Abstract
The aryl hydrocarbon receptor (AhR) is a promiscuous receptor activated by structurally diverse synthetic and natural compounds. AhR activation may lead to ligand-specific changes in gene expression despite similarities in mode of action. Therefore, differential gene expression elicited by four structurally diverse, high affinity AhR ligands (2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD; 10 nM, 30 µg/kg), 3,3′,4,4′,5-pentachlorobiphenyl (PCB126; 100 nM, 300 µg/kg), β-naphthoflavone (βNF; 10 µM, 90 mg/kg), and indolo[3,2-b]carbazole (ICZ; 1 µM)) in mouse Hepa1c1c7 hepatoma cells and C57BL/6 mouse liver samples were compared. A total of 288, 183, 119, and 131 Hepa1c1c7 genes were differentially expressed (|fold-change| ≥ 1.5, P1(t) ≥ 0.9999) by TCDD, βNF, PCB126, and ICZ, respectively. Only ~35% were differentially expressed by all 4 ligands in Hepa1c1c7 cells. In vivo, 661, 479, and 265 hepatic genes were differentially expressed following treatment with TCDD, βNF, and PCB126, respectively. Similar to Hepa1c1c7 cells, ≤34% of gene expression changes were common across all ligands. Principal components analysis identified time-dependent gene expression divergence. Comparisons of ligand-elicited expression between Hepa1c1c7 cells and mouse liver identified only 11 common gene expression changes across all ligands. Although metabolism may explain some ligand-specific gene expression changes, PCB126, βNF, and ICZ also elicited divergent expression compared to TCDD, suggestive of selective AhR modulation.
Keywords: Selective AhR modulation, Microarray, Mouse, Cross-ligand comparison
1. Introduction
The aryl hydrocarbon receptor (AhR) is a ligand-dependent basic helix-loop-helix-PER-ARNT-SIM (bHLH-PAS) transcription factor activated by structurally diverse synthetic, natural, and dietary compounds including the prototypical ligand 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) (Denison and Nagy, 2003; Denison et al., 2011). Although putative endogenous AhR ligands including Kynurenine (Opitz et al., 2011), 2-(1′H-indole-3′-carbonyl)-thiazole-4-carboxilic acid methyl ester (ITE) (Henry et al., 2010; Quintana et al., 2010) and formylindolo[3,2-b]carbazole (FICZ) (Wei et al., 1999) have been identified, its true endogenous ligand remains elusive (Denison and Nagy, 2003; Nguyen and Bradfield, 2008).
AhR activation following ligand binding causes dissociation of 90 kDa heat shock protein (HSP90), AhR-interacting protein (AIP; also known as ARA9 or XAP2), and p23, followed by translocation to the nucleus and dimerization with the AhR nuclear translocator (ARNT) (Denison and Nagy, 2003; Denison et al., 2011; Hankinson, 1995). The liganded AhR-ARNT complex then binds to dioxin response elements (DREs; core sequence 5′-GCGTG-3′) eliciting changes in gene expression (Denison and Nagy, 2003; Denison et al., 2011; Dere et al., 2011a; Hankinson, 1995). Recent evidence also suggests AhR-mediated differential gene expression independent of DREs (Beischlag et al., 2008; Denison et al., 2011; Dere et al., 2011b; Huang and Elferink, 2012; Tanos et al., 2012).
TCDD and related compounds elicit species-specific responses including teratogenicity, immunotoxicity, hepatotoxicity, and carcinogenicity (Denison et al., 2011; Hankinson, 1995; Poland and Knutson, 1982) that are mostly, if not entirely, AhR-mediated (Denison and Heath-Pagliuso, 1998; Denison et al., 2011; Gonzalez and Fernandez-Salguero, 1998). For example, AhR null mice are resistant to TCDD-mediated toxicity (Gonzalez and Fernandez-Salguero, 1998). Furthermore, while the potency of AhR ligands is determined by comparisons to TCDD, not all elicit the same effects reported with TCDD (Henry et al., 2010; McKillop and Case, 1991; Murray et al., 2010; Safe et al., 1999; Yin et al., 2012).
Selective modulation of AhR-mediated gene expression (Denison et al., 2011; Jin et al., 2012; Kremoser et al., 2007; Murray et al., 2010; Pansoy et al., 2010; Safe et al., 1999; Yin et al., 2012) is thought to be similar to selective estrogen receptor modulators (SERMs) and selective peroxisome-proliferator activated receptor modulators (SPPARMs) (Berger et al., 2003; Frasor et al., 2004; Sears et al., 2007). Selective modulation involves the induction of a ligand-dependent intermediary conformation that can range from complete inactivity to full activation (Berger et al., 2003; Denison et al., 2011; Kremoser et al., 2007). Different ligand-induced conformations change the surface topology of the activated receptor complex leading to the differential recruitment of co-activators and co-repressors in a gene-, cell-, and tissue-dependent manner that can result in ligand-specific gene expression changes (Berger et al., 2003; Brzozowski et al., 1997; Kremoser et al., 2007; Smith and O’Malley, 2004; Zhang et al., 2008). For example, the SERMs, tamoxifen and raloxifene, reposition helix 12 compared to 17β-estradiol such that they exhibit weaker agonist activity (Brzozowski et al., 1997; Levenson and Jordan, 1999). Selective AhR modulator (SAhRM) development has largely focused on immunosuppression and tumor growth inhibition (Jin et al., 2012; Murray et al., 2010; Safe et al., 1999; Yin et al., 2012). More specifically, alkyl polychlorinated dibenzofurans inhibit mammary tumor growth through an AhR-dependent mechanism absent of Cyp1a1 induction and toxicity (Safe et al., 1999) while 1-allyl-3-(3,4-dimethoxyphenyl)-7-(trifluoromethyl)-1H-indazole (SGA360) elicits AhR-mediated immunosuppression independent of DREs without Cyp1a1 induction (Murray et al., 2010). Meanwhile, TCDD, 1,2,3,7,8-pentachlorodibenzo-p-dioxin (PeCDD), 2,3,7,8-tetrachlorodibenzofuran (TCDF), 2,3,4,7,8-pentachlorodibenzofuran (PeCDF), and 3,3′4,4′-pentachlorobiphenyl (PCB126) exhibit different ligand-dependent co-activator recruitment to the AhR with varying Cyp1a1 induction efficacy (Zhang et al., 2008), consistent with ligand-dependent co-activator and co-repressor recruitment in a promoter-, cell-, tissue- and species-specific manner indicative of selective modulation.
The ability of various chemicals to selectively modulate the AhR could have important implications for risk assessment as current approaches assume a common mode of action using toxic equivalency factors (van den Berg et al., 2000). In order to examine if AhR ligands which activates the canonical AhR pathway demonstrate selective modulation, differential gene expression elicited TCDD, PCB126, β-naphthoflavone (βNF), and indolo-[3,2b]-carbazole (ICZ) was examined in mouse Hepa1c1c7 cells and C57BL/6 liver samples. Although each ligand exhibits high AhR binding affinity, Cyp1a1 mRNA induction and the induction of aryl hydrocarbon hydroxylase activity (Boobis et al., 1977; Chen et al., 1995, 2010; Denison and Nagy, 2003; Denison et al., 2011; Kopec et al., 2008; Pohjanvirta et al., 2002), they are structurally diverse with different metabolism kinetics. Therefore, global gene expression profiles were compared not only to identify conserved differential expression but also to investigate divergent and ligand-specific gene expression changes suggestive of SAhRM activity.
2. Materials and methods
2.1. In vitro treatment
All in vitro studies were performed as previously described (Dere et al., 2006). Briefly, Hepa1c1c7 cells (Dr. O. Hankinson, University of California, Los Angeles, CA) were cultured in phenol-red free DMEM/F12 media (Invitrogen, Carlsbad, CA) supplemented with 5% fetal bovine serum (FBS; Hyclone, Logan, UT), 2.5 µg/mL amphotericin B (Invitrogen), 50 µg/mL gentamycin (Invitrogen), 100 U/mL penicillin (Invitrogen), and 100 µg/mL streptomycin (Invitrogen). Cells were maintained under standard culture conditions, 5% CO2 at 37 °C. Treatment with either 10 nM TCDD (Dere et al., 2006), 100 nM PCB126, 10 µM βNF, 1 µM ICZ, or DMSO vehicle control was conducted for 1, 2, 4, 8, 12, 24, or 48 h. Concentrations of TCDD, βNF or ICZ were chosen to elicit maximal Cyp1a1 induction in concentration-response studies (unpublished results) while PCB126 concentration was chosen based on its toxic equivalency factor (TEF) of 0.1. Cells for three biological replicates were collected in 2.0 mL TRIzol Reagent (Invitrogen) for RNA isolation in all in vitro studies.
