Abstract
Osteosarcoma is a neoplasm of mesenchymal origin with features of osteogenic differentiation. Patients with recurrent or metastatic disease have a very poor prognosis. To define the landscape of somatic mutations in pediatric osteosarcoma, we performed whole-genome sequencing of DNA from 20 osteosarcoma tumor samples and matched normal tissue (obtained from 19 patients) in the discovery cohort as well as 14 samples from 13 patients in the validation cohort. Our results demonstrate that pediatric osteosarcoma is characterized by multiple somatic chromosomal lesions, including structural variations (SVs) and copy number alterations (CNAs). Moreover, single nucleotide variations (SNVs) exhibit a pattern of localized hypermutation called “kataegis” in 50% of the tumors. Despite these regions of kataegis across the osteosarcoma genomes, we detected relatively few recurrent SNVs, and only when SVs were included did we identify the major pathways that are mutated in osteosarcoma. We identified p53 pathway lesions in all 19 patient’s tumors in the discovery cohort, 9 of which were translocations in the first intron of the TP53 gene, leading to gene inactivation. This mechanism of p53 gene inactivation is unique to osteosarcoma among pediatric cancers. In an additional cohort of 32 patients, TP53 gene alterations were identified in 29 of those tumors. Beyond TP53, the RB1, ATRX and DLG2 genes showed recurrent somatic alterations (SNVs and/or SVs) in 29–53% of the tumors. These data highlight the power of whole-genome sequencing in identifying recurrent somatic alterations in cancer genomes that may be missed using other methods.
INTRODUCTION
Osteosarcoma is the most common malignant bone tumor in children and adolescents, with approximately 400 new cases each year in the United States(Ottaviani and Jaffe, 2009). Although most cases are sporadic, the risk of osteosarcoma is increased in those with various genetic diseases, including hereditary retinoblastoma, Li Fraumeni syndrome, and germ line mutations of RecQL4(Hicks et al., 2007; Kleinerman et al., 2005; McIntyre et al., 1994). Current multimodal therapies that incorporate surgical excision and combination chemotherapy (i.e., doxorubicin, methotrexate, and cisplatin) cure approximately 70% of patients (Meyers et al., 2005). However, clinical outcomes and therapeutic strategies have remained virtually unchanged over the past 20 years (Smith et al., 2010). Prognosis depends on the presence of metastatic disease, completeness of surgical resection, and histologic response to preoperative chemotherapy (Bielack et al., 2002). Patients with either metastatic disease at diagnosis or recurrent disease have a poor prognosis (Goorin et al., 2002; Kempf-Bielack et al., 2005; Meyers et al., 2005). A detailed whole-genome sequence (WGS) analysis of osteosarcoma has not been performed, and the results could offer valuable insights into the molecular pathology of this disease.
Previous genomic studies have demonstrated that osteosarcoma has some of the highest rates of chromosomal mutations, i.e., copy number variations (CNVs) and structural variations (SVs), in human cancer (Bridge et al., 1997; Sadikovic et al., 2009; Selvarajah et al., 2008; Squire et al., 2003). These lesions have been attributed, in part, to mutations in the RB1 and TP53 tumor suppressor genes, which are believed to be required for maintenance of genomic stability (Manning and Dyson, 2012; Meek, 2009). Indeed, the targeted deletion of p53 and Rb1 in murine osteoblasts is sufficient to induce metastatic osteosarcoma with molecular, cellular, and genomic features of the human disease (Walkley et al., 2008). Tumors with missense mutations in the TP53 gene may have higher levels of genomic instability (Liu et al., 2010) and may be more aggressive than those with mutations that eliminate p53 protein expression (Muller et al., 2011). Unfortunately, the high rates of CNVs and SVs in osteosarcoma have made it difficult to distinguish driver mutations from passenger mutations, and this has slowed the development of molecular-targeted therapies for this disease. Moreover, it is not known why some tumors fail to respond to therapy and metastasize.
In this study, we characterized the genomic landscape of osteosarcoma by performing WGS on 34 osteosarcoma tumor and matched non-tumor tissue samples from 32 patients. Our results demonstrated that pediatric osteosarcomas had one of the highest rates of SVs of any pediatric cancer sequenced to date (Downing et al., 2012b), but had relatively few recurrent SNVs. However, when SVs and SNVs were combined, inactivating mutations were identified in the p53 pathway in ~95% of tumors, with a majority of the TP53 mutations being translocations or focal deletions. Recurrent SVs were also identified in RB1, ATRX and DLG2. Taken together our results provide novel insights into the molecular pathology of pediatric osteosarcoma and demonstrate that comprehensive WGS is required to elucidate the complete genetic landscape of osteosarcoma.
RESULTS
Whole-Genome Sequencing of Primary and Metastatic Osteosarcoma
Using a paired-end sequencing approach, we generated 10,265 Gb of sequence data for DNA in 20 tumor specimens and matched normal DNA from 19 patients with osteosarcoma in our discovery cohort and 14 tumor specimens and matched normal DNA from 13 patients in validation cohort (Suppl. Table 1); 9,671 Gb (94%) were successfully mapped to the reference genome (Suppl. Table 2). In the discovery cohort, the samples included 17 pretreatment diagnostic samples (16 primary, 1 metastatic), 1 recurrent metastatic sample (SJOS001), and two tumor specimens from the same patient (SJOS010). This patient had metachronous osteosarcoma: the original tumor was in the right femur; 4 years later, osteosarcoma was found in the left femur; 3 years thereafter, osteosarcoma was detected in the left tibia. There was no evidence of lung metastases at the time of surgical resection of the tumor specimen (SJOS010_D) from the left tibia. The second tumor specimen (SJOS010_M) from this patient was a metastatic lung lesion that was removed 2 years after the first sample was obtained from the left tibia (Suppl. Table 1).
All 20 tumor samples in the discovery cohort were conventional high-grade intramedullary osteosarcoma. The average genome coverage was 44×, and the average exon coverage was 39×; 99% of SNPs detected across all 39 genomes showed concordance with their corresponding SNP array genotype calls, at the same genomic positions (Suppl. Table 2). Validation was carried out using custom liquid capture for all SNVs, SVs, and insertions or deletions (indels) identified in the original sequence data. Combining the discovery and validation cohorts, we identified 50,426 validated somatic sequence mutations and 10,806 SVs (Suppl. Table 3 and Suppl. Materials and Methods). These included 856 nonsilent tier-1 mutations in genes, 4,651 tier-2 mutations in evolutionarily conserved regions of the genome, and 43,782 tier-3 mutations in nonrepetitive regions of the genome that are not part of tiers 1 or 2 (Suppl. Table 3). The average number of sequence mutations was 1483.1 per case (range, 610–5,178), with 25.2 mutations per case (range, 5–103) resulting in amino acid changes (Suppl. Table 3). The estimated mean mutation rate was 1.15×10−6 per base (range 4.90×10−7–3.99×10−6). Among the validated SVs, 377 were predicted to produce an in-frame fusion protein (Suppl. Table 3). Good quality RNASeq data of were available for 5 tumors with 64 predicted fusion SVs. Among them, 15 SVs (23%) were expressed (Suppl. Table 4).
