Abstract
Halobacterium sp. NRC-1 is a wild-type extremophilic microbe that is naturally tolerant to high levels of ionizing radiation. Mutants of strain NRC-1 with even higher levels of resistance to ionizing radiation, named RAD, were previously isolated after selecting survival to extremely high-doses of ionizing radiation. These RAD mutants displayed higher transcription levels for the rfa3 operon, coding two subunits of the RPA-like putative single-stranded binding protein, rfa3 and rfa8, and a third downstream gene, ral. In order to bioengineer cells with increased tolerance to ionizing radiation and further explore the genetic basis of the RAD phenotype, we placed the rfa3 operon under control of the gvpA promoter in a Halobacterium expression plasmid, pDRK1. When pDRK1 was introduced into the wild-type NRC-1 strain, overproduction of the Rfa3 and Rfa8 proteins was observed by Western blotting and proteomic analysis. The Halobacterium strains expressing Rfa3 and Rfa8 also displayed improved survival after exposure to ionizing radiation, similar to the RAD mutants, when compared to wild-type strain NRC-1. The Rfa3 and Rfa8 proteins co-purified by affinity chromatography on single-stranded DNA-cellulose columns, confirming the ability of the proteins to bind to single-stranded DNA as well as their relative abundance in the wild-type, RAD mutants, and rfa3 operon overexpression strains. These results clearly establish that overexpression of haloarchaeal RPA promotes ionizing radiation resistance in Halobacterium sp. NRC-1 and that the Rfa3 and Rfa8 subunits bind single-stranded DNA. Bioengineering cells with increased levels of ionizing radiation resistance may have potential value in medical and environmental applications.
Keywords: Halophile, DNA repair, Gene expression, Archaea
Introduction
Halophilic Archaea (also called Haloarchaea) represent some of the most extremophilic microorganisms known. Haloarchaea inhabit hypersaline habitats with 3–5 M sodium chloride concentrations and contain equally high concentration of salts in their cytoplasm (DasSarma and DasSarma 2012). In addition, Haloarchaea may be found in diverse habitats, including both hot and cold hypersaline environments, such as in hot deep sea brines in the Red Sea, and in Deep Lake of Antarctica. Some Haloarchaea are polyextremophilic, displaying resistance to several stressors in the environment (DasSarma 2007). Despite the substantial diversity of Haloarchaea known, all isolates characterized to date constitute members of a single family (Grant et al. 2001), one species of which, Halobacterium sp. NRC-1, has served as a model organism (DasSarma 2004).
Halobacterium sp. NRC-1 was the first in the Haloarchaea family to be sequenced and provided a convenient experimental genetic system for postgenomic studies of DNA repair and replication (DasSarma 2004). Bioinformatic analysis led to a detailed inventory of many DNA repair genes and suggested the genetic basis for the observed tolerance to ultraviolet and ionizing radiation (DasSarma et al. 2001; Capes et al. 2011). Further genetic studies provided direct evidence for the involvement of a photolyase (Phr), nucleotide excision repair system (UvrABCD), and DNA break repair (Mre11/Rad50) (McCready and Marcello 2003; Crowley et al. 2006; Kish and DiRuggerio 2008) in radiation tolerance. Genetic studies of DNA replication identified at least 10 essential genes, including genes encoding origin recognition, initiation, and elongation functions (Berquist et al. 2007). However, many other DNA repair and replication genes likely to be important for radiotolerance remain to be explored in detail.
One of the most interesting postgenomic studies of Halobacterium sp. NRC-1 was the isolation of RAD mutants with increased levels of ionizing radiation resistance (DeVeaux et al. 2007). Whereas the wild-type NRC-1 species was naturally highly ionizing radiation tolerant, with a dose of 2.9 kGy corresponding to 50% cell survival, exposure to multiple cycles of irradiation with killing doses (16–23 kGy) produced progressively more ionizing radiation resistant mutants. These RAD mutants displayed doses corresponding to 50% cell survival in the range of 7–9 kGy (DeVeaux et al. 2007). Transcriptomic analysis of these RAD mutants showed that they exhibited upregulation of the rfa3 operon coding predicted gene products with homology to subunits of a mammalian replication protein A or RPA-type single-stranded DNA binding protein. Three genes were implicated, including rfa3 and rfa8, coding proteins with oligonucleotide binding (OB) folds, and a third gene, ral, with unknown function, but conserved in all sequenced Haloarchaea (Fig. 1) (DeVeaux et al. 2007; Capes et al. 2012).
Fig. 1.
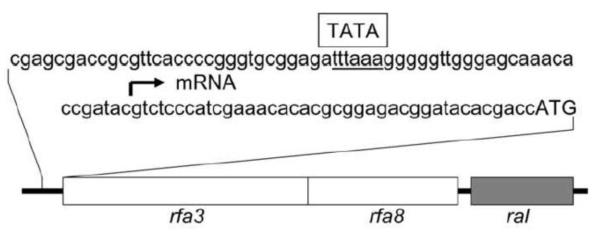
The rfa3 operon region. Sequence of the promoter region is shown above, with location of mRNA start site marked by an arrow and putative TATA-box labeled. Below, boxes indicate the relative location and sizes of the rfa3, rfa8, and ral genes.
The mammalian RPA is a heterotrimeric protein containing RPA71 (large subunit), RPA32 (medium subunit), and RPA14 (small subunit), which function together in DNA replication and repair. In Halobacterium sp. NRC-1, there are five genes coding proteins with homology to the RPA large subunit, each containing at least one OB-fold (Cubeddu and White 2005; Capes et al. 2012). This class includes the Rfa3 protein, which contains one OB-fold and one nucleic acid-binding Zn-finger motif (Chen et al. 2005; DeVeaux et al. 2007). There are two homologs to the medium subunit of RPA, including the Rfa8 protein which contains one OB-fold. Orthologous genes coding for Rfa3 and Rfa8-like proteins (with similar genetic organization and 66–70 % amino acid sequence identity) were found in the genomes of all other sequenced Haloarchaea. Similar genes and operons are also present in members of the euryarchaeal families Thermococcales and Methanosarcinales (Komori and Ishino 2001).
In the current work, we sought to explore the basis of rfa3 operon involvement in more detail. In particular, we wanted to know if cells with higher radiation tolerance could be bioengineered simply by overexpressing the RPA genes. For this goal, we also developed a new high-level expression system for Halobacterium sp. NRC-1 and used it for purification of the RPA protein and establishing its single-stranded DNA binding activity.
Materials and methods
Enzymes and chemicals
Restriction enzymes, T4 DNA ligase, and Taq DNA polymerase were purchased from New England Biolabs (Beverly, MA, USA). Most other chemicals were purchased from Sigma (St. Louis, MO, USA).
