Abstract
In diploid organisms, the frequency and nature of sexual cycles have a major impact on genome-wide patterns of heterozygosity. Recent population genomic surveys in the budding yeast, Saccharomyces cerevisiae, have revealed surprising levels of genomic heterozygosity in what has been traditionally considered a highly inbred organism. I review evidence and hypotheses regarding the generation, maintenance, and evolutionary consequences of genomic heterozygosity in S. cerevisiae. I propose that high levels of heterozygosity in S. cerevisiae, arising from population admixture due to human domestication, coupled with selfing during rare sexual cycles, can facilitate rapid adaptation to novel environments.
1 Introduction
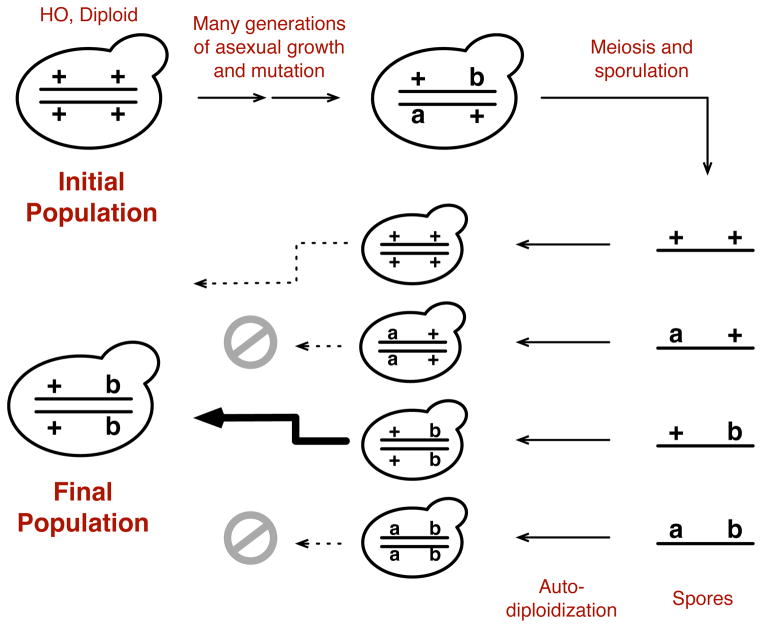
In 1994 the pioneering yeast geneticist Robert Mortimer proposed the “Genome Renewal Hypothesis” to explain patterns of genetic variation observed in the budding yeast Saccharomyces cerevisiae (Mortimer et al., 1994; Mortimer, 2000). Mortimer and colleagues observed that most yeast strains isolated from vineyards were diploid and heterozygous at one or more loci. The vast majority were also homothallic, meaning that haploid cells produced from these strains were capable of undergoing mating-type switching followed by mother-daughter mating. This process, known as autodiplodization or haploselfing, leads to diploid cells that are homozygous at all but the mating type locus. Mortimer documented a negative correlation between the number of detectable heterozygosities in vineyard isolates and the percentage of viable spores produced; homozygous isolates had nearly 100% spore viability while heterozygous isolates showed clear evidence for deleterious or sometimes lethal alleles. Finally, isolates that were homozygous were inferred to have been derived from heterozygous backgrounds via autodiploidization. Mortimer and colleagues proposed that these observations could be explained by an evolutionary scenario involving long periods of clonal reproduction in which diploid strains accumulated recessive, primarily deleterious alleles in a heterozygous state. They posited that rare sexual cycles involving meiosis followed by mating type switching and autodiploidization would facilitate the loss of deleterious alleles and fix beneficial alleles, thus leading to “Genome Renewal” (Figure 1).
Fig. 1.
A schematic illustration of Mortimer’s Genome Renewal Hypothesis. This figure illustrates the key features of the scenario Mortimer described for Genome Renewal, starting from a homothallic (HO) diploid background. The pluses indicate wild-type alleles, while ‘a’ and ‘b’ indicate recessive alleles arising during periods of asexual propagation. In this example, ‘a’ is a deleterious allele, while ‘b’ is a beneficial.
Recently, Masel and Lyttle (2011) developed a mathematical model of evolution under a life history regime like that proposed by Mortimer. This model, which assumed a small number of heterozygous sites and additivity of allelic effects, considered the effect of different mating strategies on heritable genetic variation for different selection regimes and for a range of dominance coefficients. Masel and Lyttle showed that clonal reproduction coupled with rare selfing can lead to an epistatic increase in heritable phenotypic variation per sexual episode, relative to an out-crossing strategy. They concluded that clonal expansion coupled with rare selfing can thus act as a type of ‘evolutionary capacitor’, allowing cryptic genetic variation to build up during periods of clonality, and exposing that variation to selection following periods of environmental stress that are severe enough to induce sexual cycles (Masel and Lyttle, 2011).
The Genome Renewal hypothesis is based on the assumption that the ‘ground state’ is genomic homozygosity, and the number of heterozygous loci that accumulate between periods of selfing should therefore be modest. However, recent genome sequencing of environmental isolates of S. cerevisiae has revealed that many strains harbor abundant polymorphism in the form of thousands of heterozygous sites across the genome (Magwene et al., 2011; Borneman et al., 2011). In the following pages I re-examine the Genome Renewal hypothesis in light of this discovery, focusing in particular on the implications of extensive heterozygosity coupled with homothallism with respect to adaptation to new niches. I argue that, for highly heterozygous homothallic strains, the adaptive evolutionary landscape has a high degree of “accessibility” because offspring that sample large regions of genotypic and phenotypic space can be generated rapidly from a single founding individual.
2 Evidence for the Genome Renewal Hypothesis
Since Mortimer and colleagues first put forth the Genome Renewal Hypothesis, a large number of studies have contributed to an increasingly detailed portrait of population genetic and genomic variation in S. cerevisiae (e.g. Fay and Benavides, 2005; Gresham et al., 2006; Liti et al., 2009; Schacherer et al., 2009; Skelly et al., 2009). Below I touch on only a fraction of this literature, that which bears most directly on the Genome Renewal Hypothesis. For a more exhaustive overview of yeast population genetics and genomics I refer the reader to several recent reviews (Liti and Schacherer, 2011; Sipiczki, 2011; Hittinger, 2013).