Primary and metastatic osteosarcomas had high rates of validated SVs (Fig. 1a, Suppl. Fig. 1 and Suppl. Table 1). The number of SVs and CNVs, background mutation rate, and number of nonsilent tier-1 mutations were significantly higher in osteosarcoma compared to medulloblastoma and T-ALL (Robinson et al., 2012; Zhang et al., 2012) (Fig. 1b). However, only the number of SVs was significantly higher in osteosarcoma compared to another pediatric solid tumor with high rates of somatic alterations (embryonal rhabdomyosarcoma (Chen et al., 2013)) (Fig. 1b). SVs and CNVs in cancer can be caused by sequential accumulation of distinct lesions or acute events such as chromothripsis (Stephens et al., 2011). Global patterns of SV and CNV from WGS analysis of osteosarcoma suggest that the majority of them were generated by sequential accumulation of SVs (Fig. 1c, d) but chromothripsis was detected at specific genomic regions in 4 samples (chr14 in SJOS002_D, chr17q in SJOS003_D, chr6q in SJOS005_D and chr13 in SJOS010_M, Suppl. Table 5 and Suppl. Materials and Methods). We used a modified version of GISTIC analysis (Suppl. Materials and Methods) to identify regions of the osteosarcoma genome with recurrent copy number alterations in the discovery cohort. The TP53, RB1, MYC and PTEN pathways were recurrently mutated as well as ATRX, LSAMP-AS3, CCNE1 and a genomic region on chromosome 16 containing COPS3, PMP22, MAPK7, NCOR1 and UBB (Suppl. Fig. 1c). Among SNVs with sufficient coverage in both SJOS010 samples (20X), 673 SNVs were validated in both samples, 1686 in diagnostic only and 1408 in metastasis only, indicating that these 2 tumors shared limited amount of common mutations and were divergent early in the progression.
Figure 1. Whole genome sequencing of osteosarcoma.
a) Representative CIRCOS plots of validated mutations and chromosomal lesions in a diagnostic and metastatic osteosarcoma tumor from different patients. Loss of heterozygosity (orange), gain (red), and losses (blue) are shown. Intrachromosomal translocations (green lines) and interchromosomal translocations (purple lines) are indicated. Sequence mutations in Refseq genes included silent single nucleotide variants (SNVs, green), nonsense and missense SNVs (brown), splice-site mutations (dark blue), and insertion/deletion mutations (red). An additional track was added to the innermost ring of the plot showing the density of SNVs to highlight regions adjacent to SVs characteristic of kataegis. b) Boxplots of validated basal mutation rate (BMR), number of non-silent single nucleotide variations (SNVs), total structural variations (SVs) and number of total copy number variations (CNVs) in the ERMS and ARMS tumors in the discovery cohort. The (*) represents statistical significance of p<0.001 as compared to the osteosarcoma genomes. c) Representative plot of sequence reads on chromosome 12 for the matched germline (green) and tumor (red) sample. Several distinct regions of copy number change are identified (arrows) spanning the MDM2 gene consistent with sequential chromosomal lesions. d) MDM2 FISH of SJOSO18 showing amplification (red) relative to the probe for chromosome 12 (green). Abbreviations: ERMS, embryonal rhabdomyosarcoma; TALL, T-cell acute lymphoblastic leukemia; MB, medulloblastoma.
Using the functional point mutations (including SNVs and indels) and structural variations, the GRIN(Pounds et al., 2013) package identified TP53 (FDR =3.6E-51, mutated in 28/34 samples) RB1 (FDR = 1.1E-5, mutated in 10/34 samples), ATRX (FDR = 2.4E-4, mutated in 10/34 samples) and DLG2 (FDR = 0.044, mutated in 18/34 samples) as significantly mutated genes. All genes excepted DLG2 were mutated by point mutations (9 for TP53, 3 for RB1 and 5 for ATRX) and SVs in multiple tumors (18 for TP53, 7 for RB1 and 5 for ATRX). DLG2 was exclusively mutated by SVs.
Osteosarcoma Tumor Purity and Tumor Heterogeneity
As a first step to analyze intratumor heterogeneity, the tumor purity (ratio of tumor cells to all cells) was estimated from the WGS data (Suppl. Materials and Methods). For each tumor, regions of the genome that had copy number alterations (CNAs) and corresponding changes in their loss of heterozygosity (LOH) were analyzed to identify the maximum proportional representation of a dominant somatic lesion in the tumor. The estimated tumor purity ranged from 44% to 89% (Suppl. Fig. 2). Next, we analyzed intratumor heterogeneity using the purity adjusted mutant allele frequency (MAF) derived from deep sequencing of all SNVs by capture enrichment and Illumina sequencing (Suppl. Materials and Methods). Eleven tumors (SJOS001_M, SJOS004, SJOS005, SJOS008, SJOS012, SJOS013, SJOS015, SJOS001103_D1, SJOS001105_D1, SJOS001123_D1, SJOS001125_D1) were excluded from quantitative heterogeneity analysis due to insufficient number of SNVs in copy-neutral regions. Statistical modeling demonstrated that 61% (14/23) of osteosarcomas in this group had evidence of multiple clones including metastatic samples SJOS010_M, SJOS001107_M1 and SJOS001107_M2 (Suppl. Fig. 2).
Kataegis in Osteosarcoma
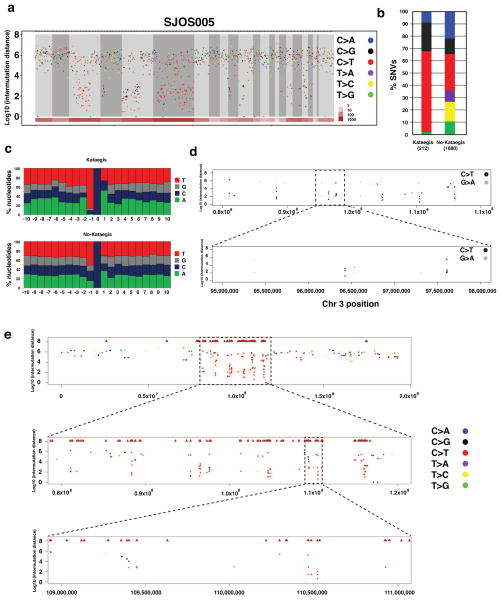
The background sequence mutation rate and the number of SVs in osteosarcomas are among the highest of the 15 pediatric cancers that have been analyzed by whole-genome sequencing to date (Downing et al., 2012a). To determine if there was any relationship between the SVs and location, distribution or type of SNV in the osteosarcoma genomes, we plotted the validated SVs and SNVs for each sample and analyzed the intermutation distance (Suppl. Fig. 3). Hypermutable regions with the 5 hallmarks of kataegis (Nik-Zainal et al., 2012) were identified in 17 of the osteosarcoma tumors (Fig. 2a and Suppl. Materials and Methods). The 5 characteristics of kataegis include: 1) enriched C->T and C->G substitutions at TpCpX trinucleotides (Figure 2b, c), 2) the same class of nucleotide mutation occurring for contiguous stretches before switching to a different class (Figure 2d), 3) mutations within short stretches of the genome occurring on the same parental chromosome (Suppl. Fig. 3a), 4) clustering of heavily mutated short stretches of the genome at multiple scales (shown at whole chromosome scale, 40 Mb scale and 2 Mb scale in Figure 2e), and 5) association of the hypermutated region with SV breakpoints (Fig. 2e and Suppl. Information). The regions of the genome with kataegis were not recurrent in our cohort nor were they associated with recurrently mutated genes in osteosarcoma (Suppl. Fig. 3b). Tier1 SNVs in kataegis regions were not significantly associated with the expression status (p=0.16 by Fisher’s exact test).
