Abstract
Nonlinear dimensionality reduction is essential for the analysis and the interpretation of high dimensional data sets. In this manuscript, we propose a distance order preserving manifold learning algorithm that extends the basic mean-squared error cost function used mainly in multidimensional scaling (MDS)-based methods. We develop a constrained optimization problem by assuming explicit constraints on the order of distances in the low-dimensional space. In this optimization problem, as a generalization of MDS, instead of forcing a linear relationship between the distances in the high-dimensional original and low-dimensional projection space, we learn a non-decreasing relation approximated by radial basis functions. We compare the proposed method with existing manifold learning algorithms using synthetic datasets based on the commonly used residual variance and proposed percentage of violated distance orders metrics. We also perform experiments on a retinal image dataset used in Retinopathy of Prematurity (ROP) diagnosis.
Keywords: Machine Learning, Manifold Learning, Nonlinear Dimensionality Reduction
1. Introduction
Due to the recent advances, acquisition of large volumes of high dimensional data has become more common in every aspect of daily life: stock market, social media, medical data, etc. Analysis and interpretation of such data requires finding meaningful low-dimensional structures in these huge data sets. Manifold learning attempts to accomplish such data explorations and dimensionality reductions.
Manifold learning can be regarded as identifying a nonlinear mapping from the original higher dimensional data space to a lower dimensional representation. Existing methods can be classified into three categories: global methods that tend to preserve global properties in the low-dimensional representation, local methods that aim to preserve the local geometry in the embedded space and techniques based on global alignment of multiple linear models [1]. Multidimensional scaling (MDS) is one of the global methods that finds a projection of the original data while preserving the pairwise Euclidean distances [2]. In the literature, various techniques are proposed to minimize MDS cost function [3, 4, 5, 6]. Similarly, in Isomap, one uses a geodesic distance estimation to use with MDS [7]. Different variations of Isomap have been proposed in the literature: landmark and conformal Isomap [8]. On the other hand, local methods [9, 10, 11, 12, 13, 14, 15] constructs the lower dimensional data using the local linear relations in the original space. Local tangent space alignment (LTSA) [9] represents the local geometry of the manifold with local tangent spaces that are learned through the neighborhood of each sample. Similarly, local linear embedding (LLE) [10] aims to preserve local neighborhood information, while Semidefinite Embedding (SDE) [11] involves preserving local isometries on a k-nn graph. Coifman et al. [14] presents a method that constructs local coordinates by learning a family of diffusion maps (DM). Another use of local geometry is by locally smooth manifold learning (LSML) [12, 13] which is based on learning a warping function, that takes any sample in the manifold and generates its neighbors. Stochastic neighborhood embedding (SNE) [15] and its variations [16, 17] are among probabilistic approaches that construct the neighborhood relations based on Gaussian kernels. Although local approaches have computational advantages, they might have limitations in preserving global geometry, especially if the data is sparse. Other methods that are based on global alignment of linear models aim to combine the local and global techniques by fitting a number of local linear models and merging them with a global alignment. Local linear coordination (LLC) [18] and manifold charting [19] methods fall into this category.
In this manuscript, we propose a nonlinear dimensionality reduction method that extends the basic idea used by the MDS and its variations. Although the ultimate goal is to preserve the distance orders, MDS algorithm only focuses on minimizing the mean-squared error between the input and output distances [2]. There is no explicit constraint on the distance orders during the solution of this optimization problem. Moreover, this minimization results in a linear relationship between distance spaces. Linear fit assumption between the distances in the original and low-dimensional projection spaces is very restricted and embedding is achieved in this restrictive family. To address these two important issues, we first generalize the mean squared error cost function to include more general relations between distance spaces. Instead of assuming a predefined relationship, we propose to also learn the relationship between distances while we project the data from the original space. Our only assumption on the relationship is to have a monotonic non-decreasing function in order to preserve the distance relationships observed in the original space in the projected space. Then, we develop a constrained optimization problem by incorporating the distance orders as inequality constraints. As a solution of this problem, we not only learn the data in the projected space but also learn the non-linear relation between distance spaces. The final form of the proposed optimization problem is a generalization of the existing global MDS-based manifold learning algorithms such that the existing methods are approximate solutions of the simplified version of this problem. In this manuscript, we focus on the formulation and theoretical aspects of the problem. Possible acceleration of the proposed method by convex relaxations and further approximations to analyze real data will be part of our future research.
Another commonly used manifold learning algorithm which has a nonlinear mapping between distance spaces is Sammons mapping [20]. Sammon’s map, a nonlinear extension of MDS, first maps the input data to a nonlinear predefined feature space and tries to preserve the distances in this feature space. Different than Sammon’s mapping, we assume an unknown nonlinear relationship between the input and output distances while preserving the distance orders from the original space.
The rest of the paper is organized as follows: We first define the notation used throughout the paper in section 2.1. Next, problem formulation is presented in section 2.2. The solution of the optimization problem and performance evaluation metrics are explained in section 2.3 and section 2.4 respectively. In section 3, we report the experiments and results and the paper is concluded in section 4.
2. Learning Algorithm
In this section, we describe the proposed method for manifold learning. We first define the data model and notations to be used throughout the manuscript. Then using this model we formulate the desired manifold learning problem and develop our algorithm. We derive an optimization problem that solves the manifold learning algorithm, starting with the commonly used cost function, mean-squared error minimization, and demonstrate that this cost function can be extended to include different distance relations between the original and projection space data points, and explicit constraints that preserve distance orders in the projected space.
2.1. Data Model and Notation
We represent the original and the projected data spaces as 𝒳 and 𝒴, respectively. Then, xi ∈ 𝒳 and yi ∈ 𝒴 with i = 1, …, N are the data points. In this representation, x and y are vectors and N is the number of the data points. We assume that dim(𝒳) = d ≥ dim(𝒴) = d̃. Moreover, we have and as the distances between the ith and jth data points in the original and the low-dimensional data spaces, ‖ · ‖ represents the L2-norm of a vector.
2.2. Problem Formulation
We formulate the manifold learning algorithm as a constrained optimization problem. Our approach restricts the minimum mean-squared error solutions used by some existing manifold learning algorithms [2, 7]. Specifically, these aim to minimize the difference between the distances of any two points in the original and projected spaces. That is, the difference between and for ∀i, j = 1, …, N is minimized, which on average results in a linear relationship between each and pair (as a result of the least-square solution).
| (1) |
where is the Euclidean distance and is the distance between the ith and jth points. Note here that the minimization is performed over the data points yi in the projected space.