2.2. In vivo exposures
Animal studies were performed as previously described (Boverhof et al., 2005; Kopec et al., 2008). In short, immature female C57BL/6 ovariectomized (ovx) mice (post natal day 25, Charles River Laboratories, Portage, MI) were housed at 23 °C with 30–40% humidity and 12-h light/dark cycle. Mice were fed Harlan Teklad 22/5 Rodent Diet 8640 (Madison, WI) ad libitum and had free access to deionized water. Mice were acclimated for 3 days, and then orally gavaged once with 30 µg/kg TCDD (Boverhof et al., 2005), 300 µg/kg PCB126 (Kopec et al., 2008), 90 mg/kg βNF, or sesame oil (vehicle). The PCB126 dose was chosen based on its toxic equivalency factor (TEF) of 0.1, while doses ≥80 mg/kg βNF elicit maximal hydroxylase activity (Boobis et al., 1977). Animals were sacrificed by cervical dislocation at 2, 4, 8, 12, 18, 24, 72, 120 or 168 h post-dose. Liver samples (~70 mg) for three biological replicates were removed, flash frozen in liquid nitrogen, and stored at −80 °C until RNA isolation.
2.3. RNA isolation
Total RNA for three biological replicates per time-point was isolated as previously described (Boverhof et al., 2005; Dere et al., 2006; Kopec et al., 2008). Briefly, TRIzol Reagent was added to samples and homogenized (Mixer Mill 300, Retsch, Germany) for in vitro and in vivo isolation, respectively. Total RNA was isolated according to manufacturer’s instructions with an additional phenol:chloroform extraction, and re-suspended in RNA storage solution (Ambion Inc., Austin, TX). Quantity and quality was assessed spectrophotometrically (A260/A280) and by denaturing gel inspection.
2.4. Microarray annotation and experimental design
Custom mouse cDNA microarrays containing 13,361 features were used. Published datasets for C57BL/6 mice dosed with TCDD (Boverhof et al., 2005), PCB126 (Kopec et al., 2008), and for Hepa1c1c7 cells treated with TCDD (Dere et al., 2006) were used, and complemented with unpublished datasets for βNF in C57BL/6 mice and βNF, PCB126, or ICZ treated Hepa1c1c7 cells, all using the same cDNA microarray within a 4 year period. Annotation was updated using the National Center for Biotechnology Information (NCBI) standalone blast (release 2.2.27+). Briefly, cDNA probe sequences were extracted from TIMS dbZach database (Burgoon and Zacharewski, 2007) and individually blasted to a custom NCBI Reference Sequence (RefSeq) database limited to verified and modeled transcripts (NM_, NR_, XM, and XR_ RefSeq identifiers) using default parameters and a bit score ≥200 threshold. RefSeq annotated probes were then matched to Entrez Gene IDs for 13,361 features identifying 8846 unique Entrez Gene IDs. Probe sequences are available online at dbzach.fst.msu.edu.
Custom microarrays comprised PCR amplified cDNAs printed onto epoxy-coated glass slides (Schott-Nexterion, Duryea, PA) at Michigan State University Research and Support Facility (http://www.genomics.msu.edu/) using an Omnigrid arrayer (GeneMachines, San Carlos, CA) equipped with 48 (4 × 12) Chipmaker 2 pins (TeleChem, Sunnyvale, CA). Post-processing consisted of 30 min in a humidity chamber followed by 60 min at 120 °C, and washing to remove unbound DNA or buffer. Total RNA (30 µg) from three biological replicates was reverse transcribed using Cy3- or Cy5-deoxyuridine triphosphate (dUTP) (Amersham, Piscataway, NJ). Labeled cDNA was purified using a Qiagen PCR purification kit (Qiagen, Valencia, CA), and Cy3- and Cy5-labeled samples were mixed, vacuum dried, and re-suspended in hybridization buffer. The re-suspended mixture was heated at 95 °C for 3 min and hybridized on the array under a 22 × 60 mm LifterSlip (Erie Scientific Company, Portsmouth, NH) at 42 °C for 18–24 h. All hybridizations involved samples compared to time-matched vehicle controls with two independent labelings (dye-swap) as previously described (Boverhof et al., 2005; Dere et al., 2006; Kopec et al., 2008). Microarrays were washed, centrifuged, and scanned at 635 nm (Cy5) and 532 nm (Cy3) on an Affymetrix 428 (Affymetrix, Santa Clara, CA) or GenePix 4100A (Molecular Devices, Union City, CA) scanner. Feature and background intensities were extracted using GenePix Pro (Molecular Devices, Sunnyvale, CA). Microarray datasets will be made available on the Gene Expression Omnibus (GEO). More detailed microarray printing, labeling, hybridization, and washing protocols can be found at dbzach.fst.msu.edu.
2.5. Microarray normalization and analysis
Datasets were normalized using a semiparametric approach (Eckel et al., 2005) in SAS v9.1 (SAS Institute Inc., Cary, NC). Posterior probability P1(t) values were calculated on a per-gene and time point basis using an empirical Bayes method (Eckel et al., 2004) implemented in R v1.8.1 (http://www.r-project.org). Differentially expressed genes were identified using a |fold-change| ≥ 1.5 and P1(t) ≥ 0.9999 as in previous studies (Boverhof et al., 2005; Dere et al., 2006; Kopec et al., 2008). Principal components analysis (PCA) was performed in Rv2.15.0 using all cDNA probes with available data across all treatments and time points. Gene expression at each time point for each ligand was used for PCA analysis while 80% confidence level ellipses were determined using the companion to applied regression (car2.0-15) R package (Fox and Weisberg, 2011).
2.6. Functional annotation
Database for Annotation, Visualization, and Integrated Discovery (DAVID; david.abcc.ncifcrf.gov) was used. All differentially expressed genes (|fold-change| ≥ 1.5, P1(t) ≥ 0.9999) were used as a background list for the assessment of enrichment of in vitro or in vivo conserved regulated genes. Functional terms were limited to Gene Ontology (GO) biological processes (GOTERM_BP_FAT) and molecular functions (GOTERM_MF_FAT). Enrichment scores represent the geometric mean (−log scale) of EASE p-values determined for individual members of annotation clusters.
3. Results
3.1. In vitro microarray analysis
Time-dependent changes elicited by 10 nM TCDD (Dere et al., 2006), 10 µM βNF, 100 nM PCB126, and 1 µM ICZ were evaluated in Hepa1c1c7 hepatoma cells. A total of 288 (130 induced and 158 repressed) TCDD-, 183 (125 induced and 58 repressed) βNF-, 119 (91 induced and 28 repressed) PCB126-, and 131 (78 induced and 53 repressed) ICZ-elicited gene expression changes (|fold-change| ≥ 1.5 and P1(t) ≥ 0.9999) were identified (Fig. 1). Cyp1a1 showed the greatest induction while Alb was most repressed by all ligands, although there were differences in efficacy (Supplementary Figs. S2 and S3). For example, TCDD elicited a ~40-fold induction of Cyp1a1 while PCB126 elicited a ~10-fold-change in Hepa1c1c7 cells. Similar induction profiles by all ligands were observed with AhR battery genes including Nqo1 and Aldh3a1 (Nebert et al., 2000) but with more similar efficacies.
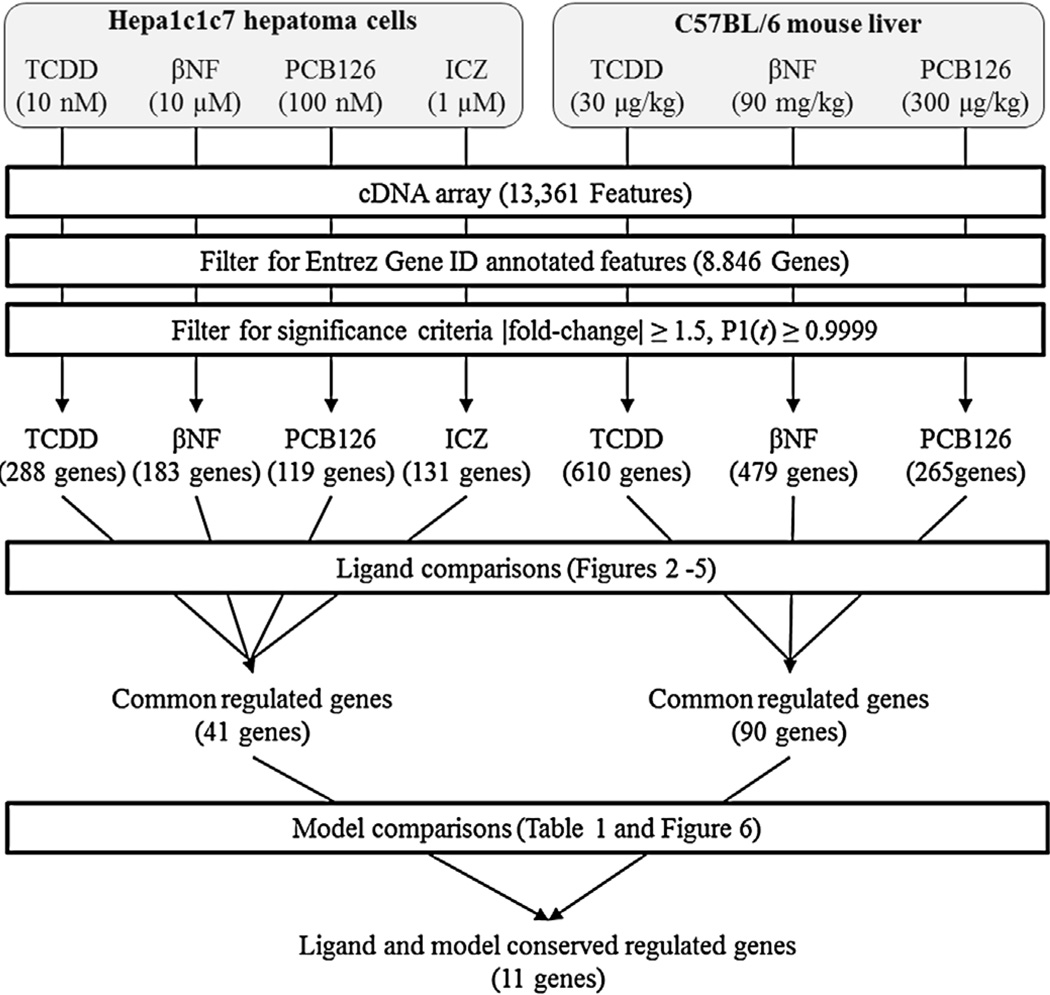
Fig. 1.
