Abstract
Purpose:
Anaplastic lymphoma kinase (ALK) rearrangements are present in an important subset of non-small-cell lung cancer (NSCLC) and predict for response to the tyrosine kinase inhibitor crizotinib. In this study, we evaluated the yet unknown frequency and functional role of ALK splicing isoforms in NSCLC.
Experimental Design:
We analyzed 270 cases of NSCLC for ALK kinase domain splicing aberrations, and in addition generated constructs with full length EML4-ALK (E13;A20) and a splicing isoform.
Results:
Splicing isoforms of the kinase domain of ALK - including complete skipping of exon 23 (ALKdel23, ALK p.I1171fs*42) and exon 27 (ALKdel27, ALK p.T1312fs*0) - were identified in 11.1% (30/270 cases) of NSCLC, and these changes co-existed with ALK rearrangements, KRAS mutations and EGFR mutations. ALK splicing isoforms were observed with full length EML4-ALK in crizotinib-naïve and treated NSCLCs. ALK T1312fs*0 was unable to render cells solely dependent on ALK signaling. Unlike EML4-ALK and EML4-ALK p.L1196M, EML4-ALK T1312fs*0 did not autophosphorylate ALK or other phospho-tyrosine sites. Co-expression of equal amounts of EML4-ALK T1312fs*0 and EML4-ALK did not result in resistance to crizotinib, while co-expression of EML4-ALK L1196M with EML4-ALK resulted in resistance to inhibition of ALK by crizotinib.
Conclusions:
ALK kinase splicing isoforms were present in NSCLC and even if translated seemed to be non-functional variants of ALK.
Keywords: lung cancer, non-small-cell lung cancer, tyrosine kinase, kinase inhibitor, anaplastic lymphoma kinase, ALK, crizotinib, alternative splicing, exon skipping, exon 27, exon 23
INTRODUCTION
Anaplastic lymphoma kinase (ALK) rearrangements, either inversions or translocations, are present in approximately 5% of all non-small-cell lung cancers (NSCLCs) at the time of diagnosis 1. The most frequent inversions event occur within the short arm of chromosome 2 and result in the fusion of the echinoderm microtubule-associated protein-like 4 (EML4) with ALK 2,3. The most common EML4-ALK fusion variants are E13;A20 and E20;A20 4.
EML4-ALK variants are transforming tyrosine kinases that activate the phosphatidylinositol-3-kinase (PI3K) and mitogen-activated protein kinase (MAPK) downstream pathways 5,6. Tumors with EML4-ALK and other ALK rearrangements are oncogene addicted to ALK signaling and can be inhibited by ALK tyrosine kinase inhibitors (TKIs) in preclinical models 5,7,8. In the clinic, NSCLCs with ALK rearrangements are sensitive to the multitargeted ALK TKI crizotinib 9 and this drug received approval by the Food and Drug Administration (FDA) in 2011 to be used in this NSCLC subtype 1. The reported response rates exceed 60% with a mean progression-free survival of over 7-10 months prior to acquisition of resistance to crizotinib monotherapy 10,11.
ALK kinase domain mutations and non-kinase domain splicing variants, which are present in a subset of neuroblastomas 12, have not been reported in ALK TKI-naïve NSCLCs. Our group reports herein the identification and characterization of splicing isoforms of the kinase domain of ALK.
MATERIALS AND METHODS
Patient and tumor characteristics, plus statistical methods
268 Chinese patients from the Queen Mary Hospital (University of Hong Kong) and 2 patients from the Beth Israel Deaconess Medical Center with NSCLC were studied. Approval of this study was obtained from ongoing Institutional Review Board (IRB) approved protocols at these institutions. Clinical, pathologic and molecular characteristics were obtained retrospectively from extraction of medical records 13. Fisher’s exact test was performed to compare categorical variables and Student’s t-test for continuous variables.
Sequencing of ALK
Total DNA and RNA were extracted using standard techniques. The expression of ALK kinase domain was initially screened by reverse transcriptase PCR using forward (5’-GACCATCATGACCGACTACAA-3’) and reverse (5’-AGTGGACCATATTCTATCGGC-3’) primers flanking ALK exons 20 and 29, respectively. Cases showing splicing variations were confirmed by sequencing. Epidermal growth factor receptor (EGFR) mutations and KRAS mutations were analyzed as previously published 14,15. ALK rearrangements were identified using ALK reverse transcriptase PCR primers or ALK FISH 9,16.
Cell culture
COS-7 cells were maintained in Dulbecco's Modified Eagle's Medium (Mediatech, Manassas, VA) supplemented with 10% fetal bovine serum (FBS). Ba/F3 cell lines were maintained in RPMI-1640 supplemented with 10% FBS and 5% WEHI-conditioned medium as the source of IL-3.
