Abstract
Regulation of neural stem cell (NSC) fate decisions is critical during the transition from a multicellular mammalian forebrain neuroepithelium to the multi-layered neocortex. Forebrain development requires coordinated vascular investment alongside NSC differentiation. Vascular endothelial growth factor A (Vegf) has proven to be a pleiotrophic gene whose multiple protein isoforms regulate a broad range of effects in neurovascular systems. To test the hypothesis that the Vegf isoforms (120, 164, and 188) are required for normal forebrain development, we analyzed the forebrain transcriptome of mice expressing specific Vegf isoforms, Vegf120, VegfF188, or a combination of Vegf120/188. Transcriptome analysis identified differentially expressed genes in embryonic day (E) 9.5 forebrain, a time point preceding dramatic neuroepithelial expansion and vascular investment in the telencephalon. Meta-analysis identified gene pathways linked to chromosome-level modifications, cell fate regulation, and neurogenesis that were altered in Vegf isoform mice. Based on these gene network shifts, we predicted that NSC populations would be affected in later stages of forebrain development. In the E11.5 telencephalon, we quantified mitotic cells [Phospho-Histone H3 (pHH3)-positive] and intermediate progenitor cells (Tbr2/Eomes-positive), observing quantitative and qualitative shifts in these populations. We observed qualitative shifts in cortical layering at P0, particularly with Ctip2-positive cells in layer V. The results identify a suite of genes and functional gene networks that can be used to further dissect the role of Vegf in regulating NSC differentiation and downstream consequences for NSC fate decisions.
Keywords: neural development, forebrain, VEGF, Pax6, Tbr2, neurogenesis
Introduction
The neocortex is a highly ordered six-layered structure, the development of which is dependent upon an outward expansion of forebrain neuroepithelium accompanied by blood vessel investment from the outer pial surface. It is from the neural stem cell (NSC) populations within the neuroepithelium that the majority of neural and glial derivatives of the central nervous system (CNS) arise. Identification of the key molecular regulators of these developmental transitions continues to be an ongoing area of research, particularly with regard to the multi-layered cortex (Pierani and Wassef, 2009; Borello and Pierani, 2010). The angiogenesis that accompanies the expansion of the neuroepithelium is as tightly regulated, spatially and temporally, as the generation of NSCs (Vasudevan and Bhide, 2008; Vasudevan et al., 2008; James and Mukouyama, 2011). Essential to the coordination of both neurogenesis and angiogenesis is the regulation of cell survival, cell proliferation, polarity, differentiation, and migration, as directed by a reservoir of growth factor and extracellular matrix (ECM) signals within the cellular microenvironment (Sobeih and Corfas, 2002; Hynes, 2009). One growth factor that has been directly implicated in both angiogenesis and neurogenesis is Vascular endothelial growth factor A (Vegf). In recent years, an expanded role for Vegf has been suggested based on its potential to directly affect neuronal survival and proliferation in vitro and in vivo (Jin, 2002; Zhu et al., 2003; Sun et al., 2006), and migrating neural progenitors in the cerebellum (Ruiz de Almodovar et al., 2010).
One of the most intriguing aspects of Vegf as a dual regulator of both neurogenesis and angiogenesis in the developing CNS is the fact that Vegf A transcripts can be alternatively spliced to generate a variety of isoforms that differ in their biochemical and signaling properties (Matsumoto and Claesson-Welsh, 2001; Krilleke et al., 2009; Koch et al., 2011; Mackenzie and Ruhrberg, 2012). While several splice variants have been identified, the predominant isoforms in the mouse brain are 120, 164, and 188 (Ng et al., 2001; Ruhrberg et al., 2002; Ruiz de Almodovar et al., 2010; Darland et al., 2011). These isoforms differ in amino acid number and composition resulting in a differential ability to bind heparin and heparan sulfate proteoglycans (HSPG) within the microenvironment as well as their ability to associate with the ECM. The Vegf120 isoform lacks the HSPG binding domain normally coded for by exons 6 and 7, and is thus freely diffusible throughout the extracellular microenvironment (Krilleke et al., 2009). In contrast, the Vegf188 isoform contains the full HSPG binding domain and is locally retained within the microenvironment. Vegf164, the primary Vegf isoform in the brain, contains a partial HSPG binding domain and has the potential to be locally retained as well as diffusible (Houck et al., 1992; Park et al., 1993; Ferrara, 1999). Vegf protein has been localized to radial glia, neurons, and vascular cells of developing human fetal telencephalon (Virgintino et al., 2003), supporting a role for this multipotent factor at several levels in developing cortical neuroepithelium. Less is known, however, about the role of Vegf in the differentiation, proliferation, and survival of NSCs, such as radial glia, a cell type gaining appreciation for its role as a neural precursor (Gotz et al., 2002; Heins et al., 2002; Gotz and Barde, 2005; Mori et al., 2005).
Given the contextual overlap between neurogenesis and vascular development, it is critical to investigate the regulatory role of Vegf, a factor potentially impacting all the cell types involved. Lines of mice expressing individual or combinations of Vegf isoforms have been established that display abnormalities in a range of developing organ systems (Carmeliet et al., 1999; Ng et al., 2001; Stalmans et al., 2002; Ishida et al., 2003; Ruiz de Almodovar et al., 2010; Darland et al., 2011). Taking advantage of these mice, we examined neural stem cell differentiation in the presence of altered Vegf isoform profiles in mice expressing only Vegf120 (120/120), only Vegf188 (188/188), or both Vegf120 and 188 (120/188). Since the Vegf164 isoform is the predominant form in the early forebrain (Darland et al., 2011), these Vegf isoform mice represent both loss of function (in the form of the Vegf164 allele) as well as altered localization based on the differential distribution of the Vegf isoforms in the microenvironment. The Vegf120/188 mice represent a putative functional rescue as the combination of isoforms provides both a local source of Vegf, the non-diffusible Vegf188, and a diffusible source in the non-HSPG binding Vegf120. We have previously shown that Vegf120 mice, lacking Vegf isoforms Vegf164 and Vegf188, have decreased Pax6 mRNA expression at embryonic day (E) 9.5 in conjunction with decreased proliferation in neuroepithelial cells along the ventricular zone border (Darland et al., 2011). However, we did not observe a quantitative reduction in Tbr2-positive cells at E11.5, suggesting that Vegf120 alone is sufficient to support formation of this early neuronal source population. Tbr2-positive cell populations contribute directly to formation of the upper neocortical layers (Hevner et al., 2001; Waclaw and Campbell, 2009; Bedogni et al., 2010). Changes affecting cell fate choice, whether to differentiate or proliferate, at any of these critical points could have profound effects on later neocortical layering specification (Hevner et al., 2001; Sessa et al., 2008).
In the current study, we have used mice expressing single or combinations of Vegf isoforms to investigate the effects of altered Vegf localization on developing cortical neuroepithelium. In essence, we used these mice to change the microenvironmental distribution of Vegf isoforms and assess the impact of these changes on the transcriptome profile as well as the subsequent effects on proliferation and differentiation in forebrain neuroepithelium. We propose that Vegf isoforms play a key role in modulating the coordinated investment of vasculature in the CNS along with the differentiation and proliferation of NSCs in the forebrain.
Materials and Methods
Generation of mouse embryos and genotyping
Vegf188, Vegf120, and Vegf120/188 transgenic mouse lines have been described in (Carmeliet et al., 1999; Ng et al., 2001; Stalmans et al., 2002). Vegf120/188 embryos were generated by crossing Vegf188 heterozygotes or homozygotes with heterozygous Vegf120 mice. Timed-pregnant mice (plug date, day 0.5) were used to generate embryos at E9.5, and E11.5. Postnatal day zero mice (P0.5) were collected within 12 hours of birth. Genotype was determined using purified genomic DNA obtained from tail cuts and standard PCR protocols described in (Carmeliet et al., 1996; Carmeliet et al., 1999; Darland et al., 2011). This study followed the NIH recommended guidelines for the care and use of animals in research and was approved by the University of North Dakota Institutional Animal Care and Use Committee (#0807-1c; #1204-3c).
Microarray and Differential Gene Expression Analysis
The two dorsal anterior bulges of the E9.5 telencephalon were dissected away from the forebrain with the top of the presumptive nasal process as the most anterior anatomical marker and the forward process of the diencephalon as the most posterior anatomical marker. The optic cup was not included in the dissected material and pial vessels were peeled away where possible. Total RNA was purified from the microdissected telencephalons as described in (Darland et al., 2011). Samples from two wild type, four Vegf188, and three Vegf120/188 were submitted to Genome Explorations Inc. (Memphis, TN) for processing and preliminary analysis. The RNA concentration was determined from the OD260/280 ratio and quality determined by capillary electrophoresis on an RNA 6000 Nano Lab-on-a-Chip kit and the Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). 15 µg of biotinylated cRNA were hybridized for 16 hr at 45°C on the GeneChip 430.2 mouse array (Affymetrix; GPL1261). GeneChips were washed and stained with streptavidin-phycoerythrin using the Affymetrix Fluidics Station 450, according to the manufacturer's protocol. Hierarchical clustering analysis was conducted using the PLIER (Therneau and Ballman, 2008) values for genes that were differentially changed at least 0.5 fold with t-test values ≤ 0.05. The log2-transformed values were mean centered prior to clustering analysis by the farthest neighbor method with Euclidean distance and Pearson Correlation as the similarity metrics. Previously published gene array data for wild type (n=4) and Vegf120 (n=4) E9.5 mice were also included in the overall analysis and batch adjusted to account for temporal variation in runs [(Darland et al., 2011); GEO record GSE30767].
Microarray Meta-Analysis
Data from six E9.5 and four E11.5 wild types microarrays .CEL files, from Hartl et al., 2008, were downloaded from the Gene Expression Omnibus (GEO record GSE8091) and were compared against our microarrays (6 E9.5 wild types, 4 E9.5 Vegf120, 4 E9.5 Vegf188, and 3 E9.5 Vegf120/188 mice; 4 of the E9.5 wild types and the 4 Vegf120 arrays were taken from [(Darland et al., 2011); GEO record GSE30767]. The E9.5 microdissection described by Hartl and coworkers (Hartl et al., 2008) was comparable to our method and the resulting transcriptomic analysis showed similar arrays of genes expressed in the multiple batches of E9.5 wild type samples. Principal Components Analysis revealed a batch variation effect, so raw expression values from all arrays were PLIER normalized and Log2 transformed. A MAS5 algorithm (Hubbell et al., 2002) was used to generate Presence/Absence calls for each gene, and probes with less than two “presence” hits were removed from further analysis. Samples were batch normalized using a non-parametric Empirical Bayes approach with a multivariate model (Batch, Age, and Genotype) using the R-script ComBat.R (Johnson et al., 2007). The BAMarray freeware (Ishwaran et al., 2006) was used to run a Bayesian-modeled ANOVA to test for significance using the E9.5 wild types as a baseline for comparison. Genes detected as significantly different from wild type were based on an adaptive test statistic that compared the increasing distance of the Zcut from zero as well as the posterior variance as it approached one. The Affymetrix annotation numbers for genes that were differentially expressed among the Vegf isoform samples relative to wild type were uploaded onto the Database for Annotation, Visualization, and Integrated Discovery (DAVID) and the enriched clusters identified with the mouse genome as the population background (Huang da et al., 2009; Huang da et al., 2009). The geometric mean of all the enrichment values for each gene per cluster is expressed as the enrichment score. The Expression Analysis Systematic Explorer (EASE) score is expressed as a p value (determined from a modified Fisher’s exact test) (Hosack et al., 2003) and reflects the significance of the enrichment score as a representation of a functional gene group relative to the expected representation based on the population background. Gene functional categories and individual networks of interest were identified based on high stringency settings and the EASE score (Hosack et al., 2003). Differentially expressed genes (DEGs) from key clusters were run through the Search Tool for the Retrieval of Interacting Genes (STRING 9.0; http://string-db.org) (Szklarczyk et al., 2011) to build interaction networks linking functional groups as well as individual genes.
