) expression consistently precedes major morphogenetic movements of the FCs. DE-Cad is expressed in both the germline and the somatic follicle cells and is required for anchoring the somatic stem cell in the germarium niche (Song and Xie, 2002) and positioning the oocyte in the posterior of the egg chamber (Godt and Tepass, 1998; Gonzalez-Reyes and St Johnston, 1998). DE-Cad is also expressed in the border cells during stage 9 before border cell migration (Niewiadomska et al., 1999; Oda et al., 1997) (Fig. 1A1, arrowhead).
DE-Cad shifts from being expressed in an anterior band, corresponding to the centripetal migrating follicle cells (CMFCs) at stage 10A, to strong expression in the midline cells and weak expression in the floor and roof cells during stage 10B, when these cells apically constrict and intercalate (Fig. 1A2-5; James et al., 2002). The pattern continues to show differential DE-Cad expression in the cells forming the dorsal anterior eggshell structures during stages 11 and 12. DE-Cad is also expressed in the posterior FCs, which form the aeropyle (Fig. 1A5, inset).
2. Cadherin-N/DN-Cad
DN-Cad is expressed during early oogenesis in the FCs (Fig. 1B1). The adherens junctions contain both DN-cadherin and DE-cadherin until stage 10 (Tanentzapf et al., 2000), which is unusual because DE-Cad and DN-Cad are not usually co-expressed (Horne-Badovinac and Bilder, 2005). DN-Cad expression is not detected in the FCs after stage 9 (Fig. 1B2-6).
3. fat2
fat2 is expressed during the early stages of oogenesis, but the resolution of the assay is not sufficient to determine if it is the germline or the FCs (Fig. 1C1). Later at stage 10A, fat2 shifts from being expressed uniformly in all MBFCs and nurse cells to being expressed very strongly in the floor cells and weakly in the roof cells (Fig. 1C2-C6) with loss of expression in the other MBFCs during stages 10B-14. The transcript is found in the apical region of FCs. Fat2 shares similarity to Drosophila Fat in the extracellular domain, but lacks the PCP signaling domain found in the intracellular domain of Fat (Castillejo-Lopez et al., 2004; Saburi et al., 2008). fat2 has been implicated in tube formation throughout embryogenesis, including the trachea, salivary glands, proventriculus, and hindgut (Castillejo-Lopez et al., 2004). Based on cultured cell studies and in vivo loss-of-function analysis, Castillejo-Lopez et al. propose that fat2 does not play a role in adhesion but, perhaps due to its size, acts as a spacer for the tube lumen (Castillejo-Lopez et al., 2004).
4. calsyntenin-1 (cals)
calsyntenin-1 shows expression during stages 7-9 in the MBFCs and in the border cells during migration (Fig. 1D1 and inset). Calsyntenin-1(cals) expression is uniform during stage 10A (Fig. 1D2), but is restricted to the dorsal region at stage 10B (Fig. 1D3) with stronger expression in midline in stages 11/12 (Fig 1D4-D5). Expression is also detected in the posterior FCs during the late stages (Fig. 1D4, inset).
5. Cad74A
The expression patterns for Cad74A (Fig. 1E1-6) was published previously (Zartman et al., In Press). Cad74A is expressed in the polar cells during early oogenesis (Fig. 1E1, arrows). During late oogenesis, Cad74A is expressed in all FCs contacting the oocyte in stage 10B, except for the roof cells where high levels of the zinc-finger transcription factor Broad (Deng and Bownes, 1997; Dorman et al., 2004; Tzolovsky et al., 1999; Ward and Berg, 2005) are sufficient to repress Cad74A expression. Overexpresssion of Cad74A in the roof cells results in short, flattened DAs due to the hindered migration of roof cells, suggesting a possible role in the remodeling of the adherens junctions or modulation of the apical membrane. The Cad74A null allele shows reproducible but incompletely penetrant DA defects (Zartman et al., In Press).