In the proposed algorithm, we compute as the estimated geodesic distance between the ith and jth data points. We follow the method described in [7] to compute the geodesic distances in the original space. First, the Euclidean distance between every pair of data points in the original space (data pairwise distance matrix) is computed. Then, a k-nearest neighbor (knn) graph or ε-ball graph is generated. That is, k-nearest neighbors of a data point or neighbors within ε distance for each datum is taken, and the edge lengths from points outside these areas to the reference datum are set to be infinite, and the pairwise distance matrix is updated accordingly. Finally, Floyd algorithm is applied over this matrix to find approximate geodesic distances between the data pairs [7]. Floyd’s algorithm, an example of dynamic programming, finds the shortest path between each pair of vertices in a weighted graph [21].
In our algorithm, we propose to generalize the minimum mean-squared error approach in (1) to include a broader relationship between the distances in the original and the low-dimensional projection space. For that purpose we rewrite (1) as
| (2) |
where h(·) is a monotonic nondecreasing function. We represent the derivative of h(·) as
| (3) |
where k(η) is a translation-invariant kernel function and wkl ’s are the multiplicative coefficients. We force wkl ≥ 0 to have h(·) as a nondecreasing function. That is, we represent the derivative of h(·) as a nonnegative weighted sum of kernel functions. Monotonic nondecreasing functions h(·) will guarantee that we preserve the order of original distances in the projected space.
Then, we have
| (4) |
where
| (5) |
Kernel functions are positive semidefinite and hence are appropriate to represent the derivative of a monotonic nondecreasing function h(·) [22]. A translation-invariant kernel k(η) is chosen such that 0 ≤ k(η) < ∞, , and limη→0 k(η) = δ(η). We specifically use Gaussian radial basis functions (rbf), since they were shown to universally approximate any function with any desired accuracy [23]. Then, we rewrite (5) as
| (6) |
where Φ(·) is the Gaussian cumulative distribution function, and σ is the kernel width (standard deviation), which is estimated using Silverman’s rule [24].
From (3), we have different non-negative wkl coefficients. We form the M × 1 size vector w as
| (7) |
We also define as the N d̃ × 1 size data vector in the projected space. Using the definitions for w and yd, and the specifications provided for the function h(·), we then formulate the manifold learning algorithm as the following optimization problem.
| (8) |
where ⪰ represents an element-wise inequality, such that we require each element of w is greater than or equal to zero. Note that in (8), we aim to avoid the global minimum solution where all yi, i = 1, …, N are the same and w = 0, which is not a valid projection solution. Therefore, we update the problem as
| (9) |
The existing manifold learning algorithms do not explicitly enforce constraints to preserve the distance relationships observed in the original space. In our method, we propose to consider the following constraint: If T distance pairs in the original space are ordered as , we require to have . This is an approximation to preserving the order relation-ship between every distance pair in the original space. Selecting T achieves a reduction in complexity, but also may reflect preference about which distance orders are more important to preserve for the user. Then, we have the following optimization problem
| (10) |
Depending on the data under consideration, feasibility in (10) could be difficult to achieve, it might not even be possible to satisfy all the inequality constraints; therefore, to obtain a solution to the proposed constrained optimization problem, we further update it by perturbing (relaxing) the distance inequality constraints to
| (11) |
where ξ = [ξ1 ⋯ ξT]T is the vector of the perturbation slack variables which store the deviation from the nonlinear inequality constraints representing the order relationship between the distance pairs in the projection space, γ is the penalty factor forcing the problem to satisfy as many order relationships as possible, and α’s demonstrate the importance of different ξ’s such that preserving some inequalities could be more important than others.
In order to use in 2.3, we follow the above discussions, and define
θ = [ydT, wT, ξT]T is an (N d̃ + M + T) × 1 vector,
,
A = [0 I] as (M + T) × (M + T + N d̃) size matrix,
,
.
Note that we improve the problem in (11) with the condition , and note also that due to the quadratic dependency of the cost function to w, we can always find a scaled w to obtain the minimum of the cost function at , and hence we define . Then, we rewrite the problem as
| (12) |
In our implementation, we always used a chain of T = M − 1 inequalities but one can choose arbitrary pairs of inequalities. In the following we also refer as , then .
2.3. Solution of the Optimization Problem
In this section, we explain the solution method that we employ to solve the constrained optimization problem proposed in (12). We first redefine the problem using extra slack variables as
| (13) |
Then the Lagrangian of the problem is computed as
| (14) |
where q and z = [z1, …, z3M−2]T are the Lagrange multipliers.
Using the proposed problem in (13), and the Lagrangian in (14), we compute the Karush-Kuhn-Tucker (KKT) conditions for this problem [25]
| (15a) |
| (15b) |
| (15c) |
| (15d) |
| (15e) |
where
is the gradient of the cost function f (θ) with respect to θ,
∇θceq = 1, a vector of ones,
D(θ) is the Jacobian of the vector of functions c(θ), such that DT (θ) = [∇θc1(θ) ⋯ ∇θc3M−2(θ)],
S is a diagonal matrix with the s vector in the diagonal, such that S = diag(s).
Interior-point methods were shown to outperform active set [26] and augmented Lagrangian methods [27] in large scale problems [28, 29]. We therefore solve the problem in (13) using interior-point methods such that we define pθ, ps, pq, and pz as the step lengths in the variables θ, s, q, and z, respectively, and apply the Newton’s method to compute the primal-dual update matrix as
| (16) |
where
is the Hessian of the Lagrangian, and
Z = diag(z).
We solve the system in (16) using the algorithm described in [30, 31, 32]. We approximate the Hessian of the Lagrangian using limited-memory Broyden-Fletcher-Goldfarb-Shanno (lBFGS) method [33]. lBFGS is a limited memory approximation of the BFGS method to be used in large scale optimization problems [25]. We provide the derivations of gradient ∇θf(θ), Jacobian D(θ), and Hessian in the appendix. The dimension of unknowns, n, in our problem is O(N2) and, we know that lBFGS takes O(n2) time per iteration, hence the run time for the optimization algorithm per iteration is O(N4). The optimization method converges to a local minima due to the non-convexity of the proposed problem. Convex relaxations to this problem and convergence analysis is a topic of our future work.