Strains, media, and culture conditions
Escherichia coli DH5α transformants were grown at 37 °C in Luria-Bertani (LB) medium supplemented with 100 μg/ml ampicillin. Halobacterium sp. strains NRC-1 (ATCC number 700922 and JCM number 11081), LH5, LH7a, and derivatives were cultured in CM+ medium, and mevinolin-resistant (Mevr) transformants were selected by culturing in CM+ containing 20 μg of mevinolin/ml (a gift from Merck, Rahway, N.J., USA) for the derivatives at 42 °C with shaking as previously described (DasSarma et al. 1995; Berquist et al. 2006, 2007). For solid media, 2% (w/v) agar was added.
Construction of the rfa3 operon expression plasmids, pRK2 and pDRK1
To facilitate Rfa3, Rfa8 and Ral expression in Halobacterium sp. NRC-1, an overexpression vector was constructed. We used the β-galactosidase (bga) pMC2 (Karan et al. 2013) expression plasmid as the backbone. The bga gene was excised and replaced with PCR amplified Halobacterium sp. NRC-1 rfa3 operon (VNG2160, 2162, 2163; see NCBI accession number AE004437.1) by cloning via the NdeI and BamHI sites, to generate the pRK2 expression plasmid. Next, the gvpA promoter was PCR amplified and used to replace the KpnI - NdeI fragment containing the cspD2 promoter, resulting in the pDRK1 expression plasmid (Fig. 2). The constructs were validated by DNA sequencing. Primers used for PCR amplification and DNA sequencing are listed in Table 1.
Fig. 2.
Map of Halobacterium sp. NRC-1 expression vector pDRK1. Relative size, location, and transcriptional orientation of bla gene (coding β-lactamase for ampicillin resistance), mev gene (coding HMG-CoA reductase for mevinolin resistance), rfa3 operon, and rep gene (coding Halobacterium pGRB1 replicase) are shown with arrows. Position of the gvpA promoter is indicated by an arrow and labeled P. Locations of KpnI, NdeI and BamHI restriction sites used for construction are indicated.
Table 1.
Oligonucleotides used in this study as primers for PCR and DNA sequencing
| Primer | Sequence (5'→3') | Application |
|---|---|---|
| SD4.NdeI.S | GGAATTCCATATCAGCGACCTTTCAG | rfa3 operon amplification |
| SD4.BamH1A | ACGCGGATCCGACCGCGCCAGCCGG | rfa3 operon amplification |
| gvpA-NdeI | CTCAAGGTATACCACTAGACCCTAAT | gvpA promoter amplification |
| gvpA-KpnI | ACTCATGGTACCTACTTCTCTCCAGT | gvpA promoter amplification |
| 2160F | ATGACGGCGTCCTCGCCGAA | rfa3 operon sequencing |
| 2160R | TCAGATCGACCTCGCGCGGA | rfa3 operon sequencing |
| 2162F | ATGAGCGGCGCACCCACCCGC | rfa3 operon sequencing |
| 2162R | TCACTCCGGCGCGTCGAACT | rfa3 operon sequencing |
| 2163F | ATGGGGAACAAGAACAAGAC | rfa3 operon sequencing |
| 2163R | CTATCCGAGCTGGTAGGGTT | rfa3 operon sequencing |
| pKJ-cspD2F | GCTGGACTGCCTTTTCTTCG | gvpA promoter sequencing |
| pKJ-BamH1-3'160R | GTTACTCCACCGTCATTCAG | gvpA promoter sequencing |
Expression of the rfa3 operon in Halobacterium sp. NRC-1 and protein preparation
Halobacterium sp. NRC-1 was transformed with pDRK1, using the EDTA-PEG method (DasSarma et al. 1995) and transformants were selected on CM+ agar plates supplemented with 20 μg/ml mevinolin. The Halobacterium sp. NRC-1 (pDRK1) strain was grown to late log phase (OD600 of 0.9–1.0) at 42 °C in CM+ medium supplemented with 20 μg/ml mevinolin and further incubated at 15 °C for 72 h to induce Rfa3, Rfa8 and Ral production.
For protein preparations, cells were harvested by centrifugation (6,000×g, 4 °C, 10 min) in a Sorvall RC-5B centrifuge and disrupted in buffer containing 50 mM Tris-HCl (pH 8.0), 2 M KCl, 10% v/v glycerol, 0.1 mM ZnCl2, 50 mM MgCl2, 1 mM EDTA, and 100 μg PMSF/ml using a sonicator (Model CL-18, Thermo Fisher Scientific, Waltham, USA). Cell debris was removed by centrifugation (25,000×g, 4 °C, 10 min) in an Eppendorf 5417C centrifuge to obtain the cell-free supernatant. The cell-free supernatant after filtering through a Nalgene membrane filter (pore size, 0.2 μm) was used as the source of crude extract.
Electrophoresis and Western blotting analysis
Western blotting analysis of cell lysates or purified protein was done essentially as described previously (Halladay et al. 1993; Shukla and DasSarma 2004). Briefly, the electrophoretically fractionated cell lysates or pure protein were electroblotted onto 0.45 μm Immobilon-P polyvinylidene difluoride (PVDF) membranes (Millipore Corp., Boston, MA) for 1 h at 100 volts using a Bio-Rad mini gel blotter. After electroblotting, the membranes were washed twice for 5 min with PBS buffer, blocked for 1 h with 3 % bovine serum albumin (BSA, Sigma Corp., St. Louis, MO) in PBS buffer. The membranes were incubated for 1 h at room temperature or overnight at 4 °C with gentle shaking with Rfa3 antibodies diluted 1:1000. The Rfa3-specific rabbit polyclonal antibodies were raised against a synthetic peptide at the C-terminus of Rfa3 (RPDTESVLIRARSI) (Thermo Scientific, Rockford, IL, USA).
The membranes were then washed 5 times each for 5 min with 20 ml PBS buffer containing 0.1% Tween 20, and incubated for 1 h with a 1:2500 dilution of goat anti-rabbit secondary antibodies-alkaline phosphatase conjugate (Sigma, St. Louis, MO, USA) in a solution containing 3 % BSA in PBS buffer. For chromogenic detection of the protein bands, membranes were incubated in nitro-blue tetrazolium and 5-bromo-4-chloro-3'-indolyphosphate substrate (Thermo Scientific, Rockford, IL USA) according to the manufacturer's specification.
X-ray irradiation and survival
For irradiation, each strain was grown in liquid culture to stationary phase (OD660nm~1.3), divided into aliquots (50 μl), and subjected to a predetermined set of doses (0 Gy, 1 kGy, 2 kGy, 3 kGy) using a Pantek X-ray machine (250 kV, 13 mA) at a dose rate of 7.8 Gy/min. Immediately after irradiation, the aliquots were diluted in basal salts and plated in duplicate or triplicate. The average colony forming units (cfu/ml) were compared to cfu for unirradiated samples to determine survival at each dose.