Amplicon sub-cloning and Quantitative Real-Time PCR
Quantitative real time PCR was performed with SybrGreen detection on a StepOne Real Time PCR System (Applied Biosystems). Primer pairs were designed for each target and optimized for GC content (45–50%), base pair length (20–26 oligonucleotides), melting temperature (54–60°C), and amplicon size (75–450bp). Primer pairs were designed for each target gene and optimized using Primer Express Software (Applied Biosystems) and were selected based on minimal hairpin and dimerization secondary structures using Oligo Analyzer (Integrated DNA Technologies, Coralville, IA). The mRNA FASTA sequences were taken from the Entrez Gene website (http://www.ncbi.nlm.nih.gov/sites/entrez) and compared against those found on Ensembl Mouse Gene Viewer (http://www.ensembl.org/Mus_musculus/index.html); see Supplemental Table 1 for Primers). Target product was amplified from E9.5 and E11.5 wild-type neuroepithelium cDNA and subcloned into TOPO-TA-Sequencing (Invitrogen). Cloned products were sequence confirmed using BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) on an ABI Prism 3100 Genetic Analyzer. Validated products were amplified off the plasmid, gel purified (Qiagen) and quantified with a Nanodrop spectrophotometer. The analysis approach was based on the previously published protocol (Rhen et al., 2007) with modifications as described. Purified DNA product for each primer pair was used to generate 7-step standard curves plus a no-template control (only correlation coefficients of 0.98–0.99 were used) and the amplification efficiency was determined from the slope of the standard curve using the forumula: log10[−1/slope] – 1. The efficiencies for the primers used ranged from 85–100%. The CT of the product was corrected using the efficiency value determined for each primer pair and a melting curve was included in the run to check for uniform product formation based on the sequence-confirmed amplicon. This approach was used to calculate the amount of target cDNA, relative to the standard curve, transcribed from the mRNA for a target gene and expressed in attograms of DNA per 2.5 ng total RNA loaded into the original cDNA synthesis reaction. Amplification of 18S rRNA and glyceraldehyde-3-phosphate dehydrogenase (Gapdh) or 18S rRNA alone were run for each cDNA sample in parallel and run as a covariate in the analyses to control for variation in transcription efficiency and sample-to-sample variation. For comparison of qPCR and microarray data, we used the log2 values for the fold change relative to wild type and did linear regression for comparison. General statistical analyses were conducted using R (http://www.R-project.org), JMP (SAS Institute, Inc., Cary, NC), and GraphPad Prism 4.0 (GraphPad Software, Inc., La Jolla, CA).
Immunolabeling
Embryos were fixed in buffered 3.7% paraformaldehyde and equilibrated to 30% sucrose. Cryosectioning of Neg50 (Fisher Scientific/Thermo Scientific, Pittsburgh, PA) embedded tissue was cut with a Leica HM550 cryostat and immunolabeling completed as described (Darland et al., 2011). In brief, sections were blocked and permeabilized in 3% donkey serum, 2% goat serum (Vector Laboratories, Burlingame, CA), 0.1% Triton X-100, and 1% bovine serum albumin (BSA) in phosphate-buffered saline (PBS) overnight at 4°C. The primary antibody incubation was 2 hours at room temperature or overnight at 4°C. The absence of primary antibody or use of species-matched immunoglobulins were used as negative controls. Rabbit polyclonal antibodies were phospho-histone H3 (pHH3, 1:200, Upstate Biotechnology, Lake Placid, NY), Tbr2, and Ctip2 (1:400, Abcam, Cambridge, MA). For colormetric detection, biotinylated secondary antibodies (Goat anti-rabbit~biotin, 1:200; Jackson Laboratories) were incubated for 45 minutes at room temperature, followed by Vectastain Elite ABC horseradish peroxidase staining as per the manufacturer’s protocol. The immunolabeled sections were counterstained with methyl green. For immunofluorscent detection of primary antibodies, fluorochrome-coupled secondary antibodies were incubated in block solution for one hour at room temperature (cy3, 1:200; Jackson Immunoresearch Laboratories, West Grove, PA). Nuclei were labeled with DAPI (Sigma Chemical Company, St. Louis, MO).
Design-based Stereology
Design-based stereology was used to quantify pHH3- or Tbr2-positive nuclei as described (Darland et al., 2011). In brief, sections from six E11.5 wild type, four Vegf120, three Vegf188, and six Vegf120/188 mice were serially sectioned in the parasagittal plane at 30 microns with a ten-section interval. Primary antibody positive nuclei and total nuclei were counted using an Olympus BX51WI microscope with a motorized XYZ stage. Quantification was done using the Optical Fractionator workflow in StereoInvestigator 9.0 (Microbrightfield, Inc., Wiliston, VT). For the pHH3 counting, the contour outlined the neuroepithelium within 100 µm of the ventricular surface of the forebrain. For the Tbr2 counting, the contour outlined the pial and ventricular surface of the forebrain. Positive nuclei and total nuclei were counted in every 10th section in systematically-selected frames based on optical dissector frames and grid sizes empirically determined for each counting paradigm so that the coefficient of error was less than 0.1 (10%). The total numbers were estimated with the optical fractionator formula as previously described (Darland et al., 2011). Neuroepithelial volume was measured based on the total tracing area, the actual z-plane measured section thickness (to account for tissue shrinkage), and the section interval. The statistical analyses were performed using GraphPad Prism (GraphPad Software, Inc., LaJolla, CA) or JMP (SAS Institute Inc., Cary, NC). Comparisons between wild type, Vegf120, Vegf188, and Vegf120/188 mice were run using standard Analysis of Variance (ANOVA) with Dunnett’s post hoc test for comparisons of Vegf isoforms against a wild type baseline. Data shown are the mean +/− the standard error of the mean (SEM) with the number of replicates indicated in the figure legend.
Protein Quantification and ELISA Assays
Forebrain (presumptive telencephalon) and midbrain (presumptive diencephalon, including the ganglionic eminence) tissue from E11.5 embryos was microdissected and triturated in modified RIPA lysis buffer (25 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS) containing a cocktail of protease and phosphatase inhibitors (Sigma) and passed through 18 and 23 gauge needles to disrupt cells and shred genomic DNA. Non-soluble material was separated from the lysed material with a 10-minute centrifugation at 20,000 rcf and samples were stored at −80°C. Total protein from E11.5 brain tissue was determined with a Bio-Rad RCDC Protein Assay and loaded at the appropriate concentration for each ELISA detection assay. For detection of VEGF protein, 50 µg of total protein was loaded in a VEGF ELISA kit (R & D Systems) with the assay conducted following manufacturer’s instructions. The VEGF levels in E11.5 lysate samples were determined using a VEGF standard curve starting at 500 pg diluted to 1:1 seven times. The r2 for the standard curve was 0.9992 and the slope used to calculate the unknown concentrations within the linear range of the assay. Values are expressed as pg Vegf/µg total protein in the assay.
Western Blot
Protein lysates (10 µg) from the forebrain of E11.5 embryos were run on 4–20% SDS-PAGE gels (BioRad) and transferred to a nylon membrane with total protein determined by BioRad Protein Assay. The membranes were blocked overnight at 4°C with 5% non-fat dry milk, 0.05% Tween 20, in Tris-buffered saline (TBS). Membranes were incubated with Caspase 3 (1:1000, Catalog # 9662; Cell Signaling Tech, Beverly, MA) primary antibody diluted in TBS-tween for 1–2 hours at room temperature. Membranes were incubated with an alkaline phosphatase conjugated 2° antibody (Sigma) for one hour at RT. Bound antibody was visualized using CDPStar (Fisher Scientific, Pittsburgh, PA) as the enzyme substrate and emitted light captured with the UVP Imaging System (BioRad). Band pixel density was determined using ImageJ software (freeware available through National Institutes of Health website).
Results
Previous studies have established that Vegf and its isoforms are expressed throughout the neuroepithelium in early cortical development. In situ hybridization studies have localized Vegf expression in the neuroepithelium throughout embryonic cortical development (Dumont et al., 1995; Ferrara et al., 1996; Breir et al., 1995). Mice expressing the lacZ gene under the control of a Vegf promoter demonstrated nuclei positive for Vegf expression throughout the forebrain neuroepithelium in E9.5 mice (Darland et al., 2011). Expression of the Vegf isoforms, Vegf120, Vegf164, and Vegf188 has been detected by qPCR in the forebrain at E7.5, E9.5, and E11.5 (Darland et al., 2011) with Vegf164 as the primary isoform expressed at E9.5. We used Vegf ELISAs to compare total levels of Vegf protein in the forebrains of wild type, Vegf120, Vegf188, and Vegf120/188 mice at E11.5. We found that there was no statistical difference in total Vegf protein among any of the Vegf isoform mice relative to wild type (Supplemental Figure 1).
In order to clarify the role of Vegf and its predominant isoforms in early cortical development, we took the approach of analyzing transcriptome-level changes associated with altered Vegf isoform expression in the forebrain. This allowed us first to determine which genes were expressed in E9.5 neuroepithelium and then to identify possible regulatory networks developmentally linked downstream of altered Vegf expression. To this end, we isolated early neuroepithelium, removing the majority of surrounding pial vasculature from E9.5 wild type mice as well as mice expressing the Vegf188 isoform only, or a combination of the Vegf120 and Vegf188 isoforms (Vegf120/188). We chose the E9.5 time point in order to look at shifts in gene expression attributable to the changes in available Vegf isoform profile independent of vascular investment within the neuroepithelium of the forebrain. At E9.5 the pial vessels have elaborated on the surface of the developing forebrain, but significant sprouting into the primitive telencephalon does not occur until E11.5 (Bar, 1980; Risau, 1997; Vasudevan and Bhide, 2008). Therefore, the E9.5 time point represents a period during which the cells within the neuroepithelium are relatively homogenous, as the NSCs have yet to begin the rapid differentiation that occurs at later time points and the periventricular vasculature has not been elaborated. We did not detect shifts in VegfR2 (Kdr) or VegfR1 (Flt-1) in the microarray.
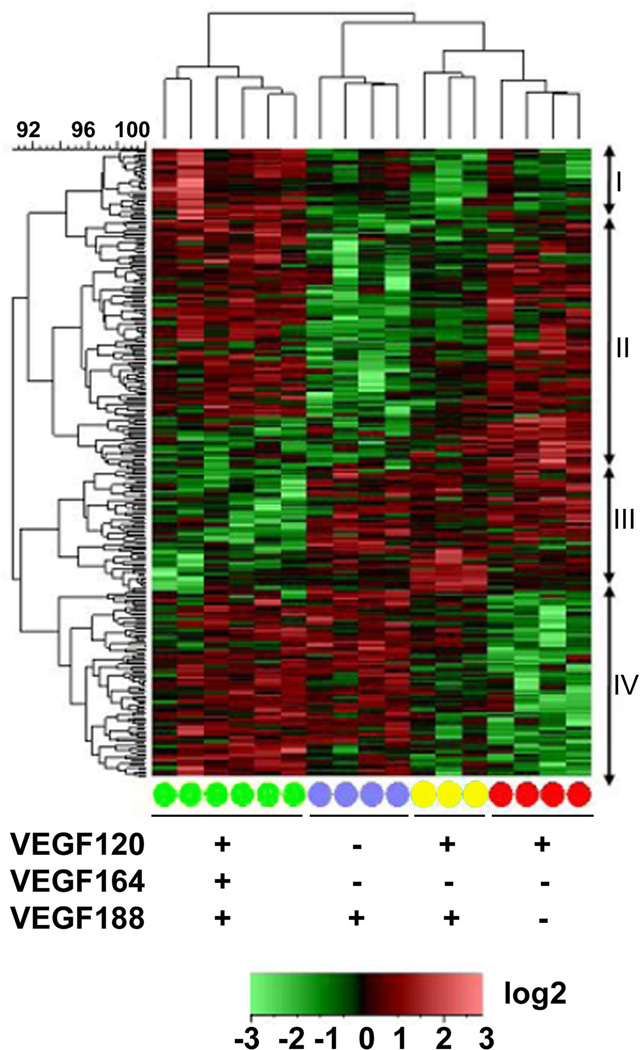
To our knowledge these microarrays represent the first transcriptome-wide assessment of the effects of Vegf misexpression in the primitive forebrain with the Vegf188 and Vegf120/188 isoform mice. To be as comprehensive as possible, we included previously published transcriptome data from Vegf120 and wild type arrays [(Darland et al., 2011); GEO record GSE30767] in our analysis. We conducted ANOVA analysis on PLIER- and batch-normalized array data sets from wild type and Vegf isoform mice to generate a heat map quantifying differentially-expressed genes (DEG) (Figure 1). With this analysis approach, genes were identified based on a Log2 fold change greater than 0.5 and a p-value of 0.05, or less. Based on this preliminary analysis we identified genes that were differentially expressed between wild type and Vegf120 (112 genes), Vegf188 (140 genes), and Vegf120/188 (152 genes). The annotated gene lists and their fold change relative to wild type in the array are provided in Supplemental Table 2. The patterns of gene expression were quite distinct among samples from mice differentially expressing Vegf isoforms, with different gene clusters down-regulated in the Vegf120 and Vegf188 mice, relative to the wild type mice. The pattern for the Vegf120/188 mice appeared intermediate between the Vegf120 and Vegf188 mice rather than a recapitulation of the wild type expression pattern.
Figure 1. Transcriptional profiling of embryonic forebrains from E9.5 Vegf isoform mice reveals differential expression patterns for single versus dual expressing isoforms.