Generation of constructs/vectors with EML4-ALK variants and reagents
The pCDNA3.1 EML4-ALK E13;A20 was a kind gift of Dr. Henry Koon (Case Western Reserve University, Cleveland, OH). The pCDNA3.1-Myc-His-Tag (Invitrogen, Carlsbad, CA) vector was used as the template for the introduction of EML4-ALK E13;A20 (i.e., EML4-ALK variant 1) and mutated constructs for COS-7 experiments. The derived constructs EML4-ALK ALK p.L1196M, EML4-ALK ALK p.T1312fs*0 and ALK p.1312fs*0 were generated by site-directed mutagenesis. Retroviral infection and selection of Ba/F3 cells for IL-3 independence were performed using previously described methods 17. Crizotinib was purchased from LC Laboratories (Woburn, MA), dissolved in dimethyl sulfoxide (DMSO) and stored at −80°C.
Western blotting, immunoprecipitation and antibodies
Cells were lysed and protein extracts obtained for analysis 17. For immunoprecipitation, protein G agarose/sepharose beads were used for pre-clearing the cell lysates and capturing the immunocomplex after incubation with the immunoprecipitating antibody (Myc). Phospho-ALK (pTyr1096), ALK, phospho-Tyrosine (pTyr100) and Myc-Tag antibodies were purchased from Cell Signaling Technology (Beverly, MA). β-actin was purchased from Santa Cruz Biotechnology (Dallas, TX). Phospho-ALK was diluted 1:500 and all other primary antibodies were diluted 1:1000, while secondary antibodies were diluted 1:10000.
RESULTS
ALK splicing variants with skipping of exons 23 or 27 in NSCLC
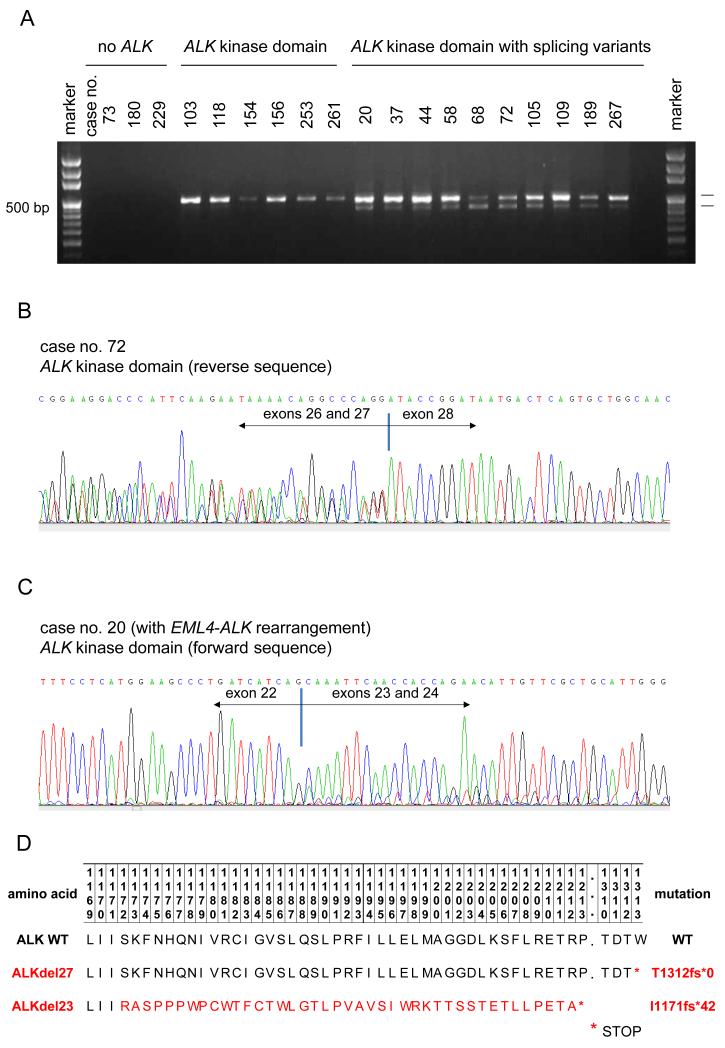
We attempted to identify ALK kinase domain splicing abnormalities in NSCLC samples by analyzing cDNA derived from 270 cases of NSCLC (Table 1). Interestingly, the ALK kinase domain was solely expressed in 113 of 270 (41.85%) tumors (Figure 1A). ALK kinase domain (i.e., exons 20 to 28) splicing variants were noted in 11.1% (30/270) of cases. The major changes observed were either complete skipping of exon 27 of ALK (ALKdel27) in 11 cases or complete skipping of exon 23 (ALKdel23) of ALK in 19 cases (Figures 1B and C, respectively). Both splicing isoforms have a nucleotide sequence that leads to early stop codons (Figure 1D): ALKdel27 to the protein ALK p.T1312fs*0, and ALKdel23 to ALK p.I1171fs*42.
TABLE 1.