Comparative microarray analysis of differential gene responses induced by AhR ligands. Custom cDNA microarrays were filtered for differentially expressed features (|fold-change| ≥ 1.5, P1(t) ≥ 0.9999) and Entrez Gene ID annotation. Datasets were compared to identify conserved, ligand- and model-specific differentially expressed genes.
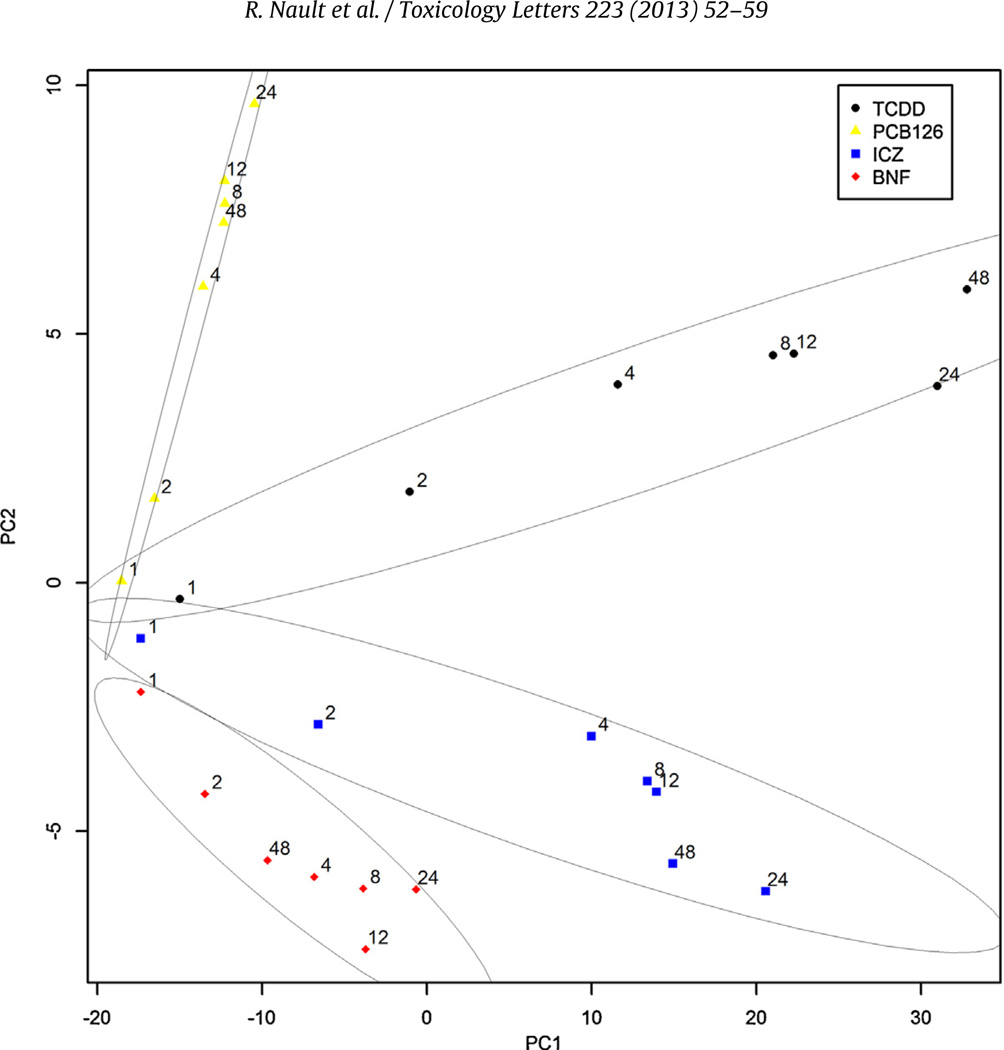
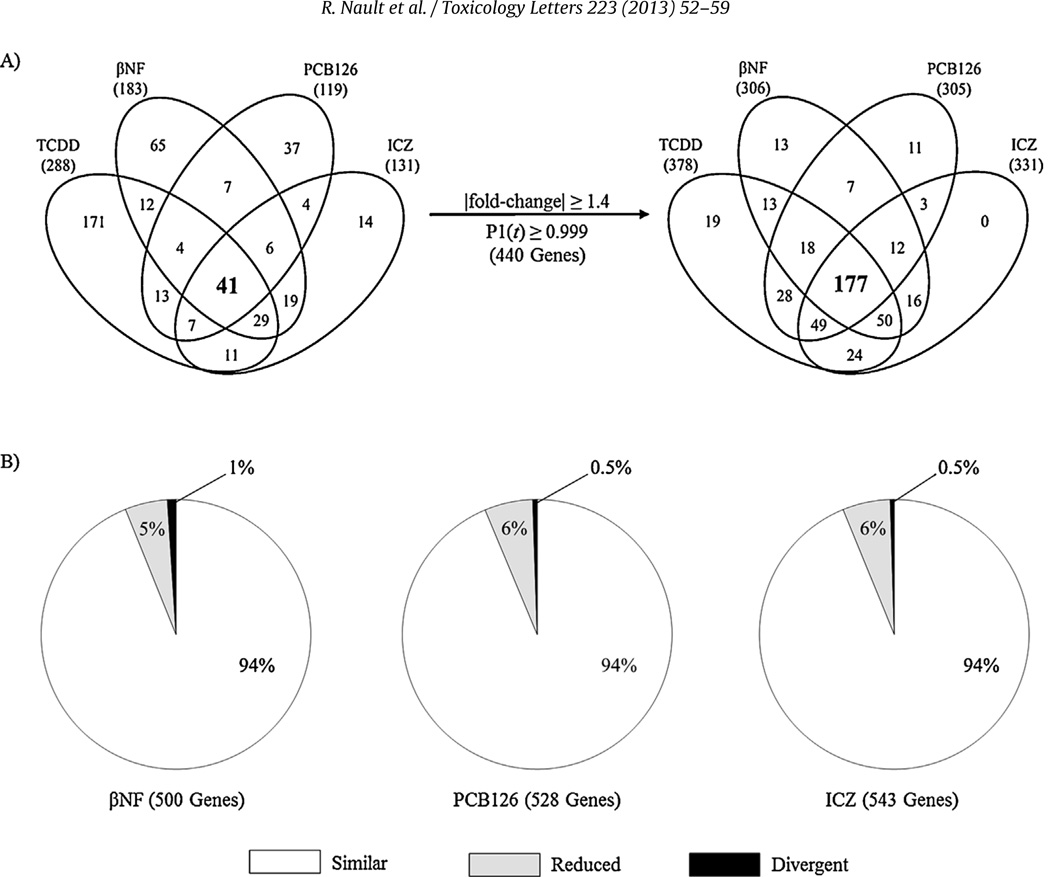
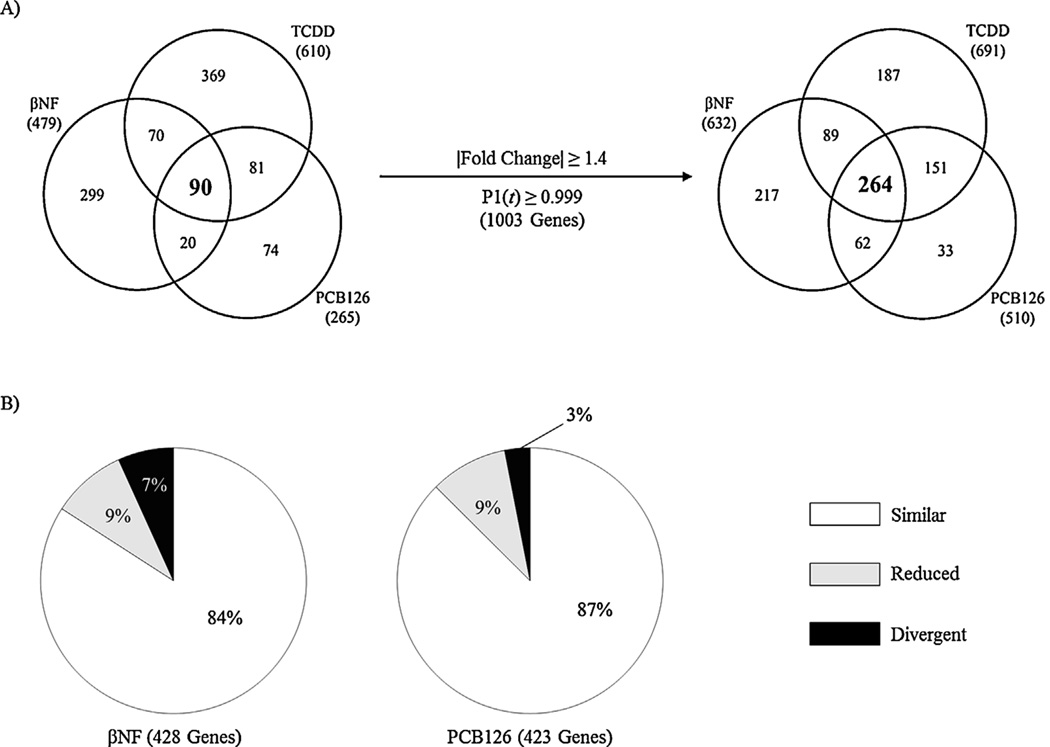
PCA suggests time-dependent divergence of gene expression. In Hepa1c1c7 gene expression, PC1 and PC2 accounted for 76% of the variance and revealed separation of TCDD and PCB126 from βNF and ICZ along PC1 with time while expression at individual time points diverged along both PC1 and PC2 (Fig. 2). Consistent with these results, expression profile comparisons suggest the majority of genes exhibit ligand-specific differential expression (Fig. 3A). However, relaxation of the selection criteria from |fold-change| ≥1.5 and P1(t) ≥ 0.9999 to |fold-change| ≥1.4 and P1(t) ≥ 0.999 for a union of 440 differentially expressed genes across all treatments increased the overlap >4-fold between TCDD, βNF, PCB126 and ICZ (41 to 177 genes), suggesting that many common differentially expressed genes were originally eliminated due to the stringent criteria. Under relaxed criteria, ≤5% of differentially expressed genes exhibit ligand-specific regulation with no ICZ-specific gene expression (Fig. 3B). Moreover, examination of fold-change similarity relative to TCDD revealed comparable expression profiles, with ≤ 1% (3 genes; Col1a1, Cdca5, and Zfp219) of βNF-, PCB126- and ICZ-elicited gene expression changes exhibiting divergent expression (i.e., induced by one ligand but repressed by another; Fig. 3B).
Fig. 2.
Principal component analysis (PCA) of TCDD-, PCB126 (PCB)-, ICZ-, or βNF-elicited differential gene expression in treated Hepa1c1c7 cells. Numbers above points represent duration of exposure to AhR ligand for TCDD (●), PCB126 (▲), ICZ (■), or βNF (◆). Principal components 1 and 2 account for 76% of the total variance for cDNA probes with available data across all treatments and time-points (12,104 clone IDs) used for this analysis. Gray ellipses representing 80% confidence levels were determined using the companion to applied regression (car2.0-15) R package (Fox and Weisberg, 2011).