Differentially expressed genes from Vegf188 (blue circles), Vegf120/188 (yellow circles), and Vegf120 (red circles) E9.5 forebrains were compared to wild type (green circles) samples using the Affymetrix mouse genomic chip 430.2 array. The log2-transformed PLIER values were row mean centered and the Complete Linkage algorithm, based on a Euclidean Distance similarity metric, was used to generate the heatmap for relative signals. Differentially expressed genes were identified based on a Log2 fold-change ≥ 0.5 and ANOVA p-value ≤ 0.05. The individual genotype samples were clustered based on a Pearson Correlation. Vegf120/188 (yellow, n=3) forebrain transcriptomes were clustered between the Vegf120 and Vegf188 transcriptomes, implying that the Vegf120/188 expression profile differs significantly from wild type (green, n=6) and was more closely related to the other Vegf isoform mice. The Vegf120/188 transcriptome phenotype appears to be a combination of the Vegf120 (red, n=4) and the Vegf188 (purple, n=4) transcriptomes, indicating that the presence of both a diffusible and non-diffusible isoform is not sufficient to rescue the wild-type transcriptome phenotype in the absence of the primary Vegf164 isoform. Cluster I shows increased expression in the wild type mice compared to the three isoform mice (32 probe sets). Cluster II showed a general decrease in expression in the Vegf188 and Vegf120/188 mice (111 probe sets). Cluster III showed a decrease in gene expression among the wild-type mice (55 probe sets). Cluster IV showed a decrease in expression amongst the Vegf120 and Vegf120/188 mice (83 Probe sets). Presence (+) or absence (−) of a Vegf isoform in a particular group is indicated below the heat map. Annotated gene information can be found in Supplemental Table 2.
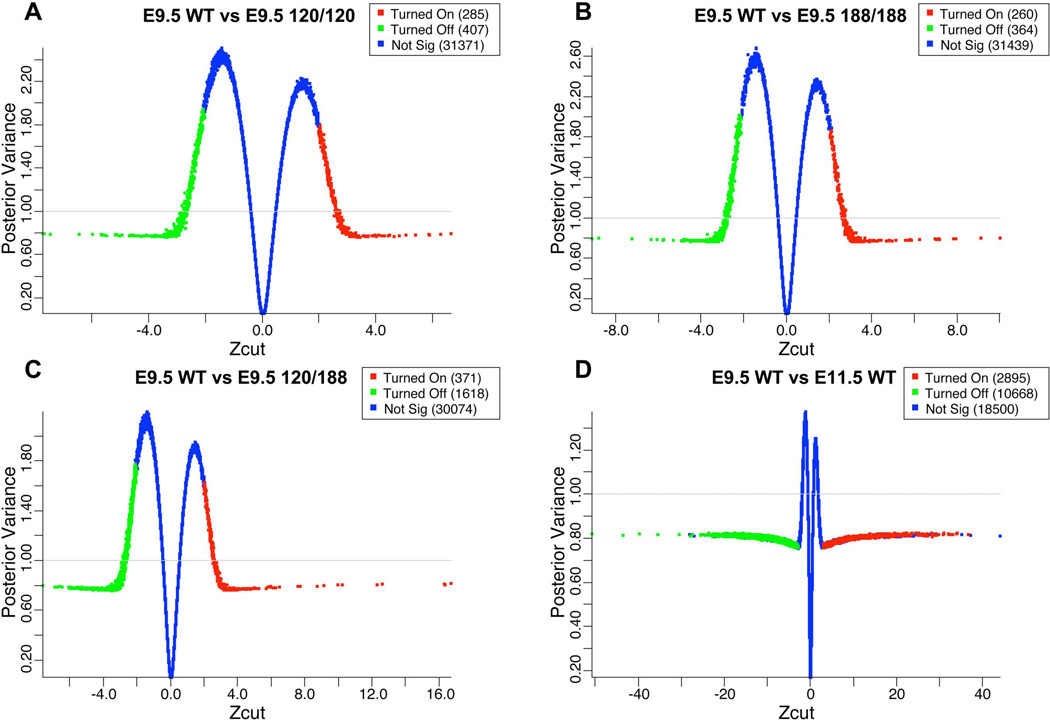
In a separate, more in-depth approach to analyzing the microarray data, we conducted a Bayesian analysis on batch- and PLIER-normalized data sets to potentially elicit additional differential gene expression patterns and enriched gene functional clusters. We chose this approach based on its ability to reduce the false discovery rate while maintaining acceptable statistical power in a highly dimensional data set with multiple batches (Ishwaran and Sunil Rao, 2008), as well to increase our statistical confidence in shifts in gene expression with low fold change. We compared our E9.5 wild type samples to each of the E9.5 Vegf isoform samples, as well as E9.5 and E11.5 wild types that had been previously published for a comprehensive early forebrain transcriptomic analysis by Hartl and coworkers (Hartl et al., 2008). This comparison included our E9.5 wild types, Vegf188 and Vegf120/188 samples (n=2, n=4 and n=3 respectively), and previously published E9.5 wild types, E11.5 wild types, and E9.5 Vegf120 samples (n=10, n=4, and n=4 respectively). The shrinkage plots generated by BAMArray analysis show differential gene expression as a Zcut (modified z-score) relative to the posterior variance of the comparison (Figure 2A–D). In the Vegf120 mice we observed 285 genes that were up-regulated and 407 genes that were down-regulated with respect to the E9.5 wild type samples. The Vegf188 mice showed the fewest genes statistically changed with 260 up-regulated genes and 364 down-regulated. The Vegf120/188 had the greatest number of genes shifted with 371 up-regulated and 1618 down-regulated. The E11.5 wild type mice had 2,895 genes up-regulated and 10,667 genes down-regulated, a reflection of the dramatic developmental changes occurring from E9.5 to E11.5 in the forebrain. As a control we compared the batch normalized E9.5 wild types from the three different technical batches, including our own (Darland et al., 2011) and that of Hartl and coworkers (Hartl et al., 2008) and found no differentially expressed probes (data included as Supplemental Figure 2D), demonstrating proof of principal for our technical and analytical approaches.
Figure 2. Meta-analysis of transcriptome shifts of embryonic forebrain in Vegf isoform mice compared against an E9.5 wild type background.
Microarray data from two previously published articles were downloaded from the Gene Expression Omnibus and incorporated into our analysis (Hartl et al., 2008); Darland et al, 2011). Unprocessed .CEL files were downloaded from GEO [6 E9.5 wild types, 4 E11.5 wild types from (Hartl et al., 2008), and 4 E9.5 wild type and 4 E9.5 Vegf120 from (Darland et al., 2011)] and analyzed with our microarray runs (2 E9.5 wild types, 4 E9.5 Vegf188, and 4 E9.5 Vegf120/188). All runs were PLIER and batched normalized, Presence/Absence filtered with MAS5, Log2 transformed, and tested for significance using a Bayesian modeled ANOVA run with BAMarray software and expressed as Zcuts relative to the E9.5 wild types. The genes that were significantly up-regulated (red) or down-regulated (green) were selected based on an adaptive test statistic that used the increasing distance of the Zcut from zero and the approach of the posterior variance as it neared one. The shrinkage plots shown are with the Zcut values graphed relative to the posterior variance. Significance is a reflection of distance from the 0 point for E9.5 wild type versus Vegf120 (A), Vegf188 (B), Vegf120/188 (C), and E11.5 wild type (D). Genes that were not significantly changed (blue, Not Sig) center around a Zcut of zero and an increasing posterior variance away from one. Supplemental Table 3 contains the specific information on the individual probes and their scores.
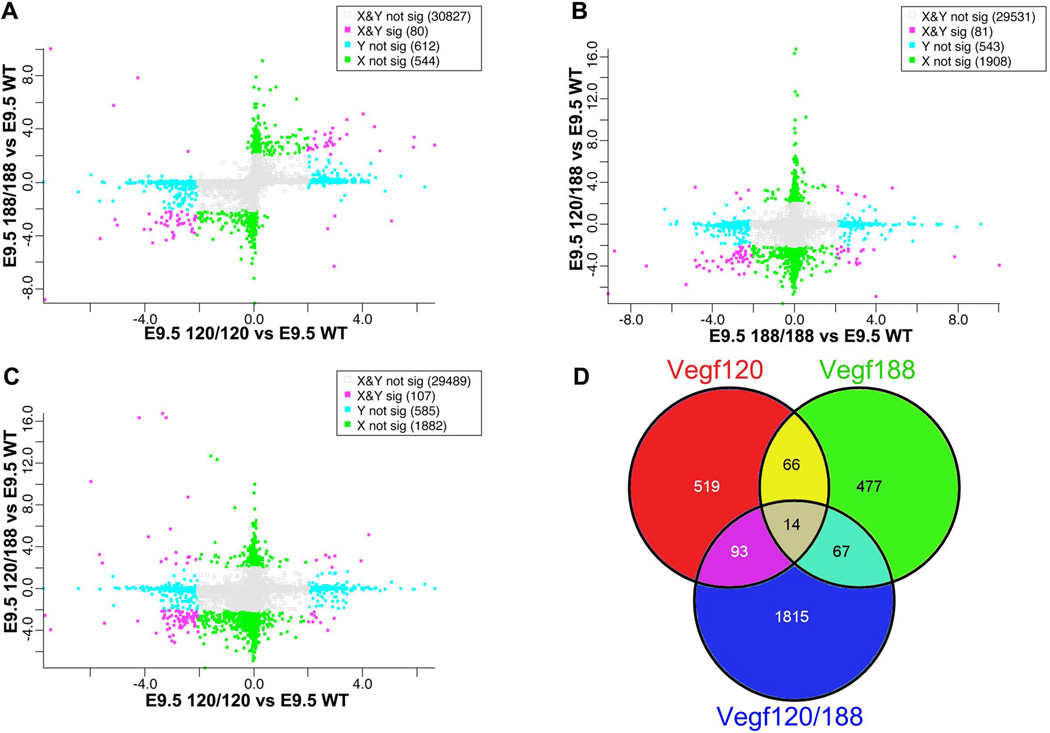
In addition to quantifying the differential gene expression pattern between each of the isoform mice and the wild type forebrain samples, we wanted to compare changes in gene expression profiles generated between the isoform mice relative to the critical shifts that occurred relative to E9.5 wild types to this end, we generated scatter plots of the Zcut values generated by group for each of the Vegf isoform mice relative to wild type. We then compared each group to each other (Figure 3 A–C). The Vegf120 and Vegf188 mice shared 80 DEGs. The Vegf120 and Vegf120/188 shared 107 DEGs while the Vegf188 and Vegf120/188 shared 81 genes (Figure 3D). When the Vegf isoform mice were compared to the E11.5 mice a large proportion of genes were shifted in the same direction (Vegf120 = 219 genes, Vegf188 = 92 genes, and Vegf120/188 = 662) (Supplemental Figure 2A–C). The differentially expressed genes identified by this approach are listed in Supplemental Table 3 along with their Zcut score and the direction of change in the array.
Figure 3. Cross-comparisons of transcriptional shifts between Vegf isoform forebrains.
Zcuts from different Vegf isoform group comparisons versus wild type were plotted on the X and Y axis. Scatter plots compare how pairs of Vegf isoform forebrains shift relative to E9.5 wild type (A–C). Genes are sorted based on being significant in one group but not the other (blue and green boxes), significant in both groups (red boxes), or not significant in either (grey boxes). Genes found in the upper-right and lower-left quadrants are shifted in the same direction relative to the E9.5 wild type base line. A Venn diagram shows differentially expressed genes (DEG) unique to the Vegf120 (red, 519 DEGs), Vegf188 (green, 477 DEGs), and Vegf120/188 (blue, 1815 DEGs), as well as DEGs common between the Vegf isoform mice independent of direction of change relative to wild type (D).
To validate our microarray analysis we performed qPCR on a subset of genes. Genes were selected for qPCR analysis based on the criteria that they were either detected as significantly changed in the microarray analysis, or have a well-established role in neurogenesis based on evidence in the literature. We collected total RNA from the neuroepithelial forebrains of wild type, Vegf120, Vegf188, and Vegf120/188 mice using the same dissection as described for the microarray collections. We quantified expression for two reference genes, Gapdh and 18S rRNA; three regulators of NSC proliferation and neurogenesis, Pax6, Neurogenin 2 (Neurog2), and Fez Family Zinc Finger 2 (Fezf2); one neural tube patterning morphogen, Sonic hedgehog (Shh); and two epigenetic modifiers, Suppressor of Zeste 12 (Suz12) and DNA (cytosine-5-)-methyltransferase 3 alpha (Dnmt3a). ANOVAs using the reference genes as covariates followed by a Tukey’s post test were used to test for significance. The group signal means, p-values, and fold changes versus wild type are consolidated in Supplemental Table 4. Gapdh and 18S rRNA were selected as reference genes to use as a basis for comparison in covariate analyses of selected genes of interest. One-way ANOVAs of 18S rRNA and Gapdh demonstrated that there were no significant shifts in expression among any group.