Baseline patient, tumor and molecular characteristics of non-small-cell lung cancers with or without anaplastic lymphoma kinase (ALK) kinase domain splicing variants
| Total (n=270) | |||
|---|---|---|---|
|
ALK kinase domain splicing variant expression (n=30) |
Lack of ALK
kinase domain splicing variant expression (n=240) |
p-value |
|
| Age median(range) |
61 (29-81) |
64 (33-79) |
0.110 |
| Women n (%) | 13 (43.3%) | 123 (51.2%) | 0.444 |
| Race n (%) Asian White |
29 (96.7%) 1 (3.3%) |
239 (99.6%) 1 (0.4%) |
0.210 |
| Smoking status n (%) Current smoker Former smoker Never smoker |
12 (40.0%) 4 (13.3%) 14 (46.7%) |
77 (32.1%) 33 (13.8%) 130 (54.1%) |
0.660 |
| Stage n (%) I II III IV |
16 (53.3%) 2 (6.7%) 9 (30.0%) 3 (10.0%) |
138 (57.5%) 45 (18.8%) 52 (21.7%) 5 (2.1%) |
0.033 |
| Histology n (%) Adenocarcinoma Squamous cell carcinoma Other (NOS) |
24 (80.0%) 4 (13.3%) 2 (6.7%) |
189 (78.8%) 30 (12.5%) 21 (8.7%) |
0.925 |
|
EGFR mutation n (%) Positive/Mutated Negative/Wild-type |
9 (30%) 21 (70%) |
116 (48.3%) 124 (51.7%) |
0.079 |
|
KRAS mutation n (%) Positive/Mutated Negative/Wild-type |
3 (10%) 27 (90%) |
19 (7.9%) 221 (92.1%) |
0.721 |
|
ALK rearrangement n (%) Positive Negative |
4 (13.3%) 26 (86.6%) |
15 (6.2%) 225 (93.8%) |
0.244 |
|
ALK splicing variant n (%) exon 23 skipping exon 27 skipping |
19 (63.3%) 11 (36.7%) |
0 (0%) 0 (0%) |
NA |
n, number; EGFR, epidermal growth factor receptor; KRAS, v-ki-ras2 Kirsten rat sarcoma viral oncogene homolog; NOS, not otherwise specified
FIGURE 1.
Screening for ALK kinase domain splicing variants in non-small-cell lung cancer (NSCLC).
A. Reverse transcriptase PCR using primers that flank exons 20 to 29 of ALK (encompass the kinase domain). Cases with or without ALK kinase domain expression, and those expressing ALK slicing variants;
B. Sequence chromatogram of a tumor specimen with complete skipping of exon 27 of ALK (ALKdel27). Representative sequences from cDNA isolated from case no. 72 highlighting the ALK exon 26-exon 27 and ALK exon 26-exon 28 co-existing sequences;
C. Sequence chromatogram of a tumor specimen with complete skipping of exon 23 of ALK (ALKdel23). Representative sequences from cDNA isolated from case no. 20 highlighting the ALK exon 22-exon 23 and ALK exon 22-exon 24 co-existing sequences. Reference ALK gene sequence (NM_004304.3; homo sapiens ALK mRNA);
D. Proposed amino acid sequence of wild type (WT) ALK, ALKdel27 and ALKdel23. The latter truncated proteins generate early stop codons. *denotes a STOP codon sequence.
The clinical, pathological and molecular characteristics of NSCLCs with or without splicing abnormalities of ALK were similar (Table 1). ALK splicing variants were observed in cases with or without ALK rearrangements, KRAS mutations and EGFR mutations (Table 1). Out of the 4 cases with ALK rearrangements and ALK splicing variants, one had EML4-ALK with ALKdel27 and 3 had EML4-ALK with ALKdel23 (Table 1). Only the EML4-ALKdel27 case received crizotinib. Out of the cases with EML4-ALKdel23, 2 remain free of disease after curative surgical resection and 1 had recurrence 29 months after surgical resection and died prior to crizotinib therapy.
EML4-ALK positive NSCLC with ALKdel27
The EML4-ALK rearranged NSCLC that harbored ALKdel27 had crizotinib-resistant malignant lymphatic tissue analyzed in our group’s attempt to sequence the ALK kinase domain of crizotinib-naïve and crizotinib-resistant NSCLCs. The patient had attained significant symptomatic response and minor tumor regression of his intra-thoracic disease best classified as stable disease for 8 months prior to development of crizotinib-resistant lesions 18. cDNA derived from crizotinib-resistant EML4-ALK positive left axillary node tissue was sequenced to encompass exons 20-28 of ALK. We identified an overlapping sequence amidst the wild-type ALK kinase domain of this EML4-ALK fusion, which corresponded to complete skipping of exon 27 of ALK (Figure 2A). cDNA isolated from non-malignant lymphoid tissue from the same patient only contained ALK and not EML4-ALK; and ALK T1312fs*0 was not present (data not shown). Interestingly, the crizotinib-naïve tumor from the patient also disclosed EML4-ALK with ALK T1312fs*0. Sequence of the exon-intron junction of exons 26-27 and 27-28 using DNA isolated from the crizotinib-resistant sample did not yield abnormal sequences.