Fig. 3.
Comparison of ligand-dependent differential gene expression changes in Hepa1c1c7 cells. (A) Ligand-dependent differential gene expression identified under stringent criteria (left; |fold-change| ≥ 1.5, P1(t) ≥ 0.9999) revealed increased overlap following relaxation of filtering criteria (right; |fold-change| ≥ 1.4 and P1(t) ≥ 0.999). (B) βNF, PCB126, or ICZ datasets (not filtered for |fold-change| or P1(t) value) were compared to TCDD regulated genes (|fold-change| ≥ 1.5, P1(t) ≥ 0.9999). Genes were classified as similar (|fold-change| ≥ 70% of TCDD mediated fold-change; white), reduced (|fold-change| ≥ 1.5 and ≤70% of TCDD mediated fold-change; gray), or divergent (|fold-change| ≥ 1.5 but in opposite direction compared to TCDD; black) based on the maximal fold-change for each gene elicited by TCDD.
3.2. In vivo microarray analysis
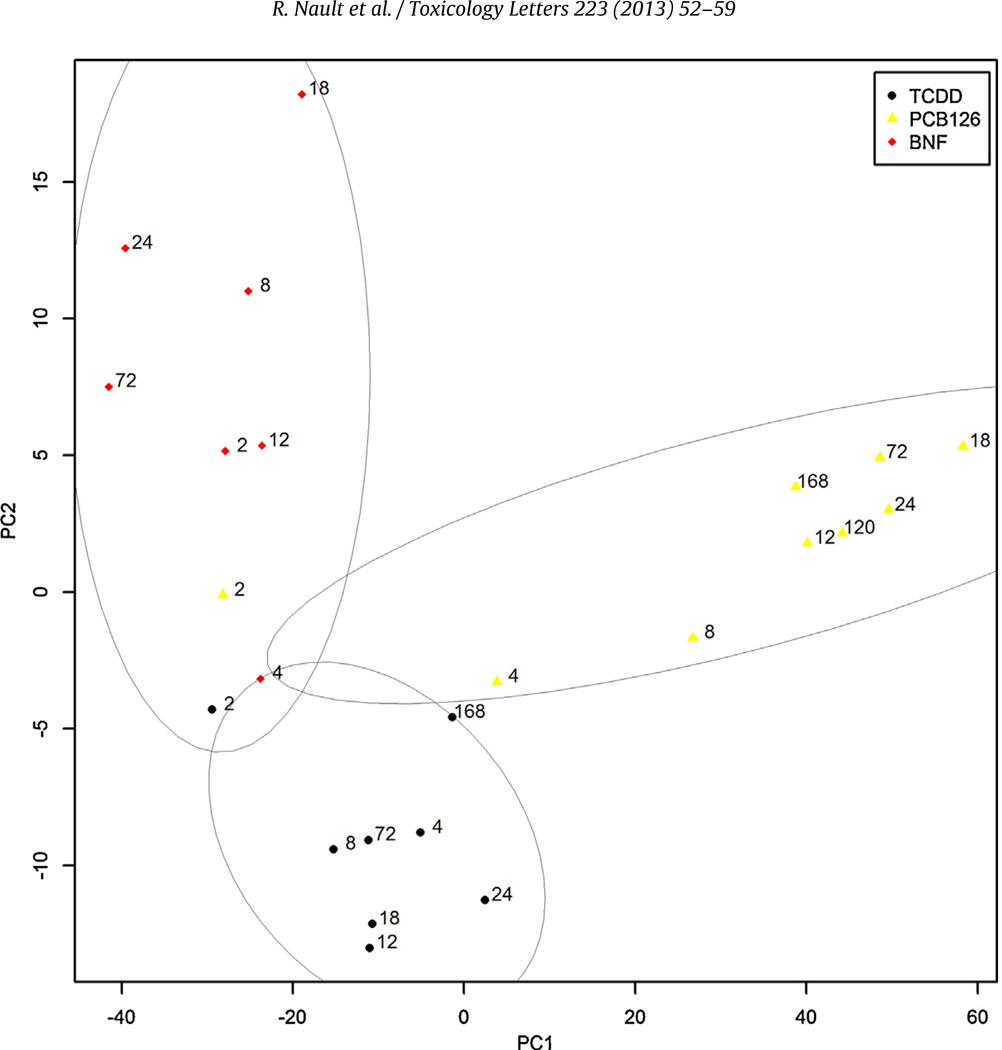
We have previously reported the hepatic gene expression effects of TCDD- and PCB126- in orally gavaged C57BL/6 mice (Boverhof et al., 2005; Kopec et al., 2008). In this study, comparative analysis was extended to include the effects of 90 mg/kg βNF on hepatic gene expression using the same dosing regimen, animal model, and data analysis approach. TCDD, βNF, and PCB126 elicited the differential expression of 610 (331 induced and 279 repressed), 479 (416 induced and 63 repressed) and 265 (166 induced and 99 repressed) genes, respectively (Fig. 1). Similar to Hepa1c1c7 cells, PCA revealed time-dependent divergent gene expression between TCDD and βNF with separation along PC1 while PCB126 largely separated along PC2. PC1 and PC2 accounted for 84% of the variance (Fig. 4). Ligand-specific differential gene expression in vivo (Fig. 5A) was reduced ~3-fold with relaxed selection criteria (overlap increased from 90 to 264 genes; Fig. 5A). However, ligand-specific differential expression only decreased 27% (82 genes) for βNF compared to 49% (182 genes) and 55% (41 genes) for TCDD and PCB126, respectively, revealing some ligands are more similar than others as suggested by PCA analyses. Moreover, 3% (13 genes) of PCB126- and 7% (29 genes) of βNF-elicited differential gene expression exhibit divergent regulation (i.e., induced by one ligand but repressed by another) relative to TCDD (Fig. 5B).