In assessing the DEGs we first quantified changes in Shh as it has been identified as critical to the dorsal-ventral patterning of the telencephalon (Rash and Grove, 2007; Komada et al., 2008; Li et al., 2009; Ferri et al., 2013) and was identified as shifted in the meta-analysis. In general, Shh is a ventralizing factor in the forebrain and its actions are mediated by the Gli family of transcription factors and Smoothened (Smo) [reviewed in (Hebert and Fishell, 2008)]. Our qPCR analysis demonstrated that the Vegf188 mice had a roughly fifty percent decrease in expression levels versus wild type and the Vegf120 mice (p < 0.1; Supplemental Table 4). No other groups were statistically different, matching what we observed with the microarray analysis. One of the mechanisms of Shh regulation of dorsal-ventral patterning is through the down-regulation of Pax6 (Ericson et al., 1997; Kioussi et al., 1999).
Pax6 is a well-established marker of neural stem cells in the ventricular zone of the developing cortex, and expression of this key transcription factor is linked to maintenance of the neural stem cell phenotype, regulation of intermediate progenitor genesis, and neurogenesis (Ericson et al., 1997; Osumi, 2001; Heins et al., 2002; Jones et al., 2002; Englund et al., 2005; Bayatti et al., 2008; Osumi et al., 2008; Sakurai and Osumi, 2008; Sansom et al., 2009; Suter et al., 2009). Despite the decrease in Shh expression, the microarray results demonstrated a general trend of decreased Pax6 expression in the Vegf isoform; however, the Bayesian ANOVA only detected the decrease as significant in the Vegf120/188 mice (Supplemental Table 3). The qPCR analysis corroborated the microarray values wherein Pax6 was reduced roughly 60% in all of the Vegf isoform mice relative to wild type (p < 0.05, Supplemental Table 4). Reduction in Pax6 may be indicative of a reduction in the early neural stem cell population, or a premature transition from a proliferative to a differentiative state.
NSC differentiation along a neural lineage is also regulated by Neurog2 and expression of Neurog2 is directly regulated by Pax6 (Stoykova et al., 1997; Scardigli et al., 2003). Correlating with decreased Pax6 expression, Neurog2 expression was significantly decreased in the Vegf188 and Vegf120/188 mice in the microarray. qPCR analysis confirmed the trend of decreased expression; however, it was only significantly different in the Vegf188 which had a 34% decrease relative to wild type (p < 0.05; Supplemental Table 4). Fezf2 is also a well-described regulator of differentiation, and is part of the network that can direct the differentiation of intermediate progenitors and early post-mitotic neurons in the neocortex (Shimizu et al., 2010). Both the microarray and qPCR demonstrated a trend of decreased expression in the Vegf isoform mice relative to the wild type; however, Fezf2 was not significantly changed in either analysis (Supplemental Table 3 and 4). qPCR expression levels in the Vegf120 mice were one-fold (Log2) lower than wild type (p = 0.18) and Vegf120/188 (p = 0.11).
NSC differentiation is dependent upon DNA methylation and epigenetic modification of the genome. These processes can regulate the transition from a proliferative NSC to a differentiating cell and a variety of modifying factors that impact transcription have been identified to date (reviewed in (Hu et al., 2012)). Among these is Suz12, a member of the Polycomb Repressive Complex 2 (PRC2), that serves to regulate histone methylation (Pasini et al., 2004) resulting in direct consequences for transcription of target genes. Suz12 is expressed in the neocortex during development, and heterozygous gene knockout results in neural tube defects and brain malformations (Miro et al., 2009). Microarray analysis showed a significant increase in Suz12 expression in the Vegf120 mice with no significant changes for the Vegf188 or Vegf120/188 mice. In support of these results, qPCR analysis indicated that Vegf120 and hVegf120/188 mean values were increased relative to wild type and Vegf188; however, the differences were not statistically significant (Supplemental Table 4). Another epigenetic modifier, Dnmt3a, regulates gene expression through de novo methylation of CpG islands. Dnmt3a works in parallel with PRC2 to regulate gene methylation during developmental neurogenesis (Wu et al., 2010). Transition from expression of Dnmt3b to Dnmt3a is an indicator of neuronal differentiation, and marks the transition of an intermediate progenitor becoming an early post mitotic neuron (Watanabe et al., 2006). Microarray analysis detected a significant decrease in Dnmt3a expression in the Vegf 188 mice, although no other groups were detected as significantly changed. qPCR quantification demonstrated that the Vegf188 mice, indeed, had a three-fold (Log2) lower expression compared to wild type, qPCR analysis also showed a 2.5 (Log2) fold lower expression the Vegf120 and Vegf120/188 (p < 0.05; Supplemental Table 4).
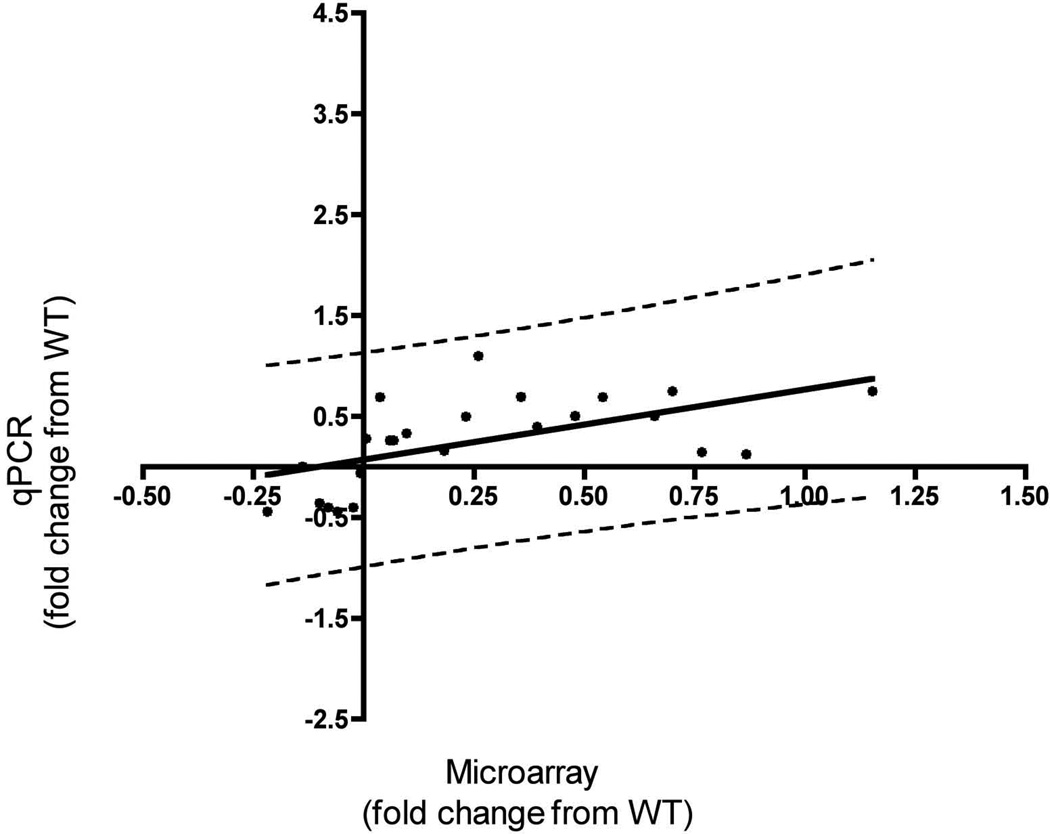
To determine whether our qPCR results were comparable to the changes observed in the microarray meta-analysis, we conducted a correlation analysis using a subset of 7 genes (Gapdh, SHH, Suz12, Dnmt3a, Pax6, Neurog2, and Fezf2) for each of the three genotype (WT versus Vegf isoform mice) comparisons. We compared Log2 fold changes of qPCR expression values (wild type versus each of the Vegf isoform mice) to their corresponding Log2 fold changes in the microarray expression values. We conducted a Spearman correlation analysis, and determined that the shift in relative expression as detected in our microarray positively correlated with the values determined by qPCR (Figure 4, Spearman’s r = 0.693, p < 0.0002, m = 0.71), confirming the qPCR validation of our array results. All the data are expressed as log2 values for comparison.
Figure 4. qPCR confirmation of the meta-array analysis.
qPCR was conducted for 7 genes and the Log2 fold change between wild type and individual Vegf isoforms (Y-Axis) was plotted against the Log2 fold change of their corresponding microarray probe expression values (X-Axis). The scale on both axes is Log2. The solid black line represents the regression line (m = 0.71) and the dashed lines indicate the 95% confidence intervals. These latter values were used to remove outliers. We ran a Spearman correlation analysis on 24 (black circles) of the 27 pairs and obtained a positive r-value of 0.693 (p < 0.0002). Quantification and statistical analysis of the qPCR runs can be found in Supplemental Table 4.
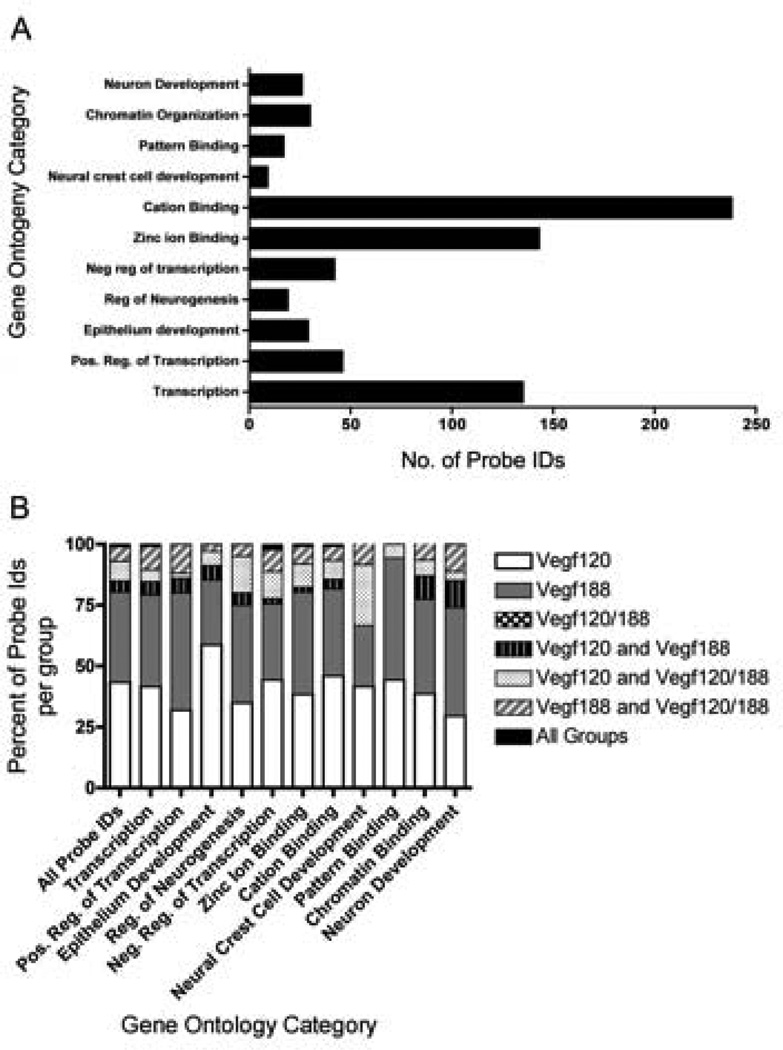
In order to characterize the functions of genes differentially expressed in the meta-array analysis among the Vegf isoform mice, we utilized the functional annotation clustering platform on the Database for Annotation, Visualization, and Integrated Discovery (DAVID) platform to identify functionally related gene groups. DAVID identifies Gene Ontology categories that are over-represented amongst a list of genes. Affymetrix Probe Ids of differentially expressed genes detected in the E9.5 isoform mice were uploaded into DAVID. Genes were identified in eleven Gene Ontology (GO) categories using an enrichment score filter of 2.0 (Figure 5A–B, Supplemental Table 5). The first functional annotation cluster identified had the GO category transcription (134 probe ids). Within this cluster there were several probes identified for their roles in cell fate choice, Notch3, and neural development, Dachshund 1 (Dach1) and Dachshund 2 (Dach2). The second annotation cluster, GO Category positive regulation of transcription (46 genes), included Meis homeobox 2 (Meis2), Distal-less homeobox2 (Dlx2), Sonic hedge hog (Shh), Sry-box 4 (Sox4), Bone morphogenetic protein 4 (Bmp4), and Neurogenin 2 (Neurog2). GO category regulation of neurogenesis (19 probes) identified many of the genes listed above, but in addition included Inhibitor of differentiation 4 (Id4), Distal-less homeobox 1 (Dlx1) and Neuropilin 1 (Nrp1). The two functional annotation clusters with the highest numbers of identified probes were zinc ion binding (143 probe ids) and cation binding (238 probe ids). Within these categories were many transcription factors, cell signaling ligands, matrix interacting proteins, and epigenetic regulators, including members of the Notch signaling pathway, Jagged 1 and Jagged 2. Several ECM and ECM-interacting proteins were identified within these clusters including Versican, and Integrin-alpha 6. The GO category neuron development (26 probe ids) contained several genes involved in neuronal migration and axon path finding, including chemokine receptor 4 (CXCR4), Eph receptor A7 (Epha7), Microtubule-associate protein 1B (Mtap1b), and Doublecortin-like kinase 1 (Dclk1). Another interesting functional annotation cluster had the GO category chromatin organization (30 probe ids). Genes in this category included histone deacetylase (Hdac9) as well as the methyl-transferases, Set domain 8 (Setd8), DNA-methyltransferase 3 alpha (Dnmt3a), and Suz12.
