Abstract
There is a “life-cycle” of pharmacodynamic (PD) biomarker assays that guides the development and clinical implementation in our laboratories. The well-recognized elements of analytical assay validation and demonstration of fitness-for-purpose of the biomarker, specimen collection, handling and assay methods are only a part of the required activities. Assay transfer across laboratories and testing on actual human clinical specimens are vital for understanding assay performance and robustness. In our experience, this patient specimen-centered approach has required assay method modifications, some unexpected, but which were critical to successful implementation in clinical trials. Additionally, dispersing assays throughout the NCI’s clinical trials network has required the development of calibrator and control materials as well as formal training courses for smooth implementation. One measure of success of this approach has been that a number of the assays developed at Frederick National Laboratory have ultimately reached the stage of commercialization, enabling wide accessibility of the PD biomarker assays by the research community.
Introduction
The development of clinical biomarkers of pharmacodynamic activity at the level of target engagement was initiated at NCI as part of the Division of Cancer Treatment and Diagnosis with the specific intent of providing an accurate, early indication of target engagement by experimental therapeutics in first in man clinical trials, focusing on tumor biopsy specimens as the preferred testing material (1). The initial project was to develop and validate an assay that could be objectively demonstrated to accurately report target engagement by veliparib. The assay readout was the quantity of enzyme product, PAR (Polyadenosyl ribose polymer) in patient tissues, previously shown to decrease in tumors and PBMCs after veliparib treatment (2). The trial objectives included demonstration of achievement of a target plasma level of drug and the inhibition of PARP1 and 2 (Polyadenosyl ribose Polymerase) and in patient biopsy specimens after administration of a single dose of veliparib (3). An additional correlative effort of the project was to measure the effect of veliparib on PARP in circulating PBMCs on the day of drug administration (4). The success of the work in demonstrating target engagement resulted in some additional, and in some respects, unanticipated findings. From this early effort with veliparib evolved an NCI effort to develop a series of pharmacodynamic markers that could be exported to the NCI clinical trials network. The aim was to move the assay beyond a simple laboratory developed test (LDT) to one that could be exported to other institutions. To achieve this goal the assays had to be not only analytically validated but also standardized so that results obtained in different institutions would have the same meaning. These assays are now available and accessible to the broader NCI extramural community (5). In the description of the NCI effort to develop pharmacodynamics markers that follows, we will highlight challenges that were encountered along the way and are summarized in Text Box 1.
Text Box 1.
Challenges encountered in the development of pharmacodynamic assays
-
Assay reproducibility – sample quantity
Small scale clinical feasibility studies on human clinical samples are critically important. Challenges for clinical samples in ELISA based assays include lower than expected levels of the PD analytes and higher than expected variation in baseline biomarker levels. Modifications in assay procedures to improve assay sensitivity were required.
-
Assay reproducibility – reagent constraints
- Reagent issues with materials purchased from commercial research vendors include lot-to-lot variability, outright inconsistency and clearly unsuitable materials
- Commercial antibodies available in large quantities either conjugated to FITC or unconjugated, and highly reproducible across lots is critical
- Reference material or quantitative reference method vital for assuring assay quality over time across laboratories
- Generation of Assay Controls and Calibrators for use in multiple laboratories is required to assure comparability of assay results.
Specimen heterogeneity.
Variable biomarker expression occurs within specimens and across diseases. This was ameliorated in immunoflourescence assays (IFAs, paraffin sections) by bounding the area of the biopsy to be tested using H&E stained slides to demonstrate presence of tumor and exclude areas of tumor necrosis.
-
Quantitation of putative drug-induced biomarker changes in biopsies.
Estimation of target concentration is problematic in IFAs. Quantifying the number of cells in the biopsy that became γH2AX positive after drug treatment and scaling the effect to a responsive xenograft tissue “reference standard” allowed determination of the fraction of cells exhibiting dsDNA breaks.
-
Measuring drug effect in individual circulating tumor cells (CTCs)
Optimal timing of the CTC collection after drug administration is not known. High drug levels immediately after IV infusion may impact CTCs already in the circulation, rather than providing a surrogate for tumor effect. For γH2AX, the 24-hour timepoint provided CTCs that were apoptotic as well as showing patterns of DNA damage.