Figure 2. Kataegis in osteosarcoma.
a) Rainfall plot showing the Log10 of the intermutation distance versus genomic position for a representative osteosarcoma sample (SJOS005) with evidence of Kataegis. The chromosomes are demarcated by gray shading and the number of SVs in each chromosome is shown in brown at the bottom. The validated SNVs are plotted and color coded by the type of mutation. b) The proportion of each type of validated SNV in osteosarcomas with evidence of kataegis versus those without kataegis. c) The distribution of each nucleotide sequence 5′ to the C mutations in tumors with kataegis and those without kataegis. d) A rainfall plot in a representative regions of chromosome 3 in SJOS005 with kataegis showing the strand of the hypermutation based on the C>T or G>A sequence clusters. f) A macrocluster of hypermutation with evidence of kataegis on chromosome 3 of SJOS005 with two sequential magnifications (boxes) showing the existence of microclusters within a single macrocluster.
Chronology of Kataegis, SVs, and Aneuploidy in SJOS005
SJOS005 has the highest proportion (11%) of kataegis SNVs in our cohort. The large number of kataegis SNVs (n=212) coupled with the accurate measurement of mutant allele fraction (MAF) of all SNVs derived from deep sequencing allowed us to analyze the chronology of kataegis in relation to other mutational events in this tumor. First, we examined MAF of SNVs in kataegis micro-clusters containing 5 or more consecutive kataegis SNVs within 10 kb. The MAF variance is relatively small (6.7% of overall variance) within a micro-cluster albeit a wide range of MAFs across micro-clusters (range: 0.142 – 0.839, median 0.364, Suppl. Fig. 3c). This pattern, along with the observation that SNVs in a microcluster occurred on the same parental chromosome, supports the hypothesis that SNVs in a kataegis microcluster originated from a single event. MAF analysis of SVs flanking “kataegis” clusters (range: 0.132 – 0.866, median 0.396) also show a significant positive correlation (p = 4.56E-5) with those of “kataegis” SNVs and there was no significant difference between them (p = 0.143 by Wilcoxon signed rank test), indicating the neighboring SVs likely arise simultaneously with kataegis SNVs (Suppl. Fig. 3d,e).
SJOS005, like most osteosarcomas, exhibits aneuploidy with copy number gains spanning more than 50% of the genome. Kataegis SNVs were significantly enriched in regions of the genome with 4 or more copies compared with non-kataegis SNVs (Suppl. Fig. 3f, p < 2.2e-16, by Fisher’s exact test). However, MAF distribution of kataegis SNVs showed a large fraction of SNVs with multiple copies of the mutant allele in amplified regions while only a single copy of mutant alleles was found in the majority of the non- kataegis SNVs (Suppl. Fig. 3g). Taken together, these data suggest that kataegis likely occur before global aneuploidy and non-kataegis SNVs occur primarily after the aneuploidy.
Structural Variations in TP53
In 1990, SVs in the first intron of the TP53 gene were detected in 10% (6/60) of osteosarcomas by using Southern blotting (Miller et al., 1990). Since that original discovery, there has been no report of SVs in the TP53 gene in other human cancers. In a more recent study, Wunder et al. sequenced exons 4–10 of the TP53 gene in 196 osteosarcoma samples and found that 19.4% (38/196) had TP53 mutations. The investigators concluded that the remaining 80.6% (158/196) had wild-type TP53 (Wunder et al., 2005). They went on to show that event-free survival of the two groups was not associated with TP53 mutation status (Wunder et al., 2005). SVs in the first intron of TP53 were not analyzed in that study, nor were other genes in the p53 pathway (e.g., MDM2 or MDMX).
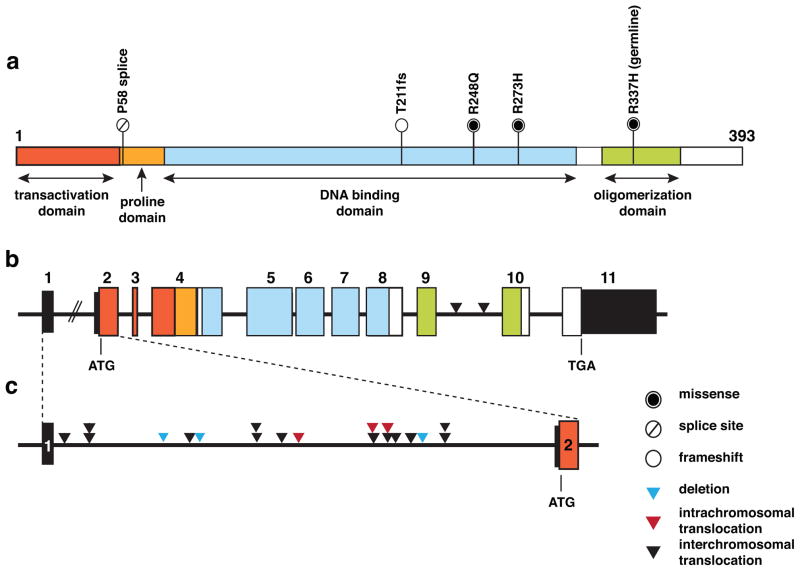
We discovered that the p53 pathway was mutated in all 20 tumor samples from the 19 patients in our discovery cohort. The majority (95%; 19/20) had either sequence mutations or structural variations in the TP53 gene, and one (SJOS018) had an MDM2 amplification (see Fig. 1c, d and Suppl. Table 4). Surprisingly, 55% of the tumors (11/20) had SVs in the TP53 gene, and the majority of those were translocations with breakpoints that were confined to the first intron of the gene (90%, 19/21 SV breakpoints, Fig. 3a–c and Suppl. Table 6). Based on our calculation of soft-clipped reads supporting the SV breakpoints, these loss-of function SVs were clonal. Indeed, some tumors had rearrangements in both alleles of TP53, resulting from two or more independent translocations (Suppl. Table 6). One patient’s tumor (SJOS006) had a germline SNV (R337H), one (SJOS012) had a somatic splice site mutation and two (SJOS004 and SJOS010) had somatic missense SNVs (Fig. 3a–c and Suppl. Table 6). The remaining 4 patients had tumors that harbored indels in the TP53 gene. Loss of heterozygosity (LOH) at the TP53 locus was evident in 40% (8/20) of the osteosarcoma tumors. Each of the lesions in TP53 was validated by custom liquid capture, along with all the other SNVs, SVs, and indels. In total, 15 tumors had biallelic inactivation of TP53, four had monoallelic inactivation of TP53, one had MDM2 amplification (Suppl. Table 6, Figure 1c).