Figure 5. Enriched Gene Ontology Categories in Vegf isoform mice.
DAVID analysis identified GO categories that were overly represented amongst the differentially expressed genes. Affymetrix probe identification numbers (Ids) of DEGs that were detected as significantly changed by BAMarray in the Vegf120, Vegf188, and Vegf120/188 microarray meta-analyses were uploaded as a single list into DAVID. Functional Annotation Clustering was run on the high setting with the Bonferroni adjustment and GO Terms with an Enrichment Score within a range of 2 to 6 and p-values less than 0.01 were reported. 417 unique differently expressed genes were identified by DAVID with GO categories that were statistically over-represented. The bar graphs represent the number of probe Ids for each functional category. Specific probe Ids, as well as, the DAVID statistics are located in Supplemental Table 5 (A). The gene contribution from each group of Vegf isoform mice into the various GO categories were broken down into their relative proportions. Of these genes roughly 40% were found changed in either the Vegf188 mice only, or Vegf120 mice only. No genes were identified by DAVID as being unique to the Vegf120/188 mice (B).
Quantitative differences in NSC of developing Vegf isoform mice
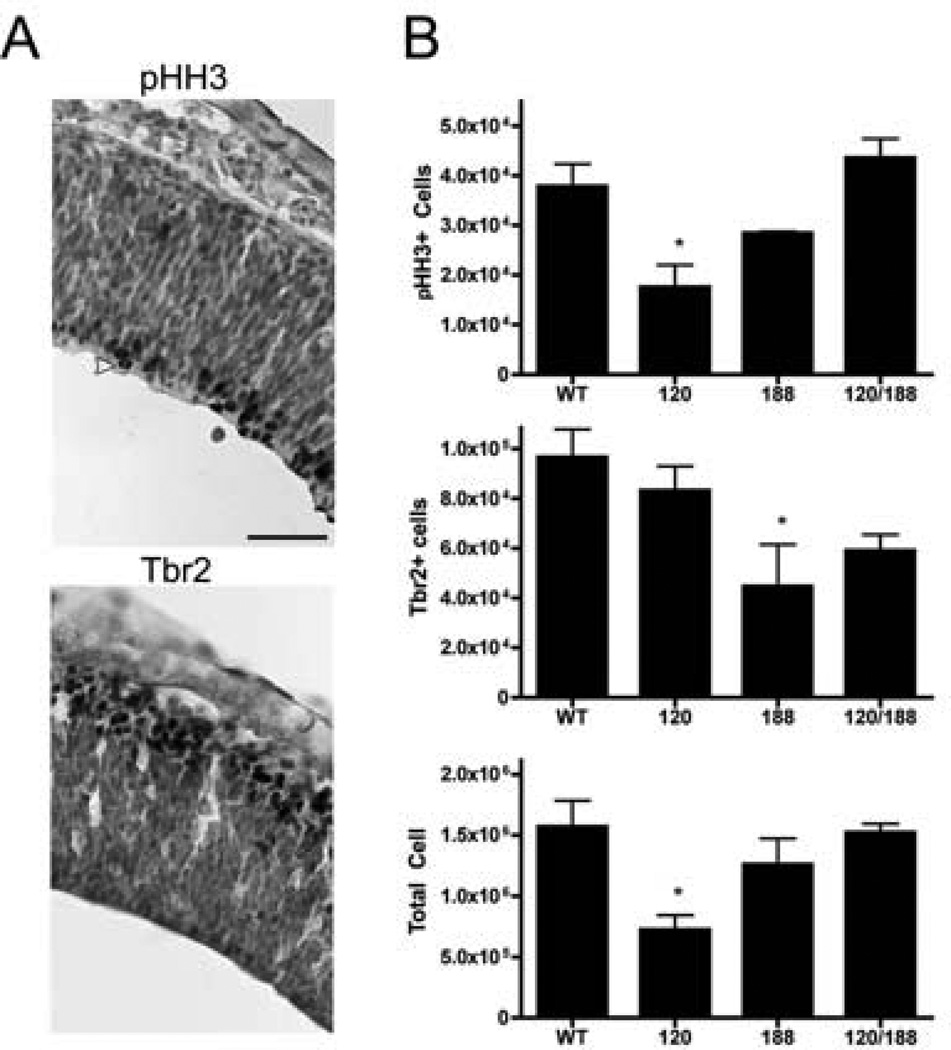
Because our data mining efforts from early forebrain sources had yielded a significant number of genes that had previously been associated with cell fate choices in NSC, we wanted to determine the consequence of a shift in Vegf isoform profile on subsequent neuroepithelial development. Therefore, we quantified proliferating cells in the ventricular zone in parallel with the early differentiating cells at E11.5. We immunolabeled mitotic cells within the ventricular zone using pHH3 and identified the early differentiating populations of NSCs using Tbr2 (Figure 6). At this stage, Tbr2 marks cells that have differentiated into either early post-mitotic neurons and Cajal-Retzius cells of the pre-plate, or early intermediate progenitors of the subventricular zone (Hodge et al,. 2013; Hevner et al., 2002; Englund et al., 2005; Kowalczyk et al., 2009). In either case Tbr2 marks cells that have transitioned from a Pax6-positive cell dividing along the ventricular surface to a cell that has differentiated and migrated towards the pial surface. We labeled all nuclei with a methyl-green counterstain and quantified volume and total cell number. Forebrain volume and total cell number were decreased in the Vegf120 mice as previously reported, but was not significantly changed in the other Vegf isoform mice (Darland et al., 2011). We counted proliferating NSCs within the putative Pax6-positive ventricular zone in E11.5 forebrains from the Vegf isoform mice. (Osumi, 2001; Darland et al., 2011). We previously reported that the Vegf120 mice had a decreased pHH3-positive NSC population as well as a decrease in total neuroepithelial cells (Darland et al., 2011). Despite this decrease in overall cell numbers, the Vegf120 mice maintained an equivalent number of Tbr2-positive cells compared to wild type. Therefore, additional wild types (n=6) and Vegf120 (N=4) were counted as well as Vegf188 (N=3) and Vegf120/188 mice (n=6). Our results were consistent with previous stereological counts for Vegf120 mice, with a decrease in overall neuroepithelial cells, and pHH3-positive cells, compared to wild type (P < 0.05). The Vegf120 isoform mice had decreased pHH3-positive cells compared to the Vegf120/188 mice (P < 0.05). The Vegf188 showed no significant changes in the total number of neuroepithelial cells or pHH3-positive cells, but the Vegf188 did have a significant decrease in Tbr2-positive cells compared to wild type (P < 0.05). The Vegf120/188 mice were not statistically different from the wild type mice in pHH3-positive, Tbr2-positive cells, or total neuroepithelial cell number.
Figure 6. Vegf regulation of NSC proliferation and differentiation.
E11.5 wild types, Vegf120, Vegf188, and Vegf120/188 forebrains were sectioned in the parasagittal plane and immunolabeled with either pHH3 (A) or Tbr2 (B) and counter-stained with methyl-green. Representative images of a rostral portion of the telencephalon stained with pHH3 and Tbr2 are shown here in the wild type mouse forebrain (400× Magnification, Scalebar = 25 um). The E11.5 forebrains showed no major differences in the distribution of pHH3- or Tbr2-positive cells among the Vegf isoform mice or wild type (Data not shown). A yellow arrowhead indicates a NSC dividing along the ventricular surface, and the red arrowhead shows a basal mitosis of a cell dividing away from the ventricular surface. Counts of pHH3-positive, Tbr2-positive cells, and total neuroepithelial cells of the forebrain were completed using Stereoinvestigator (B). Groups were tested for significant differences using a 1-way ANOVA followed by a Dunnett’s post-test using wild types as the control. The Vegf120 mice showed a significant reduction in total neurolepithelial cells and pHH3+ cells versus wild type. (p < 0.05, wild type n = 6, Vegf120 n = 4) The Vegf188 mice showed reduced numbers of Tbr2+ cells compared to wild type (p < 0.05, Vegf188 n = 3). The Vegf120/188 mice were not statistically significant from wild type in any category (p > 0.05, n = 6). Errors bars represent standard error of the mean (Red error bars were included in the pHH3 graph for clarity).
To determine if the decrease in total neuroepithelial cell number in the Vegf120 forebrain was due to an increase in cell death at E11.5, we examined protein levels of cleaved and uncleaved Caspase 3 using western blots. Cleavage of Caspase 3 is associated with the initiation of apoptotic pathways (Janicke et al., 1998; Hemachandran et al., 2002; Ouyang et al., 2012). We found that there was no difference in the ratio of cleaved Caspase 3 to Pro-Caspase 3 among or between any of the wild type or Vegf isoform mice (Supplemental Figure 4) at E11.5.
Given the changes we saw at E11.5 in the proliferating and differentiating NSC populations we wanted to determine if there were any overt consequences to early cortical layering. We conducted a histological survey at P0, an embryonic time point at which early cortical layers V and VI have been initiated but the boundaries for the upper layers are not yet clearly defined (Price et al., 2006). Analysis of hematoxylin-stained P0 sections revealed marked differences between the wild type and Vegf isoform mice and a representative series is shown (Supplemental Figure 5). At the caudal-most end of the lateral ventricle, just prior to the emergence of the hippocampus and dentate gyrus, we observed a qualitative reduction in several of the presumptive cortical layers in the Vegf isoform mice compared to wild type. The Vegf120 mice had increased cerebrovascular hemorrhage associated with their decreased vascular integrity and early postnatal lethality as has been previously reported (Carmeliet et al., 1996; Ruhrberg et al., 2002; Stalmans et al., 2002).
In order to look more specifically at the changes in cortical layering we tracked expression of Coup-TF interacting protein 2 (Ctip2), a factor that has been associated with cell fate choice in upper cortical layers (Molyneaux et al., 2007; Fishell and Hanashima, 2008). Ctip2 immunolabeling of the caudate putamen revealed comparable patterns of distribution with positive nuclei clustered ventral and medial to the developing cortical layers (Supplemental Figure 6). In contrast, the Ctip2-positive nuclei of cortical layer V were distributed across a wider band in the VEGF188 mice and VEGF120/188, relative to the wild type (Supplemental Figure 6).
Discussion
Our results support the hypothesis that the composition of Vegf isoforms present in the microenvironment of the forebrain contributes to early cortical neurogenesis. Vegf and its receptors are expressed at the proper times and places to coordinate the establishment of the vasculature and development of NSCs in the forebrain (Darland et al., 2011). We were particularly interested in the E9.5 developmental stage as a crux point for gene expression changes responsible for laying the groundwork for subsequent NSC differentiation and eventual neocortical layering. An advantage of this early time point is that it occurs before extensive vascularization of forebrain neuroepithelium. The blood vessels at the pial surface have not significantly invested the neuroepithelium at E9.5, which, in turn, reduces the confounding vascular variable for interpreting the gene expression results.
The expression shifts detected in the Vegf isoform mice transcriptomes provided evidence that early forebrain neuroepithelium is responsive to altered Vegf bioavailability. The neuroepithelial transcriptome expression patterns are unique amongst the different Vegf isoform mice, demonstrating that each Vegf isoform has the capacity for eliciting different downstream effects on gene expression. The transcriptome expression patterns in the Vegf120 and Vegf188 mice were unique. The Vegf120/188 mice displayed a transcriptome profile that was a blend of the Vegf120 and Vegf188 expression patterns, rather than a recapitulation of wild type expression. This evidence implies that Vegf120/188 mice are not a rescue of the wild type phenotype, at the transcriptome level, but rather represent a ‘loss of function’ phenotype with respect to the primary Vegf164 isoform. The Vegf120/188 does rescue the Vegf120 phenotype, in that the Vegf120/188 mice are overtly healthy and viable, whereas their Vegf120 counterparts are early post-natal lethal. Therefore, addition of a single allele of Vegf188 codes for sufficient protein to allow post-natal development to proceed in these mice.