Single-stranded DNA-cellulose affinity chromatography
The cleared lysate, prepared as described above, was loaded onto a single-stranded DNA-cellulose column (St. Louis, MO, USA) equilibrated with a buffer containing 20 mM Tris-HCl (pH 7.4), 2 M NaC1, 1 mM EDTA, and 10% (w/v) glycerol. The column was washed with the same buffer until all unbound proteins were removed, and bound proteins were eluted by decreasing the salt concentration (1 M and 0.1 M NaCl) in the same buffers in a stepwise fashion.
Proteomic analysis
Aliquots of whole cell lysates containing 200 μg of proteins were digested with sequencing-grade modified trypsin [50:1 (w/w)] (Promega, Madison, WI, USA) in a final volume of 200 μL of 50 mM ammonium bicarbonate (pH 8.3) at 37 °C for 16 h, followed by purification using a C18 spin column (Pierce, Rockford, IL, USA) according to the manufacturer's suggested protocol. The trypsinized samples were then lyophilized and stored at −20 °C for further LC-MS/MS analysis.
Approximately 2 μg of trypsinized peptides in 0.1% (v/v) formic acid were analyzed using an Agilent 1200 series nano-HPLC system (Agilent Technologies, Palo Alto, CA, USA) coupled to a hybrid linear ion trap-Orbitrap (LTQ-Orbitrap Discovery) mass spectrometer (Thermo Scientific, San Jose, CA, USA). Each sample was analyzed with five MS/MS runs in which the MS survey scans were acquired over a full m/z range of 400–2000 and by gas-phase fractionation (GPF) of four restricted windows (400–521, 516–690, 685–968, and 963–2000 m/z), as described previously (Chu et al. 2011a; Yi et al 2002). The six most intense precursor ions within each MS survey scan were sequentially isolated for collision-induced dissociation using the data-dependent mode with the isolation width: 1 m/z, dynamic exclusion duration of 45 s, and an exclusion list size of 150.
MS/MS raw data files in Thermo XCalibur (version 2.0.7) binary format were converted to the mzXML open data format using the ReAdW (version 4.6.0) program (Pedrioli et al. 2004). COMET algorithm was applied to search the spectra against the combined forward and reversed (decoy) protein sequences of Halobacterium sp. NRC-1 (2,427 proteins; EMBL-EBI Integr8, www.ebi.ac.uk/integr8/EBI-Integr8-HomePage.do) (Elias and Gygi 2007; Keller et al. 2005; Ng et al. 2000).
The search parameters included a precursor mass tolerance of 2.1 Da, fragment bin tolerance 1.0005, fragment bin offset 0.4, a minimum of ten matched peaks, up to 3 possible oxidation of the methionine (+15.9949Da) residue, and default settings of other parameters as suggested by the software developers. The cleaving agent was not specified for non-constraint database searches. COMET results were further processed using the Trans Proteomic Pipeline (Elias and Gygi 2007; Keller et al. 2002; 2005; Nesvizhskii et al. 2003) as previously described (Chu at al 2011a; 2011b). Peptides and decoy sequences with a PeptideProphet probability above 0.9 were considered as significant hits. False discovery rate (FDR) = (number of decoy peptide matches/number of peptide matches) × 100%. The MS proteomic data have been deposited in the PeptideAtlas (Desiere et al. 2005) with the dataset identifier PASS00325 (http://www.peptideatlas.org/PASS/PASS00325).
Liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis
LC-MS/MS was used to identify proteins purified by single-stranded DNA-cellulose affinity chromatography. The eluted proteins were subjected to 12 % SDS-PAGE and Coomassie stained. Protein bands were excised, destained, and subjected to reduction (with DTT) and alkylation (with iodoacetamide) and subsequently proteolyzed with trypsin (Promega, Madison, WI, USA). Digested peptides were desalted by C18 stage-tip (3M emphore , C18 Octadecyl extraction disks) and then eluted with 0.1% TFA in 60% acetonitrile and re-suspended in 7 μL 0.1% formic acid for LC-MS/MS analysis. Protein identification of peptides was performed on a Q-Exactive instrument (Thermo Scientific, Rockford, IL, USA) interfaced with a Proxion nanoflow LC system. Peptides were fractionated by reverse-phase HPLC on a 75 μm × 15 cm PicoFrit column with a 15 μm emitter (PF3360-75-15-N-5, New Objective, Woburn, MA, USA) in-house packed with Bruker Michrom (Auburn, CA, USA) Magic C18AQ (5 μm, 120Å) using 0–60% acetonitrile/0.1% formic acid gradient over 70 min at 300 nl/min. Eluting peptides were sprayed directly into Q-exactive at 2.0 kV. Survey scans (full ms) were acquired from 350–1,800 m/z with up to 15 peptide masses (precursor ions) individually isolated with a 1.9 Da window and fragmented (MS/MS) using a collision energy of 29 and 30 s dynamic exclusion. Precursor and the fragment ions were analyzed at 70,000 and 17,500 resolution, respectively. Peptide sequences were identified from isotopically resolved masses in MS and MS/MS spectra extracted with and without deconvolution using Thermo Scientific MS2 processor and Xtract software. Data was searched against Halobacterium sp. NRC-1 from the NCBI nr 2012 database. Oxidation on methionine, deamidation on residues N and Q, phospho S/T/Y, (as different variable modifications) and carbamidomethyl on cysteine (fixed), as modifications using Sequest software interfaced in the Proteome Discoverer 1.4 workflow. Mass tolerances on precursor and fragment masses were 20 ppm and 0.05 Da, respectively.
Results
Construction of an rfa3 operon overexpression strain
An expression vector, pMC2, capable of shuttling between Halobacterium sp. NRC-1 and E. coli was recently constructed using the pGRB1 miniplasmid, the mevinolin resistance gene, and pUC18 E. coli vector (Karan et al. 2013). Plasmid pMC2 also contained the H. lacusprofundi bga gene under the control of the cspD2 promoter of NRC-1. In order to express the rfa3 operon, we first replaced the bga gene, via the flanking NdeI and BamHI sites, with the PCR amplified rfa3 operon (Table 1). Next, we PCR amplified the gvpA promoter region of NRC-1, which was selected on the basis of prior transcriptomic data indicating that it was a stronger promoter (Coker et al. 2007), and replaced the cspD2 promoter region via flanking KpnI and NdeI sites. The resulting construct, named pDRK1, shown in Fig. 2, contains the rfa3 operon downstream of the gvpA promoter, the mevinolin-resistance selectable marker for Halobacterium, the ampicillin-resistance marker for E. coli, and replication ability for both microorganisms. To construct a Halobacterium rfa3 operon overexpression strain, we transformed pDRK1 into the wild-type strain to produce the bioengineered strain Halobacterium sp. NRC-1 (pDRK1).