Figure 3. Validated mutations in TP53.
a) Structure of the TP53 gene showing the transactivation, proline, DNA binding and oligomerization domains with splice site, frameshift and missense mutations in the 19 patient’s tumors in the discovery cohort. b) Structure of the genomic locus of the TP53 gene showing the exon boundaries color coded in accordance with the protein domains shown in (a). Sites of interchromosomal translocations are shown with black arrowheads between exons 9 and 10. The sizes of the introns and exons are scaled proportionally except for intron 1, which is much larger than the other introns in human TP53. c) A magnified view of intron 1 of TP53 showing the deletions (blue arrowheads), intrachromosomal (red arrowheads) and interchromosomal translocations (black arrowheads).
The majority (90%) of the TP53 associated SVs (18/20) did not have a fusion gene partner and were predicted to generate null alleles (Suppl. Table 6). Of the 2 SVs that have a fusion partner, one generated a fusion gene where the intact TP53 coding region (without Exon 1) was fused with the promoter and the first exon of ASIC2 in SJOS015_D (Suppl. Table 6). The other SV generated a fusion protein of TP53_SFSWAP (Suppl. Figure 4) in SJOS007_D.
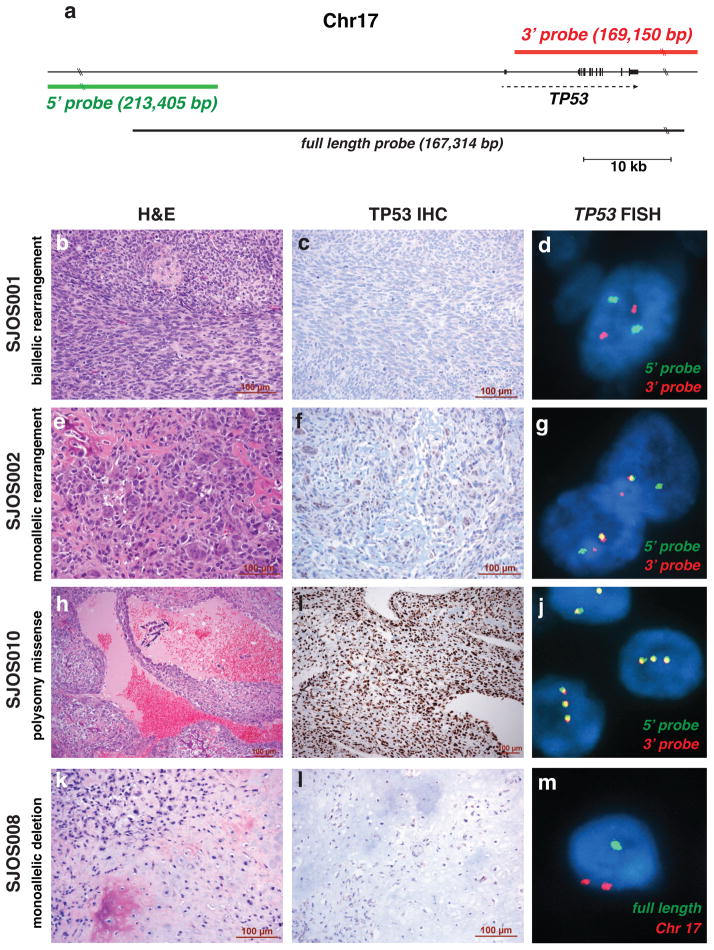
To further validate the translocations in the TP53 gene identified by WGS, we developed a break-apart fluorescence in situ hybridization (FISH) assay with separate probes spanning the 5′ and 3′ regions of the gene (Fig. 4a). We also developed a FISH assay with a probe spanning the entire TP53 gene (Fig. 4a) to assess ploidy and determine if the gene was deleted. To complement the FISH analysis, we performed P53 immunostaining to verify that the tumors with missense mutations had accumulated high levels of nuclear p53 protein. We successfully performed FISH in 18 of 20 tumors and p53 immunostaining on all 20 tumors (Suppl. Table 7). Among the 18 samples in which FISH was successfully performed, 10 showed evidence of TP53 gene rearrangement (Fig. 4b–m and Suppl. Table 7). Of the 3 tumors harboring a deletion in TP53 by WGS, one showed gene rearrangement by the TP53 break-apart FISH (Suppl. Table 7). Using the TP53 full-length FISH probe, one of the two showed gene deletion, whereas the other showed a polysomy pattern. Overall, there was perfect concordance between the WGS data and the FISH data. All 3 tumors (SJOS004, SJOS006, and SJOS010) with strong nuclear immunostaining for P53 (3+) had missense mutations in the gene. One tumor with structural change of p53 (SJOS007_D, with a predicted TP53_SFSWAP fusion) had weak staining (1+); the other osteosarcomas with gene translocations or deletion scored rare or negative for p53 immunostaining (Suppl. Table 7).
Figure 4. FISH and IHC for TP53 in Osteosarcoma.
a) Genomic location of the 5′ (green) and 3′ (red) break-apart FISH probes showing their position relative to the TP53 gene. The full-length probe used to identify deletions at the TP53 locus is shown in black. b–d) Images of H&E, TP53 IHC and break apart FISH for SJOS001 with biallelic rearrangement of the TP53 gene. e–g) Images of H&E, TP53 IHC and break apart FISH for SJOS002 with monoallelic rearrangement of the TP53 gene. h–j) Images of H&E, TP53 IHC and break apart FISH for SJOS010 with polysomy and a missense mutation leading to elevated accumulation of nuclear TP53 protein. k–m) Images of H&E, TP53 IHC and FISH using the full length probe (green) for SJOS008 with monoallelic deletion of TP53.
To extend the data on TP53 inactivation in osteosarcoma, we performed TP53 and MDM2 FISH, TP53 immunostaining, and TP53 exon sequencing on tumor samples from an additional 32 patients (Suppl. Table 8). We found that 50% (16/32) of the patients had TP53 rearrangements, 22% (7/32) had missense mutations, 16% (5/32) had nonsense mutations, 6% (2/32) had a TP53 deletion, and 3% (1/32) had an MDM2 amplification (Suppl. Table 8). There were three patients with tumor with no evidence of a p53 pathway mutation using these assays. Taken together, our data from the discovery and validation cohorts suggest that SVs in TP53 are the most common mechanism of p53 pathway inactivation in osteosarcoma.