6. Cad86C
Cad86C is not expressed (Fig. 1F1, 2) until stage 10B, when it is strongly detected in the roof cells and in a ventral band, corresponding to high Broad levels (Fig. 1F3-6) (Dorman et al., 2004). During stages 11 and 12, a strong hybridization signal is detected in the roof cells and the midline cells separating the roof primordia. Overexpression of Cad86C in the imaginal discs is sufficient to cause apical constriction (Schlichting and Dahmann, 2008), suggesting that it may be involved in apical constriction of the roof cells during dorsal appendage formation.
7. Cad87A
Identified as a possible vertebrate Cadherin23 homolog (Fung et al., 2008), Cad87A is expressed in the oocyte during the early stages of oogenesis (Fig. 1G1). During stage 10B, Cad87A is expressed uniformly in the FCs, with stronger expression in the dorsal anterior (Fig. 1G3). During stages 11/12, expression is particularly strong in the dorsal region except for the floor cells (Fig 1G4, arrow).
8. Cad88C
Cad88C expression is not detectable in the follicle cells up to stage 10A (Fig. 1H1, 2). In stage 10B, all follicle cells contacting the oocyte express Cad88C (Fig. 1H3). In stages 11-12, expression is confined to dorsal-anterior cells and posterior cells (Fig. 1H4-5).
9. Cad99C
The expression and function of Cad99C was previously reported and is required for microvilli morphology and the proper secretion of the vitelline membrane (D'Alterio et al., 2005; Schlichting et al., 2006). Cad99C, a homolog of Protocadherin 15 and required for microvilli morphology, is expressed in stages 4-8 in the anterior and posterior follicle cells and in the follicle cells that migrate to contact the oocyte at stage 9 (D'Alterio et al., 2005; Schlichting et al., 2006) (Fig. 1I1). However, we also find that Cad99C is repressed in the roof cells at stages 11/12 (Fig. 1I4).
1.3 Towards a spatial atlas of morphogenesis
Implementation of the morphogenetic program in several contexts has been found to be robust to the single loss-of-function perturbations of many effector proteins (Kleve et al., 2006; Lovegrove et al., 2006; Schlichting and Dahmann, 2008; Zartman et al., In Press). In the case of our recent study on Cad74A (Zartman et al., In Press), the lack of a strongly penetrant phenotype led us to hypothesize that perhaps other cadherins are expressed in similar expression patterns. In an unbiased pilot screen for expression patterns of morphogenesis genes, we found that a significant fraction (9/17) of the Drosophila cadherin superfamily is differentially expressed during Drosophila oogenesis. Our analysis of mRNA expression addresses the variety of spatial patterns but does not account for gene or protein activity, which must be examined to understand the lack of visible phenotypes in loss-of-function perturbations.
Remarkably, cadherin expression patterns are especially diverse during the later stages of oogenesis (stages 10-14). To provide abstractions of patterns, we have recently proposed an annotation system which compactly describes gene expression patterns during stages 10-12 of oogenesis (Figure 2A, B; Yakoby et al, under review). The geometric annotation terms are derived from the underlying signaling dynamics of the EGFR and BMP signaling pathways and summarize all known FC expression patterns during mid/late oogenesis (Goentoro et al., 2006; Lembong et al., In Press; Yakoby et al., under review). All expression patterns are summarized as Boolean combinations of seven primitive shapes: uniform (U), dorsal (D), midline (M), anterior (A), roof (R), floor (F), and posterior (P) (Fig. 2A). Three principle operations are performed to construct more complex patterns representing the integrate output of EGFR and DPP signaling: union (+), difference (\) or intersection (∩) (Table 2; Yakoby et al., under review).
Table 2.
