Abstract
Mutations in NPHS2, the gene that encodes podocin, are well-established causes of both familial and sporadic steroid-resistant focal segmental glomerulosclerosis (FSGS) in the pediatric population, but have not been well-characterized in late-onset disease. To investigate the role of NPHS2 polymorphisms in sporadic cases of late-onset FSGS, we studied 377 biopsy-confirmed FSGS cases and 919 controls. We identified 18 single nucleotide polymorphisms (SNPs) by resequencing a subgroup of cases and controls, and subsequently genotyped African-American and European-American cases and controls for five missense SNPs, three SNPs within introns, and four SNPs in the 3′ untranslated region. No homozygotes or compound heterozygotes were observed for any missense mutation. R138Q carriers were more frequent among FSGS cases relative to controls (OR = 4.9, P = 0.06), but heterozygosity for the other four missense mutations was equally distributed among FSGS cases and controls. Finally, a common haplotype of noncoding SNPs carried by 20% of African-Americans, but not observed in European-Americans, was strongly associated with a 50% reduction in risk for sporadic FSGS (OR = 0.5, P = 0.001). These results indicate that genetic variation or mutation of NPHS2 may play a role in late-onset sporadic FSGS.
Focal segmental glomerulosclerosis (FSGS) is the leading cause of primary nephrotic syndrome in adults and is an important cause of ESRD in adults and children.1,2 Mutations in the NPHS2 gene, encoding podocin, account for 20 to 40% of familial, autosomal recessive, pediatric steroid-resistant nephrotic syndrome (SRNS) (reviewed by Pollak3 and Niaudet4) and approximately 10% of idiopathic childhood SRNS.4 When histology is available, most of the cases represent FSGS. Nearly all patients with NPHS2 homozygous or compound heterozygous mutations commonly present before the age of 6 yr.4,5 Only five families in which one or more affected people presented after the age of 21 have been reported, and each of the families was segregatingR229Q along with a second mutation as complex heterozygotes with FSGS.6,7 Podocin, expressed exclusively in the glomerular podocyte, is an integral membrane protein located on the foot processes adjacent to the slit diaphragms that play a critical role in regulating hydraulic flow and protein filtration from the plasma space into the urinary space.8 Podocin interacts with other podocyte proteins, including recruiting nephrin to plasma membrane lipid rafts,9 and CD2AP.10
In familial autosomal recessive SRNS with childhood onset, pathogenic podocin mutations occur as homozygotes and/or as compound heterozygotes. In a large study of mostly Europeans and Northern Arabs,11 the NPHS2 mutation rate among families showing autosomal recessive patterns of transmission was 43% with an average age of onset of 4.8 yr. The mutation prevalence among sporadic pediatric SRNS cases with an average age of onset of 8.6 yr was reported to be 10.5%.11 In a survey of 165 families who had SRNS with one or more children with congenital or early-onset SRNS, homozygotes or compound heterozygotes for missense mutations were observed in 26% of families, whereas individuals with only a single pathogenic mutation were observed in 2% of families.11
More than 30 distinct NPHS2 mutations, occurring either in homozygosity or compound heterozygosity, have been associated with FSGS.12 R138Q is the most frequently observed pathogenic mutation occurring either as a homozygote or as a compound heterozygote in 21 of 165 families and 19 of 253 unrelated children with FSGS, respectively.5,6 Podocin R138Q is retained within the endoplasmic reticulum and therefore cannot interact normally with nephrin.13 The R229Q amino acid substitution was the second most commonly observed podocin mutation in patients with SRNS; however, its pathogenic role in SRNSis not clear because it is observed with similar frequencies in patients with SRNS and steroid-sensitive nephrotic syndrome as well as normal control subjects (7, 6, and 9%, respectively).6,11 This common polymorphism is almost always observed as a compound heterozygote together with a pathogenic mutation in early-onset familial and sporadic SRNS, suggesting that R229Q may increase susceptibility only in association with a second NPHS2 mutation. One study14 reported that R229Q carriers in the general population are at increased risk for micro albuminuria. Support for a functional role for R229Q comes from in vitro studies showing decreased binding between purified nephrin and in vitro–translated R229Q podocin.7
Most studies have addressed the role of NPHS2 mutations in young children with SRNS or FSGS.15–17 The role of NPHS2 mutations in adults with FSGS, however, has not been well characterized. Tsukaguchi et al.7 genotyped 91 adult patients with FSGS from the north eastern United States for the R229Q mutation and found that 11 (12%) were heterozygotes for R229Q. Further NPHS2 mutational screening of these 11 individuals revealed a second mutation, R291W, in two patients. The allele frequencies for the R229Q polymorphism in control populations was 1.6% for 32 individuals of African ancestry or origin, 3.1% for 49 Brazilian individuals, and 3.6% for 124 people of European ancestry compared with 6% in the 91 adult patients with sporadic FSGS.
Idiopathic FSGS is four-fold more common in African Americans (AA)2 and HIV-1-associated FSGS is 18- to 50-fold more common among AA compared with European Americans (EA).18,19 The lifetime risk for FSGS has been estimated to be approximately 1 in 139 for AA, compared with 1 in 588 for EA.2,19 Freedman et al.20 found that compared with patients with ESRD and without HIV-1, patients with ESRD and HIV-1 infection were more likely to have relatives with ESRD, suggesting that a genetic predisposition to renal injury might predispose to multiple forms of kidney disease. AA are also at increased risk for diabetic nephropathy and hypertensive nephrosclerosis. The genetic basis underlying the predilection of AA to develop kidney disease in general and adult-onset, sporadic FSGS in particular has not been defined. A genome-wide scan of nondiabetic ESRD in AA sibling pairs implicated the NPHS2 gene. An insertion, IVS3 + 9insA, in the third intron was significantly elevated in AA patients without diabetes and with ESRD compared with normal control subjects (0.018 versus 0.002), suggesting that infrequent noncoding variants may be associated with nondiabetic ESRD in AA.21 The role of NPHS2 variation in AA with adult-onset sporadic FSGS or with HIV-1–associated FSGS has not yet been investigated.