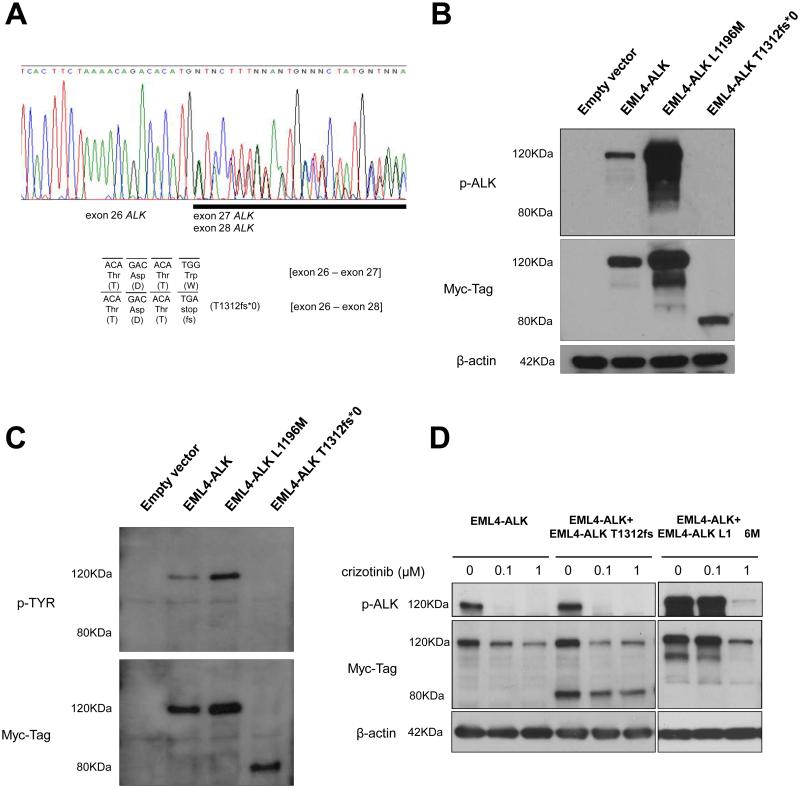
FIGURE 2.
Evaluation of EML4-ALK T1312fs*0 in preclinical models.
A. Sequence chromatogram of an EML4-ALK positive tumor specimen. Representative sequences from cDNA isolated from the aforomentioned lymph node. Nucleotide areas corresponding to anaplastic lymphoma kinase (ALK) exon 26, 27 and 28 are highlighted. Reference ALK gene sequence (NM_004304.3; homo sapiens ALK mRNA); the expected nucleotide and amino acid sequences for the wild-type ALK exon 26-exon 27 junction, and for the splicing variant with ALK exon 26-exon 28 junction are shown. The latter generates the early stop codon sequence p. T1312fs*0 (n.TGG->TGA).
B. Western blot analysis of protein extracts from COS-7 cells transfected with EML4-ALK constructs and empty vector. EML-ALK E13;A20 constructs with a 3' Myc-His-Tag were cloned into pcDNA3.1. COS-7 cells were transfected with 1 μg of DNA and protein extracts were collected 24 hours later. The radiograph shows the expression of phosphorylated ALK (p-ALK Tyr1096) and EML4-ALK detected by using a Myc-Tag antibody. EML4-ALK T1312fs*0 results in an 80 KDa protein, different from the expected size - 117 kDa - of EML4-ALK WT and EML4-ALK containing the L1196M crizotinib-resistant mutation.
C. Western blot analysis of protein extracts from COS-7 cells transfected with EML4-ALK-Myc-His-Tag constructs or empty vector and immunoprecipitated with Myc-Tag antibody. The upper figure shows the expression of phospho-tyrosine (p-TYR) and the lower figure shows the expression of EML4-ALK detected by using a Myc-Tag antibody. EML4-ALK T1312fs*0 results in an 80 KDa protein that is not phosphorylated.
D. Western blot analysis of protein extracts from COS-7 cells co-transfected with different DNA amounts of EML4-ALK constructs and empty vector and treated with crizotinib or vehicle (DMSO). The left panel with E13;A20 EML4-ALK (0.4 μg) + empty vector (0.6 μg); the middle with EML4-ALK (0.4 μg) + empty vector (0.2 μg) + EML4-ALK T1312fs*0 (0.4 μg) in which EML4-ALK T1312fs*0 does not result in resistance to crizotinib as verified by ALK phosphorylation inhibition using Tyr1096 phospho-ALK antibody; the right with EML4-ALK (0.4 μg) + empty vector (0.2 μg) + EML4-ALK L1196M (0.4 μg) in which the co-transfection of EML4-ALK L1196M is shown in the right panel and discloses resistance to inhibition of phospho-ALK by crizotinib. Blots were cropped to highlight the referenced molecular weight.
EML4-ALKdel27 generates the truncated EML4-ALK T1312fs*0 protein with inability to auto-phosphorylate ALK
Since both ALKdel27 and ALKdel23 would potentially lead to truncated proteins lacking the full kinase domain of ALK (Figure 1F), we wanted to understand the biological properties of one of these splicing variants; if translated into protein. We decided to study the skipping of exon 27 of ALK (T1312fs*0) in the context of EML4-ALK, and to this end generated EML4-ALK E13;A20 (EML4-ALK) constructs with or without ALK T1312fs*0, and in addition EML4-ALK with ALK L1196M. The latter is known to represent a crizotinib-resistant gatekeeper mutation 19.