Geometric annotations of cadherin patterns in the MBFCs
| Gene | Stage 10 A | Stage 10 B | Stage 11/12 |
|---|---|---|---|
| DE-Cad | A | M ∪ D ∪ P | A ∪ D ∪ P |
| DN-Cad | - | - | - |
| fat2 | U | R ∪ F | R ∪ F |
| cals | U | U ∪M | M ∪ D |
| Cad74A | - | U \ R | U \ R |
| Cad86C | - | R | D \ D ∩ A |
| Cad87A | - | U ∪ D | U ∪ D \ F |
| Cad88C | U | D ∪P | D ∪ P |
| Cad99C | A ∪ U | A ∪ P | M ∪ U \ R |
Of particular interest, several cadherins show complex overlapping dynamic expression patterns in the dorsal anterior FCs: midline, floor and roof cells, which are responsible for forming the operculum and DAs (Fig. 2B). Remarkably, no two cadherin genes share an identical expression profile, suggesting that the robustness of morphogenesis is not due to an absolute redundancy in cadherin expression.
Cad74A (stages 10B-12) and Cad99C (stages 11/12) show repression in the roof cells relative to expression in the MBFCs (Fig. 2B). DE-Cad, fat2 and cals show high expression in the floor cells and graded expression in the roof cells. Cad86C initially shows strong expression only in the roof cells whereas Cad87A shows relative repression in the floor cells. Cad88C also shows specific expression to the dorsal anterior cells. Dissecting the regulatory network that leads to such spatial pattern diversity will provide insight into how roof, floor and midline cell identities are maintained during late oogenesis. In particular, the relationship between previously mentioned cadherins and Broad, which is a zinc finger transcription factor highly expressed in the roof cells (Deng and Bownes, 1997; Dorman et al., 2004; Ward and Berg, 2005; Yakoby et al., 2008), will need to be investigated further and compared to the regulation model of Cad74A as well as the expression of other classes of effector molecules (Kleve et al., 2006; Laplante and Nilson, 2006; James et al., 2002; Zartman et al., In Press).
The expression patterns found in this study adds further weight to the possibility that a “combinatorial code” of cadherin expression may play a role in segregating and maintaining different cell sub-populations in the FCs (Foty and Steinberg, 2005; Fung et al., 2008; Lovegrove et al., 2006; Schlichting and Dahmann, 2008). For example, similar overlapping patterns for the non-classical cadherins are found in other developmental contexts (Lovegrove et al., 2006; Schlichting and Dahmann, 2008). Based on previous reports and the new patterns identified in this study, we hypothesize that the partially overlapping syn-expression of a subset of adhesion genes ensures robust morphogenesis of the dorsal appendages, providing built-in degeneracy to the epithelial folding code. Deciphering and parsing the spatial grammar of morphogenesis during DA formation will require manipulating the relative expression of sets of adhesion genes. Comparison of the relative overlaps between cadherin expression patterns in other models of morphogenesis will provide a necessary test for the universality of an epithelial folding code that converts spatial patterns of gene expression into mechanical properties and a final morphology.
3. Experimental procedures
Whole-mount in situ hybridization
cDNA clones for fat, fat2, Ret, Cad74A, Cad87A, Cad96Ca and Cad99C were gifts of M. Halfon. cDNA for dachsous came from the Drosophila Genomics Resource Center (Bloomington, IN). Primers used to amplify the remaining cadherin genes from cDNA obtained from the ovary are listed in Table 3. Products from PCR amplification were cloned using a StrataClone PCR Cloning Kit (Stratagene). For Cad88C, the underlined restriction sites (Table 3) for BamHI and Asp718 were used for cloning the PCR product into pBluescript and the clone was sequenced. Clones were designed to be between 900 – 1200 bases. Clones were then sequenced (GeneWiz) and BLASTed against the D. melanogaster genome (FlyBase) to confirm clone identity and orientation. Digoxygenin-labeled RNA probes were made from the cDNA clones. During the in situ experiments, a previously tested control probe was used to ensure the quality of in situ hybridization assay. The in situ hybridization protocol is optimized for assaying gene expression in the follicle cells and was the same as previously described (Wang et al., 2006; Yakoby et al., 2008) without an RNase treatment step. Ore R and y w were used for the in situ hybridization experiments.
Table 3.