Given the importance of podocin in the glomerular filtration barrier and the causal role of NPHS2 mutation in childhood SRNS and FSGS, we investigated the role of NPHS2 variation in sporadic, biopsy-proven FSGS in a case-control study that included both HIV-1–infected and uninfected AA and HIV-1–uninfected EA. The goals of this study were (1) to determine the extent of NPHS2 open reading frame variation in patients with FSGS and the general population for EA and AA by resequencing; (2) to assess the effects of NPHS2 variation in AA with HIV-1–associated, biopsy-proven FSGS and in AA and EA with sporadic FSGS; and (3) to assess the pathologic consequences of P20L, G42R, R138Q, R229Q, and A242V in a well-powered, population-based study that included >900 control subjects.
RESULTS
Polymorphisms in NPHS2
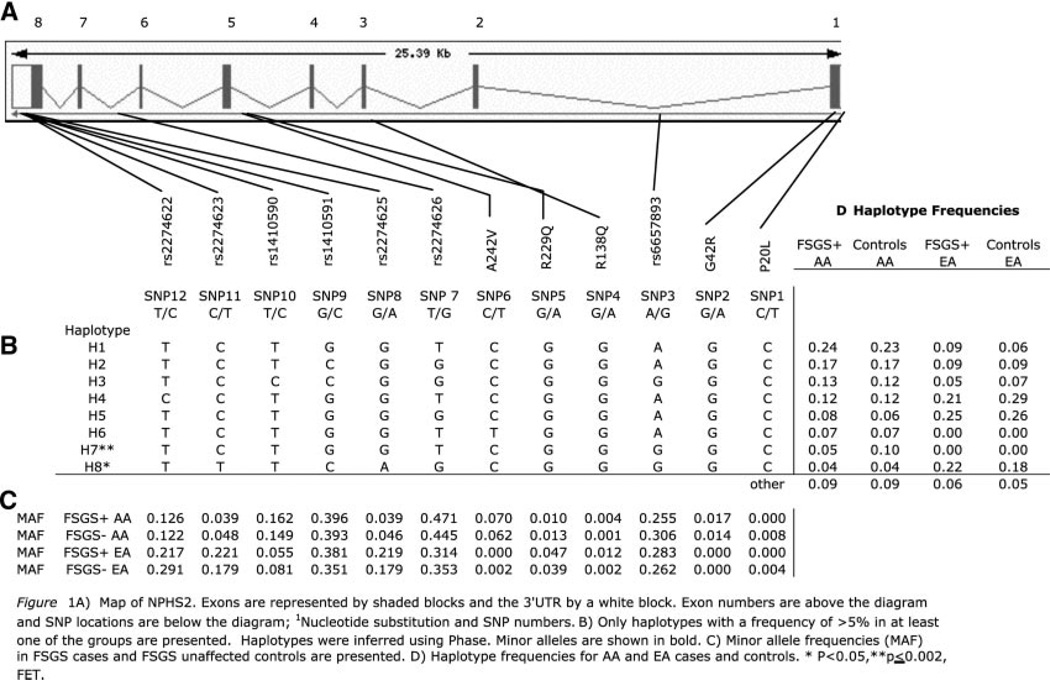
The human NPHS2 gene spans approximately 25 kb on chromosome 1q25.2 and has eight exons and seven introns (Figure 1A). DNA resequencing of the exons and intron-exon junctions for a subset of 265 patients and 179 control subjects was used to detect open reading frame mutations. Five nonsynonymous, codon-changing variants and five synonymous (silent) single-nucleotide polymorphisms (SNP) were identified. The missense mutations were P20L, G42R (exon 1), R138Q (exon 3), R229Q, and A242V (exon 5). The synonymous changes were G34G (exon 1) and S96S (exon 2) and A297A, A318A, and L346L (exon 8). In addition, two noncoding SNP were observed: −52C/G and −51G/T in the 5′ untranslated region. No nonsynonymous SNP were observed in exons 2, 4, 7, or 8.
Figure 1.
(A) Map of NPHS2. Exons are represented by shaded blocks and the 3′ untranslated region by a white block. Exon numbers are above the diagram, and SNP locations are below the diagram. (1) Nucleotide substitution and SNP numbers. (B) Only haplotypes with a frequency >5% in at least one of the groups are presented. Haplotypes were inferred using Phase. Minor alleles are shown in bold. (C) Minor allele frequencies (MAF) in patients with FSGS and control subjects are presented. (D) Haplotype frequencies for AA and EA patients and control subjects. *P < 0.05, **P ≤ 0.002, Fisher exact test.
The five nonsynonymous SNP were genotyped in the patients and control subjects for assessment of their genotypic frequencies. In addition, National Center for Biotechnology Information database (db) SNP with available TaqMan assays were genotyped: SNP 3 (intron 1), SNP 7 (intron 6), SNP 8 (intron 7), and SNP 9 to 12 in the 3′ untranslated region (Figure 1A). The minor allele frequencies for the noncoding SNP were quite different between AA and EA; this was particularly striking for SNP 8, 10, 11, and 12, for which there was a two- to three-fold difference in minor allele frequencies between AA and EA control subjects (Figure 1C). Among the nonsynonymous SNP, SNP2 (G42R) was observed only in AA, SNP5 (R229Q) was four-fold more frequent in AA control subjects (4%) compared with in EA control subjects (1%), and SNP6 (A242V) was 30-fold more frequent in EA control subjects (6%) compared with AA control subjects (0.2%). Similar differences in haplotype frequencies were also observed (Figure 1, C and D).