2.1 Most Environmental Isolates of S. cerevisiae are Diploid and Homothallic
Saccharomyces cerevisiae has a haplo-diploid life cycle, and can propagate asexually as either haploid or diploid cells. Despite the ability to grow vegetatively in a haploid state, S. cerevisiae is predominantly isolated from the environment as diploid cells, though aneuploid and polyploid isolates are not uncommon (Guijo et al., 1997). For example, Cubillos et al. (2009) found that approximately 95% of the 200+ wine strains they examined had a DNA content consistent with diploidy. Muller and McCusker (2009), based on a diverse sample of 170 isolates, estimated that 70–80% of their strains were diploid, with the remaining 20–30% of strains being triploid or tetraploid. Similarly, Sniegowski et al. (2002) show that each of the ten S. cerevisiae strains they isolated from an oak forest were diploid.
With respect to homothallism, Mortimer (2000) found that approximately 89% of the wine isolates he analyzed were homothallic, and Cubillos et al. (2009) similarly found that the majority of the wine strains they characterized were homothallic. All of the oak isolates studied by Sniegowski et al. (2002) were homothallic. The survey by Muller and McCusker (2009), which included strains from a wider variety of environmental contexts, paints a somewhat more complicated picture of homothallism. 27 of the 28 non-clinical isolates they examined were homothallic, but four of the eight clinical isolates the examined were heterothallic. Muller and McCusker suggested that an increased frequency of heterothallism in clinical isolates might result from indirect selection associated with a selective advantage for heterozygosity in clinical environments.
2.2 Patterns of Heterozygosity in S. cerevisiae
Mortimer’s assessments of heterozygosity in wine isolates was based on segregation of phenotypic traits such as the ability to grow on different carbon sources, growth rate mutations, etc. (Mortimer et al., 1994). Mortimer and colleagues found that roughly 65% of the more than 200 strains analyzed were heterozygous at one or more loci, as determined by tetrad analysis.
Subsequent studies that have reported data on heterozygosity in environmental isolates of S. cerevisiae have primarily focused on molecular genotyping. For example, Fay and Benavides (2005) analysis of 81 strains indicated that approximately 40% of the strains they characterized were heterozygous for at least one of five loci. Muller and McCusker (2009), based on data from 12 microsatellite markers, found that approximately 80% of their strains were heterozygous for at least one locus, with clinical isolates having higher average heterozygosity than non-clinical strains. Similarly, Diezmann and Dietrich (2009) undertook a population genetic survey of 103 S. cerevisiae strains at five loci and found that between 33% and 88% of strains from human associated environments (clinical, brewery, fruit) were heterozygous at one or more loci. In contrast all of the soil or bark isolates were homozygous at all loci examined, leading them to conclude that the soil isolates represent lineages that have experienced no or little outcrossing in contrast to the more clearly recombinant strains associated with agricultural and clinical settings.
These studies, based on a modest number of loci, established that heterozygosity is relatively common in S. cerevisiae isolates. However, the initial yeast genome (Goffeau et al., 1996; Wei et al., 2007) and population genomic (Gresham et al., 2006; Schacherer et al., 2009; Liti et al., 2009) studies used strains derived from monosporic derivatives, thus making it impossible to characterize heterozygosity on a genome-wide scale. Therefore the genomic extent of heterozygosity wasn’t appreciated until the first studies describing the sequencing of unmanipulated diploid genomes were published. The first report of this kind was the characterization of the genome of a diploid strain used in bioethanol production (Argueso et al., 2009). Argueso et al. (2009) arrived at an estimate of ~2 heterozygous SNPs per Kb, corresponding to at least ~24,000 heterozygous sites genome wide. Shortly thereafter the first population genomic surveys of unmanipulated diploids demonstrated that many S. cerevisiae strains isolated from both industrial and non-industrial contexts were highly heterozygous, many possessing greater than 30,000 heterozygous sites across the genome (Magwene et al., 2011; Borneman et al., 2011). For example, Magwene et al. (2011), based on whole genome sequencing of 11 diploid isolates plus genotyping of nine loci in 18 additional strains, concluded that approximately 60% of strains they examined had modest (> 5,000 sites) to extensive (> 15,000 sites) genomic heterozygosity. Subsequent genome sequencing of additional environmental isolates has led to similar findings (Akao et al., 2011; Babrzadeh et al., 2012; Hyma and Fay, 2013).
Several authors have noted that strains with high levels of heterozygosity are preferentially isolated from human associated environments (Diezmann and Dietrich, 2009; Muller and McCusker, 2009; Magwene et al., 2011). Clinical and industrial isolates stand out in this regard, but a number of strains isolated from agricultural contexts, such as fruit trees, also have high levels of heterozygosity (Diezmann and Dietrich, 2009; Hyma and Fay, 2013; Magwene et al., 2011). By contrast, S. cerevisiae isolated from relatively undisturbed environments such as oak forests in both North America and Asia have very low levels of heterozygosity (Kuehne et al., 2007; Wang et al., 2012). Similarly, isolates of the undomesticated sister species, Saccharomyces paradoxus, also exhibit very little genomic heterozygosity (Tsai et al., 2008). These data suggest that for budding yeast homozygosity may be the rule in the absence of human domestication.