2.4. Performance Analysis
In this section, we describe a metric that we define to demonstrate the performance of the proposed method. The metric we consider is the percentage of the pairwise inequalities violated in the low-dimensional space:
| (17) |
where Tt = M (M − 1)/2 is the total number of distance inequalities since we always choose to employ a chain of M − 1 inequalities and Tυ is the number of distance relationships violated in the projected space. In the proposed method, we compute Tt considering the estimated geodesic distance relationships observed in the original space. Recall that in our algorithm, for example if the geodesic distance is larger than , we propose to preserve such relationships approximately for every data points indexed with i, j, k, l ∈ [1, …, N] in the projected space. We then accordingly compute Tυ by counting violated order relationships. According to the definition in (17), the lower the τ is, the more accurate the subspace projection is. We refer to this metric as constraint violation percentage (CVP).
Another commonly used performance metric is the residual variance between the estimated geodesic distances in the original space and the Euclidean distances in the projected space [7]. Defining and as two M × 1 vectors, the residual variance is computed using
| (18) |
where
, with d̅y as the mean,
, with d̅x as the mean,
.
3. Numerical Examples
We demonstrate the performance of the proposed method on synthetic datasets and compare with the following existing methods: Isomap [7], LTSA [9], LLE [10], SDE [11], LSML [12, 13], DM [14]. Penalty factor γ in (11) is taken as 1 and the scalar ec that bounds the sum of w’s in (12) is selected as 10 in our experiments. α coefficient that indicate the importance of preserving an order constraints is taken as 1 for all order constraints. The variables that are being optimized are initialized randomly. The geodesic distance estimation is done with a knn-graph where k = 5. Similarly, neighborhood information is represented through knn-graphs with k = 5 in Isomap, LTSA, SDE and LSML methods. The number of neighbors for LLE is set as k = 5. We use the default values for all other parameters in the methods, that we compare against the proposed technique.
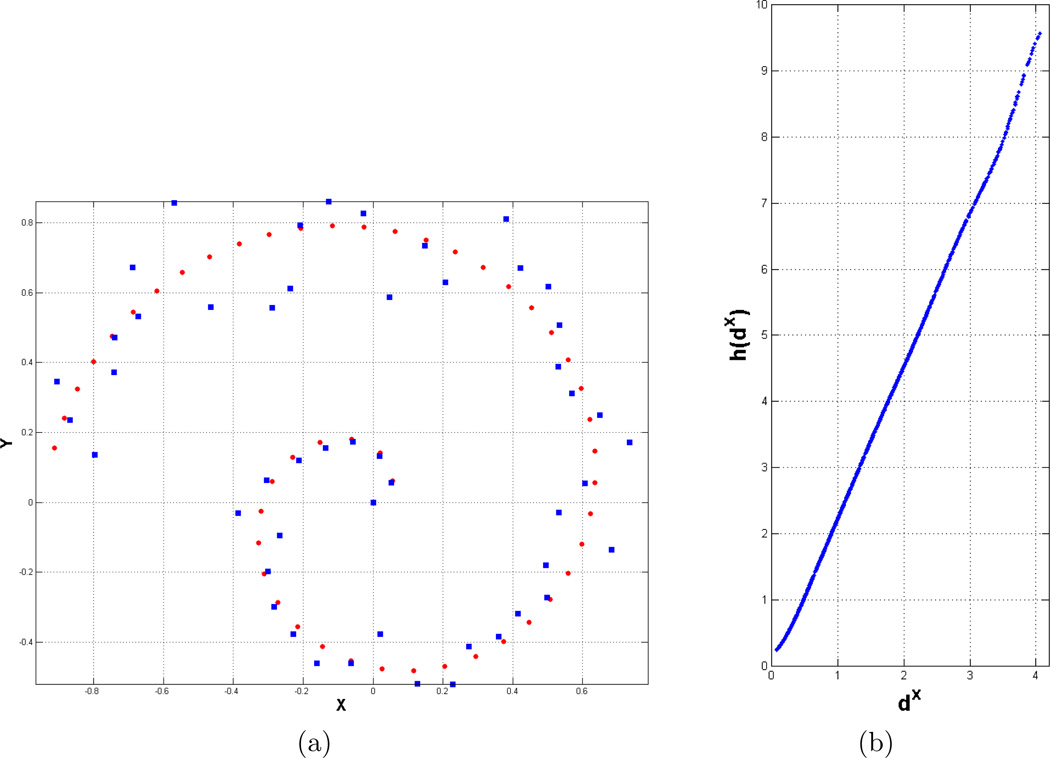
In the first set of experiments, we perform noise analysis on the synthetically generated growing band (GB) dataset with N = 50 samples, which contains a single dimensional manifold (see Figure 2(a)). For a given noise variance σ2, the two dimensional dataset is generated by the following model
| (19) |
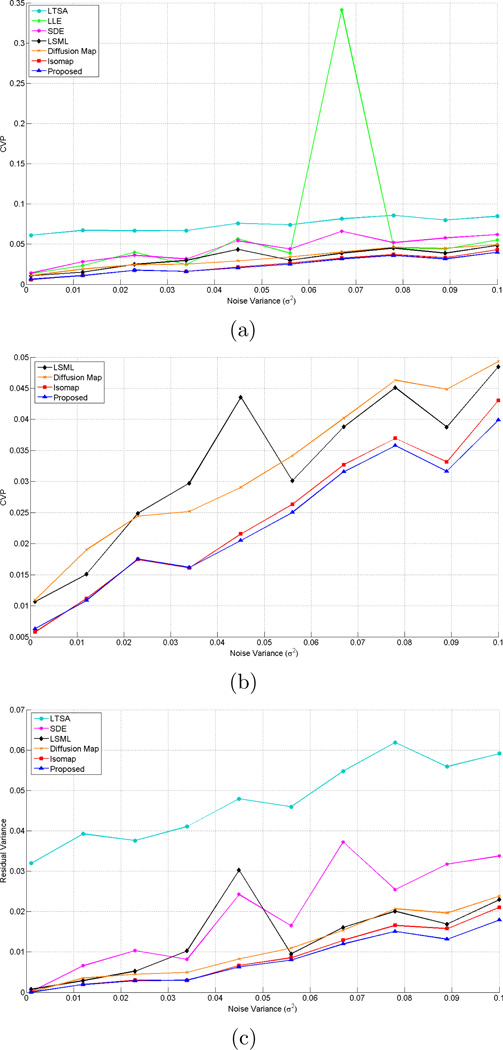
where ρ is selected as 10 and β(t) = 𝒩 (0, σ2t) is a Gaussian random variable with mean 0 and variance σ2t. For this data set we use different noise levels, σ2 changing from 0.001 to 0.1. In this experiment, our goal is to explore possible non-linear relationship between the distances in the original and the projection spaces. The performance metrics we use are constraint violation percentage (CVP) and residual variance as described in Section 2.4. In Figure 2, and Table 1, we report noise analysis results. Figure 2 displays the CVP and residual variance for different values of the noise variance using the proposed technique and all other techniques. Figure 2(b) is a zommed in version of Figure 2(a) displaying the results for LSML, DM, Isomap and the proposed methods. The residual variance for LLE is omitted in Figure 2(c) for better visualization, since it produces very high values compared to the other methods. As can be seen the proposed method shows a better performance than the other methods with respect to both evaluation criteria. The method that perfoms similar to the proposed method is Isomap. All the other methods are local and they perform poorly due to the sparsity of the data. The performance difference between Isomap and the proposed method increases as the noise variance increases, because for low noise variances, the data resembles to a line more and the distance relations are close to being linear. Notice that the residual variance shows the same trend with CVP, since both metrics aim to quantify the consistency between distances, see also Table 1 for the summary of performance analysis results. Figure 1(b) shows the h(·) function as a result of the proposed optimization scheme for σ2 = 0.1. The resulting fit does not show a linear relation, instead it maps the small distances to even smaller distances and the large distances to even larger ones. Our analysis shows that this kind of relationship outperforms the linear fit assumed by MDS.