Genotype frequencies were within Hardy-Weinberg expectations, with the exception of SNP 8, 9, and 11, for which there was an excess of homozygotes for the minor alleles in the EA FSGS case group (for SNP 8 and 11, observed 12 of 128 versus expected 6 of 128; P = 0.00005). In EA, SNP 8 and 11 are in absolute linkage disequilibrium (D′ = 1) and highly correlated (r2 = 1). SNP 9, which occurs on three haplotypes, is also highly correlated (D′ = 1; r2 >0.4) with SNP 8 and 11 on haplotype 8 (Figure 1B). The concordance and similar genotype frequencies among these three tightly linked SNP indicate that the elevated frequency of homozygotes for these three alleles in the EA FSGS group was not due to genotyping errors. Furthermore, all SNP were genotyped using independent assays. These data suggest that homozygosity for SNP 8, 9, and 11 may be associated with increased FSGS risk in EA.
Association of SNP Alleles with FSGS
Because of the differences in FSGS risk, allele frequencies, and haplotype structure between the two racial groups, we analyzed AA and EA separately for SNP and haplotype associations. We also analyzed the AA HIV-associated and HIV-1–uninfected groups separately, but because the results for the two groups were similar, we combined the AA HIV-1–associated and HIV-1–uninfected groups to increase power.
For the nonsynonymous SNP specifying P20L, G42R, R138Q, R229Q, and A242V, heterozygotes were observed in both patients and control subjects in AA and EA, and, with the exception of R138Q, these SNP were equally distributed among patients and control subjects in both groups (Table 1). We also observed no differences in distribution of the nonsynonymous SNP between patients and control subjects when we stratified the analysis by HIV-1 status (data not shown). There was a trend for R138Q carriers to be more frequent among patients with FSGS relative to control subjects: EA 2 versus 0.4% (odds ratio [OR] 4.4; P = 0.23) andAA1 versus 0.2% (OR 5.2; P = 0.19). In a combined analysis of EA and AA, the OR of FSGS among R138Q carriers was 4.9 (P = 0.06). The two EA and two AA FSGS heterozygotes for R138Q received a diagnosis of FSGS at ages 14, 24, 38, and 39. No compound heterozygotes or homozygotes were observed for any of the codon-changing SNP in the FSGS-affected patients. The polymorphism P20L was observed in the homozygous state in a single control subject. We obtained urine protein-to-creatinine ratios for 44% of EA and 88% of AA identified with a codon-changing mutation or polymorphism; none of these individuals with urine protein results had proteinuria, including one of two individuals who were heterozygous for R138Q and 24 of 37 individuals who were heterozygous for R229Q.
Table 1.
Genotype frequencies for nonsynonymous sites in patients with FSGS and control subjectsa
| Mutation | AA | EA | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MAF | Genotype, number (frequency) | Total | MAF | Genotype, number (frequency) | Total | |||||
| P20L | C/C | C/T | T/T | C/C | C/T | T/T | ||||
| FSGS | 0 | 242 (1.00) | 0 | 0 | 242 | 0.001 | 128 (1.00) | 0 | 0 | 128 |
| control | 0.008 | 621 (1.00) | 8 (0.013) | 1 (0.002) | 630 | 0.004 | 279 (0.99) | 2 (0.007) | 0 | 281 |
| G42R | G/G | G/A | A/A | G/G | G/A | A/A | ||||
| FSGS | 0.017 | 234 (0.97) | 8 (0.033) | 0 | 242 | 0 | 128 (1.00) | 0 | 0 | 128 |
| control | 0.014 | 616 (0.977) | 18 (0.03) | 0 | 634 | 0 | 280 (1.00) | 0 | 0 | 280 |
| R138Q | G/G | G/A | A/A | G/G | G/A | A/A | ||||
| FSGS | 0.004 | 245 (0.99) | 2 (0.01) | 0 | 247 | 0.012 | 127 (0.98) | 2 (0.02) | 0 | 129 |
| control | 0.001 | 636 (1.00) | 1 (0.002) | 0 | 637 | 0.002 | 280 (1.00) | 1 (0.004) | 0 | 281 |
| R229Q | G/G | G/A | A/A | G/G | G/A | A/A | ||||
| FSGS | 0.01 | 242 (1.00) | 5 (0.002) | 0 | 247 | 0.047 | 117 (0.91) | 12 (0.09) | 0 | 129 |
| control | 0.013 | 618 (0.975) | 16 (0.002) | 0 | 634 | 0.039 | 250 (0.92) | 21 (0.08) | 0 | 271 |
| A242V | C/C | C/T | T/T | C/C | C/T | T/T | ||||
| FSGS | 0.07 | 209 (0.864) | 32 (0.13) | 1 (0.004) | 242 | 0 | 128 (1.00) | 0 | 0 | 128 |
| control | 0.062 | 531 (0.881) | 69 (0.11) | 3 (0.005) | 603 | 0.002 | 280 (1.00) | 1 (0.004) | 0 | 281 |
Minor allele frequency.
Six of seven intronic SNP were associated with FSGS among either EA or AA, with the OR ranging from 0.46 to 2.50 (0.01≥ P ≤ 0.05), as shown in Table 2. The strongest association was with SNP3 among AA. AA with the SNP3 G allele were under-represented in the HIV-1–positive FSGS (OR 0.46; P = 0.01), HIV-1–negative FSGS (OR 0.75; P = 0.11), and the combined FSGS (OR 0.67; P = 0.01) patient groups relative to their control groups, with the strongest association observed for patients with HIV-1 and FSGS. Significant associations were also observed for SNP 7 and 12 (Table 2). In EA, the highly correlated SNP 8 A/A and SNP 11 T/T homozygotes (OR 2.5; P = 0.04) were overrepresented and SNP 12 C was underrepresented (OR 0.65; P = 0.05) in patients with FSGS.
Table 2.