PAR Immunoassay Development
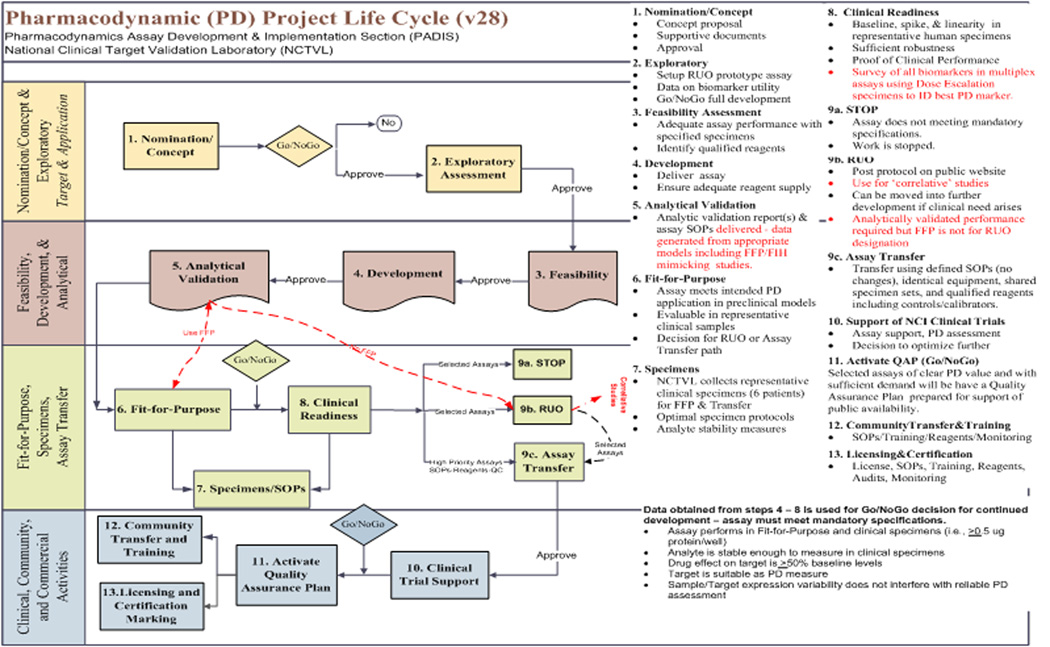
Readying PD assays for clinical implementation involves development and validation of clinically transferable standard operating procedures for the entire process from collection and stabilization of the specimen, sample preparation, assay performance through analysis, interpretation and reporting of the data (Figure 1). Our goal is to develop and optimize procedures that are sufficiently accurate and precise and that have a suitable dynamic range and sensitivity for the clinically available materials (biopsies, PBMCs, CTCs) and the anticipated drug response level. A critical assay performance parameter for PD studies is assay reproducibility. The total imprecision from all sources must be sufficiently low in order to make it possible for the desired drug effect measurement to reach statistical significance (1). Preclinical modeling not only validates the intended assay procedures, but even more importantly, helps guide the design of the clinical trial by providing information regarding the dose necessary to achieve a reproducibly measurable effect on the PD marker and the window of time for tumor biopsy after drug administration that is most likely to yield a PD signal.
Fig. 1.
Functional Flow Chart of the PD assay development, implementation and support strategy used in the Division of Cancer Treatment and Diagnosis for support of early stage clinical trials.
Substantial effort is expended to mirror the intended clinical sampling and assay procedures in fit-for-purpose preclinical studies, however experience in our program has confirmed that it is also critical to perform a small scale clinical feasibility study on human clinical samples and fine tune assay procedures when required prior to analyzing clinical trial specimens. Some of the challenges that we have been faced with upon initial evaluation of clinical samples in ELISA based assays, are lower than expected levels of the PD analytes and higher than expected variation in baseline biomarker levels. One unexpected finding during the early implementation of the PAR IA measurement on core biopsies was that the protein yield from an 18 gage needle biopsy was significantly lower and more variable compared to the protein yield that had been obtained from core needle biopsies of human tumor xenograft models during fit-for-purpose studies. This required larger volumes of lysate to be loaded into the assay well in order to achieve the desired protein load for the assay measurement, requiring assay method modifications. A similar problem arose during early implementation of an immunoassay for Total Topoisomerase I (Top1). Baseline levels of Total Top1 were found to be lower than expected in human biopsies as compared to xenograft models. Additionally, the baseline level of this biomarker is highly variable (6). In the case of this assay, the lysis buffer and procedure were modified to allow for a larger volume of lysate to be loaded into the assay well without interference. A new procedure was quickly established and was shown to significantly improve assay sensitivity on clinical biopsies as part of the ongoing evaluation of indenoisoquinolines class of investigational Top1 inhibitors, and subsequently was demonstrated to measure a decrease of total TOP1 in response to TOP1 inhibitors therapy (7).