Primers used for mRNA probe generation
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| DE-Cad | GCCCAATGGTCACAAGAA CT |
CTCGTTTTGGAGCACAGTGA |
| DN-Cad | GTCAATCGTCCGGTTCAG TT |
CACCGATTTTCCCTCAGTGT |
| CadN2 | TGCCAATCTCAGCGTTAC AG |
CCATCATCCAGCGTTTTCTT |
|
Starry night/ Flamingo |
CTTTTCGTCTCCGTCAAAG C |
GATCGAGGGAGGCATATTGA |
| Calsyntenin-1 | TGGCAACACCGATAATGA AA |
CTCCTATGCATTGCGACAGA |
| Cad88C | CGGGATCCCTTTAGCATT CGAGAGATCG |
GGGGTACCTTTAGCCACTGAT GCTGCTC |
| Cad89D | CAAGCCCAATACACCGAA CT |
ACTTTGCTAGGTCCCGGTTT |
| Cad86C | CACTGAATCTGGACGCTG AA |
GCCAGAAGAGCACCTTGTTC |
| Cad96Cb | AATCTCGAGCGGATTCCT TT |
GAGGGCCATGCTACTACTGG |
Acknowledgements
We thank M. Halfon for providing cDNA clones for several cadherin genes and the DGRC (Bloomington, IN) for cDNA clones for ds and C. Bristow for helpful comments on the manuscript. J.J.Z. is supported by the Fannie and John Hertz Foundation and the Princeton Wu fellowship. This work has been supported by the following NIH grants: P50 GM071508 and R01 GM078079 to S.Y.S.
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrescia C, Sjöstrand D, Kjaer S, Ibáñez CF. Drosophila RET contains an active tyrosine kinase and elicits neurotrophic activities in mammalian cells. FEBS Letters. 2005;579:3789–3796. doi: 10.1016/j.febslet.2005.05.075. [DOI] [PubMed] [Google Scholar]
- Becam IE, Tanentzapf G, Lepesant J-A, Brown NH, Huynh J-R. Integrin-independent repression of cadherin transcription by talin during axis formation in Drosophila. Nat Cell Biol. 2005;7:510–516. doi: 10.1038/ncb1253. [DOI] [PubMed] [Google Scholar]
- Berg CA. The Drosophila shell game: patterning genes and morphological change. Trends in Genetics. 2005;21:346–355. doi: 10.1016/j.tig.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Castillejo-Lopez C, Arias WM, Baumgartner S. The fat-like gene of Drosophila is the true orthologue of vertebrate fat cadherins and is involved in the formation of tubular organs. J Biol Chem. 2004;279:24034–24043. doi: 10.1074/jbc.M313878200. [DOI] [PubMed] [Google Scholar]
- D'Alterio C, Tran DD, Yeung MW, Hwang MS, Li MA, Arana CJ, Mulligan VK, Kubesh M, Sharma P, Chase M, et al. Drosophila melanogaster Cad99C, the orthologue of human Usher cadherin PCDH15, regulates the length of microvilli. J Cell Biol. 2005;171:549–558. doi: 10.1083/jcb.200507072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis F, Habermann B, Dahmann C. PDZ-domain-binding sites are common among cadherins. Development Genes and Evolution. 2006;216:737–741. doi: 10.1007/s00427-006-0097-0. [DOI] [PubMed] [Google Scholar]
- Deng WM, Bownes M. Two signalling pathways specify localised expression of the Broad-Complex in Drosophila eggshell patterning and morphogenesis. Development. 1997;124:4639–4647. doi: 10.1242/dev.124.22.4639. [DOI] [PubMed] [Google Scholar]
- Dorman JB, James KE, Fraser SE, Kiehart DP, Berg CA. bullwinkle is required for epithelial morphogenesis during Drosophila oogenesis. Dev Biol. 2004;267:320–341. doi: 10.1016/j.ydbio.2003.10.020. [DOI] [PubMed] [Google Scholar]
- Duffy JB. GAL4 system in Drosophila: a fly geneticist's Swiss army knife. Genesis. 2002;34:1–15. doi: 10.1002/gene.10150. [DOI] [PubMed] [Google Scholar]