Genotype frequencies and OR for SNP showing significant associations with FSGS in AA or EA
| SNP | HIV Status | Group | n (Frequency) | Total | Dominant Model | Recessive Model | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | P | |||||||
| African Americans | ||||||||||
| SNP3 | A/A | A/G | G/G | |||||||
| rs6657893 | HIV+ | FSGS | 35 (0.660) | 16 (0.302) | 2 (0.038) | 53 | 0.46(0.25to0.85) | 0.015 | 0.69 (0.15to3.16) | 1.0 |
| Control | 114 (0.471) | 115 (0.475) | 13 (0.054) | 242 | ||||||
| HIV− | FSGS | 104 (0.547) | 68 (0.358) | 18 (0.095) | 190 | 0.75(0.53to1.06) | 0.11 | 0.89 (0.50to1.60) | 0.77 | |
| Control | 186 (0.474) | 165 (0.421) | 41 (0.105) | 392 | ||||||
| Combined | FSGS | 139 (0.572) | 84 (0.346) | 20 (0.082) | 243 | 0.67(0.50to0.90) | 0.01 | 0.96 (0.56to1.65) | 1.00 | |
| Control | 300 (0.473) | 280 (0.442) | 54 (0.085) | 634 | ||||||
| SNP7 | T/T | T/G | G/G | |||||||
| rs2274626 | HIV+ | FSGS | 21 (0.404) | 20 (0.385) | 11 (0.211) | 52 | 0.62(0.34to1.15) | 0.14 | 1.28 (0.61to2.70) | 0.55 |
| Control | 72 (0.296) | 129 (0.531) | 42 (0.173) | 243 | ||||||
| HIV− | FSGS | 53 (0.276) | 90 (0.469) | 49 (0.255) | 192 | 1.08(0.74to1.59) | 0.70 | 1.44 (0.95to2.17) | 0.08 | |
| Control | 114 (0.292) | 201 (0.515) | 75 (0.192) | 390 | ||||||
| Combined | FSGS | 74 (0.303) | 110 (0.451) | 60 (0.246) | 244 | 0.96(0.70to1.32) | 0.80 | 1.44 (1.01to2.05) | 0.05 | |
| Control | 186 (0.294) | 330 (0.521) | 117 (0.185) | 633 | ||||||
| SNP8 | G/G | G/A | A/A | |||||||
| rs2274625 | HIV+ | FSGS | 51 (0.962) | 2 (0.038) | 0 (0.000) | 53 | 2.69(0.61to11.78) | 0.27 | ||
| Control | 218 (0.905) | 23 (0.095) | 0 (0.000) | 241 | ||||||
| HIV− | FSGS | 175 (0.911) | 17 (0.088) | 0 (0.000) | 192 | 1.09(0.59to2.02) | 0.75 | Zero cell | 0.55 | |
| Control | 360 (0.918) | 29 (0.074) | 3 (0.077) | 392 | ||||||
| Combined | FSGS | 226 (0.922) | 19 (0.077) | 0 (0.000) | 245 | 0.88(0.51to1.52) | 0.79 | Zero cell | 0.56 | |
| Control | 578 (0.913) | 52 (0.082) | 3 (0.047) | 633 | ||||||
| SNP11 | C/C | C/T | T/T | |||||||
| rs2274623 | HIV+ | FSGS | 50 (0.943) | 3 (0.056) | 0 (0.000) | 53 | 0.54(0.16to1.86) | 0.44 | ||
| Control | 216 (0.900) | 24 (0.100) | 0 (0.000) | 240 | ||||||
| HIV− | FSGS | 176 (0.917) | 16 (0.083) | 0 (0.000) | 192 | 0.98(0.53to1.83) | 1.00 | Zero cell | 0.55 | |
| Control | 357 (0.915) | 30 (0.777) | 3 (0.077) | 390 | ||||||
| Combined | FSGS | 226 (0.922) | 19 (0.077) | 0 (0.000) | 245 | 0.84(0.49to1.45) | 0.59 | Zero cell | 0.56 | |
| Control | 573 (0.909) | 54 (0.086) | 3 (0.048 | 630 | ||||||
| SNP12 | T/T | T/C | C/C | |||||||
| rs2274622 | HIV+ | FSGS | 33 (0.623) | 18 (0.340) | 2 (0.038) | 53 | 2.04(1.08to3.82) | 0.04 | 1.87 (0.35to9.93) | 0.61 |
| Control | 188 (0.770) | 51 (0.209) | 5 (0.020) | 244 | ||||||
| HIV− | FSGS | 155 (0.803) | 36 (0.186) | 2 (0.010) | 193 | 0.83(0.54to1.27) | 0.40 | 1.00 (0.18to5.55) | 1.0 | |
| Control | 300 (0.771) | 85 (0.218) | 4 (0.010) | 389 | ||||||
| Combined | FSGS | 188 (0.764) | 54 (0.219) | 4 (0.016) | 246 | 1.05(0.75to1.49) | 0.79 | 1.15 (0.35to3.76) | 0.76 | |
| Control | 448 (0.771) | 136 (0.215) | 9 (0.014) | 633 | ||||||
| European Americans | ||||||||||
| SNP3 | A/A | A/G | G/G | |||||||
| rs6657893 | HIV− | FSGS | 68 (0.535) | 46 (0.362) | 13 (0.102) | 127 | 1.09(0.71to1.66) | 0.75 | 1.31 (0.64to2.69) | 0.45 |
| Control | 153 (0.556) | 100 (0.364) | 22 (0.080) | 275 | ||||||
| SNP7 | G/G | G/T | T/T | |||||||
| rs2274626 | HIV− | FSGS | 61 (0.473) | 55 (0.426) | 13 (0.101) | 129 | 0.87(0.57to1.32) | 0.52 | 0.67 (0.34to1.29) | 0.27 |
| Control | 122 (0.439) | 116 (0.417) | 40 (0.144) | 278 | ||||||
| SNP86 | G/G | G/A | A/A | |||||||
| rs2274625 | HIV− | FSGS | 84 (0.656) | 32 (0.250) | 12 (0.094) | 128 | 1.12(0.72to1.74) | 0.65 | 2.49 (1.07to5.81) | 0.04 |
| Control | 188 (0.681) | 77 (0.279) | 11 (0.039) | 276 | ||||||
| SNP11 | C/C | C/T | T/T | |||||||
| rs2274623 | HIV− | FSGS | 84 (0.651) | 33 (0.256) | 12 (0.093) | 129 | 1.14(0.74to1.78) | 0.57 | 2.50 (1.07to5.82) | 0.04 |
| Control | 190 (0.681) | 78 (0.280) | 11 (0.039) | 279 | ||||||
| SNP12 | T/T | T/C | C/C | |||||||
| rs2274622 | HIV− | FSGS | 80 (0.620) | 42 (0.326) | 7 (0.054) | 129 | 0.65(0.43to1.00) | 0.05 | 0.53 (0.22to1.25) | 0.18 |
| control | 143 (0.516) | 107 (0.386) | 27 (0.097) | 277 | ||||||
Multiplicative-type corrections such as Bonferroni corrections for correlated genetic factors and tests are highly conservative. We therefore present the q value, a measure of the false discovery rate (FDR; i.e., false positive) expected for a given P value.22 For nearly all noncoding and coding SNP, the q values were NS. Only for SNP3 in AA did q values approach significance, with values of 10% FDR in the HIV-1–associated group and 12% combining all FSGS patients and control subjects.