Figure 2.
Noise analysis on GB dataset: (a) CVP on datasets with different noise variances using LTSA [9], LLE [10], SDE [11], LSML [12, 13], DM [14], Isomap [7] and the proposed methods, (b) Zoomed in version of (a) with LSML [12, 13], DM [14], Isomap [7] and the proposed methods for better visualization, (c) Comparative residual variance results. Residual variance for LLE is omitted since it gives very high values.
Table 1.
Performance analysis on GB dataset with different noise variances (σ2) (top), on spiral dataset (middle) and on Swiss roll dataset (bottom).
| σ2 | LTSA | LLE | SDE | LSML | DM | Isomap | Proposed | |
|---|---|---|---|---|---|---|---|---|
| # Violated Constraints | 0.001 | 45856 | 10085 | 10500 | 7999 | 8204 | 4325 | 4738 |
| 0.012 | 50324 | 17520 | 21111 | 11305 | 14250 | 8369 | 8140 | |
| 0.023 | 49974 | 29876 | 27048 | 18646 | 18306 | 13077 | 13154 | |
| 0.034 | 50137 | 18397 | 23712 | 22296 | 18881 | 12074 | 12126 | |
| 0.045 | 56904 | 42447 | 40638 | 32656 | 21800 | 16168 | 15380 | |
| 0.056 | 55594 | 29117 | 33005 | 22574 | 25582 | 19745 | 18766 | |
| 0.067 | 61210 | 255814 | 49481 | 29075 | 30130 | 24515 | 23671 | |
| 0.078 | 64284 | 34232 | 38870 | 33809 | 34714 | 27706 | 26834 | |
| 0.089 | 59952 | 32911 | 43207 | 29071 | 33610 | 24856 | 23735 | |
| 0.1 | 63429 | 41436 | 46387 | 36335 | 36975 | 32244 | 29907 | |
| Residual Variance | 0.001 | 0.0320 | 0.9640 | 0.0002 | 0.0007 | 0.0001 | 0.00003 | 0.00004 |
| 0.012 | 0.0392 | 0.9503 | 0.0066 | 0.0029 | 0.0035 | 0.0019 | 0.0019 | |
| 0.023 | 0.0376 | 0.9863 | 0.0103 | 0.0052 | 0.0045 | 0.0030 | 0.0029 | |
| 0.034 | 0.0411 | 0.9597 | 0.0082 | 0.0103 | 0.0049 | 0.00297 | 0.00299 | |
| 0.045 | 0.0479 | 0.9826 | 0.0242 | 0.0303 | 0.0082 | 0.0066 | 0.0063 | |
| 0.056 | 0.0460 | 0.9768 | 0.0165 | 0.0095 | 0.0110 | 0.0086 | 0.0080 | |
| 0.067 | 0.0548 | 1.0 | 0.0372 | 0.0161 | 0.0153 | 0.0129 | 0.0120 | |
| 0.078 | 0.0619 | 0.9611 | 0.0255 | 0.0201 | 0.0207 | 0.0166 | 0.0151 | |
| 0.089 | 0.0559 | 0.9626 | 0.0317 | 0.0169 | 0.0197 | 0.0158 | 0.0132 | |
| 0.1 | 0.0591 | 0.9838 | 0.0338 | 0.0230 | 0.0238 | 0.0210 | 0.0179 |
| LTSA | LLE | SDE | LSML | DM | Isomap | Proposed | |
|---|---|---|---|---|---|---|---|
| #Violated Constraints | 114430 | 210611 | 17138 | 16814 | 231680 | 8729 | 8519 |
| CVP | 0.1526 | 0.2809 | 0.0229 | 0.0224 | 0.3090 | 0.0116 | 0.0114 |
| Residual Variance | 0.2151 | 0.9063 | 0.0040 | 0.0049 | 0.78 | 0.0013 | 0.0010 |
| LTSA | LLE | SDE | LSML | DM | Isomap | Proposed | |
|---|---|---|---|---|---|---|---|
| #Violated Constraints | 1151299 | 440909 | 262700 | 183962 | 1281232 | 97043 | 68838 |
| CVP | 0.1807 | 0.0692 | 0.0412 | 0.0289 | 0.2011 | 0.0152 | 0.0108 |
| Residual Variance | 0.3646 | 0.0386 | 0.0136 | 0.0094 | 0.4917 | 0.0019 | 0.0011 |
Figure 1.
(a) Growing band dataset for noise variance σ2 = 0.045. The 2D data samples are indicated with blue squares. X coordinates of red dots illustrate the original 1D manifold. (b) Resulting h(·) function on GB dataset with σ2 = 0.1.