Rfa3 protein levels measured by Western blotting analysis
To measure the Rfa3 protein levels in Halobacterium strains, we carried out Western blotting analysis of wild-type strain NRC-1, two RAD strains (LH5 and LH7a), which had previously been shown to contain high levels of rfa3 operon transcripts, and the bioengineered strain Halobacterium sp. NRC-1 (pDRK1). A rabbit antiserum which had been raised against a synthetic peptide of Rfa3 was used as probe (see Materials and Methods). The Western blotting results are shown in Fig. 3A, and quantification results with densitometry, using total cell proteins for normalization, shown in Fig 3B. The Rfa3 protein levels were 10 to 16-fold higher for the RAD strains compared to the wild-type strain, and even higher (23-fold) for the bioengineered Halobacterium sp. NRC-1 (pDRK1) overexpression strain.
Fig. 3.
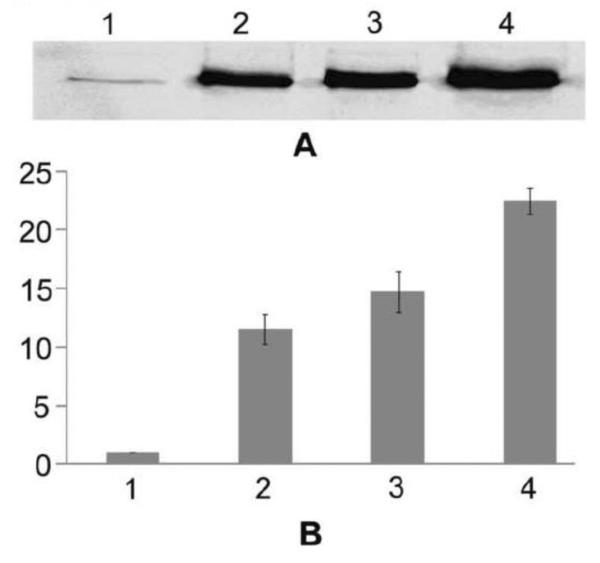
Western blotting analysis of Halobacterium sp. NRC-1 and derivative strains using rabbit Rfa3 antiserum. A. Equal quantities of total cell lysate protein samples were electrophoresed on a 12% polyacrylamide-SDS gel, transferred to PVDF membrane, and probed with Rfa3 antibody followed by a secondary goat anti-rabbit antibody-alkaline phosphatase conjugate and chromogenic substrate. Lane 1: Halobacterium sp. NRC-1, Lane 2: Halobacterium sp. LH5, Lane 3: Halobacterium sp. LH7a, Lane 4: Halobacterium sp. NRC-1 (pDRK1). B. Relative intensities of the bands analyzed by ImageJ software. Vertical axis shows the relative band intensity for the same strains as in part A in the horizontal axis. Values are the average of triplicate experiments with standard deviation shown with error bars.
Rfa3 and Rfa8 protein levels measured by proteomic analysis
In order to further assay the rfa3 operon gene products, we conducted proteomic analysis of wild-type NRC-1 strain, the two RAD strains, and the bioengineered Halobacterium sp. NRC-1 (pDRK1) overexpression strain. Total cell proteins were extracted after hypotonic lysis and proteolytic digestion with trypsin. Protein fragments were analyzed by LC-MS/MS followed by bioinformatic analysis to identify the number of tryptic peptides observed. When the results were normalized and peptide fragments compared for relative abundance, the four Halobacterium strains showed the expected levels of Rfa3 protein (Table 2), as well as Rfa8 protein. NRC-1 had the lowest Rfa3 and Rfa8 levels, the RAD strains had intermediate levels (8 to 16-fold higher), and the bioengineered Halobacterium sp. NRC-1 (pDRK1) strain had the highest levels (5–12 fold higher than the RAD mutants). In contrast, the level of Ral protein was not significantly changed, although in this case, only a few diagnostic peptides were detected.
Table 2.
Normalized relative spectral counts of rfa3 operon proteins in Halobacterium sp. strains whole cell lysates (FDR < 0.6)
| Halobacterium sp. strains | ||||
|---|---|---|---|---|
| Protein | NRC-1 | LH5 | LH7a | pDRK1 |
| Rfa3 | 1 | 8.2 | 13.2 | 67.6 |
| Rfa8 | 1 | 11.2 | 15.5 | 188.8 |
| Ral | 1 | 0.6 | 0.9 | 2.8 |
Improved ionizing irradiation resistance of strains overexpressing Rfa3 and Rfa8
In order to test the effect of the Rfa3 and Rfa8 protein levels on ionizing radiation tolerance, we used a Pantek X-ray machine to irradiate stationary phase Halobacterium cultures of strain NRC-1, the two RAD strains, and the bioengineered Halobacterium sp. NRC-1 (pDRK1) strain. Each strain was subjected to 0, 1, 2, or 3 kGy X-ray doses and percent survival was plotted (Fig. 4). Interestingly, the bioengineered Halobacterium sp. NRC-1 (pDRK1) strain, like the two RAD mutants, was found to be significantly more resistant to ionizing radiation compared to the parent wild-type strain. The Rfa3 and Rfa8 overproducing strains, both selected (RAD) and engineered (pDRK1), survived better by more than 1 order of magnitude higher at the higher ionizing radiation doses tested (2 or 3 kGy). These findings are consistent with our previous comparative transcriptomics results that overexpression of the rfa3 operon is responsible for the increased ionizing radiation resistance of Halobacterium strains. Moreover, ionizing radiation used in the present experiments is biologically approximately two times more effective than high energy electrons used in the previous study. These results also extended our understanding of the protective effect from β-particles observed previously to X-rays (DeVeaux et al. 2007).
Fig. 4.
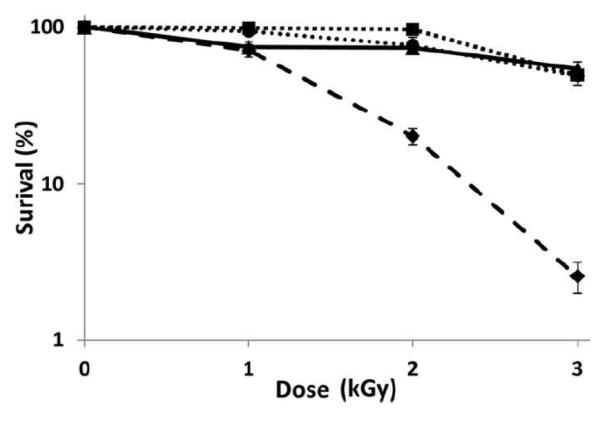
Surviving fraction of Halobacterium sp. NRC-1 and derivative strains after irradiation with ionizing radiation. Survival curves are shown for Halobacterium sp. NRC-1 (dashed ◆), LH5 (round dots, •), LH7a (square dots, ■), and pDRK1 (solid, ▲) exposed to different doses (0, 1, 2 and 3 kGy) of ionizing radiation (percent surviving in log scale (vertical axis) versus radiation dose in kGy(horizontal axis). Values are the average of triplicate experiments with standard deviation shown with error bars.