Fig. 4.
Principal component analysis (PCA) of hepatic gene expression fold-changes for TCDD-, PCB126 (PCB)-, or βNF-treated mice. Numbers represent duration of exposure to AhR ligand for TCDD (●), PCB126 (▲), or βNF (◆). PC1 and PC2 account for 84% of the variance for cDNA probes with available data across all treatments and time-points used for this analysis (11,165 clone IDs). 80% confidence level ellipses were determined using the companion to applied regression (car2.0-15) R package (Fox and Weisberg, 2011).
Fig. 5.
Comparison of ligand-dependent differential gene expression changes in C57BL/6 mouse liver. (A) Ligand-regulated genes identified under stringent criteria (left; |fold-change| ≥ 1.5, P1(t) ≥ 0.9999) showed a dramatic increase in overlap following relaxation of filtering criteria (right; |fold-change| ≥ 1.4 and P1(t) ≥ 0.999). (B) βNF or PCB126 datasets (not filtered for |fold-change| or P1(t) value) were compared to TCDD regulated genes (|fold-change| ≥ 1.5, P1(t) ≥ 0.9999). Genes were classified as similar (|fold-change| ≥ 70% of TCDD mediated fold-change; white), reduced (|fold-change| ≥ 1.5 and ≤70% of TCDD mediated fold-change; gray), or divergent (|fold-change| ≥ 1.5 but in opposite direction compared to TCDD; black) based on the maximal fold-change for each gene elicited by TCDD.
3.3. Conserved and model-specific ligand-mediated responses
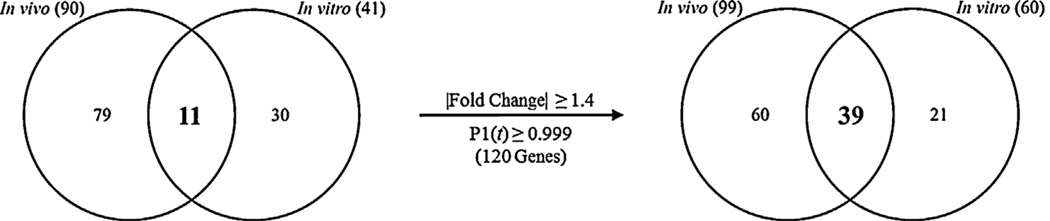
A total of 41 and 90 genes were identified as differentially expressed by all AhR ligands in Hepa1c1c7 cells and C57BL/6 liver samples, respectively (Figs. 3A and 5A). Functional annotation of the 41 Hepa1c1c7 genes identified over-represented functions associated with oxidation and reduction (enrichment score 1.02) and vasculature development (enrichment score 0.8). Glutathione (enrichment score 1.36) and lipid metabolism (enrichment score 1.23) were identified as the most enriched functions among the 90 common C57BL/6 genes. Only 11 genes were identified as differentially expressed by TCDD, βNF, PCB126 and ICZ in both Hepa1c1c7 cells and C57BL/6 mouse liver samples (Fig. 6; Supplementary Table 1). Not surprisingly, these include genes commonly used as markers of AhR ligand exposure such as Cyp1a1, Tiparp and Nqo1. Similar to in vitro and in vivo comparisons, relaxation of filtering criteria (|fold-change| ≥ 1.4, P1(t) ≥ 0.999) increased the overlap from 11 to 39 genes (Fig. 6).
Fig. 6.
Comparison of in vitro and in vivo conserved TCDD-, βNF-, PCB126-, and ICZ-differentially expressed genes. The Venn analysis for a union of 120 genes showed more overlap for differentially expressed genes following treatment with TCDD, βNF, PCB126, and ICZ increased when stringent criteria (left; |fold-change| ≥ 1.5, P1(t) ≥ 0.9999) were relaxed (right; |fold-change| ≥ 1.4 and P1(t) ≥ 0.999).
Further filtering identified 21 genes differentially expressed by TCDD, βNF, PCB126 and ICZ only in Hepa1c1c7 cells (Supplementary Table 1). Interestingly, all of the ligands repressed the expression of the hepatocyte marker Alb, which showed the greatest down-regulation. Alb repression was exclusively in vitro suggesting AhR activation may alter the functional status of Hepa1c1c7 cells. Other in vitro specific genes include the induction of several lipid and carbohydrate metabolism genes including Aldh3a1, Cyp2s1, H6pd, Pla2g4a, and Rbp4. Similarly, 60 genes exhibited differential expression by TCDD, βNF and PCB126 exclusively in C57BL/6 mouse liver (Supplementary Table 1). Numerous lipid metabolism genes showed in vivo specific AhR ligand expression including the induction of Cd36, Lrp2, Acot7, and Pla2g2c, and the down-regulation of Srebf1, Gpd2, and Dak.
4. Discussion
In this study, TCDD-, PCB126-, βNF- and ICZ-elicited differential gene expression was compared in Hepa1c1c7 cells and C57BL/6 liver samples. Although these ligands are structurally diverse with different metabolism and elimination pharmacokinetics, all bind the AhR with high affinity (Bohonowych and Denison, 2007) and elicit AhR-mediated differential gene expression. TCDD and PCB126 are metabolized slowly, or not at all, with in vivo elimination rates ranging from days to weeks in rodents (Gasiewicz et al., 1983; Kopec et al., 2013; Pohjanvirta et al., 1990). In contrast, βNF and ICZ are rapidly metabolized with almost complete elimination of βNF in mouse liver within 6 days (Boobis et al., 1977; Chen et al., 1995; Pohjanvirta et al., 2002). These pharmacokinetic differences are reflected in time-dependent differential gene expression with PCA showing divergent gene expression changes at later time points, particularly TCDD comparisons to PCB126, βNF or ICZ. This is consistent with previous reports of time-dependent divergence of gene expression for the rapidly metabolized AhR ligand 2-(1′H-indole-3′-carbonyl)-thiazole-4-carboxylic acid methyl ester (ITE) compared to TCDD in mouse lung fibroblasts (Henry et al., 2010), and may be indicative of secondary responses elicited by metabolites rather than the parent compound. However, the majority of PCB126-, βNF-, and ICZ-elicited gene expression changes were in common with TCDD and therefore, considered AhR-mediated. More specifically, ≥87% PCB126-, ≥84% βNF- and ≥94% ICZ-elicited differentially expressed genes exhibited fold-changes ≥70% of the TCDD-elicited fold-change. Consequently, divergent responses cannot be explained by metabolism and/or elimination alone.
Selective receptor modulators (SRMs) are ligands that elicit agonistic and/or antagonistic activity in a gene-, cell-, and/or tissue-specific manner (Frasor et al., 2004; Kremoser et al., 2007; Smith and O’Malley, 2004). Tamoxifen, the prototypical selective estrogen receptor modulator (SERM), exhibits antagonist activity in breast tissue with partial agonist activity in the uterus and bone, with no significant estrogen-like activity in the central nervous system (Levenson and Jordan, 1999; Levenson et al., 2002; Smith and O’Malley, 2004). Moreover, two other SERMs, raloxifene and trans-hydroxytamoxifen exhibit both partial agonism, and antagonize estrogen regulated genes in MCF-7 human breast cancer cells (Frasor et al., 2004). Comparisons of tamoxifen and 17α-ethynylestradiol elicited gene expression identified several ligand-specific gene expression differences (Fong et al., 2007; Kwekel et al., 2009). These studies demonstrate that SRMs can elicit unique gene expression profiles even in the same model. SRMs differ by inducing unique ligand-binding domain conformations that affect the recruitment and interaction with available co-activators and co-repressors within a model (Kremoser et al., 2007; Levenson and Jordan, 1999; Smith and O’Malley, 2004; Zhang et al., 2008). Therefore, it is not surprising that SRMs for the same receptor elicit conserved as well as ligand-specific gene expression changes within the same model resulting in some receptor-mediated responses that differ in potency and/or efficacy. For example, Gadd45b in vitro and Fabp5 in vivo were induced ~4-fold by TCDD but only ~2-fold by PCB126, βNF and ICZ in each model. Such effects may be due to specific ligand-induced binding domain conformations that affect co-activator recruitment. Other genes exhibit divergent profiles (i.e., induced by one ligand but repressed by another) such as in Hepa1c1c7 cells where Col1a1 was induced by TCDD but repressed by βNF, Cdca5 was induced by TCDD but repressed by PCB126, and Zfp219 was induced by TCDD but repressed by both βNF and ICZ. Co-activator and co-repressor availability contributing to cell- and tissue-specific ligand activity (Smith and O’Malley, 2004) may also explain the divergent AhR-mediated gene expression reported between Hepa1c1c7 cells and mouse liver samples following TCDD treatment (Dere et al., 2006) and is further supported by our PCB126 and βNF data. However, divergent expression could also be due to the formation of metabolites with unique or altered SRM efficacy and potency and/or other off-target effects. For example, although tamoxifen elicits antiestrogenic activity in breast tissue, 4-hydroxytamoxifen has greater binding affinity for the estrogen receptor and is considered the more active agent.