2.3 What Generates and Maintains Heterozygosity?
The nature and extent of genomic heterozygosity that sequencing of diploid strains has revealed is surprising given that previous studies (Ruderfer et al., 2006) had suggested that sex in yeast primarily involves inbreeding via intratetrad mating (Tsai et al., 2008). Inbreeding of any type quickly leads to loss of heterozygosity and mating type switching by halplo-selfing immediately homozygoses the entire genome except at the mating type locus (Kirby, 1984). Magwene et al. (2011) concluded that the extensive heterozygosity seen in many strains was most likely resulted from out-crossing between genetically diverse lineages. This seems to be at odds with studies that have estimated that outcrossing occurs only about once every 50,000 – 100,000 mitotic generations in yeast (Ruderfer et al., 2006; Tsai et al., 2008). However, population genomic analyses demonstrate quite clearly that there has been significant admixture between S. cerevisiae lineages (Liti et al., 2009; Schacherer et al., 2009), and a recent study suggests that outcrossing in S. cerevisiae may be considerably higher than previous estimates (Kelly et al., 2012). Regardless of the actual rates of outcrossing, the very high levels of heterozygosity observed in clinical, industrial, and many agricultural isolates begs the question of what contributes to the maintenance of heterozygosity? Magwene et al. (2011) suggested that differences between heterozygous and homozygous strains might be the result of alternate life history strategies that favor different frequencies of sexual cycles, and proposed that heterozygous strains represent lineages that are less likely to undergo sexual cycles, thus preserving heterozygosity. Heterothallism should also tend to favor the preservation of heterozygosity; as noted above, clinical isolates of S. cerevisiae, which tend to have among the highest levels of genome-wide heterozygosity, show a trend towards a greater frequency of heterothallism (Muller and McCusker, 2009).
2.4 Heterozygosity and Spore Viability
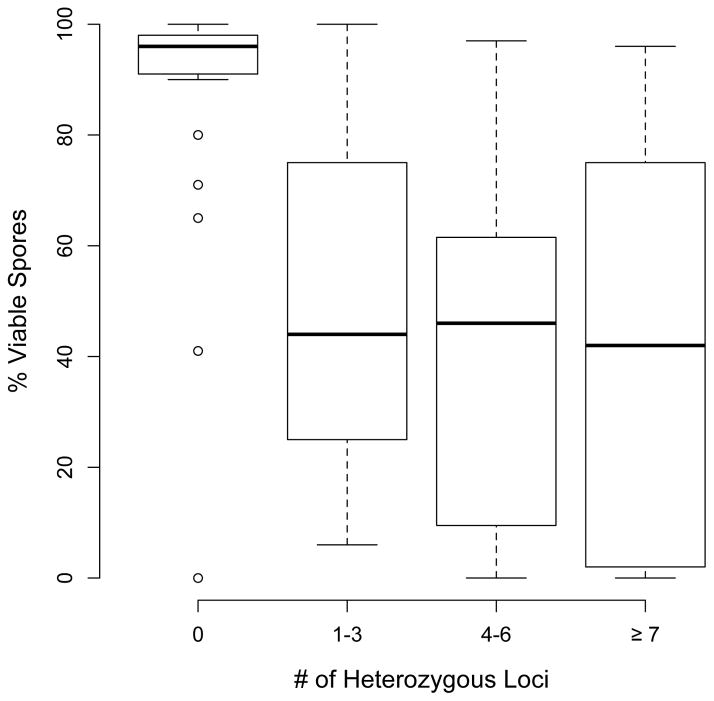
One of the findings that motivated the Genome Renewal hypothesis was the observation that spore viablity tended to decrease with increased heterozygosity (Mortimer et al., 1994). Mortimer found that strains that were completely homozygous sporulated at high frequency and had spore viabilities near 100%. By contrast approximately 47% of strains that were heterozygous at one or more loci were also heterozygous for lethal or deleterious alleles (Mortimer, 2000). In Figure 2 I present a re-analysis of data from Muller and McCusker (2009) that supports the pattern suggested by Mortimer; spore viability is negatively correlated with the number of heterozygous loci (Figure 2; Spearman rank correlation ρ = −0.50, p < 0.0001 by permutation test).
Fig. 2.
Relationship between the number of heterozygous loci and spore viability for 108 diploid strains, based on data from Muller and McCusker (2009). The rank correlation between heterozygosity and percent spore viability is −0.5.
Some of this reduction of spore viability is likely due to recessive deleterious or lethal alleles as suggested by Mortimer, however this may also reflect incompatible genetic combinations arising under reproductive isolation between the backgrounds that contributed to the formation of the heterozygotes (Cubillos et al., 2011). Such incompatibilities may result from both large scale genomic changes (e.g. aneuploidy) between strain backgrounds or may involve single nucleotide changes, either neutral or adaptive, that arise under reproductive isolation. For example, Demogines et al. (2008) identified naturally segregating allelic variation in two genes, MLH1 and PMS1, involved in DNA mismatch repair. Particular combinations of alleles at these two loci result in a low-fitness mutator phenotype (Demogines et al., 2008).
3 Molecular and Phenotypic Consequences of Heterozygosity
High levels of heterozygosity are likely to have a significant impact on molecular interactions. For example, the strain EM93 is the primary ancestor of the standard reference strain S288c and has more than 24,000 heterozygous sites (Magwene et al., 2011; Esberg et al., 2011). Table 1 shows the predicted impact of this heterozygosity on the proteome; approximately 37% of proteins in EM93 are present as two different peptide sequences. While it is hard to know how much of this protein polymorphism has functional effects, this nevertheless represents a very large pool of variation present within a single strain background. There is also abundant heterozygosity in non-coding regions, which has the potential to affect regulatory networks through effects such as allele specific gene expression (Gagneur et al., 2009) and differential protein-DNA interactions (Zheng et al., 2010). At the level of cellular phenotypes, extensive heterozygosity might to contribute to cell-to-cell heterogeneity in clonal populations, through mechanisms such as allele specific gene and protein expression (Levy et al., 2012) and differential regulation of epigenetic silencing (Halme et al., 2004).
Table 1.