- Foty RA, Steinberg MS. The differential adhesion hypothesis: a direct evaluation. Developmental Biology. 2005;278:255–263. doi: 10.1016/j.ydbio.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Fung S, Wang F, Chase M, Godt D, Hartenstein V. Expression profile of the cadherin family in the developing Drosophila brain. J Comp Neurol. 2008;506:469–488. doi: 10.1002/cne.21539. [DOI] [PubMed] [Google Scholar]
- Godt D, Tepass U. Drosophila oocyte localization is mediated by differential cadherin-based adhesion. Nature. 1998;395:387–391. doi: 10.1038/26493. [DOI] [PubMed] [Google Scholar]
- Goentoro LA, Reeves GT, Kowal CP, Martinelli L, Schupbach T, Shvartsman SY. Quantifying the Gurken morphogen gradient in Drosophila oogenesis. Dev Cell. 2006;11:263–272. doi: 10.1016/j.devcel.2006.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Reyes A, St Johnston D. The Drosophila AP axis is polarised by the cadherin-mediated positioning of the oocyte. Development. 1998;125:3635–3644. doi: 10.1242/dev.125.18.3635. [DOI] [PubMed] [Google Scholar]
- Halbleib JM, Nelson WJ. Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes Dev. 2006;20:3199–3214. doi: 10.1101/gad.1486806. [DOI] [PubMed] [Google Scholar]
- Hill E, Broadbent ID, Chothia C, Pettitt J. Cadherin superfamily proteins in Caenorhabditis elegans and Drosophila melanogaster. J Mol Biol. 2001;305:1011–1024. doi: 10.1006/jmbi.2000.4361. [DOI] [PubMed] [Google Scholar]
- Hinton HE. Respiratory systems of insect egg shells. Annu Rev Entomol. 1969;14:343–368. doi: 10.1146/annurev.en.14.010169.002015. [DOI] [PubMed] [Google Scholar]
- Horne-Badovinac S, Bilder D. Mass transit: epithelial morphogenesis in the Drosophila egg chamber. Dev Dyn. 2005;232:559–574. doi: 10.1002/dvdy.20286. [DOI] [PubMed] [Google Scholar]
- Hynes RO, Zhao Q. The evolution of cell adhesion. J Cell Biol. 2000;150:F89–96. doi: 10.1083/jcb.150.2.f89. [DOI] [PubMed] [Google Scholar]
- James KE, Dorman JB, Berg CA. Mosaic analyses reveal the function of Drosophila Ras in embryonic dorsoventral patterning and dorsal follicle cell morphogenesis. Development. 2002;129:2209–2222. doi: 10.1242/dev.129.9.2209. [DOI] [PubMed] [Google Scholar]
- Kleve CD, Siler DA, Syed SK, Eldon ED. Expression of 18-wheeler in the follicle cell epithelium affects cell migration and egg morphology in Drosophila. Dev Dyn. 2006;235:1953–1961. doi: 10.1002/dvdy.20820. [DOI] [PubMed] [Google Scholar]
- Laplante C, Nilson LA. Differential expression of the adhesion molecule Echinoid drives epithelial morphogenesis in Drosophila. Development. 2006;133:3255–3264. doi: 10.1242/dev.02492. [DOI] [PubMed] [Google Scholar]
- Lembong J, Yakoby N, Shvartsman SY. Spatial regulation of BMP signaling by patterned receptor expression Tissue Engineering. 14 doi: 10.1089/ten.tea.2008.0098. In Press. [DOI] [PubMed] [Google Scholar]
- Lovegrove B, Simões S, Rivas ML, Sotillos S, Johnson K, Knust E, Jacinto A, Hombría JC-G. Coordinated Control of Cell Adhesion, Polarity, and Cytoskeleton Underlies Hox-Induced Organogenesis in Drosophila. Current Biology. 2006;16:2206–2216. doi: 10.1016/j.cub.2006.09.029. [DOI] [PubMed] [Google Scholar]
- Montell DJ, Rorth P, Spradling AC. slow border cells, a locus required for a developmentally regulated cell migration during oogenesis, encodes Drosophila C/EBP. Cell. 1992;71:51–62. doi: 10.1016/0092-8674(92)90265-e. [DOI] [PubMed] [Google Scholar]