Gain of function mutations in p53 can prevent ATM-dependent DNA repair in some contexts (Liu et al., 2010; Song et al., 2007). We did not find any significant difference in CNV (p=0.20 by Wilcoxon rank sum test), SV (p=0.85), SNV (p=0.43), non-silent tier 1 mutations (p=0.66) or background mutation rate (p=0.43) in the osteosarcoma samples with mutant p53 versus those with inactivating (nonsense, deletion and truncation) mutations in TP53. Clinical follow-up data was available for 44 patients in our combined cohort. At the time of analysis, 30 of the 44 patients were alive with a median follow-up time of 46.6 months (range 6.6–167.6 months). Survival analysis, including event-free survival and overall survival, did not show a significant difference in outcome for the patients whose tumors carried TP53-missense mutations (10 patients) versus those with TP53-truncating mutations (34 patients) with log-rank test p values of 0.88 and 0.64, respectively.
RB1, ATRX and DLG2 is Recurrently Mutated in Osteosarcoma
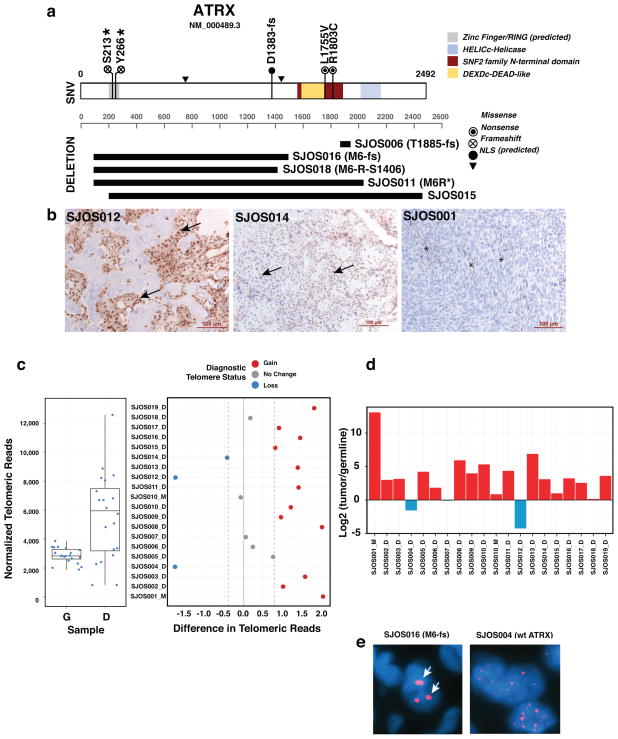
ATRX is part of a multiprotein complex that regulates chromatin remodeling, nucleosome assembly, and telomere maintenance. It has recently been shown that ATRX mutations in neuroblastoma are associated with age at diagnosis (Cheung et al., 2012). Most neuroblastomas with ATRX mutations have evidence of alternative lengthening of telomeres, as measured by WGS, telomere FISH, and telomere qPCR (Cheung et al., 2012). In our osteosarcoma discovery cohort, we identified five tumors (SJOS001, SJOS002, and SJOS007, SJOS001112-M2, SJOS001117-D1) with point mutations in ATRX and five with focal deletions or SVs affecting the coding region of the gene (Fig. 5a and Suppl. Table 9). There was no significant gender bias in ATRX mutations (p=0.25 by Fisher’s exact test) even though it is located on the X chromosome. There was no association between ATRX mutation and the type of TP53 mutation (missense versus inactivating mutations). By immunohistochemistry, 31% (6/19) of the tumors in the discovery cohort were ATRX negative (Fig. 5b and Suppl. Table 9). The sample with a missense mutation (SJOS007-R1803C) and one with a SV (SJOS018) were heterogeneous for ATRX protein expression. Analysis of telomere sequence reads from the WGS data and qPCR of telomeres showed that the majority of osteosarcomas had longer telomeres (Fig. 5c,d). Telomere FISH was successfully performed on 14 samples. Evidence of alternative lengthening of telomeres was found in 85% (12/14), including the 7 samples with ATRX mutations (Suppl. Table 9).
Figure 5. ATRX mutations correlate with ALT in osteosarcoma.
a) Diagram of the 5 single-nucleotide variations, 4 deletions and 1 inter-chromosome SV found in the ATRX genes of the osteosarcoma cohort. 3 of the samples (SJOS006, SJOS018 and SJOS011) with ATRX SVs had matching RNAseq data. SJ006 has a short deletion at exon 23 and the RNASeq data confirmed a read-through event that would result in a T1885 frameshift. For SJOS011, the RNAseq and WGS supported a junction connecting exon 1 to exon 28 creating a nonsense mutation (M6R*). For SJOS018, the RNASeq and WGS data supported a deletion connecting exon 1 to exon 13 thereby creating an in frame fusion protein (M6RS1406). The WGS for SJOS016 predicts a deletion that connects exon 1 to exon 16 producing a frameshift (M6fs). b) Representative IHC for ATRX showing nuclear ATRX in a sample with intense staining and wild type ATRX (SJOS012), a sample with fainter nuclear localized ATRX (SJOS014) and a sample with a nonsense mutation (SJOS001). Arrows indicate representative nuclei stained positive for ATRX. Asterisks indicate ATRX immunopositive vascular endothelial cells among the tumor cells that are negative for ATRX IHC. c,d) Relative telomere length in the osteosarcomas compared to that in the matched germline DNA, as analyzed by WGS and qPCR. e) Representative telomere FISH showing characteristics of ALT (arrow) in an osteosarcoma with an ATRX deletion.
Beyond TP53 and ATRX, other significant recurrent genetic lesions that were validated were RB1 (10/34, FDR q = 1.1E-5) and DLG2 (18/34, FDR q = 0.044). DLG2 encodes a multi-PDZ domain protein that is involved in epithelial polarity during cell division and has been implicated in cancer cell invasion. In Drosophila, DLG is a tumor suppressor, but a clear tumor suppressor function has not yet been confirmed for DLG2 in human cancer. All DLG2 lesions were SVs, consistent with the possibility that the genomic instability in osteosarcoma leads to recurrent genetic lesions in important cancer pathways that may contribute to disease progression. Additional studies on a larger cohort will be required to achieve any association with response to therapy or overall survival.
Structural Variations in Cancer Genes
Ostersarcoma has the highest number of SVs in the pediatric cancers analyzed so far and SVs breakpoints alone contributed 91% (9605/10523) of all functional genetic lesions in the combined discovery and validation WGS cohorts. To further evaluate the possibility that the genomic instability in osteosarcoma leads to mutations in important cancer genes, we tested whether the observed SV breakpoints were enriched in COSMIC cancer genes. In total, 122 cancer genes had at least one SV breakpoint in the WGS cohort (Suppl. Table 10). All but one tumor (SJOS001118_D1) had at least one breakpoint (range from 1 to 40) in a cancer gene. There were a total of 415 SV breakpoints across all cancer genes among all WGS tumors, ranging from 0 to 38 (TP53), which was highly enriched (p = 2.5E-11) estimated by the GRIN software. SV breakpoint enrichment in the cancer genes was highly significant even when we excluded TP53 from the list (p = 2.5E-6). Twelve of the 34 tumors (35%) achieved significant enrichment of SV breakpoints in cancer genes individually. In addition, some tumors have “fold-back intrchromosomal translocations” (Campbell et al., 2010) to inactivate tumor suppressor genes (Suppl. Figure 5). These results further supported the hypothesis that genomic instability leads to lesions in various cancer genes.