Estimated number of heterozygous proteins per chromosome in the genome of the strain EM93.
| Chr | # ORFs | % ORFs |
|---|---|---|
| I | 44 | 38 |
| II | 101 | 22 |
| III | 60 | 33 |
| IV | 174 | 21 |
| V | 117 | 36 |
| VI | 75 | 53 |
| VII | 233 | 40 |
| VIII | 109 | 34 |
| IX | 119 | 49 |
| X | 192 | 48 |
| XI | 191 | 55 |
| XII | 87 | 15 |
| XIII | 223 | 44 |
| XIV | 211 | 49 |
| XV | 285 | 48 |
| XVI | 191 | 37 |
|
| ||
| Total | 2412 | 37 |
4 Evolutionary Consequences of Heterozygosity and Homothallism
Mortimer’s Genome Renewal hypothesis and related models (Masel and Lyttle, 2011; Sipiczki, 2011) primarily consider the case of accumulation of a modest number of heterozygous sites against an otherwise homozygous genomic background. However, as detailed above, many environmental isolates of S. cerevisiae are heterozygous at more than 30,000 sites across the genome. How does this degree of heterozygosity modify our view of the Genome Renewal hypothesis?
Consider the following scenario — a single heterozygous individual is introduced into a novel environment and generates a large clonal population. When nutrients become limiting the population undergoes meiosis and sporulation [most heterozygous isolates are slow to sporulate but eventually do so under extended nutrient limitation; Magwene et al. (2011)]. The meiotic products generated from this clonal population represent a sampling of a very large combinatorial space – the 2n possible allelic combinations representing the alternative alleles at heterozygous sites in the founder individual (even accounting for linkage disequilibrium, n is likely to be in the hundreds to thousands). Assuming subsequent germination of those spores, either through reintroduction of nutrients or disperal to nearby nutrient rich environments (e.g. by insect vectors; Stefanini et al. (2012)), haploids will return to diploidy either through mating type switching and autodiploidization or through intra- or intertetrad matings. Due to a high frequency of selfing and other forms of inbreeding, a large number of allelic combinations will be exposed to local selection in a primarily homozygous state, thus increasing the probability that one or more favorable genotypes will become established and thrive in the new environment.
How likely are such introductions and how much potential phenotypic variability can be exposed to selection under such a scenario? The likelihood of introductions is hard to assess given the challenges of studying yeast ecology in a natural setting, but S. cerevisiae is commonly isolated during environmental sampling in a wide variety of contexts (Hyma and Fay, 2013; McCusker et al., 1994; Naumov et al., 1998; Sweeney et al., 2004) and such introductions are likely to be facilitated, either purposefully or inadvertantly, by human activity (Goddard et al., 2010).
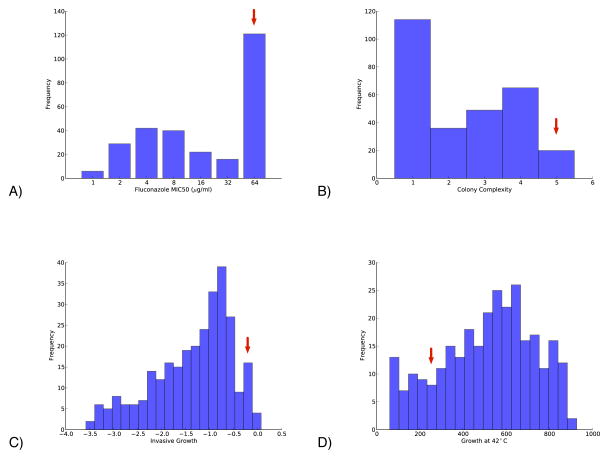
The “phenotypic potential” of heterozygous, homothallic strains can be addressed directly in the laboratory by sporulating such strains, germinating the spores at low enough density to induce autodiploidization, and assessing the phenotypes of the resulting homozygous offspring. Figure 3 shows an example of carrying out such an experiment on a highly heterozygous clinical isolate, YJM311 (McCusker et al., 1994). Each of the subfigures represents the distribution of a different phenotype of interest in homozygous offspring of YJM311. There is abundant phenotypic variation for each of the traits and the multivariate phenotypic space represented by these offspring is equally rich (Magwene, unpublished data). Illustrative of the genetic and phenotypic diversity of such strains, my laboratory has recently used such a population to map QTLs for biofilm formation in S. cerevisiae (Granek et al., 2013).
Fig. 3.
Phenotypic distributions of four traits among offspring of the highly heterozygous clinical isolate YJM311. 288 homozygous diploid offspring were generated by sporulation followed by autodiploidization. Traits assessed include: A) resistance to the antifungal drug fluconazole (MIC50 as determined in microtiter plates); B) a measure of colony biofilm complexity (Granek et al., 2013); C) a measure of invasive growth on agar substrates (logarithm of the ratio of post-wash to pre-wash colony density; (Drees et al., 2005)); and D) a measure of growth at high temperature on agar plates (square root of the mean spot density for three replicates of each segregant grown at 42°C). The red arrow in each subfigure indicates the phenotype of the parental strain YJM311.