The EML4-ALK and EML4-ALK L1196M constructs expressed proteins of their expected size of close to 117kDa (Figure 2B). However, the EML4-ALK T1312fs*0 protein was identified at a size of ~80kDa (Figure 2B). The latter results indicated that EML4-ALKdel27 could generate a truncated EML4-ALK protein.
We then decided to study the functional properties of EML4-ALK T1312fs*0. Using an antibody specific for a phosphorylation site within amino acid 1096 (tyrosine 1096) of ALK (which is still present within ALK T1312fs*0), we were only able to detect auto-phosphorylation of ALK in EML4-ALK and EML4-ALK L1196M; while EML4-ALK T1312fs*0 did not have a detectable signal (Figure 2C). We also performed an immunoprecipitation experiment to detect auto-phosphorylation of tyrosine residues within ALK or other proteins bound to EML4-ALK using a phospho-tyrosine antibody. EML4-ALK T1312fs*0 was unable to generate a measurable signal (Figure 2C). These results confirmed that EML4-ALK T1312fs*0 was unable to phosphorylate tyrosine sites and was a non-functional kinase when expressed alone. We attempted to transform a cell line system with ALK T1312fs*0 but it was ineffective in making Ba/F3 cells interleukin-3 (IL-3) independent (data not shown). Since ALK p.I1171fs*42 lacks all the amino acid sites of ALK T1312fs*0 in addition to other areas of the ALK kinase, we can foretell it will similarly be unable to transform cell line systems. EML4-ALK and EML4-ALK L1196M can make Ba/F3 cells IL-3 independent 20,21.
EML4-ALK T1312fs*0 and response to the ALK TKI crizotinib
Although we demonstrated that EML4-ALK T1312fs*0 was potentially non-functional as a “driver” oncogene, we also tested the possibility that this truncated protein could affect the exquisite sensitivity of EML4-ALK towards the ALK TKI crizotinib. When EML4-ALK was transfected alone, we were able to observe that the inhibition of phosphorylation of ALK by crizotinib occurred at doses of 0.1μM (Figure 4C). When we co-transfected equal amounts of EML4-ALK and EML4-ALK T1312fs*0 we observed that the inhibition of phosphorylation of ALK within EML4-ALK was unaffected and crizotinib continued to inhibit ALK at doses of 0.1μM (Figure 4C). When EML4-ALK and EML4-ALK L1196M were co-transfected in equal amounts, then a shift in the pattern of inhibition of phosphorylation of ALK was noted (Figure 5C); confirming that EML4-ALK L1196M is a crizotinib-resistant mutation. Taken together these results led us to speculate that EML4-T1312fs*0 was not involved in the pattern of either sensitivity or resistance of EML4-ALK to crizotinib.
DISCUSSION
We identified ALK splicing variants, involving the complete skipping of exons 23 and 27 of ALK, in samples with NSCLC. To the best of our knowledge, these results represent the first attempt to understand the biological and clinical significance of ALK kinase domain splicing variants in NSCLC. In our cohort of 270 patients, more than 10% of NSCLCs harbored either ALKdel23 (~7% of cases) or ALKdel27 (~4% of cases). However, it is noteworthy that most tumors (58.15% [157/270]) did not even demonstrate expression of the ALK kinase domain. This observation is supported by the known low to absent expression of ALK in most lung tumors that do not harbor ALK rearrangements 11,22,23. More interestingly, ALK splicing variants were noted to co-exist not only with ALK rearrangements in some samples but also with other oncogenic driver mutations (in specific EGFR and KRAS mutations). The latter may be indicative that ALK kinase domain splicing variants are “passenger” events in NSCLCs.
The other known tumor type with oncogenic ALK abnormalities that has been shown to display ALK splicing variants is neuroblastoma. In this tumor type, characterized by mutations within the tyrosine kinase of ALK in a subset of samples, truncated ALK proteins with complete skipping of exon 2-3 (ALKdel2-3) and exons 4-11 (ALKdel4-11) of ALK have been described and characterized 24,25. Different than the EML4-ALKdel27-derived protein EML4-ALK T1312fs*0 described herein by us, these truncated extracellular domain proteins led to increased constitutive kinase activity of ALK and transformation potential (i.e., ALKdel2-3 and ALKdel4-11 were putative driver events). It is possible to speculate that the opposite nature of these proteins is based on the fact that ALKdel2-3 and ALKdel4-11 retain the sequence of the active kinase domain of ALK, whereas ALKdel27 leads to a stop codon that occurs within the tyrosine kinase domain. The lack of a functional complete tyrosine kinase domain may explain the non-functional nature of the EML4-ALK T1312fs*0 truncated protein. Therefore, ALKdel27 seems to be a silent variant that indeed is a passenger alternative splicing form of ALK. ALKdel23 (ALK I1171fs*42), due to its even more significant lack of residues within the ALK kinase domain, is also expected to be a truncated and non-functional variant if translated.