SAhRMs have been developed that bind to the AhR but competitively antagonize TCDD-elicited effects. TCDD-induced enzyme expression and/or activity (AHH, EROD, and Cyp1a1 expression) (Astroff and Safe, 1989; Astroff et al., 1988; Bannister et al., 1989; Harris et al., 1989; Kim et al., 2006), AhR binding (Gasiewicz and Rucci, 1991), ER binding down-regulation (Romkes et al., 1987), and TCDD-induced porphyria (Yao and Safe, 1989), immunotoxicity (Davis and Safe, 1988; Dickerson et al., 1990), cleft palate, and hydronephrosis (Bannister et al., 1989; Biegel et al., 1989) can be antagonized by SAhRMs. More recently, SAhRMS that modulate hematopoietic progenitor expansion (Boitano et al., 2010), repress cytokine-mediated induction of complement factor genes (Murray et al., 2011), antagonize cytokine-mediated inflammatory signaling (Murray et al., 2010), and inhibit tumor growth (Jin et al., 2012; Safe et al., 1999; Yin et al., 2012) in absence of canonical DRE-elicited transcription have also been developed. Genome-wide AhR ChIP-chip and two-hybrid studies also suggest that structurally diverse ligands such as 3-methylcholanthrene (3-MC), PeCDD, PeCDF, TCDF, and PCB126 elicit DRE-dependent AhR-mediated selective modulation of gene expression (Pansoy et al., 2010; Zhang et al., 2008).
Maximal Cyp1a1 induction concentrations and doses were used for comparisons as an indication of maximal AhR activation allowing observation of any evidence of selective AhR modulation. Our study suggests, in addition to conserved responses, structurally diverse AhR ligands elicit ligand-specific in vitro and in vivo hepatic differential gene expression. For example, βNF is a commonly used non-toxic alternative to investigate AhR-mediated effects. In addition to differences in efficacy and potency compared to TCDD, βNF also had the lowest percentage (≤47%) of overlapping differentially expressed genes in Hepa1c1c7 cells and C57BL/6 mouse liver samples compared to PCB126 and ICZ. Furthermore, βNF-elicited Hepa1c1c7 gene expression was most like ICZ (≤73% overlap) while TCDD was more similar to PCB126 (≤65% overlap) in C57BL/6 mouse liver. However, relaxation of microarray filtering criteria often increased the overlap of differentially expressed genes across all ligands suggesting that several genes may be identified as ligand specific due to differences in efficacy. The conservation of a significant subset of differentially expressed genes in response to TCDD, PCB126, βNF, and ICZ suggests that potent AhR agonists elicit similar physiological responses, barring differences in metabolism. Indeed, using different dosing regimens hepatic lipid accumulation in rodents has been reported for TCDD, PCB126, βNF, and 3-MC (Boverhof et al., 2005; Bunger et al., 2008; Kopec et al., 2008)(DrugMatrix database; https://ntp.niehs.nih.gov/drugmatrix), despite pharmacokinetic differences and evidence of SAhRM activity (Pansoy et al., 2010; Zhang et al., 2008).
In summary, TCDD, PCB126, βNF, and ICZ elicit a common subset of gene expression changes with varying efficacies as well as ligand-specific differential expression in Hepa1c1c7 cells and C57BL/6 liver. The identification of a large common subset of differentially expressed genes suggests these effects are mediated by AhR. However, there were also ligand-specific gene expression changes that cannot be explained by differences in metabolism and/or elimination alone. Although ligand-specific SAhRM-like gene expression changes need to be verified (e.g., corresponding changes in protein expression, enzyme activity and/or metabolite levels) and their physiological significance determined, these results suggest structurally diverse AhR ligands exhibit some degree of SAhRM activity that, in addition to differences in pharmacodynamics, could affect potency and efficacy as well as toxicity. Therefore, in addition to differences in metabolism and/or elimination, ligand-specific SAhRM activity may have important implications for risk assessment of AhR ligands which assumes a comparable mode of action for all ligands (van den Berg et al., 2000). The significance of SAhRM activity in elucidating the mechanisms of toxicity of TCDD and related compounds, and potential implications for risk assessment remains to be determined.
Supplementary Material
HIGHLIGHTS.
Gene expression elicited by AhR ligands TCDD, PCB126, βNF and ICZ was compared.
AhR ligands TCDD, PCB126, βNF and ICZ regulate a common subset of genes.
AhR ligand elicited gene expression diverges as duration of exposure increases.
TCDD, PCB126, βNF and ICZ selectively modulate the aryl hydrocarbon receptor.
Acknowledgements
This work was supported by the National Institute of Environmental Health Sciences Superfund Basic Research Program (NIEHS SBRP P42ES04911). The authors would like to thank Dr. Darrell R. Boverhof for his support with in vivo studies and Lyle D. Burgoon for his help with the microarray analyses.
Footnotes
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Supplementary data
Supplementary Fig. S1 Correlation analysis of differential gene expression between custom cDNA and Agilent oligonucleotide microarrays.
Supplementary Figs. S2 and S3 Rank fold-change correlation of common differentially regulated genes.
Supplementary Table 1 Common and model-specific differentially regulated genes.
Supplementary Tables 2–8 Analyzed microarray datasets including previously published datasets with newly annotated probes.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.toxlet.2013.08.013.
References
- Astroff B, Safe S. 6-substituted-1,3,8-trichlorodibenzofurans as 2,3,7,8-tetrachlorodibenzo-p-dioxin antagonists in the rat: structure activity relationships. Toxicology. 1989;59:285–296. doi: 10.1016/0300-483x(89)90198-4. [DOI] [PubMed] [Google Scholar]
- Astroff B, Zacharewski T, Safe S, Arlotto MP, Parkinson A, Thomas P, Levin W. 6-Methyl-1,3,8-trichlorodibenzofuran as a 2,3,7,8-tetrachlorodibenzo-p-dioxin antagonist: inhibition of the induction of rat cytochrome P-450 isozymes and related monooxygenase activities. Mol. Pharmacol. 1988;33:231–236. [PubMed] [Google Scholar]
- Bannister R, Biegel L, Davis D, Astroff B, Safe S. 6-Methyl-1,3,8-trichlorodibenzofuran (MCDF) as a 2,3,7,8-tetrachlorodibenzo-p-dioxin antagonist in C57BL/6 mice. Toxicology. 1989;54:139–150. doi: 10.1016/0300-483x(89)90040-1. [DOI] [PubMed] [Google Scholar]
- Beischlag TV, Luis Morales J, Hollingshead BD, Perdew GH. The aryl hydrocarbon receptor complex and the control of gene expression. Crit. Rev. Eukaryot. Gene Expr. 2008;18:207–250. doi: 10.1615/critreveukargeneexpr.v18.i3.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger JP, Petro AE, Macnaul KL, Kelly LJ, Zhang BB, Richards K, Elbrecht A, Johnson BA, Zhou G, Doebber TW, Biswas C, Parikh M, Sharma N, Tanen MR, Thompson GM, Ventre J, Adams AD, Mosley R, Surwit RS, Moller DE. Distinct properties and advantages of a novel peroxisome proliferator-activated protein [gamma] selective modulator. Mol. Endocrinol. 2003;17:662–676. doi: 10.1210/me.2002-0217. [DOI] [PubMed] [Google Scholar]
- Biegel L, Harris M, Davis D, Rosengren R, Safe L, Safe S. 2,2’,4,4’,5,5’-Hexachlorobiphenyl as a 2,3,7,8-tetrachlorodibenzo-p-dioxin antagonist in C57BL/6J mice. Toxicol. Appl. Pharmacol. 1989;97:561–571. doi: 10.1016/0041-008x(89)90261-5. [DOI] [PubMed] [Google Scholar]