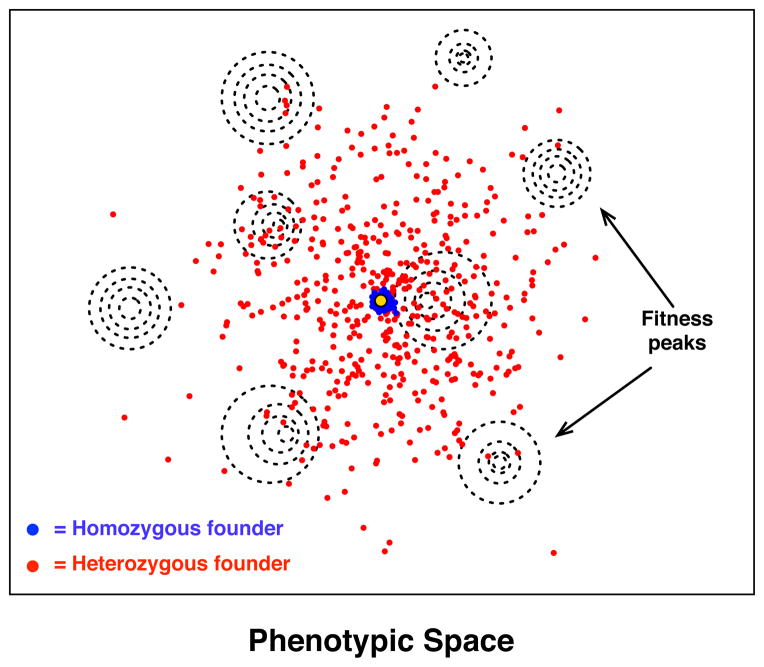
Given the very large number of allelic combinations that can be generated from a single individual, and the corresponding phenotypic variability that accompanies this genetic variation, I argue that the evolutionary landscape can be thought of as relatively “accessible” for heterozygous, homothallic strains. For highly heterozygous and homothallic strains the phenotypes of their offpsring can be radically different than their own, and the ability to generate large clonal populations means that large regions of genotypic and phenotypic space can be potentially sampled in a single sexual generation. These properties therefore seem likely to promote invasion of new niches and adaptation to novel environments. The range of phenotypes accessible from a heterozygous founder should generally increase as a function of the number of heterozygous sites and the magnitude of epistatic interactions between loci, but should be negatively correlated with the average degree of dominance (Masel and Lyttle, 2011).
5 Admixture, Heterozygosity, and Domestication
Saccharomyces cerevisiae has been at the forefront of studies of yeast population genetics but in recent years several additional species within the Saccharomyces sensu stricto complex have begun to garner similar attention (Libkind et al., 2011; Liti et al., 2009). Thus far, S. cerevisiae seems unique in the degree of admixture that has occurred between lineages. The amount of heterozygosity, and the genetic structure of geographically distinct populations in species such as Saccharomyces paradoxus seems consistent with what one would expect based on selfing followed by slow accumulation of heterozygous mutations during extended periods of asexual growth (Tsai et al., 2008).
It is likely that admixture between divergent S. cerevisiae lineages, and the resultant heterozygosity, has been both a consequence of and a contributor to domestication in S. cerevisiae. Human activity can bring divergent lineages into contact, facilitating outcrossing that establishes heterozygosity. Subsequently, the adaptive potential of such heterozygotes may have facilitated selection for traits that are desirable for brewing, baking, and industrial uses. However, these same processes may help to facilitate S. cerevisiae adaptation to new, less beneficial (from a human perspective), niches. For example, S. cerevisiae is an emerging human pathogen (McCusker, 2006), and clinical isolates are frequently highly heterozygous (Magwene et al., 2011).
It is also interesting to consider the extent to which heterozygosity and homothallism may facilitate the establishment of interspecific hybrids in the Saccharomyces genus. Prezygotic barriers to hybridzation are relatively weak and hybrids show robust vegetative growth (Morales and Dujon, 2012). However, most yeast hybrids are sterile, primarily as a result of chromosomal translocations that distinguish the different species and which lead to incomplete meiosis in hybrids (Delneri et al., 2003; Fischer et al., 2000). However, if rare compatible combinations of alleles from the two species are also homothallic (Greig et al., 2002), then the resulting heterozygosity from such interspecies crosses may play a critical role in subsequent adaptation and/or domestication. For example, it has been shown that the Saccharomyces bayanus is a complex hybrid of three species – S. eubayanus, S. uvarum and S. cerevisiae. The type strain for S. bayanus, CBS 380, has significant heterozygosity that reflects the contributions of the different parental species (Libkind et al., 2011).
6 Genome Renewal in other Fungi?
How might a modified model of genome renewal apply more broadly to other fungal lineages? Particularly interesting in this regard is recent work on the fungal pathogen Candida albicans. C. albicans is primarily isolated as a diploid, and most strains of C. albicans have extensive genomic heterozygosity (Jones et al., 2004). However, unlike S. cerevisiae, neither a true sexual cycle or haploids had been described in C. albicans until recently when Hickman et al. (2013) showed that viable haploid cells can arise from diploids, presumably through a mechanism of concerted chromosome loss. These haploid cells can mate with cells of the opposite mating type as well as autodiploidize. However, in contrast to S. cerevisiae, the homozygous diploids examined tended to have reduced fitness (though it should be noted that this observation is based on derivatives of a single diploid background). Like Mortimer, Hickman et al. (2013) propose that a temporary reduction to haploidy should help to purge recessive deleterious alleles.
7 Conclusions
The standard version the Genome Renewal hypothesis is that infrequent sexual cycles, characterized by a high degree of selfing, can help to purge deleterious alleles and fix beneficial alleles, thus helping to facilitate adaptation in yeast. However, recent discoveries from population genomic sequencing of natural S. cerevisiae strains has forced us to re-evaluate the Genome Renewal hypothesis to account for very high levels of heterozygosity observed in many environmental isolates. The hypothesis put forth here is that extensive heterozygosity coupled with clonal expansion and homothallism can act as an engine for adaptation by greatly increasing the size of the genotypic and phenotypic spaces that can rapidly explored in a single sexual generation in a population founded by a single individual. Further studies involving population genomic sequencing, experimental evolution, and microbial ecology will be needed to determine the extent to which this hypothesis holds and whether the patterns observed here are peculiar to S. cerevisiae or if they extend to other taxa that operate in a similar ecological and evolutionary milieu of clonal growth, selfing, and admixture.
Fig. 4.
A hypothetical evolutionary landscape for two different populations generated from single founding individuals. The plane of the figure represents a high dimensional phenotypic space, and the concentric dashed circles represent local fitness optima. The red population represents individuals generated from a highly heterozygous founder; the blue population reprsents derivatives of a homozygous founder. The founder phenotypes are identical for both populations (yellow dot). The populations are assumed to have been generated by initial clonal growth (with mutation) followed by a single round of meiosis and sporulation coupled with subsequent mating and/or haploselfing.