Secondly, we carry out experiments on a noisy spiral data with N = 50 samples (see Figure 3(a)). Resulting h(·) function is shown in Figure 3(b). The comparative results can be seen in Table 1. We outperform all other methods, but the performance of the proposed method is close to Isomap, and this can also be seen in h(·) function in Figure 3(b) which demonstrates an almost linear relationship. Even though we achieve an almost linear fitting, in the proposed method, we do not assume any predefined model in the algorithm, instead we learn this linear relationship as part of the solution. Other methods that give a comparable performance with the proposed method on this dataset are SDE and LSML.
Figure 3.
Spiral dataset: (a) Noisy data samples are shown with blue squares, red dots indicate the original samples before adding noise, (b) resulting h(·) function.
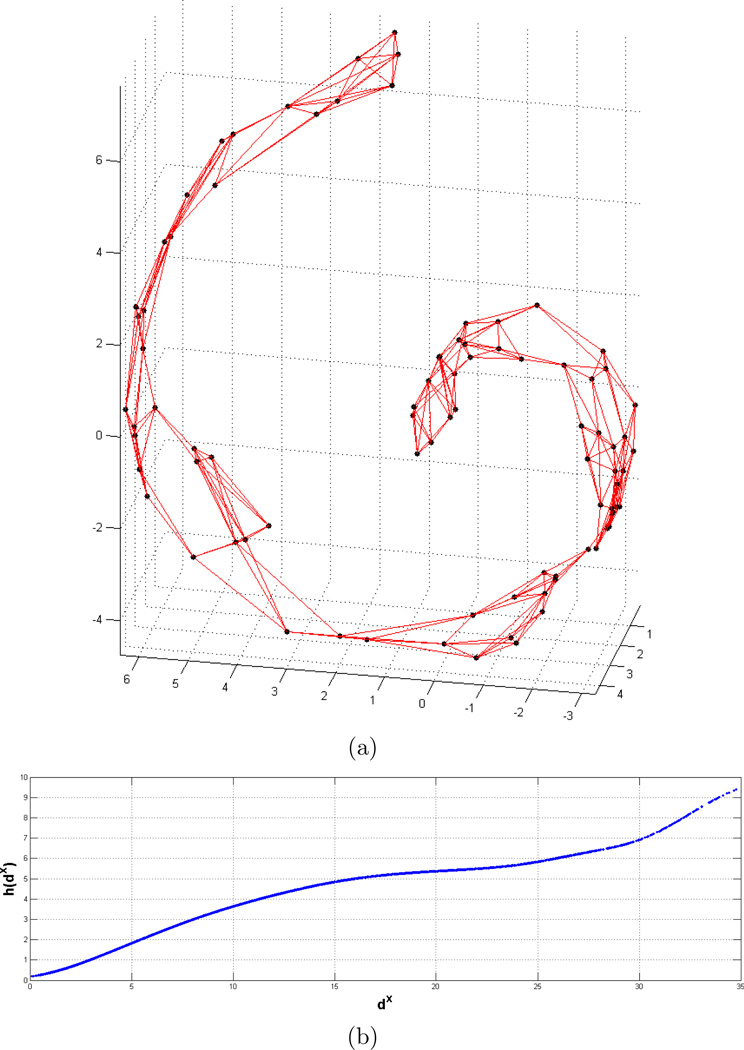
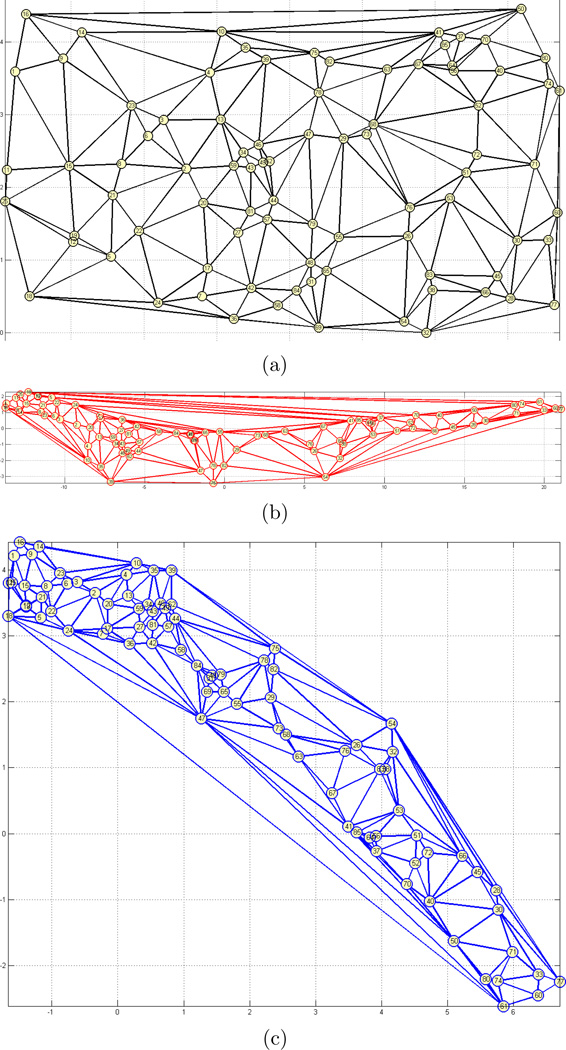
Thirdly, we perform experiments on the Swiss roll dataset with N = 85 samples (Figure 4(a)). 3D samples from the original space is projected to a 2D space in this experiment. Our goal is to demonstrate the performance of our method on a dataset having different curvature levels. Table 1 and Figure 5 report the comparative results. Also, resulting h(·) function that represents the relationship between dx and dys is reported in Figure 4(b). Since data is synthetically generated, original 2D coordinates are known and are displayed in Figure 5(a). Figures 5(b) and 5(c) show the 2D projection results with Isomap and the proposed methods, respectively. Each sample is identified with an index number and the numbers are displayed on the nodes overlayed with Delaunay triangulation edges for better visualization. In the figure, we only display the projection result for Isomap, since it has the closest performance to our method on this dataset. Although the 2D projection with the proposed method seems rotated with respect to the original data, it preserves the distance relations better than the Isomap method, and the results in Table 1 supports this observation. Note that, knn-graph does not contain any shortcuts between different sides of the Swiss roll due to the small k value. However, the motivation of the method is not to overcome the limitations of the geodesic distance estimation. Instead, we would like to solve the general optimization problem given an estimated distance matrix. With this purpose, same parameters are used for local neighborhood definitions in all of the methods.