Association of Haplotypes of NPHS2 with FSGS
Figure 1, B and D, presents the haplotype structure and the distribution of haplotype frequencies between FSGS patients and control subjects. Haplotype 7 was significantly more frequent in AA control subjects compared with AA patients with FSGS (Figure 1D). A dominant genetic model suggested that individuals carrying either one or two copies of haplotype 7 were protected against FSGS (OR 0.50; confidence interval [CI] 0.32 to 0.78; P = 0.002) compared with individuals carrying any other haplotype combination (Table 3). In EA diplotype 8 (i.e., two copies of haplotype 8) was overrepresented in EA patients with FSGS compared with control subjects (OR 2.50; CI 1.08 to 5.84; P = 0.04; Table 3). The q value for haplotype 7 in AA was 1.3%, indicating that the association is highly robust. In contrast, the q value for the association between haplotype 8 and FSGS in EA was 22%.
Table 3.
Haplotype frequencies and OR for haplotypes showing significant associations with FSGS in AA or EAa
| Haplotype | Group | N (Frequency) | Total | OR (95% CI) | P/q | ||
|---|---|---|---|---|---|---|---|
| AA (dominant model) | |||||||
| H7 | H7/H7 | H7/Hx | Hx/Hx | ||||
| FSGS | 1 (0.004) | 25 (0.10) | 221 (0.89) | 247 | 0.50 (0.32 to 0.78) | 0.002/0.013 | |
| Control | 7 (0.01) | 115 (0.18) | 516 (0.81) | 638 | |||
| EA (recessive model) | |||||||
| H8 | H8/H8 | H8/Hx | Hx/Hx | ||||
| FSGS | 12 (0.09) | 33 (0.25) | 85 (0.65) | 130 | 2.50 (1.07 to 5.84) | 0.04/0.22 | |
| Control | 11 (0.04) | 78 (0.28) | 193 (0.68) | 282 | |||
The q value indicates the FDR.
DISCUSSION
The causal role of NPHS2 mutations in familial and sporadic pediatric-onset SRNS and FSGS is well established; however, the influence of NPHS2 polymorphism and mutation on late-onset FSGS has not been fully explored in case-control studies and has not been explored at all in AA with sporadic or HIV-associated FSGS, where the incidence of FSGS is highest. In this study, we examined the role of NPHS2 variation in sporadic and HIV-associated FSGS among a mostly adult population. We first resequenced NPHS2 in 265 patients with FSGS and 179 control subjects and then genotyped coding and noncoding SNP in all 370 patients and 918 control subjects. Heterozygosity for P20L, G42R, R229Q, and A242V did not increase the risk for FSGS because the distribution of genotypes was similar in both patients and control subjects, regardless of HIV-1 status; however, R138Q heterozygosity was associated with a five-fold increased risk for FSGS, with similar effects observed in both EA and AA. This is the first study to implicate heterozygosity for R138Q as a risk factor for late-onset FSGS. No other codon-changing mutations were observed in either patients with sporadic FSGS or HIV-associated FSGS. The attributable risk of NPHS2 for adult sporadic and HIV-associated FSGS is extremely small given the low allele frequency of R138Q and the lack of involvement of other rare or common mutations.
Several studies have suggested that the missense mutations P20L and R229Q may have a role in FSGS. Caridi et al.23 reported the occurrence of P20L heterozygous single mutations in three of 120 steroid-resistant and two of 59 steroid-sensitive pediatric patients with an age of onset between 1.5 and 9.8 yr. In our study, P20L was not observed in either EA or AA patients with FSGS and was only rarely observed among EA and AA control subjects (minor allele frequency 0.04 to 0.08%, respectively), suggesting no role for P20L in late-onset FSGS, at least in our population. In the study by Caridi et al., the allele frequency was 1.3% in the patient group, but without knowledge of the allele frequency in the general population, its functional/causal role in childhood nephrotic syndrome cannot be determined. It therefore is possible that (1) P20L is a neutral polymorphism in the Italian population and has no functional consequence; (2) P20L is a causal mutation and predisposes to early-onset renal disease as a heterozygous single mutation; or (3) P20L interacts with other, as-yet-undetected mutations in NPHS2 or other gene(s), causing early-onset disease. These alternative hypotheses for P20L warrant further investigation; however, in our population of AA and EA with sporadic FSGS, P20L was not observed in any individual with FSGS, and the 10 heterozygotes and single homozygote were unaffected control subjects. Urinalysis was available for 10 of the 11 individuals with P20L, none of whom had proteinuria. These results support the first hypothesis that P20L is a neutral polymorphism.
The R229Q mutation has also been implicated in renal disease as a compound heterozygote occurring with a second NPHS2 mutation and in the heterozygous state in adult-onset FSGS.7 There is also in vitro evidence that the R229Q protein binds less efficiently to nephrin.7 In our study, R229Q occurred with near-identical frequencies in patients and control subjects and was never observed as homozygote or as a compound heterozygote. In addition, the 24 R229Q heterozygous individuals for whom urine protein results were available lacked proteinuria. We therefore conclude from this study that heterozygosity for R229Q is not a risk factor for late-onset FSGS; however, R229Q may predispose to FSGS or SRNS in the presence of a second pathologic mutation.