Acknowledgments
I thank Helen Murphy and Cliff Zeyl for discussions and feedback, as well as two anonymous reviewers for their critical comments and suggestions on the manuscript. Ludo Muller and John McCusker kindly provided data on spore viability and heterozygosity. The work was supported in part by awards from NSF (MCB-0614959) and NIH (P50GM081883-01).
Glossary
- Admixture
Interbreeding between two or more genetically distinct populations
- Autodiploidization
Mating between mother and daughter haploid cells following mating type switching. Autodiploidization leads to diploid cells that are completely homozygous across the genome, except at the mating type locus
- Heterothallic
Yeast strains that are incapable of undergoing mating type switching as haploid cells are referred as heterothallic. Such strains can be stably propagated as either haploids or diploid. Heterothallism in S. cerevisiae is usually a result of loss-of-function mutations at the HO locus
- Homothallic
Yeast strains that are capable of undergoing mating type switching and autodiploidization are referred to as homothallic. The haploid phase is usually transient in homothallic strains because they rapidly switch mating types and initiate mother-daughter cell matings, leading to diploidy
- Mating type switching
In S. cerevisiae and related yeast, mating type switching is facilitated by a high-frequency, site specific gene conversion events at the mating type (MAT) locus, induced by a site-specific endonuclease called HO, and involving silenced mating type sequence at ‘hidden’ MAT loci. in See Haber (1998) for a review of the mechanisms underlying mating type switching
- Tetrad analysis
During sporulation, haploid spores are packaged together in a structure called an ascus. The ascus, plus the four haploid spores, are referred to as a tetrad. Tetrads can be teased apart with a micromanipulator following enzymatic digestion of the ascus. Subsequent germination of spores and scoring of phenotypes facilitates genetic analyses such as linkage mapping and distinguishing between Mendelian and non-Mendelian traits
References
- Akao T, Yashiro I, Hosoyama A, Kitagaki H, Horikawa H, Watanabe D, Akada R, Ando Y, Harashima S, Inoue T, Inoue Y, Kajiwara S, Kitamoto K, Kitamoto N, Kobayashi O, Kuhara S, Masubuchi T, Mizoguchi H, Nakao Y, Nakazato A, Namise M, Oba T, Ogata T, Ohta A, Sato M, Shibasaki S, Takatsume Y, Tanimoto S, Tsuboi H, Nishimura A, Yoda K, Ishikawa T, Iwashita K, Fujita N, Shimoi H. Whole-genome sequencing of sake yeast Saccharomyces cerevisiae Kyokai no. 7. DNA Res. 2011;18(6):423–34. doi: 10.1093/dnares/dsr029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argueso JL, Carazzolle MF, Mieczkowski PA, Duarte FM, Netto OVC, Missawa SK, Galzerani F, Costa GGL, Vidal RO, Noronha MF, Dominska M, Andrietta MGS, Andrietta SR, Cunha AF, Gomes LH, Tavares FCA, Alcarde AR, Dietrich FS, McCusker JH, Petes TD, Pereira GAG. Genome structure of a Saccharomyces cerevisiae strain widely used in bioethanol production. Genome Res. 2009;19(12):2258–70. doi: 10.1101/gr.091777.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babrzadeh F, Jalili R, Wang C, Shokralla S, Pierce S, Robinson-Mosher A, Nyren P, Shafer RW, Basso LC, de Amorim HV, de Oliveira AJ, Davis RW, Ronaghi M, Gharizadeh B, Stambuk BU. Whole-genome sequencing of the efficient industrial fuel-ethanol fermentative Saccharomyces cerevisiae strain CAT-1. Mol Genet Genomics. 2012;287(6):485–94. doi: 10.1007/s00438-012-0695-7. [DOI] [PubMed] [Google Scholar]
- Borneman AR, Desany BA, Riches D, Affourtit JP, Forgan AH, Pretorius IS, Egholm M, Chambers PJ. Whole-genome comparison reveals novel genetic elements that characterize the genome of industrial strains of Saccharomyces cerevisiae. PLoS Genet. 2011;7(2):e1001287. doi: 10.1371/journal.pgen.1001287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos FA, Billi E, Zörgö E, Parts L, Fargier P, Omholt S, Blomberg A, Warringer J, Louis EJ, Liti G. Assessing the complex architecture of polygenic traits in diverged yeast populations. Mol Ecol. 2011;20(7):1401–13. doi: 10.1111/j.1365-294X.2011.05005.x. [DOI] [PubMed] [Google Scholar]
- Cubillos FA, Vásquez C, Faugeron S, Ganga A, Martínez C. Self-fertilization is the main sexual reproduction mechanism in native wine yeast populations. FEMS Microbiol Ecol. 2009;67(1):162–70. doi: 10.1111/j.1574-6941.2008.00600.x. [DOI] [PubMed] [Google Scholar]
- Delneri D, Colson I, Grammenoudi S, Roberts IN, Louis EJ, Oliver SG. Engineering evolution to study speciation in yeasts. Nature. 2003;422(6927):68–72. doi: 10.1038/nature01418. [DOI] [PubMed] [Google Scholar]