Figure 4.
Swiss roll dataset: (a) Data samples are shown with black dots overlayed with the knn-graph edges (red) where k = 5, (b) resulting h(·) function.
Figure 5.
2D representation of the Swiss roll dataset: (a) Original data in 2D, (b) Result with Isomap, and (c) Result with the proposed method.
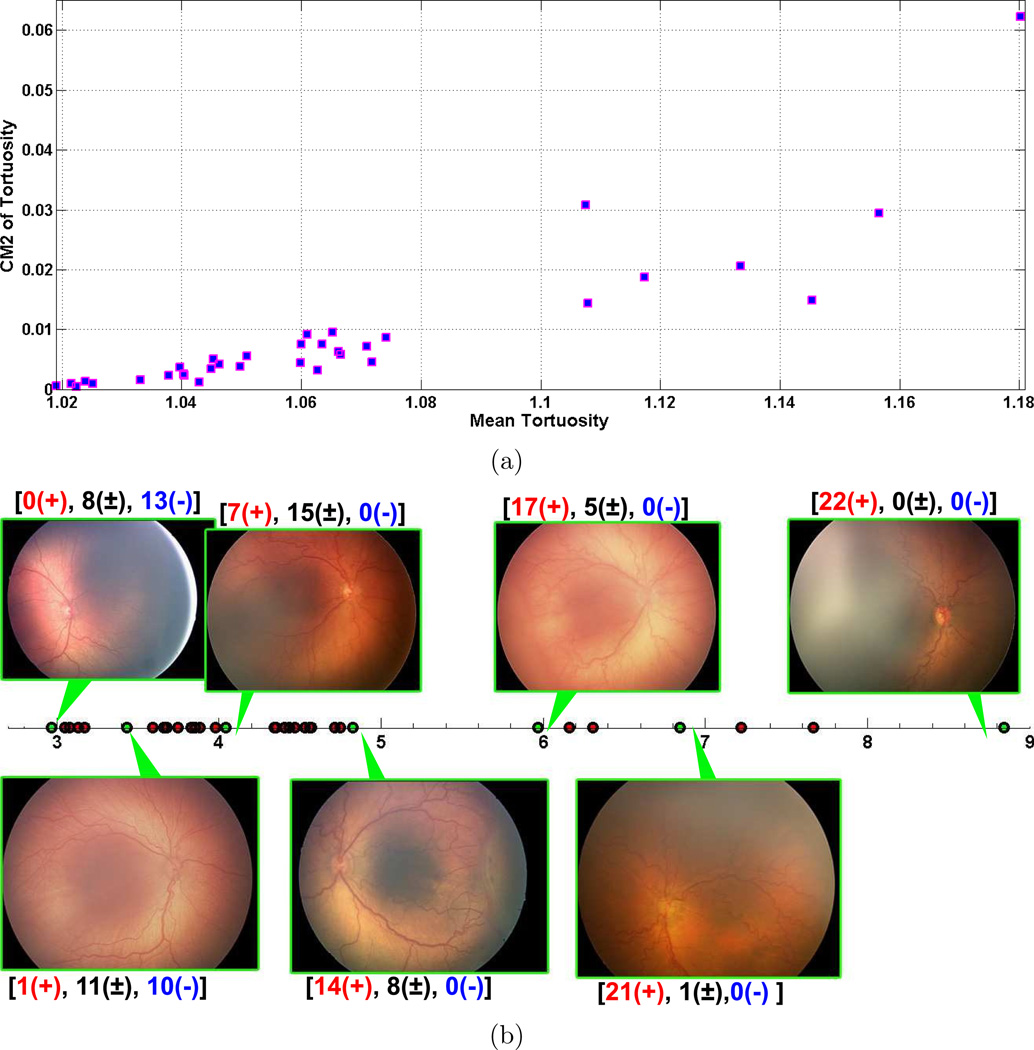
Lastly, we carry out experiments on a retinal image dataset. Retinal images are widely used by doctors to follow, treat and diagnose various diseases. Retinopathy of prematurity (ROP) is among the diseases that can be diagnosed through the use of retinal images. It is a disease affecting low-birth weight infants, in which blood vessels in the retina of the eye develop abnormally and cause potential blindness. ROP is diagnosed from dilated retinal examination by an ophthalmologist, and may be successfully treated if diagnosed appropriately [34]. According to the international classification system, clinical ROP diagnosis has three classes, plus disease, pre-plus and neither [35]. Plus disease is a critical parameter which identifies severe ROP and is characterized by tortuosity of the arteries and dilation of the veins in the retina. Pre-plus represents vascular abnormalities insufficient for plus disease but with more arterial tortuosity and venous dilation than normal. Infants with plus disease require treatment to prevent blindness, whereas those without plus disease may be monitored by serial ophthalmic examination without treatment. Studies have found that clinical plus disease diagnosis is often subjective and qualitative, and that there is significant inconsistency even among experts [36, 37]. Hence, diagnosis of ROP is a vital and challenging task. In this experiment, we use a retinal image dataset, that consists of 34 images that are diagnosed by 22 experts [38]. Vessels are manually segmented in the images. Based on manual segmentation, we compute cumulative tortuosity for each center line point of vessels and employ mean and second central moment (CM2) of these values as features [39]. For a curve, cumulative tortuosity is defined as the ratio of the curve length to the distance between the two endpoints. As can be seen in the scatter plot in Figure 6(a), the features represent a one dimensional manifold with varying noise levels as in GB dataset. Figure 6(b) shows result of our manifold learning algorithm on the dataset along with some example images. Expert diagnostic decisions are also displayed on top of each image where the numbers represent the number of experts decided plus disease (+), pre-plus (±) and neither (−) respectively. The most important observation that we make from this figure is that the amount of tortuos vessels increases as we go through the manifold. Tortuosity plays an important role for ROP diagnosis. We also observe this role by analyzing the expert diagnostic decisions on the manifold such that the number of plus-disease decisions increases, while the number of neither decisions decreases as we trace the projected points from left to right. Moreover, the number of pre-plus decisions is high in the images in the middle of the projected curve compared to the images at the left and rightmost ends. In this example, preserving distance orders during dimensionality reduction is crucial, since tortuosity is a critical factor for ROP diagnosis.