Purification and identification of Rfa3 and Rfa8 single-stranded DNA binding activity
The function of the Rfa3 and Rfa8 proteins were speculated to be in DNA binding and repair with the finding of overexpression of the rfa3 operon in the RAD mutants. To determine if these proteins bind single-stranded DNA, we employed affinity chromatography on single-stranded DNA cellulose columns. Total protein from each of the Halobacterium strains was bound to a column, washed, and bound proteins eluted. Both total and eluted proteins were profiled by SDS-polyacrylamide gel electrophoresis and Coomassie staining, and the eluted proteins were analyzed by Western blotting using Rfa3 antiserum (Fig. 5, lanes 1–3, respectively). Two major proteins were visible in the bioengineered Halobacterium sp. NRC1 (pDRK1) strain, with the heavy top band corresponding to Rfa3 based on molecular weight, estimated to be ~52 kDa. The higher apparent molecular mass was expected based on previous results with haloarchaeal proteins due to their high fraction of acidic amino acids (Halladay et al. 1993; Karan et al. 2013). Bands with the same mobility were also visible by Coomassie straining in the two RAD strains, albeit with lower intensity, suggestive of lower abundance. Western blotting showed the identity of the higher band to be Rfa3, which was also detected in wild-type strain NRC-1.
Fig. 5.

Affinity purification and Western analysis of Rfa3 and Rfa8 in Halobacterium sp. NRC-1 and derivative strains. Total protein samples from cell lysates and fractions binding to a DNA-cellulose column were electrophoresed on a 12% polyacrylamide-SDS gel, and stained with Coomassie blue, or transferred to PVDF membrane, and probed with rabbit Rfa3 antibody followed by a secondary goat anti-rabbit antibody-alkaline phosphatase conjugate and substrate. Panel A contains Halobacterium sp. NRC-1, panel B contains Halobacterium sp. LH5, panel C contains Halobacterium sp. LH7a, and panel D contains Halobacterium sp. NRC-1 (pDRK1). Lanes labeled 1 contain total cell lysate and lanes labeled 2 contain peak bound fraction from a DNA-cellulose column, Coomassie stained, and lanes labeled 3 display results of Western blots.
In order to confirm the identity of Rfa3 protein and determine the identity of the smaller, ~22 kDa protein, we excised the two bands from the bioengineered Halobacterium sp. NRC-1 (pDRK1) strain lysate from a SDS-polyacrylamide gel, and conducted LC-MS/MS analysis. The results confirmed the identities of the two proteins as Rfa3 (34.12 kDa) and Rfa8 (21.16 kDa), based on peptide sequences, molecular weight, and query coverage. The tryptic digested peptides of Rfa3 and Rfa8 from Halobacterium sp. NRC-1 (pDRK1) are tabulated in Tables 3 and 4. The peptides identified covered 91% (282/311 amino acids) of Rfa3 and 94.2% (179/190 amino acids) of Rfa8, unambiguously confirming the identity of the proteins.
Table 3.
Unique peptides of Halobacterium sp. NRC-1 Rfa3 identified by LC-MS/MS analysis
| Amino acid sequence | Number of times peptide identified | Expected mass value (Da) | Query coverag* (%) to Rfa3 |
|---|---|---|---|
| LEKLVSEYKVPVDEAR | 6 | 1875.1 | 5 |
| LVSEYK | 1 | 737.8 | 1 |
| LVSEYKVPVDEAR | 14 | 1504.7 | 4 |
| VPVDEAR | 23 | 784.8 | 2 |
| RSVENHYLDEAGLDR | 2 | 1773.8 | 4 |
| RSVENHYLDEAGLDRDDLAGGGGNDTVEVEDVDEAEQWVDITAK | 56 | 4804.0 | 18 |
| SVENHYLDEAGLDR | 6 | 1617.6 | 4 |
| SVENHYLDEAGLDRDDLAGGGGNDTVEVEDVDEAEQWVDITAK | 177 | 4647.8 | 17 |
| DDLAGGGGNDTVEVEDVDEAEQWVDITAK | 19 | 3048.1 | 9 |
| VVELWEPR | 7 | 1027.1 | 2 |
| SDSVGQVGLLGDPTGTIK | 22 | 1743.9 | 8 |
| SDSVGQVGLLGDPTGTIKFTK | 3 | 2120.3 | 9 |
| WAKSDLETLDEGQVYR | 2 | 1910.0 | 5 |
| SDLETLDEGQVYR | 83 | 1524.6 | 4 |
| LGNVVTDEYQGR | 56 | 1350.4 | 3 |
| YSVKLNK | 1 | 851.0 | 2 |
| TTSIEAVDEEFEVGDNEDEVAGALVDVQSGSGLIK | 89 | 3623.8 | 14 |
| TTSIEAVDEEFEVGDNEDEVAGALVDVQSGSGLIKR | 1 | 3780.0 | 15 |
| RCPEEDCTR | 19 | 1108.2 | 2 |
| CPEEDCTR | 16 | 952.0 | 2 |
| VLQNGRCNEHGEGDGEFDLR | 11 | 2245.3 | 6 |
| CNEHGEGDGEFDLR | 99 | 1577.6 | 4 |
| IKGVLDDGTDVHEVIFDEEATEALTGIGLEEAK | 3 | 3514.8 | 15 |
| GVLDDGTDVHEVIFDEEATEALTGIGLEEAK | 125 | 3273.5 | 14 |
| QMAKDALDTTVVADEMR | 6 | 1894.1 | 5 |
| DALDTTVVADEMR | 197 | 1435.5 | 4 |
| ATILGKYYR | 4 | 1084.2 | 2 |
| YVLANDTELLTTRPDTESVLIR | 168 | 2519.8 | 7 |
| ELLTTRPDTESVLIR | 89 | 1742.9 | 4 |
The percent of Halobacterium sp. NRC-1 Rfa3 amino acid sequence that aligns with the identified peptide sequence.
Table 4.