Interestingly, the currently approved and/or used methods of detection of ALK rearrangements in clinical practice are unable to obtain sequence reads of the introns and exons spanning the kinase domain of ALK 1,9,16. Even robust targeted next-generation sequencing techniques 26 using tumor-derived genomic DNA as a template would not be able to identify a splicing mutation or exon skipping variant within ALK. These limitations highlight the possible lack of reporting of ALK splicing variants in NSCLC.
The identification of mechanisms of resistance to crizotinib and other ALK TKIs in ALK rearranged NSCLC is of utmost importance, and to date only ALK kinase mutations and/or bypass tracks due to activation of other oncogenes (such as EGFR) have been described 11,17,27-29. ALK kinase mutations – which are present in only around one third of crizotinib-resistant samples – have been characterized in vitro as crizotinib-resistant mutations using EML4-ALK driven models 29. We were able to confirm that EML4-ALK with the gatekeeper ALK-L1196M indeed is resistant to crizotinib. Our preclinical characterization of EML4-ALKdel27 and the resulting truncated EML4-ALK T1312fs*0 mutated protein add to these prior results by annotating a non-functional and non-resistant inducing variant of EML4-ALK. The translation of these results into the clinic would inform future investigators not to alter their ALK TKI treatment strategies in the presence of ALKdel27. The patient in which we identified EML4-ALKdel27 had this variant present together with wild-type EML4-ALK in his crizotinib-naïve and resistant samples. His marked response to crizotinib 18 does not seem to have been affected by EML4-ALKdel27.
In summary, our group report that splicing isoforms of the kinase domain of ALK (either ALKdel23 or ALKdel27) were identified in 11.1% of NSCLCs, and these ALK kinase splicing variants were non-functional genomic events in NSCLC.
Acknowledgements
We would like to thank Angela Brooks for insightful comments and for sharing information on the The Cancer Genome Atlas (TCGA) project, and Dalia Ercan and Katsuhiro Okuda for their technical support.
Funding/Grant Support: This work was funded in part through fellowships from Conselho Nacional de Desenvolvimento Científico e Tecnológico (LLFP, no.237730/2012-0), the American Society of Clinical Oncology Conquer Cancer Foundation (DBC), and United against Lung Cancer (SSK); and from grants from the American Cancer Society RSG 11-186 (DBC), the National Institutes of Health CA090578 (DBC, PAJ), CA126026 (SSK), CA169259 (SSK), CA136851 (PAJ), and the University of Hong Kong CRCG 104002074 (MW).
Footnotes
Contributors: LLFP, DWW, PAJ, MPW, MPW, SSK and DBC were involved in the conception of this study; LLFP, DWW, VPT, LPC, NY, SN, HY, PAJ, MPW, SSK and DBC were involved in data acquisition, analysis and interpretation; MPW, SK, PAJ and DBC provided administrative and funding support; LLFP, NY, SN, HY, PAJ, MPW, SSK and DBC were involved in writing the report; all authors approved the final version.
Conflict of interest: DBC has received consulting fees from Pfizer, Roche and AstraZeneca. PAJ has received consulting fees from Pfizer, Roche, AstraZeneca, Boehringer Ingelheim, Sanofi Aventis and Chugai. No other conflict of interest is stated.
REFERENCES
- 1.Ou SH, Bartlett CH, Mino-Kenudson M, Cui J, Iafrate AJ. Crizotinib for the treatment of ALK-rearranged non-small cell lung cancer: a success story to usher in the second decade of molecular targeted therapy in oncology. Oncologist. 2012;17:1351–1375. doi: 10.1634/theoncologist.2012-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, Bando M, Ohno S, Ishikawa Y, Aburatani H, Niki T, Sohara Y, Sugiyama Y, Mano H. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature, 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 3.Choi YL, Takeuchi K, Soda M, Inamura K, Togashi Y, Hatano S, Enomoto M, Hamada T, Haruta H, Watanabe H, Kurashina K, Hatanaka H, Ueno T, Takada S, Yamashita Y, Sugiyama Y, Ishikawa Y, Mano H. Identification of novel isoforms of the EML4-ALK transforming gene in non-small cell lung cancer. Cancer Res. 2008;68:4971–4976. doi: 10.1158/0008-5472.CAN-07-6158. [DOI] [PubMed] [Google Scholar]