Figure 6.
Experiment on retinal images. (a) scatter plot of the features: mean tortuosity and second central moment (CM2) of tortuosity, (b) result of the proposed method along with some example images indicated with green dots. Horizontal axis represents the projected points. The numbers above each image show the number of expert decisions plus disease (+), pre-plus (±) and neither (−) respectively. Notice that, amount of curvy vessels in images increases as we go from left to right. This correlates with the expert diagnostic decisions such that the number of plus disease decisions increases from left to right, while the number of neither decisions decreases. Moreover, the number of pre-plus decisions is high in the images in the middle of the manifold and lower for images at the left and rightmost ends.
4. Conclusion
We developed a nonlinear dimensionality reduction method that is a generalization of multidimensional scaling technique. Rather than using the common mean-squared error as an unconstrained cost function, we formulated a constrained optimization problem. In this problem, we assumed an unknown monotonic nondecreasing relationship modeled by radial basis function interpolation between the distances in the original and the projected spaces to be learned as a part of the manifold learning problem. Moreover, we incorporated explicit constraints on the distance orders in the projected space. Using interior-point methods, we solved this optimization problem, and in addition to obtaining the low-dimensional representation, we also learned the nonlinear relationship between the distances. We also proposed a new performance evaluation metric based on the number of the violated distance orders. Using this metric and residual variance as the performance measures, we compared our algorithm with other popular methods on synthetic datasets. The experiments demonstrated that the proposed method outperforms the other algorithms. The local algorithms performed poorly due to the sparsity of the data. The algorithm that performed close to the proposed method was Isomap, which is global and is a special case of our method. We applied the proposed algorithm on a real dataset where preserving distance orders is crucial for correct disease diagnosis. Future work includes extending the formulation of the optimization problem with convex relaxation to obtain faster solutions for larger datasets.
highlights.
-
-
A novel manifold learning method which preserves pairwise distance order relations in the projected space
-
-
Theoretical formulation of the constrained optimization problem extending classical MDS-based mean-squared error minimization
-
-
A new performance metric that involves number of preserved distance orders
-
-
Proposed method also provides the relation between distances in original and lower dimensional spaces
Acknowledgements
This work is partially supported by NIH 1R21EY022387-01A1, NSF IIS-1118061, NSF IIS-1149570, MGHPCC 2012 Seed Funds, and Northeastern University 2013 Tier 1 Seed Funds.
Appendix
In this appendix, we compute the gradient of the cost function, the Jacobian of the inequality constraints, and the Hessian of the Lagrangian that we define in (15) and (16). We first start with the derivation of the gradient ∇θ f (θ). Following the definition of θ
| (20) |
Then rewriting and , where w is defined in (7), , and α = [α1, …, αM−1], we compute
| (21a) |
| (21b) |
Note here that , then
| (22) |
Recall that DT (θ) = [∇θc1(θ) ⋯ ∇θc3M−2(θ)], then using the definition of c(θ), we have DT (θ) = [AT ∇θ g1(θ) ⋯ ∇θ gM−1 (θ)]. Here . If and , from Section 2.2, gn (θ) = ‖ yl − yk ‖ − ‖ yi − yj ‖ + ξn, then ∇wgn (θ) = 0, ∇ξ gn (θ) = [0, …, 0, 1, 0, …, 0], a vector of zeros with 1 at the nth location, and . From the definition of gn (θ), we have ∇ym gn(θ) = 0 for m = 1, …, N, and m ≠ i, j, k, l, and
| (23) |
Recall the definition of Hessian , using this definition, we first demonstrate the computation of the Hessian of the cost function
| (24) |
Then using (21), it is easy to show that . Again from (21), we compute
| (25) |
Noticing that , we have
| (26) |
We can write , then
| (27) |
To complete the derivation of the Hessian of the Lagrangian, we continue with the computation of for m = 1, …, 3M − 2. Recalling the formula for the Jacobian of the inequality constraints we derive above, it is straightforward to show that for m = 1, …, 2M − 1, and for m = 2M, …, 3M − 2 such that
| (28) |
Then using (23) and the computation of the Jacobian matrix, we can show that , and
| (29) |
From (23) and the definition of gn (θ) recall that ∇ym gn (θ) = 0 for m = 1, …, N, and m ≠ i, j, k, l, then , for m′ = 1, …, N, also . We compute
| (30) |
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Esra Ataer-Cansizoglu, Email: ataer@ece.neu.edu.
Murat Akcakaya, Email: akcakaya@ece.neu.edu.
Umut Orhan, Email: orhan@ece.neu.edu.
Deniz Erdogmus, Email: erdogmus@ece.neu.edu.