- Bohonowych JE, Denison MS. Persistent binding of ligands to the aryl hydrocarbon receptor. Toxicol. Sci. 2007;98:99–109. doi: 10.1093/toxsci/kfm085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boitano AE, Wang J, Romeo R, Bouchez LC, Parker AE, Sutton SE, Walker JR, Flaveny CA, Perdew GH, Denison MS, Schultz PG, Cooke MP. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science. 2010;329:1345–1348. doi: 10.1126/science.1191536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boobis AR, Nebert DW, Felton JS. Comparison of beta-naphthoflavone and 3-methylcholanthrene as inducers of hepatic cytochrome(s) P-448 and aryl hydrocarbon (benzo[a]pyrene) hydroxylase activity. Mol. Pharmacol. 1977;13:259–268. [PubMed] [Google Scholar]
- Boverhof DR, Burgoon LD, Tashiro C, Chittim B, Harkema JR, Jump DB, Zacharewski TR. Temporal and dose-dependent hepatic gene expression patterns in mice provide new insights into TCDD-Mediated hepatotoxicity. Toxicol. Sci. 2005;85:1048–1063. doi: 10.1093/toxsci/kfi162. [DOI] [PubMed] [Google Scholar]
- Brzozowski AM, Pike AC, Dauter Z, Hubbard RE, Bonn T, Engstrom O, Ohman L, Greene GL, Gustafsson JA, Carlquist M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- Bunger MK, Glover E, Moran SM, Walisser JA, Lahvis GP, Hsu EL, Bradfield CA. Abnormal liver development and resistance to 2,3,7,8-tetrachlorodibenzo-p-dioxin toxicity in mice carrying a mutation in the DNA-binding domain of the aryl hydrocarbon receptor. Toxicol. Sci. 2008;106:83–92. doi: 10.1093/toxsci/kfn149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoon LD, Zacharewski TR. dbZach toxicogenomic information management system. Pharmacogenomics. 2007;8:287–291. doi: 10.2217/14622416.8.3.287. [DOI] [PubMed] [Google Scholar]
- Chen EP, Chen L, Ji Y, Tai G, Wen YH, Ellens H. A mechanism-based mathematical model of aryl hydrocarbon receptor-mediated CYP1A induction in rats using beta-naphthoflavone as a tool compound. Drug Metab. Dispos. 2010;38:2278–2285. doi: 10.1124/dmd.110.034421. [DOI] [PubMed] [Google Scholar]
- Chen YH, Riby J, Srivastava P, Bartholomew J, Denison M, Bjeldanes L. Regulation of CYP1A1 by indolo[3,2-b]carbazole in murine hepatoma cells. J. Biol. Chem. 1995;270:22548–22555. doi: 10.1074/jbc.270.38.22548. [DOI] [PubMed] [Google Scholar]
- Davis D, Safe S. Immunosuppressive activities of polychlorinated dibenzofuran congeners: quantitative structure-activity relationships and interactive effects. Toxicol. Appl. Pharmacol. 1988;94:141–149. doi: 10.1016/0041-008x(88)90344-4. [DOI] [PubMed] [Google Scholar]
- Denison MS, Heath-Pagliuso S. The Ah receptor: a regulator of the biochemical and toxicological actions of structurally diverse chemicals. Bull. Environ. Contam. Toxicol. 1998;61:557–568. doi: 10.1007/pl00002973. [DOI] [PubMed] [Google Scholar]
- Denison MS, Nagy SR. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu. Rev. Pharmacol. Toxicol. 2003;43:309–334. doi: 10.1146/annurev.pharmtox.43.100901.135828. [DOI] [PubMed] [Google Scholar]
- Denison MS, Soshilov AA, He G, DeGroot DE, Zhao B. Exactly the same but different: promiscuity and diversity in the molecular mechanisms of action of the aryl hydrocarbon (dioxin) receptor. Toxicol. Sci. 2011;124:1–22. doi: 10.1093/toxsci/kfr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dere E, Boverhof DR, Burgoon LD, Zacharewski TR. In vivo-in vitro toxicogenomic comparison of TCDD-elicited gene expression in Hepa1c1c7 mouse hepatoma cells and C57BL/6 hepatic tissue. BMC Genomics. 2006;7:80. doi: 10.1186/1471-2164-7-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dere E, Forgacs AL, Zacharewski TR, Burgoon LD. Genome-wide computational analysis of dioxin response element location and distribution in the human, mouse, and rat genomes. Chem. Res. Toxicol. 2011a;24:494–504. doi: 10.1021/tx100328r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dere E, Lo R, Celius T, Matthews J, Zacharewski TR. Integration of genome-wide computation DRE search, AhR ChIP-chip and gene expression analyses of TCDD-elicited responses in the mouse liver. BMC Genomics. 2011b;12:365. doi: 10.1186/1471-2164-12-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson R, Howie L, Davis D, Safe S. The structure-dependent effects of heptachlorodibenzofuran isomers in male C57BL/6 mice: immunotoxicity and monooxygenase enzyme induction. Fundam. Appl. Toxicol. 1990;15:298–307. doi: 10.1016/0272-0590(90)90056-p. [DOI] [PubMed] [Google Scholar]
- Eckel JE, Gennings C, Chinchilli VM, Burgoon LD, Zacharewski TR. Empirical bayes gene screening tool for time-course or dose-response microarray data. J. Biopharm. Stat. 2004;14:647–670. doi: 10.1081/BIP-200025656. [DOI] [PubMed] [Google Scholar]
- Eckel JE, Gennings C, Therneau TM, Burgoon LD, Boverhof DR, Zacharewski TR. Normalization of two-channel microarray experiments: a semiparametric approach. Bioinformatics. 2005;21:1078–1083. doi: 10.1093/bioinformatics/bti105. [DOI] [PubMed] [Google Scholar]
- Fong CJ, Burgoon LD, Williams KJ, Forgacs AL, Zacharewski TR. Comparative temporal and dose-dependent morphological and transcriptional uterine effects elicited by tamoxifen and ethynylestradiol in immature, ovariectomized mice. BMC Genomics. 2007;8:151. doi: 10.1186/1471-2164-8-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J, Weisberg S. An{R} companion to applied regressions. Thousand Oaks: Sage; 2011. [Google Scholar]
- Frasor J, Stossi F, Danes JM, Komm B, Lyttle CR, Katzenellenbogen BS. Selective estrogen receptor modulators: discrimination of agonistic versus antagonistic activities by gene expression profiling in breast cancer cells. Cancer Res. 2004;64:1522–1533. doi: 10.1158/0008-5472.can-03-3326. [DOI] [PubMed] [Google Scholar]
- Gasiewicz TA, Geiger LE, Rucci G, Neal RA. Distribution, excretion, and metabolism of 2,3,7,8-tetrachlorodibenzo-p-dioxin in C57BL/6J, DBA/2J, and B6D2F1/J mice. Drug Metab. Dispos. 1983;11:397–403. [PubMed] [Google Scholar]
- Gasiewicz TA, Rucci G. Alpha-naphthoflavone acts as an antagonist of 2,3,7, 8-tetrachlorodibenzo-p-dioxin by forming an inactive complex with the Ah receptor. Mol. Pharmacol. 1991;40:607–612. [PubMed] [Google Scholar]
- Gonzalez FJ, Fernandez-Salguero P. The aryl hydrocarbon receptor: studies using the AHR-null mice. Drug Metab. Dispos. 1998;26:1194–1198. [PubMed] [Google Scholar]
- Hankinson O. The aryl hydrocarbon receptor complex. Annu. Rev. Pharmacol. Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- Harris M, Zacharewski T, Astroff B, Safe S. Partial antagonism of 2,3,7,8-tetrachlorodibenzo-p-dioxin-mediated induction of aryl hydrocarbon hydroxylase by 6-methyl-1,3,8-trichlorodibenzofuran: mechanistic studies. Mol. Pharmacol. 1989;35:729–735. [PubMed] [Google Scholar]
- Henry EC, Welle SL, Gasiewicz TA. TCDD and a putative endogenous AhR ligand, ITE, elicit the same immediate changes in gene expression in mouse lung fibroblasts. Toxicol. Sci. 2010;114:90–100. doi: 10.1093/toxsci/kfp285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Elferink CJ. A novel nonconsensus xenobiotic response element capable of mediating aryl hydrocarbon receptor-dependent gene expression. Mol. Pharmacol. 2012;81:338–347. doi: 10.1124/mol.111.075952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin UH, Lee SO, Safe SH. Aryl hydrocarbon receptor (AhR)-active pharmaceuticals are selective AhR modulators in MDA-MB-468 and BT474 breast cancer cells. J. Pharmacol. Exp. Ther. 2012 doi: 10.1124/jpet.112.195339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Henry EC, Kim DK, Kim YH, Shin KJ, Han MS, Lee TG, Kang JK, Gasiewicz TA, Ryu SH, Suh PG. Novel compound 2-methyl-2H-pyrazole-3-carboxylic acid (2-methyl-4-o-tolylazo-phenyl)-amide (CH-223191) prevents 2,3,7,8-TCDD-induced toxicity by antagonizing the aryl hydrocarbon receptor. Mol. Pharmacol. 2006;69:1871–1878. doi: 10.1124/mol.105.021832. [DOI] [PubMed] [Google Scholar]