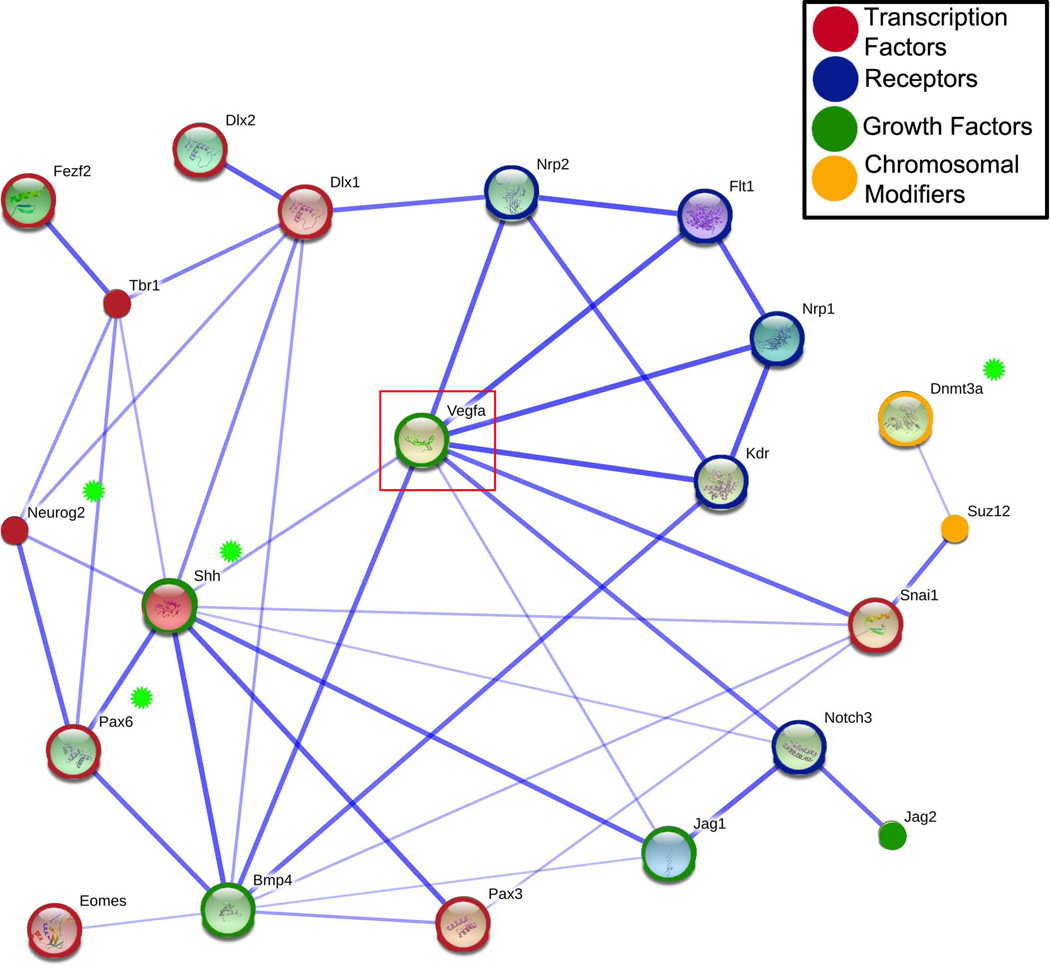
The microarray results provide evidence that altering Vegf bioavailability in the microenvironment can alter NSC fate decisions. Many of the differentially expressed genes detected in the meta-array analysis and identified as enriched by DAVID with regard to GO function can be linked directly to processes involved in regulation of cell fate choice and NSC differentiation. A predicted protein interaction network, generated with the Search Tool for the Retrieval of Interacting Genes (STRING 9.0; Figure 7) and using genes mined by DAVID clustering as well as several other genes of interest (Vegf, VegfR1, VegfR2, Tbr1, and Tbr2), provided an additional platform to identify potential pathways downstream of Vegf isoform mediated changes related to neocortical development. The STRING network demonstrated a potential role for Vegf as a bridging factor in neurovascular development by linking several factors important to both vascular patterning and neural development, such as Nrp1, Nrp2 and Notch3. Genes were identified with links to Vegf that were functionally associated with NSC specification and fate choice, migration, and epigenetic modification.
Figure 7. Predicted protein interactions network with a subset of differentially expressed genes identified in the Vegf isoform mice.
We used the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) to generate evidence-based links between and among genes identified from DAVID analysis of the top enriched gene clusters. Gene acronyms were incorporated into STRING using default settings. The components in this STRING model were simplified from a network originally constructed from genes identified as over-represented in functional categories as determined by the DAVID analysis. Blue lines indicated direct evidence linking proteins with the thickness of the line reflecting confidence of interactions based upon the number of database links. The lengths of the lines, the inner color and size of the nodes have no significance in this schematic (Small nodes indicate that no protein structure data was available). To aid in network building, we incorporated five additional genes into the STRING model: Tbr1, Tbr2/Eomes, Flt1/VegfR1, Kdr/VegfR2, and the VegfA (Vegf). Transcription factors have been circled in red, receptors in blue, growth factors in green, and chromosomal modifiers in gold. Genes of particular interest are indicated with a green star (Neurog2, Shh, Pax6, and Dnmt3a). Vegf is boxed to highlight its key position within the evidence network. The STRING network links differentially expressed genes in the Vegf isoform mice with Vegf and its receptors, providing new directions for study involving Vegf regulation of cell fate choice. Potential effectors for modifying epigenetic regulation of gene expression, Suz12 and Dmnt3a, were networked to Vegf through Snai1 or Shh. Pax6, a key regulator of NSC proliferation and neurogenesis, links to Vegf indirectly through Shh and Bmp4. Pax6 is in turn linked to regulators of differentiation Neurog2 and Dlx1, as well as markers of differentiation, Tbr1 and Tbr2.
The STRING network linked Vegf and its receptors to factors essential to forebrain patterning and specification, Bmp4 and Shh (Ericson et al., 1995; Ericson et al., 1995). Both Shh and Bmp4 are well known for their abilities to specify cell fate based on their signaling gradients and distribution in the microenvironment. These two genes directly linked to other genes involved in the regulation of NSC proliferation and differentiation (i.e Notch3, Jag1, Jag2, Pax6 and Neurog2). Pax6 is necessary for proper differentiation and stem cell fate choice in the developing forebrain (Sansom et al., 2009; Larsen et al., 2010). Reduced or lost expression of Pax6 at this early time point would potentially result in changes in forebrain patterning including establishment of the pallial-subpallial border (Georgala et al., 2011; Duan et al., 2012). Pax6-positive cells shift their expression profile to become Tbr2-positive cells as they differentiate and migrate away from the ventricular zone. These cells, in turn, express other markers of differentiation as they concomitantly migrate and stratify into the six layers of the neocortex. One such example is the differentiation of Tbr2-positve cells to Tbr1-positive cells, at E12.5–13.5, and their subsequent migration into layer VI (Hevner et al., 2001). Levels of Pax6 are particularly important in maintaining the balance between NSC proliferation, maintenance, and differentiation. Both gain of function and loss of function studies of Pax6 demonstrate that altered Pax6 results in altered neurogenesis and directly affects the number of the upper layer cortical neurons (Stoykova et al., 1997; Jones et al., 2002; Guillemot et al., 2006; Quinn et al., 2007; Sansom et al., 2009; Tuoc et al., 2009). Neurog2 is important in the differentiation of neurons and its developmental expression is directly regulated by Pax6 (Scardigli et al., 2003). Both Neurog2 and Dlx2 contribute to NSC fate choice and interact through Notch signaling (Yun et al., 2002; Scardigli et al., 2003; Ochiai et al., 2009). Pax6 and Neurog2 regulate NSC exit from the cell cycle, which in turn affects differentiation of upper layer neurons (Quinn et al., 2007; Tuoc et al., 2009). Shifts in expression of Pax6, Neurog2, Dnmt3a, Dlx1, and Dlx2 detected in the E9.5 microarrays provide evidence that the early NSC populations are vulnerable to changes in the Vegf isoform profile with regard to their cell fate decisions. Loss of Dnmt3a is linked to reduced expression of Neurog2 and Dlx2, both of which are genes critical to neurogenesis (Feng et al., 2007; Wu et al., 2010). Any factor affecting genes involved in these early stepwise NSC fate decisions would have adverse consequences on subsequent neocortical development.
Supporting this idea is the broader distribution of layer V Ctip2-positive cells at P0 in the VEGF188 and VEGF120/188 mice, implying a role for VEGF in either the establishment of layers in the neocortex by directly influencing the migration and localization of differentiating NSCs. Supporting this idea of VEGF misexpression altering post-mitotic neuronal migration is our microarray data that identified Dlx1, Dlx2, and CXCR4 as differentially expressed in the VEGF isoform mice compared to wild type. Knock-out mice for these genes have demonstrated significant changes in interneuron migration and neocortical lamination(Anderson et al., 1997; Stumm et al., 2007; Wang et al., 2011) further highlighting their critical role in cell fate decisions.
Taken together, our transcriptomic data analyses lend supporting evidence linking an altered pattern of Vegf isoform expression to shifts in genes controlling NSC specification and determination with consequences for balancing NSC differentiation and proliferation. Vegf120 mice have reduced neuroepithelial volume, total cell number, and reduced pHH3-positive proliferating cells with unaltered Tbr2-positive populations [(Darland et al., 2011), Figure 6B] with the implication that non-diffusible Vegf isoforms are necessary to maintain NSC proliferation. The Vegf188 and Vegf120/188 mice had statistically equivalent levels of pHH3-positive cells compared to wild type in agreement with the idea that a localized source of Vegf may be necessary for maintenance of NSC proliferation. The Vegf188 forebrain had reduced numbers of Tbr2-positive cells, the loss of which may be correlated with the reduced expression of Pax6, Neurog2, and Dnmt3a in the Vegf188 isoform mice. Pax6-positive cells contribute directly to the genesis of Tbr2-positive cells, so a reduced Pax6-positive population would have significant consequences for intermediate progenitor cells and differentiation of post-mitotic neurons (Quinn et al., 2007). The reduction in Tbr2-positive cells at E11.5 in the Vegf188 mice may represent a failure of Pax6-positive neural stem cells to differentiate into Tbr2-positive cells, or an early departure of the Tbr2-positive cell populations down a further differentiated cell fate choice (i.e. Tbr1-positive). The Vegf120/188 mice resembled wild type mice with regard to numbers of pHH3- and Tbr2-positive cells indicating that the presence of both diffusible and locally-retained Vegf isoforms may act as a functional rescue in animals lacking the Vegf164 isoform. However, the evidence for the Vegf120/188 mice as a rescue animal may not be that clear cut, given that the Vegf120/188 mice showed the largest numbers of altered genes in the microarray compared to the E9.5 wild types. The gene expression pattern of the Vegf120/188 isoform mice was closer to that of the Vegf120 and Vegf188 mice than it was to the wild type mice. This intermediate pattern of expression may reflect a level of phenotypic plasticity in the Vegf120/188 mice that shifts gene expression in an effort to develop the normal forebrain structure in the absence of Vegf164.
The role of Vegf in the coordination of neurovascular development is complex. Vegf and its receptors are expressed by both neuronal and vascular cell types during cortical development, and many of Vegf’s downstream effectors have been shown to play a role in both angiogenesis and neurogenesis. Observing the direct effects of Vegf on neural development is often confounded by potential indirect effects caused by the impaired vasculature. However, despite these difficulties, it is clear that the Vegf isoforms plays an important role in the development of cortex. By looking early on in development, we minimized indirect effects of abnormal vasculature and identified Vegf isoform-specific shifts in the expression of genes important to NSC differentiation independent of vascular compromise. We demonstrate that the Vegf isoform profile does affect NSC proliferation and differentiation, with the loss of diffusible isoforms (Vegf120 and Vegf164) resulting in decreased Tbr2-positive cell differentiation and the loss of non-diffusible (Vegf188 and Vegf164) isoforms causing reduce proliferation at the ventricular surface. Our findings imply that the Vegf isoforms serve to guide and fine tune NSC development by mediating the balance of their proliferation and differentiation.
Supplementary Material
Acknowledgments
The authors thank Elvira Tkach, and undergraduate students Kayla Nelson, Carol Two Hawk, Abigail Neff, and Maxwell Maltese of the UND Biology Department for their technical support. The Vegf isoform mice were a generous gift from Dr. Patricia D’Amore of the Schepens Eye Research Institute and the Departments of Ophthalmology and Pathology at the Harvard Medical School. The authors thank Dr. Joachim Klose, Director of the Proteome Research Group Professor at the Institute of Human Genetics at the Charité - University Medicine Berlin for the generous use of his transcriptomic data in our analyses (Hartle et al. 2008). The authors acknowledge the use of the University of North Dakota School of Medicine and Health Sciences Imaging core that is supported, in part, by a COBRE award through NIH/NCRR, P20RR017699. The authors wish to acknowledge the help of the faculty startup funds from the University of North Dakota Biology Department, College of Arts and Sciences and the North Dakota EPSCoR program. These studies were supported by NIH-NINDS R15 NS057807-01/-02 (DCD), 3R15NS057807-01S1 (DCD), and an American Heart Association pre-doctoral award AHA0715542Z (JTC).