It is notable that a review of the literature reveals that only five families with NPHS2 in which one or more individuals presented with FSGS as an adult (>21 yr) have been identified; NPHS2 R229Q heterozygosity was present in all five families together with a second NPHS2 mutation.6,7 Furthermore, R229Q homozygosity was never observed in any family with FSGS or individual with sporadic FSGS.6,7,11,24 These results from family studies reported by others and from this population-based study suggest that R229Q is not a highly pathogenic mutation and likely has little consequence in the heterozygote state in the absence of a second NPHS2 pathogenic mutation.
R138Q is a rare mutation that occurs slightly more frequently in EA than in AA with approximately four in 1000 AA and eight in 1000 EA carrying the mutated allele in the general population. It is unknown whether the frequency of this mutation is elevated in certain ethnicities as a result of founder effects; however, in several studies of NPHS2 mutations in early-onset SRNS, R138Q is the most common pathologic mutation. Before this study, the pathologic consequence of R138Q heterozygosity was unknown; however, we provide robust evidence that R138Q heterozygotes are at a four- to five-fold increased risk for developing FSGS and that they may be at increased risk for steroid resistance because three of the R138Q carriers were steroid resistant; the steroid responsiveness of the fourth R138Q carrier is unknown. Two control subjects were R138Q heterozygotes, but the one individual for whom urine protein determination was available lacked proteinuria. This finding suggests that family members of patients with SRNS or FSGS in whom R138Q is a contributing mutation may be at increased risk for FSGS. By inference, this finding suggests that carriers of other pathologic NPHS2 mutations may also be at increased risk for late-onset FSGS, but studies with sufficient power to address this issue will be difficult to perform.
Two haplotypes, 7 and 8, were associated with modified risk for FSGS in AA and EA, respectively. The high probability of false discovery for haplotype 8 (FDR 22%) indicates that this association is not robust; however, the FDR for haplotype 7 was only 1.3%. Approximately 20% of the AA population carry either one or two haplotype 7 chromosomes and have a 50% risk reduction for FSGS compared with those not carrying this haplotype. Haplotype 7 has no structural mutations, suggesting that the protective effect might result from an unidentified allele in a regulatory region, possibly affecting transcription rates and podocin expression. Haplotype 7 was never observed in the EA population. This finding was unexpected because FSGS incidence is higher in AA. It is therefore likely that genes other than NPHS2 are responsible for the higher rate of FSGS in the AA population.
This study shows that NPHS2 mutations and polymorphisms contribute to sporadic or HIV-1–associated FSGS in the AA and EA populations. The structural NPHS2 polymorphisms P20L, G423R, R229Q, and A242V showed no associations with sporadic FSGS in either AA or EA; however, both AA and EA who carry the R138Q allele, quite rare in the general population but relatively common in families with early-onset SRNS or FSGS, are at increased risk for developing steroid-resistant FSGS. The clinical implication of this finding is that relatives of patients who have SRNS or FSGS and in whom causal NPHS2 mutations have been identified may also be at increased risk for SRNS or FSGS if they are carriers of pathologic mutations. Further studies in large, racially diverse cohorts are needed to confirm the clinical significance of these observations.
CONCISE METHODS
Clinical Ascertainment
The study protocol was approved by the institutional review boards of all participating medical centers, and all patients gave informed consent. Inclusion criterion was diagnosis of FSGS or collapsing glomerulopathy, based on renal biopsy. Renal biopsy diagnoses were made by diverse pathologists over a period of two decades. Because the diagnostic distinction between collapsing glomerulopathy and FSGS has only recently been codified, it was not possible to make this distinction reliably in this study population; therefore, for simplicity, the diagnostic term FSGS is used for all purposes. Exclusion criteria were clinical history of post adaptive FSGS as a result of reflux nephropathy, reduced renal mass, sickle cell nephropathy, or morbid obesity. Two patients did not have a native kidney biopsy but had a presentation and course compatible with FSGS and subsequently developed FSGS in the renal transplant.
Demographics of the Study Population
Patients with renal biopsy–confirmed idiopathic FSGS or HIV-1–associated FSGS were enrolled from 22 academic medical centers in the United States as part of the NIH FSGS Genetic Study, which was initiated in 1994 and continues to accrue participants. The study enrolled 377 patients with FSGS and 919 control subjects without known kidney disease. Geographic ancestry was based on self-report. There was no evidence of population substructure among patients and control subjects.25
The study population consisted of three case groups: (1) 130 EA had biopsy-confirmed FSGS and did not have HIV-1 infection, (2) 194 AA had biopsy-confirmed FSGS and did not have HIV-1 infection, and (3) 53 AA had HIV-1–associated FSGS. All had renal biopsies that were consistent with HIV-1–associated nephropathy, with collapse of glomerular capillaries and podocyte hyperplasia. Age of renal biopsy, which was available for all cases, was taken as a proxy for age of FSGS onset. The mean age of EA and AA patients with FSGS was 35 and 34 yr, respectively. Among the AA patients, 9.7% were younger than 18 yr and 3.6% were younger than 12 yr. The percentage of EA patients who were younger than 18 and 12 yr was 19.2 and 6.1%, respectively.
The control group included 919 adults, 674 of whom were HIV-1 uninfected and 245 were HIV-1 positive. The 245 HIV-1–positive control subjects were AA patients enrolled in the Johns Hopkins AIDS Link to the Intravenous Drug Experience (ALIVE) cohort from Baltimore, MD, who did not have FSGS or evidence of nephrotic syndrome after at least 8 yr of HIV-1 infection exposure. The absence of kidney disease was defined as having normal serum creatinine (≤1.4 mg/dl) and lack of proteinuria (urine protein-to-creatinine ratio <0.5). This group represents a hypernormal sample in that they are drawn from the AA population at risk for HIV-1–associated FSGS, have been HIV-1 infected, and yet lack evidence of FSGS or proteinuria. Urine protein-to-creatinine ratios were obtained for all of the HIV-1–infected AA control subjects at the time of study enrollment.