Unique peptides of Halobacterium sp. NRC-1 Rfa8 identified by LC-MS/MS
| Amino acid sequence | Number of times peptide identified | Expected mass value (Da) | Query coverage* (%) to Rfa8 |
|---|---|---|---|
| RVFAAEFNDATFTFK | 32 | 1763.9 | 7 |
| VFAAEFNDATFTFK | 88 | 1607.7 | 7 |
| ESDDERAPLYALLPTGER | 12 | 2032.1 | 9 |
| APLYALLPTGER | 9 | 1300.5 | 6 |
| ANRVFVTGTLTETEDIGEDAEYWR | 1 | 2772.9 | 12 |
| VFVTGTLTETEDIGEDAEYWR | 128 | 2431.5 | 11 |
| VVDPTGTFFVYAGQYQPEAAAALR | 20 | 2571.8 | 12 |
| VVDPTGTFFVYAGQYQPEAAAALRDAETPTYVAVVGKPR | 1 | 4156.6 | 20 |
| DAETPTYVAVVGKPR | 12 | 1602.8 | 7 |
| TYETDDGSVNVSLRPESITMVEESVR | 77 | 2914.1 | 13 |
| TYETDDGSVNVSLRPESITMVEESVRDR | 1 | 3185.4 | 14 |
| DRWVVEAAER | 4 | 1230.3 | 5 |
| WVVEAAER | 21 | 959.0 | 4 |
| TLDRIEAFNEEGNEYAER | 22 | 2156.2 | 9 |
| TLDRIEAFNEEGNEYAERAR | 2 | 2383.5 | 10 |
| IEAFNEEGNEYAER | 65 | 1670.7 | 7 |
| IEAFNEEGNEYAERAR | 1 | 1897.9 | 8 |
| AEYGADTSAYR | 12 | 1203.2 | 5 |
| ARAEYGADTSAYR | 2 | 1430.5 | 6 |
| ARAEYGADTSAYRQAVVDALAEFDAPE | 1 | 2887.0 | 23 |
| QAVVDALAEFDAPE | 20 | 1474.5 | 7 |
The percent of Halobacterium sp. NRC-1 Rfa8 amino acid sequence that aligns with the identified peptide sequence.
Discussion
Haloarchaeal RAD mutant strains of Halobacterium sp. NRC-1 with increased ionizing radiation tolerance were previously isolated by selection with killing doses of irradiation using a linear accelerator (DeVeaux et al. 2007). Two of these independently isolated mutants showed increased transcription of the rfa3, rfa8, and ral genes, which were organized together as an operon. These findings were remarkable, given that the rfa3 and rfa8 gene products were homologs of the genes for the two larger subunits of the mammalian-type RPA, with encoded OB folds. The conclusion from this work was that increasing the expression of the archaeal RPA homolog appeared to allow cells to survive an otherwise lethal dose of ionizing radiation. However, the selection of the RAD mutants through a highly mutagenic process of irradiation left open the possibility that other unknown mutations were contributing to the observed RAD phenotype.
In this work, we have overexpressed the rfa3 operon from a newly constructed expression plasmid in a clean background of the wild-type Halobacterium sp. NRC-1 strain and found that the resulting cells have become more ionizing radiation resistant, like the RAD mutants. Based on the present data, we conclude that that increased expression of the rfa3 operon is responsible for the RAD phenotype, and that it is possible to bioengineer ionizing radiation resistant cells by this genetic perturbation. Moreover, we also found that the Rfa3 and Rfa8 proteins co-purify by affinity chromatography on single-stranded DNA columns, demonstrating that these proteins have single-stranded DNA binding activity. These findings strongly suggest that the single-stranded DNA binding activity of Rfa3 and Rfa8 together is responsible for the enhanced survival of Halobacterium sp. NRC-1 with ionizing radiation.
The evidence for upregulation of Rfa3 and Rfa8 in the RAD mutants and bioengineered Halobacterium sp. NRC-1 (pDRK1) strain comes from multiple approaches. In addition to the previously published transcriptomic results, we now have Western blotting, proteomic, and affinity chromatographic evidence. The Western blotting data showed that the Rfa3 protein levels are increased in the RAD and bioengineered strains. The proteomic data extended the evidence for increased protein levels to Rfa8 in the RAD and bioengineered strains, and confirmed the Western blotting results for Rfa3. The affinity chromatographic results also confirmed that the Rfa3 and Rfa8 protein levels are both increased in the RAD and bioengineered strains. Thus, the evidence for the involvement of Rfa3 and Rfa8 in promoting a RAD phenotype in total is compelling.
The genetic linkage of Rfa3 and Rfa8 proteins and their co-purification by the single-stranded DNA affinity chromatography suggest that they function together in the cell. In mammalian RPA, three subunits are involved in binding single-stranded DNA, RPA70, 32, and 14. Halobacterium sp. NRC-1 Rfa3 is a homolog of the mammalian large subunit, while the Rfa8 is a homolog of the medium subunit. The Rfa3 protein contains one OB-fold (positioned at 84VDI-TSI157) and also one nucleic acid-binding Zn-finger motif (189RCP-HGE209). The Rfa8 protein also contains an OB-fold (48VFV-ITM130) (Chen et al. 2005; DeVeaux et al. 2007). The degree of homology between the haloarchaeal and mammalian subunits is low, and there is no homolog of the small subunit in the Halobacterium genome, indicating that the archaeal RPA is highly diverged from its mammalian counterpart. Although the evidence for elevated levels of Rfa3 and Rfa8 in radioresistance is strong, further studies are needed to determine their precise roles in the cell in relation to each other and other proteins involved in DNA repair and/or replication.
Initially, the finding of a third gene in the rfa3 operon, ral, suggested that the encoded protein may play a role in the haloarchaeal RPA function. Subsequently, in a comparative genomic study, the ral gene was found to encode one of the haloarchaeal signature proteins, which are always present in sequenced genomes of this family, but not in any other organisms. Usually, but not always, these ral genes are genetically linked to rfa3 and rfa8, suggesting coordinate regulation (Capes et al. 2011; 2012). However, the results of proteomic analysis and affinity chromatography indicated that ral gene product is not overexpressed to the same degree as in the RAD or bioengineered strains compared to the wild-type. A possibility which has not yet been addressed is whether the ral gene product may participate directly or indirectly in controlling the levels of Rfa3 and Rfa8, as well as itself, in haloarchaeal cells. Further analysis is needed to address the degree of expression linkage between ral, and rfa3 and rfa8 in Halobacterium and the potential role of ral in the observed increases in rfa3 and rfa8.
Halobacterium sp. NRC-1 contains multiple RPA genes and several studies have examined the diversity of RPA genes in the family of Haloarchaea. Genes homologous to rfa3 (cHOG0104), of which there are 5 in NRC-1, belong to the COG1599/KOG0851/arCOG01510/1 group (Capes et al. 2011; 2012). For rfa8 (cHOG0130), there are two homologous genes in NRC-1, both belonging to the COG3390/arCOG2257 group. For ral, the signature haloarchaeal gene, there is only a single member present in each sequenced species belonging to the tucHOG0456 group. The most detailed analysis of these genes was previously conducted in the related moderate halophile, Haloferax volcanii (Stroud et al. 2012). The H. volcanii RPA3 associates with the genetically linked Rfa8 homolog in that organism (Hvo0291/RPAP3 or RpaA2), consistent with our observation of co-purification of the Rfa3 and Rfa8 in Halobacterium sp. NRC-1. Deletion of the H. volcanii rfa3 homolog (called either Hvo0292/RPA3 or RpaA1; Hartman et al. 2010; Skowyra and MacNeill 2012) results in a hypersensitive phenotype to DNA damaging agents. This finding is consistent with our results showing that increasing the Rfa3 and Rfa8 protein levels in Halobacterium enhances resistance to a DNA damaging agent. However, the difference in Rfa3 and Rfa8 protein levels between the RAD mutants and engineered overexpression strain did not translate to observable differences in the levels of radioresistance, at least at the doses tested. Additional studies aimed at higher radiation doses are needed to clarify the precise relationship between expression levels and degrees of radioresistance.