Footnotes
Competing Interests
The authors declare that they have no competing interests.
Author’s Contributions
JTC and DCD together conceived the study, carried out the experiments, completed the analyses, and wrote the manuscript. JTC conducted all the stereological counts and analyses. JTC conducted the meta-analysis on the multi-batch array data and worked with DCD to analyze and present the results. MAB helped in the original RNA purification, genotyping, and quality control for the batch microarray samples. SDS, NFC, and SAN worked with JTC and DCD to complete the qPCR assays. GJS conducted the caspase westerns as part of his undergraduate Honors Thesis and worked with DD to analyze the data. All authors made intellectual contributions to the research, edited the final version of the manuscript, and are able to defend the work.
References
- Anderson SA, Eisenstat DD, Shi L, Rubenstein JL. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997;278:474–476. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- Bar T. The vascular system of the cerebral cortex. Advances in anatomy, embryology, and cell biology. 1980;59:I–VI. 1–62. doi: 10.1007/978-3-642-67432-7. [DOI] [PubMed] [Google Scholar]
- Bayatti N, Sarma S, Shaw C, Eyre JA, Vouyiouklis DA, Lindsay S, Clowry GJ. Progressive loss of PAX6, TBR2, NEUROD and TBR1 mRNA gradients correlates with translocation of EMX2 to the cortical plate during human cortical development. The European journal of neuroscience. 2008;28:1449–1456. doi: 10.1111/j.1460-9568.2008.06475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedogni F, Hodge RD, Elsen GE, Nelson BR, Daza RA, Beyer RP, Bammler TK, Rubenstein JL, Hevner RF. Tbr1 regulates regional and laminar identity of postmitotic neurons in developing neocortex. Proceedings of the National Academy of Sciences. 2010;107:13129–13134. doi: 10.1073/pnas.1002285107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borello U, Pierani A. Patterning the cerebral cortex: traveling with morphogens. Current Opinion in Genetics & Development. 2010;20:408–415. doi: 10.1016/j.gde.2010.05.003. [DOI] [PubMed] [Google Scholar]
- Carmeliet P, Ferreira V, Breier G, Pollefeyt S, Kieckens L, Gertsenstein M, Fahrig M, Vandenhoeck A, Harpal K, Eberhardt C, Declercq C, Pawling J, Moons L, Collen D, Risau W, Nagy A. Abnormal blood vessel development and lethality in embryos lacking a single VEGF allele. Nature. 1996;380:435–439. doi: 10.1038/380435a0. [DOI] [PubMed] [Google Scholar]
- Carmeliet P, Lampugnani MG, Moons L, Breviario F, Compernolle V, Bono F, Balconi G, Spagnuolo R, Oosthuyse B, Dewerchin M, Zanetti A, Angellilo A, Mattot V, Nuyens D, Lutgens E, Clotman F, de Ruiter MC, Gittenberger-de Groot A, Poelmann R, Lupu F, Herbert JM, Collen D, Dejana E. Targeted deficiency or cytosolic truncation of the VE-cadherin gene in mice impairs VEGF-mediated endothelial survival and angiogenesis. Cell. 1999;98:147–157. doi: 10.1016/s0092-8674(00)81010-7. [DOI] [PubMed] [Google Scholar]
- Carmeliet P, Ng YS, Nuyens D, Theilmeier G, Brusselmans K, Cornelissen I, Ehler E, Kakkar VV, Stalmans I, Mattot V, Perriard JC, Dewerchin M, Flameng W, Nagy A, Lupu F, Moons L, Collen D, D'Amore PA, Shima DT. Impaired myocardial angiogenesis and ischemic cardiomyopathy in mice lacking the vascular endothelial growth factor isoforms VEGF164 and VEGF188. Nature Medicine. 1999;5:495–502. doi: 10.1038/8379. [DOI] [PubMed] [Google Scholar]
- Darland DC, Cain JT, Berosik MA, Saint-Geniez M, Odens PW, Schaubhut GJ, Frisch S, Stemmer-Rachamimov A, Darland T, D'Amore PA. Vascular endothelial growth factor (VEGF) isoform regulation of early forebrain development. Developmental Biology. 2011;358:9–22. doi: 10.1016/j.ydbio.2011.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan D, Fu Y, Paxinos G, Watson C. Spatiotemporal expression patterns of Pax6 in the brain of embryonic, newborn, and adult mice. Brain structure & function. 2012 doi: 10.1007/s00429-012-0397-2. [DOI] [PubMed] [Google Scholar]
- Englund C, Fink A, Lau C, Pham D, Daza RA, Bulfone A, Kowalczyk T, Hevner RF. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. The Journal of Neuroscience. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericson J, Muhr J, Jessell TM, Edlund T. Sonic hedgehog: a common signal for ventral patterning along the rostrocaudal axis of the neural tube. The International Journal of Developmental Biology. 1995;39:809–816. [PubMed] [Google Scholar]
- Ericson J, Muhr J, Placzek M, Lints T, Jessell TM, Edlund T. Sonic hedgehog induces the differentiation of ventral forebrain neurons: a common signal for ventral patterning within the neural tube. Cell. 1995;81:747–756. doi: 10.1016/0092-8674(95)90536-7. [DOI] [PubMed] [Google Scholar]
- Ericson J, Rashbass P, Schedl A, Brenner-Morton S, Kawakami A, van Heyningen V, Jessell TM, Briscoe J. Pax6 controls progenitor cell identity and neuronal fate in response to graded Shh signaling. Cell. 1997;90:169–180. doi: 10.1016/s0092-8674(00)80323-2. [DOI] [PubMed] [Google Scholar]
- Feng J, Fouse S, Fan G. Epigenetic regulation of neural gene expression and neuronal function. Pediatric Research. 2007;61:58R–63R. doi: 10.1203/pdr.0b013e3180457635. [DOI] [PubMed] [Google Scholar]
- Ferrara N. Role of vascular endothelial growth factor in the regulation of angiogenesis. Kidney international. 1999;56:794–814. doi: 10.1046/j.1523-1755.1999.00610.x. [DOI] [PubMed] [Google Scholar]
- Ferri A, Favaro R, Beccari L, Bertolini J, Mercurio S, Nieto-Lopez F, Verzeroli C, La Regina F, De Pietri Tonelli D, Ottolenghi S, Bovolenta P, Nicolis SK. Sox2 is required for embryonic development of the ventral telencephalon through the activation of the ventral determinants Nkx2.1 and Shh. Development. 2013;140:1250–1261. doi: 10.1242/dev.073411. [DOI] [PubMed] [Google Scholar]
- Fishell G, Hanashima C. Pyramidal neurons grow up and change their mind. Neuron. 2008;57:333–338. doi: 10.1016/j.neuron.2008.01.018. [DOI] [PubMed] [Google Scholar]
- Georgala PA, Carr CB, Price DJ. The role of Pax6 in forebrain development. Developmental neurobiology. 2011;71:690–709. doi: 10.1002/dneu.20895. [DOI] [PubMed] [Google Scholar]
- Gotz M, Barde YA. Radial glial cells defined and major intermediates between embryonic stem cells and CNS neurons. Neuron. 2005;46:369–372. doi: 10.1016/j.neuron.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Gotz M, Hartfuss E, Malatesta P. Radial glial cells as neuronal precursors: a new perspective on the correlation of morphology and lineage restriction in the developing cerebral cortex of mice. Brain Res Bull. 2002;57:777–788. doi: 10.1016/s0361-9230(01)00777-8. [DOI] [PubMed] [Google Scholar]
- Guillemot F, Molnar Z, Tarabykin V, Stoykova A. Molecular mechanisms of cortical differentiation. The European journal of neuroscience. 2006;23:857–868. doi: 10.1111/j.1460-9568.2006.04626.x. [DOI] [PubMed] [Google Scholar]
- Hartl D, Irmler M, Romer I, Mader MT, Mao L, Zabel C, de Angelis MH, Beckers J, Klose J. Transcriptome and proteome analysis of early embryonic mouse brain development. Proteomics. 2008;8:1257–1265. doi: 10.1002/pmic.200700724. [DOI] [PubMed] [Google Scholar]
- Hebert JM, Fishell G. The genetics of early telencephalon patterning: some assembly required. Nature reviews. Neuroscience. 2008;9:678–685. doi: 10.1038/nrn2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heins N, Malatesta P, Cecconi F, Nakafuku M, Tucker KL, Hack MA, Chapouton P, Barde YA, Gotz M. Glial cells generate neurons: the role of the transcription factor Pax6. Nature Neuroscience. 2002;5:308–315. doi: 10.1038/nn828. [DOI] [PubMed] [Google Scholar]
- Hemachandran M, Nijhawan R, Joshi K. Cytological grading, apoptosis, and Bcl-2 protein expression in breast cancer. Diagnostic cytopathology. 2002;26:356–359. doi: 10.1002/dc.10105. [DOI] [PubMed] [Google Scholar]
- Hevner RF, Shi L, Justice N, Hsueh Y, Sheng M, Smiga S, Bulfone A, Goffinet AM, Campagnoni AT, Rubenstein JL. Tbr1 regulates differentiation of the preplate and layer 6. Neuron. 2001;29:353–366. doi: 10.1016/s0896-6273(01)00211-2. [DOI] [PubMed] [Google Scholar]
- Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biology. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houck KA, Leung DW, Rowland AM, Winer J, Ferrara N. Dual regulation of vascular endothelial growth factor bioavailability by genetic and proteolytic mechanisms. The Journal of biological chemistry. 1992;267:26031–26037. [PubMed] [Google Scholar]
- Hu XL, Wang Y, Shen Q. Epigenetic control on cell fate choice in neural stem cells. Protein & cell. 2012;3:278–290. doi: 10.1007/s13238-012-2916-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hubbell E, Liu WM, Mei R. Robust estimators for expression analysis. Bioinformatics. 2002;18:1585–1592. doi: 10.1093/bioinformatics/18.12.1585. [DOI] [PubMed] [Google Scholar]
- Hynes RO. The Extracellular Matrix: Not Just Pretty Fibrils. Science. 2009;326:1216–1219. doi: 10.1126/science.1176009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida S, Usui T, Yamashiro K, Kaji Y, Amano S, Ogura Y, Hida T, Oguchi Y, Ambati J, Miller JW, Gragoudas ES, Ng YS, D'Amore PA, Shima DT, Adamis AP. VEGF164-mediated inflammation is required for pathological, but not physiological, ischemia-induced retinal neovascularization. The Journal of experimental medicine. 2003;198:483–489. doi: 10.1084/jem.20022027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishwaran H, Rao JS, Kogalur UB. BAMarraytrade mark: Java software for Bayesian analysis of variance for microarray data. BMC bioinformatics. 2006;7:59. doi: 10.1186/1471-2105-7-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishwaran H, Sunil Rao J. Clustering gene expression profile data by selective shrinkage. Statistics & Probability Letters. 2008;78:1490–1497. [Google Scholar]
- James JM, Mukouyama YS. Neuronal action on the developing blood vessel pattern. Seminars in Cell & Developmental Biology. 2011;22:1019–1027. doi: 10.1016/j.semcdb.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicke RU, Sprengart ML, Wati MR, Porter AG. Caspase-3 is required for DNA fragmentation and morphological changes associated with apoptosis. The Journal of biological chemistry. 1998;273:9357–9360. doi: 10.1074/jbc.273.16.9357. [DOI] [PubMed] [Google Scholar]
- Jin K. Vascular endothelial growth factor (VEGF) stimulates neurogenesis in vitro and in vivo. Proceedings of the National Academy of Sciences. 2002;99:11946–11950. doi: 10.1073/pnas.182296499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Jones L, Lopez-Bendito G, Gruss P, Stoykova A, Molnar Z. Pax6 is required for the normal development of the forebrain axonal connections. Development. 2002;129:5041–5052. doi: 10.1242/dev.129.21.5041. [DOI] [PubMed] [Google Scholar]
- Kioussi C, O'Connell S, St-Onge L, Treier M, Gleiberman AS, Gruss P, Rosenfeld MG. Pax6 is essential for establishing ventral-dorsal cell boundaries in pituitary gland development. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:14378–14382. doi: 10.1073/pnas.96.25.14378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch S, Tugues S, Li X, Gualandi L, Claesson-Welsh L. Signal transduction by vascular endothelial growth factor receptors. The Biochemical journal. 2011;437:169–183. doi: 10.1042/BJ20110301. [DOI] [PubMed] [Google Scholar]
- Komada M, Saitsu H, Kinboshi M, Miura T, Shiota K, Ishibashi M. Hedgehog signaling is involved in development of the neocortex. Development. 2008;135:2717–2727. doi: 10.1242/dev.015891. [DOI] [PubMed] [Google Scholar]
- Krilleke D, Ng YS, Shima DT. The heparin-binding domain confers diverse functions of VEGF-A in development and disease: a structure-function study. Biochemical Society transactions. 2009;37:1201–1206. doi: 10.1042/BST0371201. [DOI] [PubMed] [Google Scholar]
- Larsen KB, Lutterodt MC, Laursen H, Graem N, Pakkenberg B, Mollgard K, Moller M. Spatiotemporal distribution of PAX6 and MEIS2 expression and total cell numbers in the ganglionic eminence in the early developing human forebrain. Developmental Neuroscience. 2010;32:149–162. doi: 10.1159/000297602. [DOI] [PubMed] [Google Scholar]