- Niewiadomska P, Godt D, Tepass U. DE-Cadherin Is Required for Intercellular Motility during Drosophila Oogenesis. J Cell Biol. 1999;144:533–547. doi: 10.1083/jcb.144.3.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Uemura T, Takeichi M. Phenotypic analysis of null mutants for DE-cadherin and Armadillo in Drosophila ovaries reveals distinct aspects of their functions in cell adhesion and cytoskeletal organization. Genes to Cells. 1997;2:29–40. doi: 10.1046/j.1365-2443.1997.d01-284.x. [DOI] [PubMed] [Google Scholar]
- Pacquelet A, Rorth P. Regulatory mechanisms required for DE-cadherin function in cell migration and other types of adhesion. J Cell Biol. 2005;170:803–812. doi: 10.1083/jcb.200506131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilot F, Lecuit T. Compartmentalized morphogenesis in epithelia: from cell to tissue shape. Dev Dyn. 2005;232:685–694. doi: 10.1002/dvdy.20334. [DOI] [PubMed] [Google Scholar]
- Ray Keller LADDRS. How we are shaped: The biomechanics of gastrulation. Differentiation. 2003;71:171–205. doi: 10.1046/j.1432-0436.2003.710301.x. [DOI] [PubMed] [Google Scholar]
- Saburi S, Hester I, Fischer E, Pontoglio M, Eremina V, Gessler M, Quaggin SE, Harrison R, Mount R, McNeill H. Loss of Fat4 disrupts PCP signaling and oriented cell division and leads to cystic kidney disease. Nat Genet. 2008;40:1010–1015. doi: 10.1038/ng.179. [DOI] [PubMed] [Google Scholar]
- Schlichting K, Dahmann C. Hedgehog and Dpp signaling induce cadherin Cad86C expression in the morphogenetic furrow during Drosophila eye development. Mech Dev. 2008 doi: 10.1016/j.mod.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Schlichting K, Demontis F, Dahmann C. Cadherin Cad99C is regulated by Hedgehog signaling in Drosophila. Dev Biol. 2005;279:142–154. doi: 10.1016/j.ydbio.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Schlichting K, Wilsch-Brauninger M, Demontis F, Dahmann C. Cadherin Cad99C is required for normal microvilli morphology in Drosophila follicle cells. J Cell Sci. 2006;119:1184–1195. doi: 10.1242/jcs.02831. [DOI] [PubMed] [Google Scholar]
- Schnorr JD, Berg CA. Differential Activity of Ras1 during Patterning of the Drosophila Dorsoventral Axis. Genetics. 1996;144:1545–1557. doi: 10.1093/genetics/144.4.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schock F, Perrimon N. Molecular mechanisms of epithelial morphogenesis. Annu Rev Cell Dev Biol. 2002;18:463–493. doi: 10.1146/annurev.cellbio.18.022602.131838. [DOI] [PubMed] [Google Scholar]
- Song X, Xie T. DE-cadherin-mediated cell adhesion is essential for maintaining somatic stem cells in the Drosophila ovary. Proc Natl Acad Sci U S A. 2002;99:14813–14818. doi: 10.1073/pnas.232389399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A. In: In The development of Drosophila melanogaster. A. M., editor. Cold Spring Harbor Laboratory Press; New York: 1993. pp. 1–70. [Google Scholar]
- Sugaya R, Ishimaru S, Hosoya T, Saigo K, Emori Y. A Drosophila homolog of human proto-oncogene ret transiently expressed in embryonic neuronal precursor cells including neuroblasts and CNS cells. Mechanisms of Development. 1994;45:139–145. doi: 10.1016/0925-4773(94)90027-2. [DOI] [PubMed] [Google Scholar]