The mechanism by which the Halobacterium RPA protein enhances cell survival after high doses of ionizing radiation is not yet clear. The binding of Rfa3 and Rfa8 to single-stranded DNA and the known functions of single-stranded DNA binding proteins in both prokaryotes and eukaryotes strongly suggest that the Halobacterium RPA plays a key role in DNA replication and repair. Formation of single-stranded DNA is known to be a consequence of DNA melting in DNA replication and nucleolytic activities in DNA repair. Halobacterium RPA likely plays a role in both of these processes and an increase in its activity may help to stabilize and secure extensive DNA regions with single-stranded character. Such an activity may increase the cell's ability to repair the DNA more efficiently, possibly by promoting recombinational repair. Interestingly, proteomic analysis found that the level of RadA protein was also increased in the RAD mutants and overexpression strains, consistent with this mechanism (data not shown, see also McCready et al. 2005; Boubriak et al. 2008). However, additional roles for RPA after ionizing radiation such as replication arrest relief may also contribute to cell survival (Gygli et al. 2011).
Our ability to bioengineer increased resistance to ionizing radiation in Haloarchaea opens the possibility for a wide variety of applications to biotechnology. Radioresistant strains of Haloarchaea may have many potential uses, e.g. in bioremediation in the presence of radioactive materials, or as biosensors in space and in the food industry. Of significant interest is whether bioengineering of radioresistance will be possible in other cell types, as well, including other microorganisms and mammalian cells, through overexpression of single-strand DNA binding proteins. If radioresistance is bioengineerable generally, many other applications may be possible, including potential uses in cancer research and treatment.
Acknowledgements
This work was supported by grants from the National Aeronautics and Space Administration (NNX10AP47G), National Institutes of Health (AI107364), and the Bill and Melinda Gates Foundation (OPP1061509) to S.D. We thank the Johns Hopkins University School of Medicine Mass Spectrometry and Proteomics Facility for assistance with LC-MS/MS.
References
- Berquist BR, DasSarma P, DasSarma S. Essential and non-essential DNA replication genes in the model haloarchaeon, Halobacterium sp. NRC-1. BMC Genetics. 2007;8:31. doi: 10.1186/1471-2156-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berquist BR, Müller JA, DasSarma S. Methods in microbiology. In: Oren A, Rainey F, editors. Genetic systems for halophilic archaea. Volume 35. Elsevier/Academic Press; London: 2006. pp. 649–680. [Google Scholar]
- Boubriak I, Ng WL, DasSarma P, DasSarma S, Crowley DJ, McCready S. Transcriptional responses to biologically relevant doses of UV-B radiation in the model archaeon, Halobacterium sp. NRC-1. Saline Syst. 2008;1:13. doi: 10.1186/1746-1448-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capes MD, Coker JA, Gessler R, Grinblat-Huse V, DasSarma SL, Jacob CG, Kim JM, DasSarma P, DasSarma S. The information transfer system of halophilic archaea. Plasmid. 2011;65:77–101. doi: 10.1016/j.plasmid.2010.11.005. [DOI] [PubMed] [Google Scholar]
- Capes MD, DasSarma P, DasSarma S. The core and unique proteins of Haloarchaea. BMC Genomics. 2012;13:39. doi: 10.1186/1471-2164-13-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Kocherginskaya SA, Lin Y, Sriratana B, Lagunas AM, Robbins JB, Mackie RI, Cann IK. Biochemical and mutational analyses of a unique clamp loader complex in the archaeon Methanosarcina acetivorans. J Biol Chem. 2005;280:41852–41863. doi: 10.1074/jbc.M508684200. [DOI] [PubMed] [Google Scholar]
- Chu LJ, Chen MC, Setter J, Tsai YS, Yang H, Fang X, Ting YS, Shaffer SA, Taylor GK, von Haller PD, Goodlett DR, Ng WV. New structural proteins of Halobacterium salinarum gas vesicle revealed by comparative proteomics analysis. J Proteome Res. 2011a;10:1170–1178. doi: 10.1021/pr1009383. [DOI] [PubMed] [Google Scholar]
- Chu LJ, Yang H, Shih P, Kao Y, Tsai YS, Chen J, Huang G, Weng RR, Ting YS, Fang X, von Haller PD, Goodlett DR, Ng WV. Metabolic capabilities and systems fluctuations in Haloarcula marismortui revealed by integrative genomics and proteomics analyses. J Proteome Res. 2011b;10:3261–3273. doi: 10.1021/pr200290x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coker JA, DasSarma P, Kumar J, Müller JA, DasSarma S. Transcriptional profiling of the model Archaeon Halobacterium sp. NRC-1: responses to changes in salinity and temperature. Saline Syst. 2007;3:6. doi: 10.1186/1746-1448-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley DJ, Boubriak I, Berquist BR, Clark M, Richard E, Sullivan L, DasSarma S, McCready S. The uvrA, uvrB and uvrC genes are required for repair of ultraviolet light induced DNA photoproducts in Halobacterium sp. NRC-1. Saline Syst. 2006;2:11. doi: 10.1186/1746-1448-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubeddu L, White MF. DNA damage detection by an archaeal single-stranded DNA-binding protein. J Mol Biol. 2005;353:507–516. doi: 10.1016/j.jmb.2005.08.050. [DOI] [PubMed] [Google Scholar]
- DasSarma S. Genome sequence of an extremely halophilic archaeon. In: Fraser T, Read T, Nelson KE, editors. Microbial Genomes. Humana Press, Inc; Totowa, NJ: 2004. pp. 383–399. [Google Scholar]
- DasSarma S. Extreme microbes. American Scientist. 2007;95:224–231. [Google Scholar]
- DasSarma S, DasSarma P. Encyclopedia of Life Sciences. John Wiley & Sons, Ltd; 2012. Halophiles. [Google Scholar]
- DasSarma S, Kennedy SP, Berquist B, Ng WV, Baliga NS, Spudich JL, Krebs MP, Eisen JA, Johnson CH, Hood L. Genomic perspective on the photobiology of Halobacterium species NRC-1, a phototrophic, phototactic, and UV-tolerant haloarchaeon. Photosyn Res. 2001;70:3–17. doi: 10.1023/A:1013879706863. [DOI] [PubMed] [Google Scholar]