Blood samples from normal donors were obtained from the National Institutes of Health Clinical Center in Bethesda, MD, and the NCI-Frederick Blood Donor Program in Frederick, MD. These patients all lacked a history of kidney disease. Urine protein-to-creatinine ratios were obtained for one of two R138Q heterozygous control subjects, 24 of 37 R229Q heterozygous control subjects, 10 of 11 P20L heterozygous (nine) or homozygous (one) control subjects, 59 of 70 A242V heterozygous control subjects, and 17 of 18 G42R heterozygous control subjects.
Identification of Exonic SNP by Resequencing
Nucleotide polymorphisms in the exons of NPHS2 were identified by resequencing DNA from 265 patients with FSGS and 179 control subjects. Each of the eight exons and intron-exon boundaries were amplified using primers and conditions as described previously.24 PCR products were reacted with Big Dye Terminator Mix (Applied Biosystems [ABI], Foster City, CA) using an M13 universal primer. Sequences were determined using ABI software. Sequences were analyzed using Mutation Surveyor software (Soft Genetics, State College, PA) and by inspection by eye.
Genotyping of SNP Alleles
SNP occurring within exons were genotyped by TaqMan assays (ABI) or PCR-RFLP using the same primers and PCR conditions as were used for resequencing. All individuals included in the SNP discovery resequencing effort were also genotyped to confirm the SNP observed on the sequence traces. The results were consistent between the two methods with no discordances observed.
Seven additional dbSNP database (db) SNP from the National Center for Biotechnology Information were selected on the basis of their allele frequencies and availability as TaqMan-On-Demand assays from ABI. These SNP were genotyped according to the manufacturer’s instructions. For controlling for sample-handling or genotyping errors, 10% of the DNA samples were duplicated among plates; no discrepancies were found.
Statistical Analyses
Genetic associations between NPHS2 SNP and FSGS were assessed by comparing genotypic frequencies between patients and control subjects using Fisher exact test. The OR with 95% CI were calculated. Best haplotype reconstruction frequencies were estimated using the program PHASE 2.1.26,27 Haplotype frequencies between patients and control subjects were evaluated by Fisher exact test of one haplotype against all others. The FDR for a given P value is given by the q value22 and was calculated using the statistical package R.28 Linkage disequilibrium was calculated between all pairs of markers using the program HAPLOVIEW.29 Statistical analyses were performed with SAS 9.1 (SAS Institute, Cary, NC), unless otherwise stated.
ACKNOWLEDGMENTS
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400; and the National Institute on Drug Abuse, National Institutes of Health, under grant DA-04334. This Research was supported in part by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research and National Human Genome Research Institute.
We thank Joseph Chichetti, Randall Johnson, and Carl McIntosh for excellent technical support and the volunteers and their families for participating in this study.
Footnotes
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services; neither does mention of trade names, commercial products, or organizations imply endorsement by the US government.
DISCLOSURES
None.
REFERENCES
- 1.Schnaper HW. Idiopathic focal segmental glomerulosclerosis. Semin Nephrol. 2003;23:183–193. doi: 10.1053/snep.2003.50016. [DOI] [PubMed] [Google Scholar]
- 2.Kitiyakara C, Eggers P, Kopp JB. Twenty-one-year trend in ESRD due to focal segmental glomerulosclerosis in the United States. Am J Kidney Dis. 2004;44:815–825. [PubMed] [Google Scholar]
- 3.Pollak MR. The genetic basis of FSGS and steroid-resistant nephrosis. Semin Nephrol. 2003;23:141–146. doi: 10.1053/snep.2003.50014. [DOI] [PubMed] [Google Scholar]
- 4.Niaudet P. Podocin and nephrotic syndrome: Implications for the clinician. J Am Soc Nephrol. 2004;15:832–834. doi: 10.1097/01.asn.0000118135.00519.b0. [DOI] [PubMed] [Google Scholar]
- 5.Fuchshuber A, Gribouval O, Ronner V, Kroiss S, Karle S, Brandis M, Hildebrandt F. Clinical and genetic evaluation of familial steroid-responsive nephrotic syndrome in childhood. J Am Soc Nephrol. 2001;12:374–378. doi: 10.1681/ASN.V122374. [DOI] [PubMed] [Google Scholar]
- 6.Ruf RG, Lichtenberger A, Karle SM, Hass JP, Anacleto FE, Schultheiss M, Zalewski I, Imm A, Ruf EM, Mucha B, Bagga A, Neuhaus T, Fuchshuber A, Bakkaloglu A, Hildebrandt F Arbeitsgemeinschaft fuer Paeiatrische Nephrologie Study Group. Patients with mutations in NPHS2 (podocin) do not respond to standard steroid treatment of nephrotic syndrome. J Am Soc Nephrol. 2004;15:722–732. doi: 10.1097/01.asn.0000113552.59155.72. [DOI] [PubMed] [Google Scholar]
- 7.Tsukaguchi H, Sudhakar A, Le TC, Nguyen T, Yao J, Schwimmer JA, Schachter AD, Poch E, Abreu PF, Appel GB, Pereira AB, Kalluri R, Pollack MR. NPHS2 mutations in late-onset focal segmental glomerulosclerosis: R229Q is a common disease-associated allele. J Clin Invest. 110:1659–1666. doi: 10.1172/JCI16242. 20029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smithies O. Why the kidney glomerulus does not clog: A gel permeation/ diffusion hypothesis of renal function. Proc Natl Acad Sci U S A. 2003;100:4108–4113. doi: 10.1073/pnas.0730776100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huber TB, Koettgen M, Schilling B, Walz G, Benzing T. Interaction with podocin facilitates nephrin signaling. J Biol Chem. 2001;276:41543–41546. doi: 10.1074/jbc.C100452200. [DOI] [PubMed] [Google Scholar]