- Tanentzapf G, Smith C, McGlade J, Tepass U. Apical, lateral, and basal polarization cues contribute to the development of the follicular epithelium during Drosophila oogenesis. J Cell Biol. 2000;151:891–904. doi: 10.1083/jcb.151.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanoue T, Takeichi M. New insights into Fat cadherins. J Cell Sci. 2005;118:2347–2353. doi: 10.1242/jcs.02398. [DOI] [PubMed] [Google Scholar]
- Tepass U. Genetic analysis of cadherin function in animal morphogenesis. Curr Opin Cell Biol. 1999;11:540–548. doi: 10.1016/s0955-0674(99)00025-3. [DOI] [PubMed] [Google Scholar]
- Tepass U, Harris KP. Adherens junctions in Drosophila retinal morphogenesis. Trends in Cell Biology. 2007;17:26–35. doi: 10.1016/j.tcb.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Tepass U, Truong K, Godt D, Ikura M, Peifer M. Cadherins in embryonic and neural morphogenesis. Nat Rev Mol Cell Biol. 2000;1:91–100. doi: 10.1038/35040042. [DOI] [PubMed] [Google Scholar]
- Tzolovsky G, Deng W-M, Schlitt T, Bownes M. The Function of the Broad-Complex During Drosophila melanogaster Oogenesis. Genetics. 1999;153:1371–1383. doi: 10.1093/genetics/153.3.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt L, Schrimpf SP, Meskenaite V, Frischknecht R, Kinter J, Leone DP, Ziegler U, Sonderegger P. Calsyntenin-1, a Proteolytically Processed Postsynaptic Membrane Protein with a Cytoplasmic Calcium-Binding Domain. Molecular and Cellular Neuroscience. 2001;17:151–166. doi: 10.1006/mcne.2000.0937. [DOI] [PubMed] [Google Scholar]
- Wang X, Bo J, Bridges T, Dugan KD, Pan TC, Chodosh LA, Montell DJ. Analysis of cell migration using whole-genome expression profiling of migratory cells in the Drosophila ovary. Dev Cell. 2006;10:483–495. doi: 10.1016/j.devcel.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Ward EJ, Berg CA. Juxtaposition between two cell types is necessary for dorsal appendage tube formation. Mech Dev. 2005;122:241–255. doi: 10.1016/j.mod.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Waring GL. Morphogenesis of the eggshell in Drosophila. Int Rev Cytol. 2000;198:67–108. doi: 10.1016/s0074-7696(00)98003-3. [DOI] [PubMed] [Google Scholar]
- Wu X, Tanwar PS, Raftery LA. Drosophila follicle cells: Morphogenesis in an eggshell. Seminars in Cell & Developmental Biology. 2008;19:271–282. doi: 10.1016/j.semcdb.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakoby N, Bristow CA, Gong D, Schafer X, Lembong J, Zartman JJ, Halfon MS, Schüpbach T, Shvartsman SY. Combinatorial code for epithelial patterning in Drosophila oogenesis. Developmental Cell. doi: 10.1016/j.devcel.2008.09.008. under review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakoby N, Lembong J, Schupbach T, Shvartsman SY. Drosophila eggshell is patterned by sequential action of feedforward and feedback loops. Development. 2008;135:343–351. doi: 10.1242/dev.008920. [DOI] [PubMed] [Google Scholar]
- Zartman JJ, Yakoby N, Bristow CA, Zhou X, Schlichting K, Dahmann C, Shvartsman SY. Cad74A is regulated by BR and is required for robust dorsal appendage formation in Drosophila oogenesis. Developmental Biology. doi: 10.1016/j.ydbio.2008.07.027. In Press, Accepted Manuscript. [DOI] [PMC free article] [PubMed] [Google Scholar]