- Demogines A, Wong A, Aquadro C, Alani E. Incompatibilities involving yeast mismatch repair genes: a role for genetic modifiers and implications for disease penetrance and variation in genomic mutation rates. PLoS Genet. 2008;4(6):e1000103. doi: 10.1371/journal.pgen.1000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diezmann S, Dietrich FS. Saccharomyces cerevisiae: population divergence and resistance to oxidative stress in clinical, domesticated and wild isolates. PLoS One. 2009;4(4):e5317. doi: 10.1371/journal.pone.0005317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees BL, Thorsson V, Carter GW, Rives AW, Raymond MZ, Avila-Campillo I, Shannon P, Galitski T. Derivation of genetic interaction networks from quantitative phenotype data. Genome Biol. 2005;6(4):R38. doi: 10.1186/gb-2005-6-4-r38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esberg A, Muller LAH, McCusker JH. Genomic structure of and genome-wide recombination in the Saccharomyces cerevisiae S288C progenitor isolate EM93. PLoS One. 2011;6(9):e25211. doi: 10.1371/journal.pone.0025211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Benavides JA. Evidence for domesticated and wild populations of saccharomyces cerevisiae. PLoS Genet. 2005;1(1):66–71. doi: 10.1371/journal.pgen.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer G, James SA, Roberts IN, Oliver SG, Louis EJ. Chromosomal evolution in Saccharomyces. Nature. 2000;405(6785):451–4. doi: 10.1038/35013058. [DOI] [PubMed] [Google Scholar]
- Gagneur J, Sinha H, Perocchi F, Bourgon R, Huber W, Steinmetz LM. Genome-wide allele- and strand-specific expression profiling. Mol Syst Biol. 2009;5:274. doi: 10.1038/msb.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard MR, Anfang N, Tang R, Gardner RC, Jun C. A distinct population of Saccharomyces cerevisiae in New Zealand: evidence for local dispersal by insects and human-aided global dispersal in oak barrels. Environ Microbiol. 2010;12(1):63–73. doi: 10.1111/j.1462-2920.2009.02035.x. [DOI] [PubMed] [Google Scholar]
- Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG. Life with 6000 genes. Science. 1996;274(5287):546, 563–7. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- Granek JA, Murray D, Kayrkçi O, Magwene PM. The genetic architecture of biofilm formation in a clinical isolate of Saccharomyces cerevisiae. Genetics. 2013;193(2):587–600. doi: 10.1534/genetics.112.142067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig D, Louis EJ, Borts RH, Travisano M. Hybrid speciation in experimental populations of yeast. Science. 2002;298(5599):1773–5. doi: 10.1126/science.1076374. [DOI] [PubMed] [Google Scholar]
- Gresham D, Ruderfer DM, Pratt SC, Schacherer J, Dunham MJ, Botstein D, Kruglyak L. Genome-wide detection of polymorphisms at nucleotide resolution with a single dna microarray. Science. 2006;311(5769):1932–6. doi: 10.1126/science.1123726. [DOI] [PubMed] [Google Scholar]
- Guijo S, Mauricio JC, Salmon JM, Ortega JM. Determination of the relative ploidy in different Saccharomyces cerevisiae strains used for fermentation and ‘flor’ film ageing of dry sherry-type wines. Yeast. 1997;13(2):101–17. doi: 10.1002/(SICI)1097-0061(199702)13:2<101::AID-YEA66>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Haber JE. Mating-type gene switching in Saccharomyces cerevisiae. Annu Rev Genet. 1998;32:561–99. doi: 10.1146/annurev.genet.32.1.561. [DOI] [PubMed] [Google Scholar]
- Halme A, Bumgarner S, Styles C, Fink GR. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell. 2004;116(3):405–15. doi: 10.1016/s0092-8674(04)00118-7. [DOI] [PubMed] [Google Scholar]
- Hickman MA, Zeng G, Forche A, Hirakawa MP, Abbey D, Harrison BD, Wang YM, Su C-h, Bennett RJ, Wang Y, Berman J. The ‘obligate diploid’ Candida albicans forms mating-competent haploids. Nature. 2013;494(7435):55–9. doi: 10.1038/nature11865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hittinger CT. Saccharomyces diversity and evolution: a budding model genus. Trends Genet. 2013 doi: 10.1016/j.tig.2013.01.002. [DOI] [PubMed] [Google Scholar]
- Hyma KE, Fay JC. Mixing of vineyard and oak-tree ecotypes of Saccharomyces cerevisiae in north american vineyards. Mol Ecol. 2013 doi: 10.1111/mec.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T, Federspiel NA, Chibana H, Dungan J, Kalman S, Magee BB, Newport G, Thorstenson YR, Agabian N, Magee PT, Davis RW, Scherer S. The diploid genome sequence of Candida albicans. Proc Natl Acad Sci U S A. 2004;101(19):7329–34. doi: 10.1073/pnas.0401648101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AC, Shewmaker FP, Kryndushkin D, Wickner RB. Sex, prions, and plasmids in yeast. Proc Natl Acad Sci U S A. 2012;109(40):E2683–90. doi: 10.1073/pnas.1213449109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby GC. Breeding systems and heterozygosity in populations of tetrad forming fungi. Heredity. 1984;52:35–41. [Google Scholar]
- Kuehne HA, Murphy HA, Francis CA, Sniegowski PD. Allopatric divergence, secondary contact, and genetic isolation in wild yeast populations. Curr Biol. 2007;17(5):407–11. doi: 10.1016/j.cub.2006.12.047. [DOI] [PubMed] [Google Scholar]
- Levy SF, Ziv N, Siegal ML. Bet hedging in yeast by heterogeneous, age-correlated expression of a stress protectant. PLoS Biol. 2012;10(5):e1001325. doi: 10.1371/journal.pbio.1001325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libkind D, Hittinger CT, Valério E, Gonçalves C, Dover J, Johnston M, Gonçalves P, Sampaio JP. Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. Proc Natl Acad Sci U S A. 2011;108(35):14539–44. doi: 10.1073/pnas.1105430108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts IN, Burt A, Koufopanou V, Tsai IJ, Bergman CM, Bensasson D, O’Kelly MJT, van Oudenaarden A, Barton DBH, Bailes E, Nguyen AN, Jones M, Quail MA, Goodhead I, Sims S, Smith F, Blomberg A, Durbin R, Louis EJ. Population genomics of domestic and wild yeasts. Nature. 2009;458(7236):337–41. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G, Schacherer J. The rise of yeast population genomics. C R Biol. 2011;334(8–9):612–9. doi: 10.1016/j.crvi.2011.05.009. [DOI] [PubMed] [Google Scholar]