References
- 1.Van der Maaten L, Postma E, Van Den Herik H. Dimensionality reduction: A comparative review. Journal of Machine Learning Research. 2009;10:1–41. [Google Scholar]
- 2.Kruskal J. Multidimensional scaling by optimizing goodness of fit to a nonmetric hypothesis. Psychometrika. 1964;29(1):1–27. [Google Scholar]
- 3.Dzwinel W, Blasiak J. Method of particles in visual clustering of multidimensional and large data sets. Future Generation Computer Systems. 1999;15(3):365–379. [Google Scholar]
- 4.Pawliczek P, Dzwinel W, Yuen DA. Visual exploration of data by using multidimensional scaling on multicore CPU, GPU, and MPI cluster. Concurrency and Computation: Practice and Experience [Google Scholar]
- 5.Pawliczek P, Dzwinel W. Interactive Data Mining by Using Multidimensional Scaling. Procedia Computer Science. 2013;18:40–49. [Google Scholar]
- 6.Andrecut M. Molecular dynamics multidimensional scaling. Physics Letters A. 2009;373(23):2001–2006. [Google Scholar]
- 7.Tenenbaum J, De Silva V, Langford J. A global geometric framework for nonlinear dimensionality reduction. Science. 2000;290(5500):2319–2323. doi: 10.1126/science.290.5500.2319. [DOI] [PubMed] [Google Scholar]
- 8.Silva V, Tenenbaum J. Global versus local methods in nonlinear dimensionality reduction. Advances in Neural Information Processing Systems (NIPS) 2002;15:705–712. [Google Scholar]
- 9.Zhang Z, Zha H. Principal manifolds and nonlinear dimensionality reduction via tangent space alignment. Journal of Shanghai University (English Edition) 2004;8(4):406–424. [Google Scholar]
- 10.Roweis S, Saul L. Nonlinear dimensionality reduction by locally linear embedding. Science. 2000;290(5500):2323–2326. doi: 10.1126/science.290.5500.2323. [DOI] [PubMed] [Google Scholar]
- 11.Weinberger K, Saul L. Unsupervised learning of image manifolds by semidefinite programming. International Journal of Computer Vision. 2006;70(1):77–90. [Google Scholar]
- 12.Dollár P, Rabaud V, Belongie S. Learning to Traverse Image Manifolds. Advances in Neural Information Processing Systems (NIPS) 2006 [Google Scholar]
- 13.Dollár P, Rabaud V, Belongie S. Non-Isometric Manifold Learning: Analysis and an Algorithm; International conference on Machine Learning (ICML); 2007. [Google Scholar]
- 14.Coifman RR, Lafon S. Diffusion maps. Applied and computational harmonic analysis. 2006;21(1):5–30. [Google Scholar]
- 15.Hinton GE, Roweis ST. Stochastic neighbor embedding. Advances in Neural Information Processing Systems (NIPS) 2002:833–840. [Google Scholar]
- 16.Xie B, Mu Y, Tao D, Huang K. m-SNE: Multiview stochastic neighbor embedding, IEEE Transactions on Systems. Man, and Cybernetics. 2011;41(4):1088–1096. doi: 10.1109/TSMCB.2011.2106208. [DOI] [PubMed] [Google Scholar]
- 17.Van der Maaten L, Hinton G. Visualizing data using t-SNE. Journal of Machine Learning Research. 2008;9(2579–2605):85. [Google Scholar]
- 18.Teh Y, Roweis S. Automatic alignment of local representations. Advances in Neural Information Processing Systems (NIPS) 2002;15:841–848. [Google Scholar]
- 19.Brand M. Charting a manifold. Advances in Neural Information Processing Systems (NIPS) 2003:985–992. [Google Scholar]
- 20.Sammon J., Jr A nonlinear mapping for data structure analysis. IEEE Transactions on Computers. 1969;100(5):401–409. [Google Scholar]
- 21.Floyd R. Algorithm 97: shortest path. Communications of the ACM. 1962;5(6):345. [Google Scholar]
- 22.Hofmann T, Schölkopf B, Smola A. Kernel methods in machine learning. The annals of statistics. 2008:1171–1220. [Google Scholar]
- 23.Park J, Sandberg I. Universal approximation using radial-basis-function networks. Neural computation. 1991;3(2):246–257. doi: 10.1162/neco.1991.3.2.246. [DOI] [PubMed] [Google Scholar]
- 24.Silverman B. Density estimation for statistics and data analysis. Vol. 26. Chapman & Hall/CRC; 1986. [Google Scholar]
- 25.Nocedal J, Wright S. Numerical optimization. Springer verlag; 1999. [Google Scholar]
- 26.Boggs P, Tolle J. Sequential quadratic programming. Acta numerica. 1996;4(1):1–51. [Google Scholar]
- 27.Friedlander M, Saunders M. A globally convergent linearly constrained Lagrangian method for nonlinear optimization. SIAM Journal on Optimization. 2005;15(3):863–897. [Google Scholar]
- 28.Forsgren A, Gill P, Wright M. Interior methods for nonlinear optimization. SIAM review. 2002;44(4):525–597. [Google Scholar]
- 29.Gould N, Orban D, Toint P. Numerical methods for large-scale non-linear optimization. Acta Numerica. 2005;14:299–361. [Google Scholar]
- 30.Byrd R, Gilbert J, Nocedal J. A trust region method based on interior point techniques for nonlinear programming. Mathematical Programming. 2000;89(1):149–185. [Google Scholar]
- 31.Byrd R, Hribar M, Nocedal J. An interior point algorithm for large-scale nonlinear programming. SIAM Journal on Optimization. 1999;9(4):877–900. [Google Scholar]
- 32.Waltz R, Morales J, Nocedal J, Orban D. An interior algorithm for nonlinear optimization that combines line search and trust region steps. Mathematical Programming. 2006;107(3):391–408. [Google Scholar]
- 33.Nocedal J. Updating quasi-Newton matrices with limited storage. Mathematics of computation. 1980;35(151):773–782. [Google Scholar]
- 34.Early Treatment For ROP Cooperative Group. Revised indications for the treatment of retinopathy of prematurity; results of the early treatment for retinopathy of prematurity randomized trial. Arch Ophthalmol. 2003;121:1684–1694. doi: 10.1001/archopht.121.12.1684. [DOI] [PubMed] [Google Scholar]
- 35.The Committee for the Classification of Retinopathy of Prematurity. The international classification of retinopathy of prematurity revisited. Arch Ophthalmol. 2005;123:991–999. doi: 10.1001/archopht.123.7.991. [DOI] [PubMed] [Google Scholar]
- 36.Chiang M, Jiang L, Gelman R, Du Y. Inter expert agreement of plus disease diagnosis in retinopathy of prematurity. Arch Ophthalmol. 2007;125:875–880. doi: 10.1001/archopht.125.7.875. [DOI] [PubMed] [Google Scholar]
- 37.Wallace DK, Quinn GE, Freedman SF, Chiang MF. Agreement among pediatric ophthalmologists in diagnosing plus and pre-plus disease in retinopathy of prematurity. Journal of American Association for Pediatric Ophthalmology and Strabismus. 2008;12(4):352. doi: 10.1016/j.jaapos.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gelman R, Jiang L, Du Y, Martinez-Perez M, Flynn J, Chiang M. Plus disease in retinopathy of prematurity: pilot study of computer-based and expert diagnosis. Journal of American Association for Pediatric Ophthalmology and Strabismus. 2007;11(6):532–540. doi: 10.1016/j.jaapos.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ataer-Cansizoglu E, You S, Kalpathy-Cramer J, Keck K, Chiang M, Erdogmus D. Observer and feature analysis on diagnosis of retinopathy of prematurity. IEEE International Workshop on Machine Learning for Signal Processing (MLSP) 2012:1–6. doi: 10.1109/MLSP.2012.6349809. [DOI] [PMC free article] [PubMed] [Google Scholar]