- 10.Schwarz K, Simons M, Reiser J, Saleem MA, Faul C, Kriz W, Shaw AS, Holzman LB, Mundel P. Podocin, a raft-associated component of the glomerular slit diaphragm, interacts with CD2AP and nephrin. J Clin Invest. 2001;108:1621–1629. doi: 10.1172/JCI12849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weber S, Gribouval O, Esquivel EL, Morinière V, Tête MJ, Legendre C, Niaudet P, Antignac C. NPHS2 mutation analysis shows genetic heterogeneity of steroid-resistant nephrotic syndrome and low post-transplant recurrence. Kidney Int. 2004;66:571–579. doi: 10.1111/j.1523-1755.2004.00776.x. [DOI] [PubMed] [Google Scholar]
- 12.Franceschini N, North KE, Kopp JB, McKenzie L, Winkler CA. NPHS2 gene, nephrotic syndrome and focal segmental glomerulosclerosis: A HuGE review. Genet Med. 2006;8:63–75. doi: 10.1097/01.gim.0000200947.09626.1c. [DOI] [PubMed] [Google Scholar]
- 13.Huber TB, Simons M, Hartleben B, Sernetz L, Schnidts M, Gundlach E, Saleem MA, Walz G, Benzing T. Molecular basis of the functional podocin-nephrin complex: mutations in the NPHS2 gene disrupt nephrin targeting to lipid raft microdomains. Hum Mol Genet. 2003;12:3397–3405. doi: 10.1093/hmg/ddg360. [DOI] [PubMed] [Google Scholar]
- 14.Pereira AC, Pereira AB, Mota GF, Cunha RS, Herkenhoff FL, Pollak MR, Mill JG. NPHS2 R229q functional variant is associated with micro albuminuria in the general population. Kidney Int. 2004;65:1026–1030. doi: 10.1111/j.1523-1755.2004.00479.x. [DOI] [PubMed] [Google Scholar]
- 15.Caridi G, Bertelli R, Carrea A, Di Duca M, Catarsi P, Artero M, Carraro M, Zennaro C, Candiano G, Musante L, Seri M, Ginevri F, Perfumo F, Ghiggeri GM. Prevalence, genetics, and clinical features of patients carrying podocin mutations in steroid-resistant nonfamilial focal segmental glomerulosclerosis. J Am Soc Nephrol. 2001;12:2742–2746. doi: 10.1681/ASN.V12122742. [DOI] [PubMed] [Google Scholar]
- 16.Frishberg Y, Rinat C, Megged O, Shapira E, Feinstein S, Rass-Rothschild A. Mutations in NPHS2 encoding podocin are a prevalent cause of steroid-resistant nephrotic syndrome among Israeli-Arab children. J Am Soc Nephrol. 2002;13:400–405. doi: 10.1681/ASN.V132400. [DOI] [PubMed] [Google Scholar]
- 17.Yu Z, Ding J, Huang J, Yao Y, Xiao H, Zhang J, Liu J, Yang J. Mutations in NPHS2 in sporadic steroid-resistant nephrotic syndrome in Chinese children. Nephrol Dial Transplant. 2005;20:902–908. doi: 10.1093/ndt/gfh769. [DOI] [PubMed] [Google Scholar]
- 18.Kopp JB, Winkler C. HIV-associated nephropathy in African Americans. Kidney Int Suppl. 2003:S43–S49. doi: 10.1046/j.1523-1755.63.s83.10.x. [DOI] [PubMed] [Google Scholar]
- 19.Eggers PW, Kimmel PL. Is there an epidemic of HIV infection in the US ESRD program? J Am Soc Nephrol. 2004;15:2477–2485. doi: 10.1097/01.ASN.0000138546.53152.A7. [DOI] [PubMed] [Google Scholar]
- 20.Freedman BI, Soucie JM, Stone SM, Pegram S. Familial clustering of end-stage renal disease in blacks with HIV-associated nephropathy. Am J Kidney Dis. 1999;34:254–258. doi: 10.1016/s0272-6386(99)70352-5. [DOI] [PubMed] [Google Scholar]
- 21.Dusel JA, Burdon KP, Hicks PJ, Hawkins GA, Bowden DW, Freedman BI. Identification of podocin (NPHS2) gene mutations in African Americans with nondiabetic end-stage renal disease. Kidney Int. 2005;68:256–262. doi: 10.1111/j.1523-1755.2005.00400.x. [DOI] [PubMed] [Google Scholar]
- 22.Storey JD, Tibshirani R. Statistical significance for genome wide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caridi G, Bertelli R, Di Duca M, Dagnino M, Emma F, Muda AO, Scolari F, Miglietti N, Mazzucco G, Murer L, Carrea A, Massella L, Rizzoni G, Perfumo F, Ghiggeri GM. Broadening the spectrum of diseases related to podocin mutations. J Am Soc Nephrol. 2003;14:1278–1286. doi: 10.1097/01.asn.0000060578.79050.e0. [DOI] [PubMed] [Google Scholar]
- 24.Boute N, Gribouval O, Roselli S, Benessy F, Lee H, Fuchshuber A, Dahan K, Gubler MC, Niaudet P, Antignac C. NPHS2, encoding the glomerular protein podocin, is mutated in autosomal recessive steroid-resistant nephrotic syndrome. Nat Genet. 2000;24:349–354. doi: 10.1038/74166. [DOI] [PubMed] [Google Scholar]
- 25.Orloff MS, Iyengar SK, Winkler CA, Goddard KAB, Dart RA, Ahuja TS, Mokrzycki M, Briggs WA, Korbet SM, Kimmel PL, Simon EE, Trachtman H, Vlahov D, Michel DM, Berns JS, Smith MC, Schelling JR, Sedor JR, Kopp JB. Variants in the Wilms’ tumor gene are associated with focal segmental glomerulosclerosis in the African American population. Physiol Genomics. 2005;21:212–221. doi: 10.1152/physiolgenomics.00201.2004. [DOI] [PubMed] [Google Scholar]
- 26.Stephens M, Smith N, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stephens M, Donnelly P. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2006. Available at: http://www.R-project.org. [Google Scholar]
- 29.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of linkage disequilibrium and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]