- 4.Horn L, Pao W. EML4-ALK: honing in on a new target in non-small-cell lung cancer. J Clin.Oncol. 2009;27:4232–4235. doi: 10.1200/JCO.2009.23.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koivunen JP, Mermel C, Zejnullahu K, Murphy C, Lifshits E, Holmes AJ, Choi HG, Kim J, Chiang D, Thomas R, Lee J, Richards WG, Sugarbaker DJ, Ducko C, Lindeman N, Marcoux JP, Engelman JA, Gray NS, Lee C, Meyerson M, Janne PA. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin.Cancer Res. 2008;14:4275–4283. doi: 10.1158/1078-0432.CCR-08-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Z, Sasaki T, Tan X, Carretero J, Shimamura T, Li D, Xu C, Wang Y, Adelmant GO, Capelletti M, Lee HJ, Rodig SJ, Borgman C, Park SI, Kim HR, Padera R, Marto JA, Gray NS, Kung AL, Shapiro GI, Janne PA, Wong KK. Inhibition of ALK, PI3K/MEK, and HSP90 in murine lung adenocarcinoma induced by EML4-ALK fusion oncogene. Cancer Res. 2010;70:9827–9836. doi: 10.1158/0008-5472.CAN-10-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Settleman J. Cell culture modeling of genotype-directed sensitivity to selective kinase inhibitors: targeting the anaplastic lymphoma kinase (ALK) Semin.Oncol. 2009;36:S36–S41. doi: 10.1053/j.seminoncol.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 8.McDermott U, Iafrate AJ, Gray NS, Shioda T, Classon M, Maheswaran S, Zhou W, Choi HG, Smith SL, Dowell L, Ulkus LE, Kuhlmann G, Greninger P, Christensen JG, Haber DA, Settleman J. Genomic alterations of anaplastic lymphoma kinase may sensitize tumors to anaplastic lymphoma kinase inhibitors. Cancer Res. 2008;68:3389–3395. doi: 10.1158/0008-5472.CAN-07-6186. [DOI] [PubMed] [Google Scholar]
- 9.Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, Ou SH, Dezube BJ, Janne PA, Costa DB, Varella-Garcia M, Kim WH, Lynch TJ, Fidias P, Stubbs H, Engelman JA, Sequist LV, Tan W, Gandhi L, Mino-Kenudson M, Wei GC, Shreeve SM, Ratain MJ, Settleman J, Christensen JG, Haber DA, Wilner K, Salgia R, Shapiro GI, Clark JW, Iafrate AJ. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N.Engl.J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Camidge DR, Bang YJ, Kwak EL, Iafrate AJ, Varella-Garcia M, Fox SB, Riely GJ, Solomon B, Ou SH, Kim DW, Salgia R, Fidias P, Engelman JA, Gandhi L, Janne PA, Costa DB, Shapiro GI, Lorusso P, Ruffner K, Stephenson P, Tang Y, Wilner K, Clark JW, Shaw AT. Activity and safety of crizotinib in patients with ALK-positive non-small-cell lung cancer: updated results from a phase 1 study. Lancet Oncol, 2012;13:1011–1019. doi: 10.1016/S1470-2045(12)70344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaw AT, Engelman JA. ALK in Lung Cancer: Past, Present, and Future. J Clin Oncol, 2013;31:1105–1111. doi: 10.1200/JCO.2012.44.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y, Takita J, Choi YL, Kato M, Ohira M, Sanada M, Wang L, Soda M, Kikuchi A, Igarashi T, Nakagawara A, Hayashi Y, Mano H, Ogawa S. Oncogenic mutations of ALK kinase in neuroblastoma. Nature, 2008;455:971–974. doi: 10.1038/nature07399. [DOI] [PubMed] [Google Scholar]
- 13.Wong DW, Leung EL, So KK, Tam IY, Sihoe AD, Cheng LC, Ho KK, Au JS, Chung LP, Pik WM. The EML4-ALK fusion gene is involved in various histologic types of lung cancers from nonsmokers with wild-type EGFR and KRAS. Cancer, 2009;115:1723–1733. doi: 10.1002/cncr.24181. [DOI] [PubMed] [Google Scholar]
- 14.Costa DB, Nguyen KS, Cho BC, Sequist LV, Jackman DM, Riely GJ, Yeap BY, Halmos B, Kim JH, Janne PA, Huberman MS, Pao W, Tenen DG, Kobayashi S. Effects of erlotinib in EGFR mutated non-small cell lung cancers with resistance to gefitinib. Clin.Cancer Res. 2008;14:7060–7067. doi: 10.1158/1078-0432.CCR-08-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tam IY, Chung LP, Suen WS, Wang E, Wong MC, Ho KK, Lam WK, Chiu SW, Girard L, Minna JD, Gazdar AF, Wong MP. Distinct epidermal growth factor receptor and KRAS mutation patterns in non-small cell lung cancer patients with different tobacco exposure and clinicopathologic features. Clin Cancer Res. 2006;12:1647–1653. doi: 10.1158/1078-0432.CCR-05-1981. [DOI] [PubMed] [Google Scholar]
- 16.Wong DW, Leung EL, Wong SK, Tin VP, Sihoe AD, Cheng LC, Au JS, Chung LP, Wong MP. A novel KIF5B-ALK variant in nonsmall cell lung cancer. Cancer, 2011;117:2709–2718. doi: 10.1002/cncr.25843. [DOI] [PubMed] [Google Scholar]