Adequate functional and analytical specifications and testing procedures must be established for all critical reagents and control materials in order to identify and mitigate reagent issues with lot-to-lot variability and outright inconsistency, and to manage changes in critical reagent suppliers as R&D companies are bought and sold, and launch and then discontinue these products suppliers that are also often necessary when supporting a clinically implemented assay over the long, multi-year course cycle of clinical drug development. Early in the implementation of the PAR IA, failed lots of two different antibody reagents complicated the support of ongoing analyses. In both cases communication with the research supply company led to the identification of the process changes that were likely the root cause of the lot failures and resolution of the supply issues. Another hurdle that was faced was that the original supplier of the PAR polymer standard was acquired and the product was discontinued. Since no reference material or quantitative reference method was available for this heterogeneous analyte, an anion exchange HPLC method that was originally reported for the analysis of PAR polymer in vivo was adapted to provide a qualitative analysis of the distribution of the polymer sizes and branching structure in a given PAR standard preparation (8). This method was used to confirm the comparability of the PAR polymer from an alternate vendor. Additionally a master lot of material for the PAR polymer has been established and working lots of standard are compared to this master lot to provide continuity of assigned PAR values over time.
It was an objective of the NCI program to standardize measurement methods for tissues and then disseminate that information through the NCI clinical trials network. This in turn raised the issue of assuring assay quality over time across laboratories. In our strategy, clinically proven PD assays are transferred to the cancer research community with a training and certification program provided at the Frederick National Laboratory for Cancer Research. The training program is designed to achieve operator proficiency via intensive classroom and hands on laboratory training in the standard operating procedures for sample preparation, immunoassay measurement, data analysis and interpretation. Generation of relevant assay controls and calibrators that can be used at any of the network testing laboratories is also an important part of the strategy for achieving consistent inter-laboratory performance of an assay. These controls and calibrators vary from assay to assay, but are supported for all assays that NCI supports through community training and open access SOPs (5). We have established mechanisms to share limited amounts of assay critical reagent including controls and calibrators with newly certified PD assay operators so they can establish proficiency on an assay within their own laboratories.
As our laboratory proceeded through this process of increasing robustness and continued to generate usable data, Trevigen, the vendor of the raw materials for the validated immunoassay, elected to develop a commercial kit that has been successfully used by a number of investigators (9), and which has been offered by the company as part of a service supporting the development of PARP inhibitors by monitoring PAR in PBMCs from patients.
Phosphorylated H2Ax Assays
We next sought to develop an assay that would examine the downstream effects of PARP inhibition – DNA damage. While the PAR immunoassay measures the product of the enzyme targeted by PARP inhibitors and is therefore a direct reporter of target engagement, the pharmacological objective of PARP inhibition is to impede or prevent DNA repair. Assays to evaluate this are also needed to evaluate the efficacy combining PARP inhibitors in the clinic with drugs that cause DNA damage. Such an assay would also support the pharmacodynamic evaluation of other types of DNA damaging agents. The Division of Cancer Treatment and Diagnosis initiated a program to develop the indenoisoquinoline class of Top1 inhibitors (NSC 724998, 725776 and 706744) (10) resulting in accumulation of double strand DNA breaks. The assay implemented for this drug effect was measurement of pS139 H2Ax (called γH2AX in the literature) in formalin fixed, paraffin embedded biopsy tissues via immunofluorescence assay (IFA) using microscopy (11). Phosphorylation of histone H2AX to form γH2AX is well documented as a marker for DNA double-strand breaks and programmed cell death (11, 12, 13). Several immunological assays have been reported for evaluation of γH2AX, all based on a specific antibody that recognizes phosphorylated H2AX protein.
The work was greatly helped by the existence of a commercial monoclonal antibody (JBW301) that was available in large quantities either conjugated to FITC or unconjugated, and which was highly reproducible across lots purchased from the vendor, as established by a series of internal quality control measures including specificity testing by Western blotting and on test tissues established as positive and negative for target expression. During assay development, direct modulation of γH2AX levels by the 3 indenoisoquinoline Top1 inhibitors in mouse target tissues included skin (hair follicles become positive), intestine (crypt cells become positive), as well as in both responsive and non-responsive xenografts (13). A key outcome of this work was the demonstration of a strong correlation between tumor growth inhibition (measured xenograft volumes) and the amount of γH2Ax induced on Day 1 of the treatment cycle as a function of dose, for both Topotecan, a known Top1 inhibitor and 724998 (now called LMP400 and in clinical trials at NCI). The analytical validation, fitness-for-purpose demonstration, and inter-laboratory transfer of the tissue IFA and it’s application in the clinic demonstrated robustness of the methodology, but once again the need for assay modification arose after transfer, once testing on actual clinical specimens was initiated. Important elements that were necessary for the clinical implementation included a sampling strategy in which the area of the biopsy to be tested was bounded by use of H&E stained slides to demonstrate presence of tumor and exclude areas of tumor necrosis, then subsequently testing a series of seven, 5 micron slides, separated from each other by three sections, across the biopsy. This approach also had the advantage of providing backup slides to use for restaining if required and for additional correlative marker assessment.