MATERIALS AND METHODS
Full details of sample acquisition, molecular and biochemical procedures, informatics and whole genome sequencing are provided in the Supplementary Information. All tumors in this study were from St. Jude Children’s Research Hospital (SJCRH) patients. The SJCRH IRB approved experiments involving human subjects and informed consent was obtained from all subjects. The European Bioinformatics Institute accession number is EGAS00001000263.
DISCUSSION
Whole-genome sequencing of osteosarcoma demonstrated that the rate of SNVs was similar to that in other pediatric solid tumors, and only a few recurrent SNVs were detected. Approximately half of the osteosarcomas in our discovery cohort had a pattern of hypermutation associated with SVs called kataegis (Nik-Zainal et al., 2012). The regions of the genome with kataegis were not recurrent nor were any of the most recurrently mutated genes found in regions of kataegis. Unlike SNVs, chromosomal lesions were the major mechanism of recurrent mutations and many of the most significant chromosomal lesions were found in known cancer genes including TP53, RB1 and ATRX.
Genomic Stability and Osteosarcoma Initiation and Progression
Osteosarcomas have high rates of SVs and CNVs; thus, SVs and CNVs are a consistent hallmark of this cancer. The most frequent mutation is in TP53; by our estimates, both alleles are mutated in as many as 80% of tumors, and at least one allele is mutated in more than 90% in our discovery cohort. These data suggest that p53 mutations are a major oncogenic driver in osteosarcoma. Although this finding is not novel, what is surprising is the mechanism of inactivation. Most TP53 mutations are SVs in intron 1, which suggests that either the TP53 locus is particularly susceptible to SVs, or SVs occur at a high rate in the osteosarcoma tumor-initiating cell. Besides osteosarcomas and prostate cancers (Baca et al., 2013; Berger et al., 2011), there is no evidence of TP53 SVs in any other cancer, so the locus is probably not uniquely susceptible to chromosomal rearrangements. These data raise an intriguing possibility—genomic instability characterized by high rates of CNVs and SVs may precede TP53 inactivation and be the underlying mechanism that initiates and promotes osteosarcoma.
Many human cancers have shown evidence of genome instability thought to result from a combination of stress (i.e., replicative or oxidative stress) and genetic perturbations in cancer pathways. Our data suggest that some level of genomic instability precedes the major genetic perturbations in cancer pathways and is a probable mechanism of acquiring those mutations. Clearly, this does not preclude the possibility that genomic instability increases after mutations arise in cancer pathways, but we propose that the level of instability in the osteosarcoma tumor–initiating cell is somewhat unique.
Patients with osteosarcoma may have genetic variants or mutations that predispose their osteoblasts to a higher rate of genomic instability, which then leads to a higher risk of the cells transforming into osteosarcoma. Alternatively, osteoblasts may have a unique intracellular environment that contributes to this higher rate of SVs and CNVs. This may have to do with proliferation rate or checkpoint control, with chromosomal structure and/or segregation, or with the epigenomic landscape that somehow predisposes these tumor cells to such high rates of SVs. Consistent with this model, the osteogenic Runt-related (RUNX2) transcription factor is downregulated in osteosarcoma and appears to act as a tumor suppressor in that context. Moreover, the downregulation of RUNX2 in osteoblasts has been proposed to lead to increased DNA damage, delayed DNA repair, and genomic instability(Zaidi et al., 2007). This is just one possible mechanism to begin to explain the high-rates of chromosomal lesions present during osteosarcoma initiation.
We propose that an elevated rate of SVs leads to mutations in TP53 and other genes (e.g., RB1, DLG2 and ATRX), and then additional SVs accumulate, causing disease progression. Another model suggests that osteosarcoma cells are resistant to the normal checkpoints that prevent cells with high rates of SVs from surviving, and this allows the cells to accumulate SVs in TP53 and other genes, thereby leading to rapid progression. Thus, osteosarcoma is a unique model in which to study the regulation of genomic stability in normal proliferative progenitor cells and to advance our understanding of how those chromosomal lesions contribute to tumorigenesis.
Kataegis in Osteosarcoma
In a recent WGS study, Nik-Zainal et al. analyzed the mutational signatures of 21 breast cancers(Nik-Zainal et al., 2012). Using mathematical methods, they identified multiple signatures, some of which correlated with BRCA1 or BRCA2 mutations and distinct patterns of structural variations. One of the most striking patterns to emerge from their analysis was a distinct hypermutation phenomenon in 61% (13/21) of their breast cancer cohort that they termed “kataegis.” SNV clusters with the same 5 characteristics as those described in the study by Nik-Zainal et al. were found in 50% of the OS analyzed by WGS. Interestingly, genomic regions encoding TP53 and ATRX, the two most frequently mutated genes in osteosarcoma, do not exhibit this pattern of local hypermutation. Nor was there any association between kataegis and TP53 mutation type (i.e. SNV, indel or SV).
The mechanisms underlying kataegis in cancer and the associated SVs have not yet been elucidated. However, the specific features of kataegis provide several interesting avenues of future research that may be directly relevant to osteosarcoma initiation and progression. It has been proposed that the AID/APOBEC family of proteins may contribute to kataegis (Burns et al., 2013; Nik-Zainal et al., 2012; Roberts et al., 2012). Some of these proteins can deaminate cytosine to uracil in DNA and this in turn leads to C->T mutations. AID plays a key role in antibody diversification in B cells through local hypermutation and class-switch recombination but AID itself preferentially deaminates cytosine adjacent to a 5′ purine. However, APOBEC1 and some of the APOBEC3 proteins deaminate cytosines with 5′-T as found in kataegis hypermutated regions of breast cancer and osteosarcoma. If that process is accompanied by rearrangements, it could provide a plausible mechanism to begin to explain this unique pattern of somatic mutation in these unrelated types of cancer. Lada et al. further supported the hypothesis by showing AID/APOBEC could induce kataegis-like mutations in yeast(Lada et al., 2012). However, we did not detect differential expression in various AID/APOBEC genes (p values between 0.50 and 0.91) in the current study. To date, there is no correlation between the expression of AID/APOBEC genes and the presence of kataegis in breast cancer or osteosarcoma.
An alternative mechanism may be related to the repair of dsDNA breaks in osteosarcoma. Roberts and colleagues showed that kataegis-like hypermutations could be generated by base alkylation in long single-strand DNA at double-strand breaks (Roberts et al., 2012). The high rate of SVs in osteosarcomas compared to other pediatric cancers would therefore make this tumor type more likely to show evidence of kataegis according to this model. Additional studies will be required to determine if kataegis contributes to the high rate of SVs in osteosarcoma, if the high rates of SVs contribute to kataegis or if the two processes are unrelated.