- Magwene PM, Kaykç Ö, Granek JA, Reininga JM, Scholl Z, Murray D. Outcrossing, mitotic recombination, and life-history trade-offs shape genome evolution in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2011;108(5):1987–92. doi: 10.1073/pnas.1012544108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masel J, Lyttle DN. The consequences of rare sexual reproduction by means of selfing in an otherwise clonally reproducing species. Theor Popul Biol. 2011;80(4):317–22. doi: 10.1016/j.tpb.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker JH. Saccharomyces cerevisiae: An emerging and model pathogenic fungus. In: Heitman J, Edwards JE, Filler SG, Mitchell AP, editors. Molecular Principles of Fungal Pathogenesis. American Society for Microbiology; 2006. [Google Scholar]
- McCusker JH, Clemons KV, Stevens DA, Davis RW. Sac-charomyces cerevisiae virulence phenotype as determined with CD-1 mice is associated with the ability to grow at 42 degrees c and form pseudohyphae. Infect Immun. 1994;62(12):5447–55. doi: 10.1128/iai.62.12.5447-5455.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales L, Dujon B. Evolutionary role of interspecies hybridization and genetic exchanges in yeasts. Microbiol Mol Biol Rev. 2012;76(4):721–39. doi: 10.1128/MMBR.00022-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer RK. Evolution and variation of the yeast (saccharomyces) genome. Genome Res. 2000;10(4):403–9. doi: 10.1101/gr.10.4.403. [DOI] [PubMed] [Google Scholar]
- Mortimer RK, Romano P, Suzzi G, Polsinelli M. Genome renewal: a new phenomenon revealed from a genetic study of 43 strains of Saccharomyces cerevisiae derived from natural fermentation of grape musts. Yeast. 1994;10(12):1543–52. doi: 10.1002/yea.320101203. [DOI] [PubMed] [Google Scholar]
- Muller LAH, McCusker JH. Microsatellite analysis of genetic diversity among clinical and nonclinical Saccharomyces cerevisiae isolates suggests heterozygote advantage in clinical environments. Mol Ecol. 2009;18(13):2779–86. doi: 10.1111/j.1365-294X.2009.04234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumov GI, Naumova ES, Sniegowski PD. Saccharomyces paradoxus and Saccharomyces cerevisiae are associated with exudates of north american oaks. Can J Microbiol. 1998;44(11):1045–50. [PubMed] [Google Scholar]
- Ruderfer DM, Pratt SC, Seidel HS, Kruglyak L. Population genomic analysis of outcrossing and recombination in yeast. Nat Genet. 2006;38(9):1077–81. doi: 10.1038/ng1859. [DOI] [PubMed] [Google Scholar]
- Schacherer J, Shapiro JA, Ruderfer DM, Kruglyak L. Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature. 2009;458(7236):342–5. doi: 10.1038/nature07670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipiczki M. Diversity, variability and fast adaptive evolution of the wine yeast (Saccharomyces cerevisiae) genome—a review. Annals of Microbiology. 2011;61:85–93. doi: 10.1007/s13213-010-0086-4. [DOI] [Google Scholar]
- Skelly DA, Ronald J, Connelly CF, Akey JM. Population genomics of intron splicing in 38 saccharomyces cerevisiae genome sequences. Genome Biol Evol. 2009;1:466–78. doi: 10.1093/gbe/evp046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sniegowski PD, Dombrowski PG, Fingerman E. Saccharomyces cerevisiae and Saccharomyces paradoxus coexist in a natural woodland site in north america and display different levels of reproductive isolation from european conspecifics. FEMS Yeast Res. 2002;1(4):299–306. doi: 10.1111/j.1567-1364.2002.tb00048.x. [DOI] [PubMed] [Google Scholar]
- Stefanini I, Dapporto L, Legras JL, Calabretta A, Di Paola M, De Filippo C, Viola R, Capretti P, Polsinelli M, Turillazzi S, Cavalieri D. Role of social wasps in Saccharomyces cerevisiae ecology and evolution. Proc Natl Acad Sci U S A. 2012;109(33):13398–403. doi: 10.1073/pnas.1208362109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney JY, Kuehne HA, Sniegowski PD. Sympatric natural Saccharomyces cerevisiae and S. paradoxus populations have different thermal growth profiles. FEMS Yeast Res. 2004;4(4–5):521–5. doi: 10.1016/S1567-1356(03)00171-5. [DOI] [PubMed] [Google Scholar]
- Tsai IJ, Bensasson D, Burt A, Koufopanou V. Population genomics of the wild yeast Saccharomyces paradoxus: Quantifying the life cycle. Proc Natl Acad Sci U S A. 2008;105(12):4957–62. doi: 10.1073/pnas.0707314105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QM, Liu WQ, Liti G, Wang SA, Bai FY. Surprisingly diverged populations of Saccharomyces cerevisiae in natural environments remote from human activity. Mol Ecol. 2012;21(22):5404–17. doi: 10.1111/j.1365-294X.2012.05732.x. [DOI] [PubMed] [Google Scholar]
- Wei W, McCusker JH, Hyman RW, Jones T, Ning Y, Cao Z, Gu Z, Bruno D, Miranda M, Nguyen M, Wilhelmy J, Komp C, Tamse R, Wang X, Jia P, Luedi P, Oefner PJ, David L, Dietrich FS, Li Y, Davis RW, Steinmetz LM. Genome sequencing and comparative analysis of Sac-charomyces cerevisiae strain YJM789. Proc Natl Acad Sci U S A. 2007;104(31):12825–30. doi: 10.1073/pnas.0701291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Zhao H, Mancera E, Steinmetz LM, Snyder M. Genetic analysis of variation in transcription factor binding in yeast. Nature. 2010;464(7292):1187–91. doi: 10.1038/nature08934. [DOI] [PMC free article] [PubMed] [Google Scholar]