- Kopec AK, Boverhof DR, Burgoon LD, Ibrahim-Aibo D, Harkema JR, Tashiro C, Chittim B, Zacharewski TR. Comparative toxicogenomic examination of the hepatic effects of PCB126 and TCDD in immature, ovariectomized C57BL/6 mice. Toxicol. Sci. 2008;102:61–75. doi: 10.1093/toxsci/kfm289. [DOI] [PubMed] [Google Scholar]
- Kopec AK, Boverhof DR, Nault R, Harkema JR, Tashiro C, Sharratt B, Chittim B, Burgoon LD, Zacharewski TR. Toxicogenomic evaluation of long-term hepatic effects of TCDD in immature, ovariectomized C57bl/6 mice. Toxicol. Sci. 2013 doi: 10.1093/toxsci/kft156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremoser C, Albers M, Burris TP, Deuschle U, Koegl M. Panning for SNuRMs: using cofactor profiling for the rational discovery of selective nuclear receptor modulators. Drug Discovery Today. 2007;12:860–869. doi: 10.1016/j.drudis.2007.07.025. [DOI] [PubMed] [Google Scholar]
- Kwekel JC, Forgacs AL, Burgoon LD, Williams KJ, Zacharewski TR. Tamoxifen-elicited uterotrophy: cross-species and cross-ligand analysis of the gene expression program. BMC Medical Genomics. 2009;2:19. doi: 10.1186/1755-8794-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson AS, Jordan VC. Selective oestrogen receptor modulation: molecular pharmacology for the millennium. Eur. J. Cancer. 1999;35:1974–1985. doi: 10.1016/s0959-8049(99)00297-x. [DOI] [PubMed] [Google Scholar]
- Levenson AS, Kliakhandler IL, Svoboda KM, Pease KM, Kaiser SA, Ward JE, Jordan VC., 3rd Molecular classification of selective oestrogen receptor modulators on the basis of gene expression profiles of breast cancer cells expressing oestrogen receptor alpha. Br. J. Cancer. 2002;87:449–456. doi: 10.1038/sj.bjc.6600477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKillop D, Case DE. Mutagenicity, carcinogenicity and toxicity of beta-naphthoflavone, a potent inducer of P448. Biochem. Pharmacol. 1991;41:1–7. doi: 10.1016/0006-2952(91)90003-n. [DOI] [PubMed] [Google Scholar]
- Murray IA, Flaveny CA, Chiaro CR, Sharma AK, Tanos RS, Schroeder JC, Amin SG, Bisson WH, Kolluri SK, Perdew GH. Suppression of cytokine-mediated complement factor gene expression through selective activation of the Ah receptor with 3’,4’-dimethoxy-alpha-naphthoflavone. Mol. Pharmacol. 2011;79:508–519. doi: 10.1124/mol.110.069369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray IA, Krishnegowda G, DiNatale BC, Flaveny C, Chiaro C, Lin JM, Sharma AK, Amin S, Perdew GH. Development of a selective modulator of aryl hydrocarbon (Ah) receptor activity that exhibits anti-inflammatory properties. Chem. Res. Toxicol. 2010;23:955–966. doi: 10.1021/tx100045h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebert DW, Roe AL, Dieter MZ, Solis WA, Yang Y, Dalton TP. Role of the aromatic hydrocarbon receptor and [Ah] gene battery in the oxidative stress response, cell cycle control, and apoptosis. Biochem. Pharmacol. 2000;59:65–85. doi: 10.1016/s0006-2952(99)00310-x. [DOI] [PubMed] [Google Scholar]
- Nguyen LP, Bradfield CA. The search for endogenous activators of the aryl hydrocarbon receptor. Chem. Res. Toxicol. 2008;21:102–116. doi: 10.1021/tx7001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opitz CA, Litzenburger UM, Sahm F, Ott M, Tritschler I, Trump S, Schumacher T, Jestaedt L, Schrenk D, Weller M, Jugold M, Guillemin GJ, Miller CL, Lutz C, Radlwimmer B, Lehmann I, von Deimling A, Wick W, Platten M. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature. 2011;478:197–203. doi: 10.1038/nature10491. [DOI] [PubMed] [Google Scholar]
- Pansoy A, Ahmed S, Valen E, Sandelin A, Matthews J. 3-methylcholanthrene induces differential recruitment of aryl hydrocarbon receptor to human promoters. Toxicol. Sci. 2010;117:90–100. doi: 10.1093/toxsci/kfq096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohjanvirta R, Korkalainen M, McGuire J, Simanainen U, Juvonen R, Tuomisto JT, Unkila M, Viluksela M, Bergman J, Poellinger L, Tuomisto J. Comparison of acute toxicities of indolo[3,2-b]carbazole (ICZ) and 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in TCDD-sensitive rats. Food Chem. Toxicol. 2002;40:1023–1032. doi: 10.1016/s0278-6915(02)00067-4. [DOI] [PubMed] [Google Scholar]
- Pohjanvirta R, Vartiainen T, Uusi-Rauva A, Monkkonen J, Tuomisto J. Tissue distribution, metabolism, and excretion of 14C-TCDD in a TCDD-susceptible and a TCDD-resistant rat strain. Pharmacol. Toxicol. 1990;66:93–100. doi: 10.1111/j.1600-0773.1990.tb00712.x. [DOI] [PubMed] [Google Scholar]
- Poland A, Knutson JC. 2,3,7,8-Tetrachlorodibenzo-p-dioxin and related halogenated aromatic hydrocarbons: examination of the mechanism of toxicity. Annu. Rev. Pharmacol. Toxicol. 1982;22:517–554. doi: 10.1146/annurev.pa.22.040182.002505. [DOI] [PubMed] [Google Scholar]
- Quintana FJ, Murugaiyan G, Farez MF, Mitsdoerffer M, Tukpah AM, Burns EJ, Weiner HL. An endogenous aryl hydrocarbon receptor ligand acts on dendritic cells and T cells to suppress experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. U. S. A. 2010;107:20768–20773. doi: 10.1073/pnas.1009201107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romkes M, Piskorska-Pliszczynska J, Safe S. Effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin on hepatic and uterine estrogen receptor levels in rats. Toxicol. Appl. Pharmacol. 1987;87:306–314. doi: 10.1016/0041-008x(87)90292-4. [DOI] [PubMed] [Google Scholar]
- Safe S, Qin C, McDougal A. Development of selective aryl hydrocarbon receptor modulators for treatment of breast cancer. Expert Opin. Investig. Drugs. 1999;8:1385–1396. doi: 10.1517/13543784.8.9.1385. [DOI] [PubMed] [Google Scholar]
- Sears DD, Hsiao A, Ofrecio JM, Chapman J, He W, Olefsky JM. Selective modulation of promoter recruitment and transcriptional activity of PPARgamma. Biochem. Biophys. Res. Commun. 2007;364:515–521. doi: 10.1016/j.bbrc.2007.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CL, O’Malley BW. Coregulator function: a key to understanding tissue specificity of selective receptor modulators. Endocr. Rev. 2004;25:45–71. doi: 10.1210/er.2003-0023. [DOI] [PubMed] [Google Scholar]
- Tanos R, Patel RD, Murray IA, Smith PB, Patterson AD, Perdew GH. Aryl hydrocarbon receptor regulates the cholesterol biosynthetic pathway in a dioxin response element-independent manner. Hepatology. 2012;55:1994–2004. doi: 10.1002/hep.25571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg M, Peterson RE, Schrenk D. Human risk assessment and TEFs. Food Addit. Contam. 2000;17:347–358. doi: 10.1080/026520300283414. [DOI] [PubMed] [Google Scholar]
- Wei YD, Rannug U, Rannug A. UV-induced CYP1A1 gene expression in human cells is mediated by tryptophan. Chem. Biol. Interact. 1999;118:127–140. doi: 10.1016/s0009-2797(98)00118-5. [DOI] [PubMed] [Google Scholar]
- Yao C, Safe S. 2,3,7,8-Tetrachlorodibenzo-p-dioxin-induced porphyria in genetically inbred mice: partial antagonism and mechanistic studies. Toxicol. Appl. Pharmacol. 1989;100:208–216. doi: 10.1016/0041-008x(89)90307-4. [DOI] [PubMed] [Google Scholar]
- Yin XF, Chen J, Mao W, Wang YH, Chen MH. A selective aryl hydrocarbon receptor modulator 3,3’-Diindolylmethane inhibits gastric cancer cell growth. J. Exp. Clin. Cancer Res. 2012;31:46. doi: 10.1186/1756-9966-31-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Rowlands C, Safe S. Ligand-dependent interactions of the Ah receptor with coactivators in a mammalian two-hybrid assay. Toxicol. Appl. Pharmacol. 2008;227:196–206. doi: 10.1016/j.taap.2007.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.