- 17.Sasaki T, Okuda K, Zheng W, Butrynski J, Capelletti M, Wang L, Gray NS, Wilner K, Christensen JG, Demetri G, Shapiro GI, Rodig SJ, Eck MJ, Janne PA. The neuroblastoma-associated F1174L ALK mutation causes resistance to an ALK kinase inhibitor in ALK-translocated cancers. Cancer Res. 2010;70:10038–10043. doi: 10.1158/0008-5472.CAN-10-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costa DB, Kobayashi S, Pandya SS, Yeo WL, Shen Z, Tan W, Wilner KD. CSF concentration of the anaplastic lymphoma kinase inhibitor crizotinib. J Clin Oncol. 2011;29:e443–e445. doi: 10.1200/JCO.2010.34.1313. [DOI] [PubMed] [Google Scholar]
- 19.Choi YL, Soda M, Yamashita Y, Ueno T, Takashima J, Nakajima T, Yatabe Y, Takeuchi K, Hamada T, Haruta H, Ishikawa Y, Kimura H, Mitsudomi T, Tanio Y, Mano H. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N.Engl.J Med. 2010;363:1734–1739. doi: 10.1056/NEJMoa1007478. [DOI] [PubMed] [Google Scholar]
- 20.Sasaki T, Koivunen J, Ogino A, Yanagita M, Nikiforow S, Zheng W, Lathan C, Marcoux JP, Du J, Okuda K, Capelletti M, Shimamura T, Ercan D, Stumpfova M, Xiao Y, Weremowicz S, Butaney M, Heon S, Wilner K, Christensen JG, Eck MJ, Wong KK, Lindeman N, Gray NS, Rodig SJ, Janne PA. A novel ALK secondary mutation and EGFR signaling cause resistance to ALK kinase inhibitors. Cancer Res. 2011;71:6051–6060. doi: 10.1158/0008-5472.CAN-11-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katayama R, Khan TM, Benes C, Lifshits E, Ebi H, Rivera VM, Shakespeare WC, Iafrate AJ, Engelman JA, Shaw AT. Therapeutic strategies to overcome crizotinib resistance in non-small cell lung cancers harboring the fusion oncogene EML4-ALK. Proc.Natl.Acad.Sci.U.S.A, 2011;108:7535–7540. doi: 10.1073/pnas.1019559108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindeman NI, Cagle PT, Beasley MB, Chitale DA, Dacic S, Giaccone G, Jenkins RB, Kwiatkowski DJ, Saldivar JS, Squire J, Thunnissen E, Ladanyi M. Molecular Testing Guideline for Selection of Lung Cancer Patients for EGFR and ALK Tyrosine Kinase Inhibitors: Guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J Thorac.Oncol. 2013 doi: 10.1016/j.jmoldx.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 23.Takeuchi K, Choi YL, Togashi Y, Soda M, Hatano S, Inamura K, Takada S, Ueno T, Yamashita Y, Satoh Y, Okumura S, Nakagawa K, Ishikawa Y, Mano H. KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin.Cancer Res. 2009;15:3143–3149. doi: 10.1158/1078-0432.CCR-08-3248. [DOI] [PubMed] [Google Scholar]
- 24.Okubo J, Takita J, Chen Y, Oki K, Nishimura R, Kato M, Sanada M, Hiwatari M, Hayashi Y, Igarashi T, Ogawa S. Aberrant activation of ALK kinase by a novel truncated form ALK protein in neuroblastoma. Oncogene, 2012;31:4667–4676. doi: 10.1038/onc.2011.616. [DOI] [PubMed] [Google Scholar]
- 25.Cazes A, Louis-Brennetot C, Mazot P, Dingli F, Lombard B, Boeva V, Daveau R, Cappo J, Combaret V, Schleiermacher G, Jouannet S, Ferrand S, Pierron G, Barillot E, Loew D, Vigny M, Delattre O, Janoueix-Lerosey I. Characterization of rearrangements involving the ALK gene reveals a novel truncated form associated with tumor aggressiveness in neuroblastoma. Cancer Res, 2013;73:195–204. doi: 10.1158/0008-5472.CAN-12-1242. [DOI] [PubMed] [Google Scholar]
- 26.Wagle N, Berger MF, Davis MJ, Blumenstiel B, Defelice M, Pochanard P, Ducar M, Van Hummelen P, MacConaill LE, Hahn WC, Meyerson M, Gabriel SB, Garraway LA. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2012;2:82–93. doi: 10.1158/2159-8290.CD-11-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Costa DB, Kobayashi S. Acquired resistance to the ALK inhibitor crizotinib in the absence of an ALK mutation. J Thorac.Oncol, 2012;7:623–625. doi: 10.1097/JTO.0b013e318241daab. [DOI] [PubMed] [Google Scholar]
- 28.Katayama R, Shaw AT, Khan TM, Mino-Kenudson M, Solomon BJ, Halmos B, Jessop NA, Wain JC, Yeo AT, Benes C, Drew L, Saeh JC, Crosby K, Sequist LV, Iafrate AJ, Engelman JA. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Sci.Transl.Med. 2012;4 doi: 10.1126/scitranslmed.3003316. 120ra17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lovly CM, Pao W. Escaping ALK inhibition: mechanisms of and strategies to overcome resistance. Sci.Transl.Med. 2012;4 doi: 10.1126/scitranslmed.3003728. 120ps2. [DOI] [PubMed] [Google Scholar]