- DasSarma S, Robb FT, Place AR, Sowers KR, Schreier HJ, Fleischmann EM. Archaea: a laboratory manual - halophiles. Cold Spring Harbor Laboratory Press; New York: 1995. [Google Scholar]
- Desiere F, Deutsch EW, Nesvizhskii AI, Mallick P, King NL, Eng JK, Aderem A, Boyle R, Brunner E, Donohoe S, Fausto N, Hafen E, Hood L, Katze MG, Kennedy KA, Kregenow F, Lee H, Lin B, Martin D, Ranish JA, Rawlings DJ, Samelson LE, Shiio Y, Watts JD, Wollscheid B, Wright ME, Yan W, Yang L, Yi EC, Zhang H, Aebersold R. Integration with the human genome of peptide sequences obtained by high-throughput mass spectrometry. Genome Biol. 2005;6:R9. doi: 10.1186/gb-2004-6-1-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVeaux LC, Müller JA, Smith J, Petrisko J, Wells DP, DasSarma S. Extremely radiation-resistant mutants of a halophilic archaeon with increased single-stranded DNA-binding protein (RPA) gene expression. Radiat Res. 2007;168:507–514. doi: 10.1667/RR0935.1. [DOI] [PubMed] [Google Scholar]
- Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- Grant WD, Kamekura M, McGenity TJ, Ventosa A. Class III. Halobacteria class. nov. In: Boone DR, Castenholz RW, Garrity GM, editors. Bergey's Manual of Systematic Bacteriology. 2nd edn. Vol. 1. Springer; New York: 2001. pp. 294–334. The Archaea and the Deeply Branching and Phototrophic Bacteria. [Google Scholar]
- Gygli PE, Lockhart JS, DeVeaux LC. Role of RPA proteins in radiation repair and recovery. In: Chen C, editor. DNA Repair. Intech; Rijeka, Croatia: 2011. pp. 175–200. [Google Scholar]
- Halladay JT, Jones JG, Lin F, MacDonald AB, DasSarma S. The rightward gas vesicle operon in Halobacterium plasmid pNRC100: identification of the gvpA and gvpC gene products by use of antibody probes and genetic analysis of the region downstream of gvpC. J Bacteriol. 1993;175:684–692. doi: 10.1128/jb.175.3.684-692.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman AL, Norais C, Badger JH, Delmas S, Haldenby S, Madupu R, Robinson J, Khouri H, Ren Q, Lowe TM, Maupin-Furlow J, Pohlschroder M, Daniels C, Pfeiffer F, Allers T, Eisen JA. The complete genome sequence of Haloferax volcanii DS2, a model archaeon. PLoS One. 2010;5(3):e9605. doi: 10.1371/journal.pone.0009605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karan R, Capes MD, DasSarma P, DasSarma S. Cloning, overexpression, purification, and characterization of a polyextremophilic β-galactosidase from the Antarctic haloarchaeon Halorubrum lacusprofundi. BMC Biotechnol. 2013;13:3. doi: 10.1186/1472-6750-13-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Eng J, Zhang N, Li XJ, Aebersold R. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol Syst Biol. 2005;1:0017. doi: 10.1038/msb4100024. 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- Komori K, Ishino Y. Replication protein A in Pyrococcus furiosus is involved in homologous DNA recombination. J Biol Chem. 2001;276:25654–25660. doi: 10.1074/jbc.M102423200. [DOI] [PubMed] [Google Scholar]
- Kish A, DiRuggiero J. Rad50 is not essential for the Mre11-dependant repair of DNA double strand breaks in Halobacterium sp. str. NRC-1. J Bacteriol. 2008;190:5210–5216. doi: 10.1128/JB.00292-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCready S, Marcello L. Repair of UV damage in Halobacterium salinarum. Biochem Soc Trans. 2003;31:694–698. doi: 10.1042/bst0310694. [DOI] [PubMed] [Google Scholar]
- McCready S, Muller JM, Boubriak I, Berquist BR, Ng WL, DasSarma S. UV radiation induces homologous recombination genes in the model archaeon, Halobacterium sp. NRC-1. Saline Syst. 2005;1:3. doi: 10.1186/1746-1448-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- Ng WV, Kennedy SP, Mahairas GG, Berquist B, Pan M, Shukla HD, Lasky SR, Baliga NS, Thorsson V, Sbrogna J, Swartzell S, Weir D, Hall J, Dahl TA, Welti R, Goo YA, Leithauser B, Keller K, Cruz R, Danson MJ, Hough DW, Maddocks DG, Jablonski PE, Krebs MP, Angevine CM, Dale H, Isenbarger TA, Peck RF, Pohlschroder M, Spudich JL, Jung KW, Alam M, Freitas T, Hou S, Daniels CJ, Dennis PP, Omer AD, Ebhardt H, Lowe TM, Liang P, Riley M, Hood L, DasSarma S. Genome sequence of Halobacterium species NRC-1. Proc Natl Acad Sci U S A. 2000;97:12176–12181. doi: 10.1073/pnas.190337797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrioli PG, Eng JK, Hubley R, Vogelzang M, Deutsch EW, Raught B, Pratt B, Nilsson E, Angeletti RH, Apweiler R, Cheung K, Costello CE, Hermjakob H, Huang S, Julian RK, Kapp E, McComb ME, Oliver SG, Omenn G, Paton NW, Simpson R, Smith R, Taylor CF, Zhu W, Aebersold R. A common open representation of mass spectrometry data and its application to proteomics research. Nat Biotechnol. 2004;22:1459–1466. doi: 10.1038/nbt1031. [DOI] [PubMed] [Google Scholar]
- Shukla HD, DasSarma S. Complexity of gas vesicle biogenesis in Halobacterium sp. strain NRC-1: identification of five new proteins. J Bacteriol. 2004;186:3182–3186. doi: 10.1128/JB.186.10.3182-3186.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra A, MacNeill SA. Identification of essential and non-essential single-stranded DNA-binding proteins in a model archaeal organism. Nucleic Acids Res. 2012;40:1077–1090. doi: 10.1093/nar/gkr838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud A, Liddell S, Allers T. Genetic and biochemical identification of a novel single-stranded DNA-binding complex in Haloferax volcanii. Front Microbiol. 2012;3:224. doi: 10.3389/fmicb.2012.00224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi EC, Marelli M, Lee H, Purvine SO, Aebersold R, Aitchison JD, Goodlett DR. Approaching complete peroxisome characterization by gas-phase fractionation. Electrophoresis. 2002;23:3205–3216. doi: 10.1002/1522-2683(200209)23:18<3205::AID-ELPS3205>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]