Detailed analysis of the mutant allele frequency for one of the osteosarcoma samples in our discovery cohort with kataegis suggested a specific chronology of kataegis relative to aneuploidy and adjacent SVs. In SJOS005, kataegis SNVs along with neighboring structural rearrangements occurred first in a few genomic loci. These “kataegis” regions then underwent additional structural rearrangements, resulting in further amplification. On the other hand, aneuploidy and structural re-arrangements preceded acquisition of sequence mutations in most of the remaining regions as indicated by the single-copy mutant allele found in the non-kataegis SNVs.
TP53-Mutant or -Null Osteosarcomas
Previous studies have estimated that 20% to 70% of osteosarcomas carry mutations in the p53 pathway (Lonardo et al., 1997; Wunder et al., 2005), but our data suggest that the proportion is much higher. Recent advances in our understanding of the p53 pathway in cancer suggest that important functional differences exist between missense mutations in TP53 and inactivating mutations in TP53, with respect to tumor progression and response to therapy (reviewed in (Muller et al., 2011)). Although we found no significant difference in the overall survival between the two groups of osteosarcomas with missense and inactivating mutations, a larger cohort of genetically well-characterized osteosarcomas with a longer follow-up period may provide insight into the prognostic relevance of missense TP53 mutations versus TP53-null mutations. Also, while we did not detect any significant association of the rate of chromosomal lesions or point mutations in osteosarcomas with mutant TP53 versus those with null mutations in TP53, the cohort was relatively small and the overall proportion of tumors with mutant TP53 was limiting. A larger study with additional tumors will be required to determine if osteosarcomas with mutant TP53 show any difference in overall outcome or in their genomic landscape. The largest study to date on the relevance of TP53 mutations to the outcome of pediatric patients with osteosarcoma focused on exon sequencing and TP53 immunohistochemistry to identify missense mutations. Those tumors lacking SNVs or indels in the exons and lacking nuclear TP53 protein expression were classified and analyzed as wild-type TP53(Wunder et al., 2005). Based on the data presented here from WGS and FISH analyses of the TP53 locus, most of those samples interpreted as wild-type TP53 most likely had inactivating SVs in the TP53 gene.
Preclinical Models of Osteosarcoma
One of the best-characterized knockout mouse models of osteosarcoma was generated using an osteoblast-specific Cre transgene and Rb1Lox and Tp53Lox alleles(Walkley et al., 2008). Those mice developed metastatic osteosarcoma with many of the molecular, cellular, and histopathologic features of human osteosarcoma. Our data suggest that this model is highly relevant to human osteosarcoma because both the Rb and p53 pathways are mutated or deregulated in the majority of osteosarcomas. Genetically engineered mouse models provide a unique opportunity to study different types of genetic lesions implicated in tumorigenesis and test the effect of combinations of mutations on tumor initiation and progression. On the basis of the data presented here, we believe that testing the difference between knock-in alleles with Tp53 missense mutations(Jackson and Lozano, 2013) and those with Tp53-null mutations, as well as MDM2 overexpression, using conditional transgenic alleles may be relevant to human disease. Also, it may be useful to introduce ATRX mutations(Berube et al., 2005; Medina et al., 2009) and study the initiation and progression of osteosarcoma, as well as changes in telomeres, genome stability, and epigenetic landscape in the mouse tumors. Finally, the roles of Dlg2 have not been previously studied in osteosarcoma, and genetically engineered mouse models provide a unique opportunity to investigate the functions of those genes in osteosarcoma tumorigenesis.
Supplementary Material
Highlights.
Structural variations are the major mechanism of mutation in osteosarcoma.
The p53 pathway is mutated in 90% of osteosarcomas.
ATRX is recurrently mutated in osteosarcoma.
Approximately 50% of osteosarcomas have kataegis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, Park K, Kitabayashi N, MacDonald TY, Ghandi M, et al. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666–677. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berube NG, Mangelsdorf M, Jagla M, Vanderluit J, Garrick D, Gibbons RJ, Higgs DR, Slack RS, Picketts DJ. The chromatin-remodeling protein ATRX is critical for neuronal survival during corticogenesis. The Journal of clinical investigation. 2005;115:258–267. doi: 10.1172/JCI22329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielack SS, Kempf-Bielack B, Delling G, Exner GU, Flege S, Helmke K, Kotz R, Salzer-Kuntschik M, Werner M, Winkelmann W, et al. Prognostic factors in high-grade osteosarcoma of the extremities or trunk: an analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2002;20:776–790. doi: 10.1200/JCO.2002.20.3.776. [DOI] [PubMed] [Google Scholar]
- Bridge JA, Nelson M, McComb E, McGuire MH, Rosenthal H, Vergara G, Maale GE, Spanier S, Neff JR. Cytogenetic findings in 73 osteosarcoma specimens and a review of the literature. Cancer genetics and cytogenetics. 1997;95:74–87. doi: 10.1016/s0165-4608(96)00306-8. [DOI] [PubMed] [Google Scholar]
- Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, Leonard B, Refsland EW, Kotandeniya D, Tretyakova N, Nikas JB, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494:366–370. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, Morsberger LA, Latimer C, McLaren S, Lin ML, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Steward E, Shelat A, Qu C, Bahrami A, Hatley M, Wu G, Bradley C, McEvoy J, Pappo A, et al. RAS Pathway Mutations are Associated with High-Risk Embryonal Rhabdomyosarcoma. Cancer Cell. 2013 in press. [Google Scholar]
- Cheung NK, Zhang J, Lu C, Parker M, Bahrami A, Tickoo SK, Heguy A, Pappo AS, Federico S, Dalton J, et al. Association of age at diagnosis and genetic mutations in patients with neuroblastoma. JAMA: the journal of the American Medical Association. 2012;307:1062–1071. doi: 10.1001/jama.2012.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downing JR, Wilson RK, Zhang J, Mardis ER, Pui CH, Ding L, Ley TJ, Evans WE. The Pediatric Cancer Genome Project. Nat Genet. 2012a;44:619–622. doi: 10.1038/ng.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downing JR, Wilson RK, Zhang J, Mardis ER, Pui CH, Ding L, Ley TJ, Evans WE. The Pediatric Cancer Genome Project. Nature genetics. 2012b;44:619–622. doi: 10.1038/ng.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goorin AM, Harris MB, Bernstein M, Ferguson W, Devidas M, Siegal GP, Gebhardt MC, Schwartz CL, Link M, Grier HE. Phase II/III trial of etoposide and high-dose ifosfamide in newly diagnosed metastatic osteosarcoma: a pediatric oncology group trial. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2002;20:426–433. doi: 10.1200/JCO.2002.20.2.426. [DOI] [PubMed] [Google Scholar]