Tissue extraction assay methods have the advantage of reporting the average analyte concentration and distribution within a biopsy but have the disadvantage that the amount of signal specifically associated with tumor cells cannot be assessed. Tissue IFAs have the capability of reporting that data, but unless multiple slides are tested, actual distribution throughout the volume of the biopsy is not assessable. In addition, estimation of target concentration is problematic. We overcame these issues by quantifying the number of cells in the biopsy that became γH2AX positive after drug treatment and scaled the effect by use of a responsive xenograft tissue “reference standard” which we used to determine the fraction of cells exhibiting dsDNA breaks in a sensitive xenograft model at doses that caused tumor regression in mouse models, and at lower dose levels that did not. The ability to construct a response curve from these reference standards was enabled by the dynamic range of the assay: from 1–3% of cells positive in vehicle-treated controls to >30% positive in tumors of mice treated at the daily × 5 MTD of the test compound, which caused tumor regression.
Our preclinical experience with the robustness of γH2Ax as a PD biomarker suggested additional useful assay platforms. As a replacement for normal tissue surrogates (PBMCs, skin), we developed PD biomarker applications on the CellSearch CTC platform, already used clinically for patient monitoring (described by Yap et al. in this issue, 14) opening up the possibility of measuring drug effect on putative tumor cells in the blood instead of requiring a tumor biopsy.
We successfully developed this methodology and reported that active drugs known to induce DNA damage (topotecan, gemcitabine) also induced a strong γH2Ax signal in CTCs isolated from patients on clinical trials. In contrast, drugs that do not induce DNA damage (the kinase inhibitor fostamatinib and rapamycin (15, 16, 17)) did not induce a γH2Ax signal in CTCs. A limitation of the approach was the necessity of testing CTCs 24 hours after drug administration due to limitations within the clinic and the uncertainty around CTC half-life in the circulation (estimated to be 1 to 2 hours; 18). The concern was that the initially high blood concentration of drug during an IV infusion could cause the PD response in CTCs, rather than drug exposure inside the tumor lesions, so CTC sampling emphasized time points well after peak drug levels were reached, Although subsequent studies have not identified an immediate response in CTCs that could be associated with high plasma drug concentrations, the γH2Ax signal observed in the first clinical applications of the assay was not the punctate, focal pattern associated with the immediate response of the repair enzymes to DNA damage. Rather, the γH2Ax signal 24 hours after dosing was distributed throughout the nucleus (Figure 2), and images also showed a number of γH2Ax positive CTCs with the nuclear segmentation that is a hallmark of apoptosis (19–21).