- Li XJ, Zhang X, Johnson MA, Wang ZB, Lavaute T, Zhang SC. Coordination of sonic hedgehog and Wnt signaling determines ventral and dorsal telencephalic neuron types from human embryonic stem cells. Development. 2009;136:4055–4063. doi: 10.1242/dev.036624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie F, Ruhrberg C. Diverse roles for VEGF-A in the nervous system. Development. 2012;139:1371–1380. doi: 10.1242/dev.072348. [DOI] [PubMed] [Google Scholar]
- Matsumoto T, Claesson-Welsh L. VEGF receptor signal transduction. Sci STKE. 2001;2001:re21. doi: 10.1126/stke.2001.112.re21. [DOI] [PubMed] [Google Scholar]
- Miro X, Zhou X, Boretius S, Michaelis T, Kubisch C, Alvarez-Bolado G, Gruss P. Haploinsufficiency of the murine polycomb gene Suz12 results in diverse malformations of the brain and neural tube. Disease models & mechanisms. 2009;2:412–418. doi: 10.1242/dmm.001602. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nature reviews. Neuroscience. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- Mori T, Buffo A, Gotz M. The novel roles of glial cells revisited: the contribution of radial glia and astrocytes to neurogenesis. Current topics in developmental biology. 2005;69:67–99. doi: 10.1016/S0070-2153(05)69004-7. [DOI] [PubMed] [Google Scholar]
- Ng YS, Rohan R, Sunday ME, Demello DE, D'Amore PA. Differential expression of VEGF isoforms in mouse during development and in the adult. Dev Dyn. 2001;220:112–121. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1093>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Ochiai W, Nakatani S, Takahara T, Kainuma M, Masaoka M, Minobe S, Namihira M, Nakashima K, Sakakibara A, Ogawa M, Miyata T. Periventricular notch activation and asymmetric Ngn2 and Tbr2 expression in pair-generated neocortical daughter cells. Molecular and cellular neurosciences. 2009;40:225–233. doi: 10.1016/j.mcn.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Osumi N. The role of Pax6 in brain patterning. The Tohoku journal of experimental medicine. 2001;193:163–174. doi: 10.1620/tjem.193.163. [DOI] [PubMed] [Google Scholar]
- Osumi N, Shinohara H, Numayama-Tsuruta K, Maekawa M. Concise review: Pax6 transcription factor contributes to both embryonic and adult neurogenesis as a multifunctional regulator. Stem Cells. 2008;26:1663–1672. doi: 10.1634/stemcells.2007-0884. [DOI] [PubMed] [Google Scholar]
- Ouyang L, Shi Z, Zhao S, Wang FT, Zhou TT, Liu B, Bao JK. Programmed cell death pathways in cancer: a review of apoptosis, autophagy and programmed necrosis. Cell proliferation. 2012;45:487–498. doi: 10.1111/j.1365-2184.2012.00845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JE, Keller GA, Ferrara N. The vascular endothelial growth factor (VEGF) isoforms: differential deposition into the subepithelial extracellular matrix and bioactivity of extracellular matrix-bound VEGF. Molecular biology of the cell. 1993;4:1317–1326. doi: 10.1091/mbc.4.12.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. The EMBO journal. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierani A, Wassef M. Cerebral cortex development: From progenitors patterning to neocortical size during evolution. Development, Growth & Differentiation. 2009;51:325–342. doi: 10.1111/j.1440-169X.2009.01095.x. [DOI] [PubMed] [Google Scholar]
- Price DJ, Kennedy H, Dehay C, Zhou L, Mercier M, Jossin Y, Goffinet AM, Tissir F, Blakey D, Molnar Z. The development of cortical connections. The European journal of neuroscience. 2006;23:910–920. doi: 10.1111/j.1460-9568.2006.04620.x. [DOI] [PubMed] [Google Scholar]
- Quinn JC, Molinek M, Martynoga BS, Zaki PA, Faedo A, Bulfone A, Hevner RF, West JD, Price DJ. Pax6 controls cerebral cortical cell number by regulating exit from the cell cycle and specifies cortical cell identity by a cell autonomous mechanism. Developmental Biology. 2007;302:50–65. doi: 10.1016/j.ydbio.2006.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rash BG, Grove EA. Patterning the dorsal telencephalon: a role for sonic hedgehog? The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27:11595–11603. doi: 10.1523/JNEUROSCI.3204-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhen T, Metzger K, Schroeder A, Woodward R. Expression of putative sex-determining genes during the thermosensitive period of gonad development in the snapping turtle, Chelydra serpentina. Sexual development : genetics, molecular biology, evolution, endocrinology, embryology, and pathology of sex determination and differentiation. 2007;1:255–270. doi: 10.1159/000104775. [DOI] [PubMed] [Google Scholar]
- Risau W. Mechanisms of angiogenesis. Nature. 1997;386:671–674. doi: 10.1038/386671a0. [DOI] [PubMed] [Google Scholar]
- Ruhrberg C, Gerhardt H, Golding M, Watson R, Ioannidou S, Fujisawa H, Betsholtz C, Shima DT. Spatially restricted patterning cues provided by heparin-binding VEGF-A control blood vessel branching morphogenesis. Genes & Development. 2002;16:2684–2698. doi: 10.1101/gad.242002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz de Almodovar C, Coulon C, Salin PA, Knevels E, Chounlamountri N, Poesen K, Hermans K, Lambrechts D, Van Geyte K, Dhondt J, Dresselaers T, Renaud J, Aragones J, Zacchigna S, Geudens I, Gall D, Stroobants S, Mutin M, Dassonville K, Storkebaum E, Jordan BF, Eriksson U, Moons L, D'Hooge R, Haigh JJ, Belin MF, Schiffmann S, Van Hecke P, Gallez B, Vinckier S, Chedotal A, Honnorat J, Thomasset N, Carmeliet P, Meissirel C. Matrix-binding vascular endothelial growth factor (VEGF) isoforms guide granule cell migration in the cerebellum via VEGF receptor Flk1. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30:15052–15066. doi: 10.1523/JNEUROSCI.0477-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai K, Osumi N. The neurogenesis-controlling factor, Pax6, inhibits proliferation and promotes maturation in murine astrocytes. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2008;28:4604–4612. doi: 10.1523/JNEUROSCI.5074-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom SN, Griffiths DS, Faedo A, Kleinjan DJ, Ruan Y, Smith J, van Heyningen V, Rubenstein JL, Livesey FJ. The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self-renewal and neurogenesis. PLoS Genetics. 2009;5:e1000511. doi: 10.1371/journal.pgen.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scardigli R, Baumer N, Gruss P, Guillemot F, Le Roux I. Direct and concentration-dependent regulation of the proneural gene Neurogenin2 by Pax6. Development. 2003;130:3269–3281. doi: 10.1242/dev.00539. [DOI] [PubMed] [Google Scholar]
- Sessa A, Mao CA, Hadjantonakis AK, Klein WH, Broccoli V. Tbr2 directs conversion of radial glia into basal precursors and guides neuronal amplification by indirect neurogenesis in the developing neocortex. Neuron. 2008;60:56–69. doi: 10.1016/j.neuron.2008.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Nakazawa M, Kani S, Bae YK, Kageyama R, Hibi M. Zinc finger genes Fezf1 and Fezf2 control neuronal differentiation by repressing Hes5 expression in the forebrain. Development. 2010;137:1875–1885. doi: 10.1242/dev.047167. [DOI] [PubMed] [Google Scholar]
- Sobeih MM, Corfas G. Extracellular factors that regulate neuronal migration in the central nervous system. International journal of developmental neuroscience : the official journal of the International Society for Developmental Neuroscience. 2002;20:349–357. doi: 10.1016/s0736-5748(02)00040-0. [DOI] [PubMed] [Google Scholar]
- Stalmans I, Ng YS, Rohan R, Fruttiger M, Bouche A, Yuce A, Fujisawa H, Hermans B, Shani M, Jansen S, Hicklin D, Anderson DJ, Gardiner T, Hammes HP, Moons L, Dewerchin M, Collen D, Carmeliet P, D'Amore PA. Arteriolar and venular patterning in retinas of mice selectively expressing VEGF isoforms. The Journal of clinical investigation. 2002;109:327–336. doi: 10.1172/JCI14362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoykova A, Gotz M, Gruss P, Price J. Pax6-dependent regulation of adhesive patterning, R-cadherin expression and boundary formation in developing forebrain. Development. 1997;124:3765–3777. doi: 10.1242/dev.124.19.3765. [DOI] [PubMed] [Google Scholar]
- Stumm R, Kolodziej A, Schulz S, Kohtz JD, Hollt V. Patterns of SDF-1alpha and SDF-1gamma mRNAs, migration pathways, and phenotypes of CXCR4-expressing neurons in the developing rat telencephalon. The Journal of Comparative Neurology. 2007;502:382–399. doi: 10.1002/cne.21336. [DOI] [PubMed] [Google Scholar]
- Sun Y, Jin K, Childs JT, Xie L, Mao XO, Greenberg DA. Vascular endothelial growth factor-B (VEGFB) stimulates neurogenesis: Evidence from knockout mice and growth factor administration. Developmental Biology. 2006;289:329–335. doi: 10.1016/j.ydbio.2005.10.016. [DOI] [PubMed] [Google Scholar]
- Suter DM, Tirefort D, Julien S, Krause K-H. A Sox1 to Pax6 Switch Drives Neuroectoderm to Radial Glia Progression During Differentiation of Mouse Embryonic Stem Cells. Stem Cells. 2009;27:49–58. doi: 10.1634/stemcells.2008-0319. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, Jensen LJ, von Mering C. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic acids research. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therneau TM, Ballman KV. What does PLIER really do? Cancer informatics. 2008;6:423–431. [PMC free article] [PubMed] [Google Scholar]
- Tuoc TC, Radyushkin K, Tonchev AB, Pinon MC, Ashery-Padan R, Molnar Z, Davidoff MS, Stoykova A. Selective cortical layering abnormalities and behavioral deficits in cortex-specific Pax6 knock-out mice. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009;29:8335–8349. doi: 10.1523/JNEUROSCI.5669-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan A, Bhide PG. Angiogenesis in the embryonic CNS: A new twist on an old tale. Cell Adhesion & Migration. 2008;2:167–169. doi: 10.4161/cam.2.3.6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan A, Long JE, Crandall JE, Rubenstein JL, Bhide PG. Compartment-specific transcription factors orchestrate angiogenesis gradients in the embryonic brain. Nature Neuroscience. 2008;11:429–439. doi: 10.1038/nn2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgintino D, Errede M, Robertson D, Girolamo F, Masciandaro A, Bertossi M. VEGF expression is developmentally regulated during human brain angiogenesis. Histochemistry and cell biology. 2003;119:227–232. doi: 10.1007/s00418-003-0510-y. [DOI] [PubMed] [Google Scholar]
- Waclaw RR, Campbell K. Regional control of cortical lamination. Nature Neuroscience. 2009;12:1211–1212. doi: 10.1038/nn1009-1211. [DOI] [PubMed] [Google Scholar]
- Wang Y, Li G, Stanco A, Long JE, Crawford D, Potter GB, Pleasure SJ, Behrens T, Rubenstein JL. CXCR4 and CXCR7 have distinct functions in regulating interneuron migration. Neuron. 2011;69:61–76. doi: 10.1016/j.neuron.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe D, Uchiyama K, Hanaoka K. Transition of mouse de novo methyltransferases expression from Dnmt3b to Dnmt3a during neural progenitor cell development. Neuroscience. 2006;142:727–737. doi: 10.1016/j.neuroscience.2006.07.053. [DOI] [PubMed] [Google Scholar]
- Wu H, Coskun V, Tao J, Xie W, Ge W, Yoshikawa K, Li E, Zhang Y, Sun YE. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 2010;329:444–448. doi: 10.1126/science.1190485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun K, Fischman S, Johnson J, Hrabe de Angelis M, Weinmaster G, Rubenstein JL. Modulation of the notch signaling by Mash1 and Dlx1/2 regulates sequential specification and differentiation of progenitor cell types in the subcortical telencephalon. Development. 2002;129:5029–5040. doi: 10.1242/dev.129.21.5029. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Jin K, Mao XO, Greenberg DA. Vascular endothelial growth factor promotes proliferation of cortical neuron precursors by regulating E2F expression. FASEB J. 2003;17:186–193. doi: 10.1096/fj.02-0515com. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.