- Hicks MJ, Roth JR, Kozinetz CA, Wang LL. Clinicopathologic features of osteosarcoma in patients with Rothmund-Thomson syndrome. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2007;25:370–375. doi: 10.1200/JCO.2006.08.4558. [DOI] [PubMed] [Google Scholar]
- Jackson JG, Lozano G. The mutant p53 mouse as a pre-clinical model. Oncogene. 2013 doi: 10.1038/onc.2012.610. [DOI] [PubMed] [Google Scholar]
- Kempf-Bielack B, Bielack SS, Jurgens H, Branscheid D, Berdel WE, Exner GU, Gobel U, Helmke K, Jundt G, Kabisch H, et al. Osteosarcoma relapse after combined modality therapy: an analysis of unselected patients in the Cooperative Osteosarcoma Study Group (COSS) Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2005;23:559–568. doi: 10.1200/JCO.2005.04.063. [DOI] [PubMed] [Google Scholar]
- Kleinerman RA, Tucker MA, Tarone RE, Abramson DH, Seddon JM, Stovall M, Li FP, Fraumeni JF., Jr Risk of new cancers after radiotherapy in long-term survivors of retinoblastoma: an extended follow-up. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2005;23:2272–2279. doi: 10.1200/JCO.2005.05.054. [DOI] [PubMed] [Google Scholar]
- Lada AG, Dhar A, Boissy RJ, Hirano M, Rubel AA, Rogozin IB, Pavlov YI. AID/APOBEC cytosine deaminase induces genome-wide kataegis. Biology direct. 2012;7:47. doi: 10.1186/1745-6150-7-47. discussion 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu DP, Song H, Xu Y. A common gain of function of p53 cancer mutants in inducing genetic instability. Oncogene. 2010;29:949–956. doi: 10.1038/onc.2009.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonardo F, Ueda T, Huvos AG, Healey J, Ladanyi M. p53 and MDM2 alterations in osteosarcomas: correlation with clinicopathologic features and proliferative rate. Cancer. 1997;79:1541–1547. [PubMed] [Google Scholar]
- Manning AL, Dyson NJ. RB: mitotic implications of a tumour suppressor. Nature reviews Cancer. 2012;12:220–226. doi: 10.1038/nrc3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre JF, Smith-Sorensen B, Friend SH, Kassell J, Borresen AL, Yan YX, Russo C, Sato J, Barbier N, Miser J, et al. Germline mutations of the p53 tumor suppressor gene in children with osteosarcoma. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 1994;12:925–930. doi: 10.1200/JCO.1994.12.5.925. [DOI] [PubMed] [Google Scholar]
- Medina CF, Mazerolle C, Wang Y, Berube NG, Coupland S, Gibbons RJ, Wallace VA, Picketts DJ. Altered visual function and interneuron survival in Atrx knockout mice: inference for the human syndrome. Human molecular genetics. 2009;18:966–977. doi: 10.1093/hmg/ddn424. [DOI] [PubMed] [Google Scholar]
- Meek DW. Tumour suppression by p53: a role for the DNA damage response? Nature reviews Cancer. 2009;9:714–723. doi: 10.1038/nrc2716. [DOI] [PubMed] [Google Scholar]
- Meyers PA, Schwartz CL, Krailo M, Kleinerman ES, Betcher D, Bernstein ML, Conrad E, Ferguson W, Gebhardt M, Goorin AM, et al. Osteosarcoma: a randomized, prospective trial of the addition of ifosfamide and/or muramyl tripeptide to cisplatin, doxorubicin, and high-dose methotrexate. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2005;23:2004–2011. doi: 10.1200/JCO.2005.06.031. [DOI] [PubMed] [Google Scholar]
- Miller CW, Aslo A, Tsay C, Slamon D, Ishizaki K, Toguchida J, Yamamuro T, Lampkin B, Koeffler HP. Frequency and structure of p53 rearrangements in human osteosarcoma. Cancer research. 1990;50:7950–7954. [PubMed] [Google Scholar]
- Muller PA, Vousden KH, Norman JC. p53 and its mutants in tumor cell migration and invasion. The Journal of cell biology. 2011;192:209–218. doi: 10.1083/jcb.201009059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- Pounds S, Cheng C, Li S, Liu Z, Zhang J, Mullighan C. A genomic random interval model for statistical analysis of genomic lesion data. Bioinformatics. 2013;29:2088–2095. doi: 10.1093/bioinformatics/btt372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, Sterling J, Thompson C, Harris S, Mav D, Shah R, Klimczak LJ, Kryukov GV, Malc E, Mieczkowski PA, et al. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol Cell. 2012;46:424–435. doi: 10.1016/j.molcel.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L, Phoenix TN, Hedlund E, Wei L, Zhu X, et al. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488:43–48. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadikovic B, Yoshimoto M, Chilton-MacNeill S, Thorner P, Squire JA, Zielenska M. Identification of interactive networks of gene expression associated with osteosarcoma oncogenesis by integrated molecular profiling. Human molecular genetics. 2009;18:1962–1975. doi: 10.1093/hmg/ddp117. [DOI] [PubMed] [Google Scholar]
- Selvarajah S, Yoshimoto M, Ludkovski O, Park PC, Bayani J, Thorner P, Maire G, Squire JA, Zielenska M. Genomic signatures of chromosomal instability and osteosarcoma progression detected by high resolution array CGH and interphase FISH. Cytogenetic and genome research. 2008;122:5–15. doi: 10.1159/000151310. [DOI] [PubMed] [Google Scholar]
- Smith MA, Seibel NL, Altekruse SF, Ries LA, Melbert DL, O’Leary M, Smith FO, Reaman GH. Outcomes for children and adolescents with cancer: challenges for the twenty-first century. J Clin Oncol. 2010;28:2625–2634. doi: 10.1200/JCO.2009.27.0421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Hollstein M, Xu Y. p53 gain-of-function cancer mutants induce genetic instability by inactivating ATM. Nature cell biology. 2007;9:573–580. doi: 10.1038/ncb1571. [DOI] [PubMed] [Google Scholar]
- Squire JA, Pei J, Marrano P, Beheshti B, Bayani J, Lim G, Moldovan L, Zielenska M. High-resolution mapping of amplifications and deletions in pediatric osteosarcoma by use of CGH analysis of cDNA microarrays. Genes, chromosomes & cancer. 2003;38:215–225. doi: 10.1002/gcc.10273. [DOI] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkley CR, Qudsi R, Sankaran VG, Perry JA, Gostissa M, Roth SI, Rodda SJ, Snay E, Dunning P, Fahey FH, et al. Conditional mouse osteosarcoma, dependent on p53 loss and potentiated by loss of Rb, mimics the human disease. Genes Dev. 2008;22:1662–1676. doi: 10.1101/gad.1656808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunder JS, Gokgoz N, Parkes R, Bull SB, Eskandarian S, Davis AM, Beauchamp CP, Conrad EU, Grimer RJ, Healey JH, et al. TP53 mutations and outcome in osteosarcoma: a prospective, multicenter study. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2005;23:1483–1490. doi: 10.1200/JCO.2005.04.074. [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Pande S, Pratap J, Gaur T, Grigoriu S, Ali SA, Stein JL, Lian JB, van Wijnen AJ, Stein GS. Runx2 deficiency and defective subnuclear targeting bypass senescence to promote immortalization and tumorigenic potential. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19861–19866. doi: 10.1073/pnas.0709650104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.