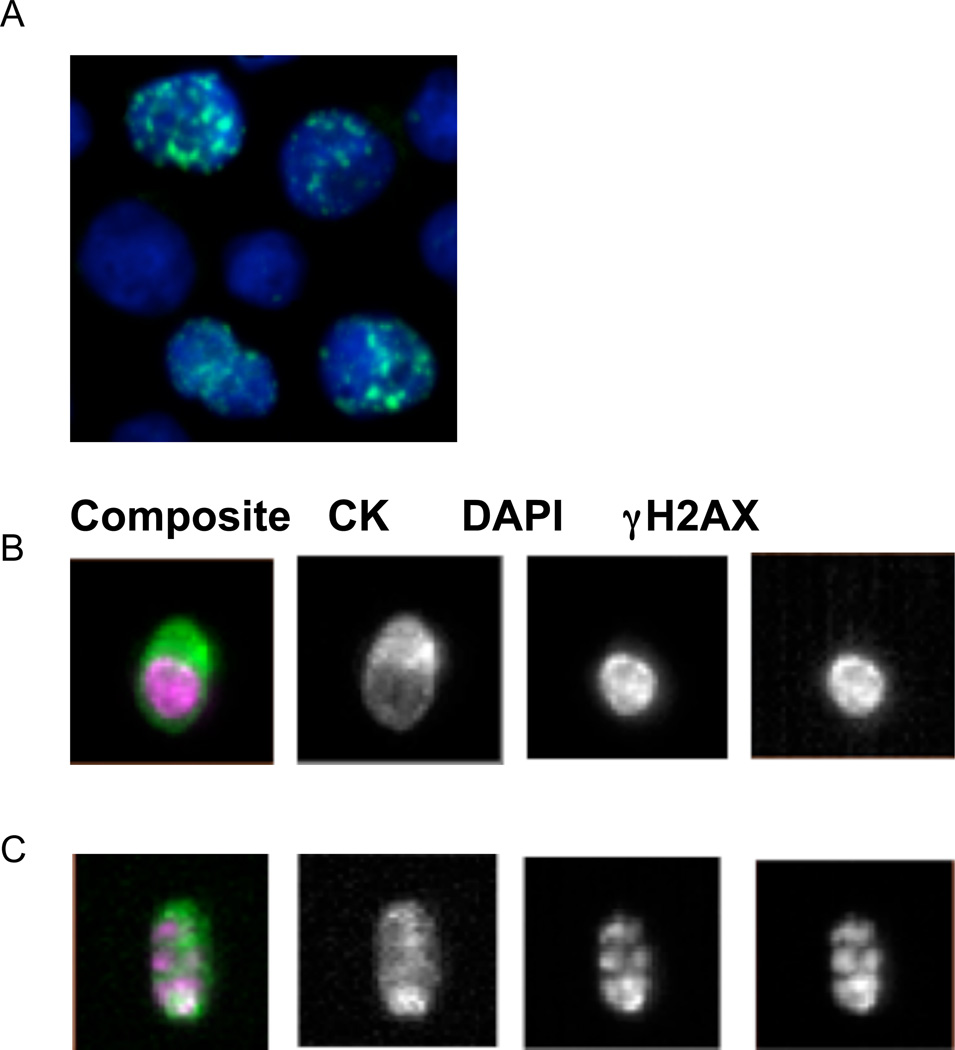
Figure 2.
γH2AX detection in CTC
A, Photomicrograph of γH2AX-postive MCF-7 breast cancer cells used as the positive control. Cells were treated with 1 εmol/L topotecan for 2 hours and stained with γH2AX-AF488 (green) and DAPI (blue). Image capture was conducted using Leica 5000 DM. Final Magnification 200x.
B & C, Images of γH2AX-postive CTCs from a breast cancer patient treated with ABT-888 combined with Cytoxan. The blood samples were obtained from the patient in cycle one on (B) day 2, 24 hours after drug administration and (C) day 5, 24 hours after drug administration. The EpCAM-enriched CTCs were stained with conjugated antibodies for CK-PE, CD45-APC, DAPI, and γH2AX-AF488 and analyzed on the CellSearch system.
The original IFA with drug response calibrators for use with tumor biopsies and the CellSearch CTC-based assay are now part of the NCI training program for clinical trial laboratories, and assay controls and validation methods are also taught and distributed. Veridex currently offers γH2AX testing for clients in its CLIA laboratory.
Assay selection for specific translational research is largely dependent on study intent, assay sensitivity and specificity, intra- and inter-assay variability, and quantitative/dynamic range, and labor and equipment costs. Western blotting and Dot blotting assays are used for detecting γH2AX in cell lysates, while conventional or confocal microscopy based IFAs and flow cytometry based fluorescence-activated cell sorting (FACS) are used for quantifying γH2AX signal from intact cells (2, 22), although the latter are much more costly and have lower throughput. With these limitations in mind, a third assay was developed and reported (23), using the same tissue extraction and two-site EIA technique developed for measuring PAR. This γH2AX immunoassay employs one unique pair of high affinity anti-γH2AX antibodies as a sandwich bracketing the γH2AX antigen, rather than one antibody, which improves assay specificity compared to a single antibody detection step. A mouse monoclonal antibody to phosphorylated H2AX is immobilized onto each well of plate as capture antibody. A rabbit polyclonal antibody against total H2AX of both phosphorylated and non-phosphorylated protein is used for detection. Anti-rabbit HRP conjugate is subsequently used as the reporter in the presence of chemiluminescent substrates for γH2AX quantitation. Synthetic phosphorylated H2AX peptide standard at 8 different concentrations from 1 to 128 pM is used as quantitative calibrators, and well-characterized extracts of the cancer cell line MCF-7 at 3 known concentrations are used for assay controls. γH2AX quantitation is normalized based on cell numbers or total protein in lysate. A detailed assay SOP is being posted on the NCI-DCTD website (5), and Trevigen has developed and commercialized a quantitative sandwich EIA to measure γH2AX in cell and tissue extracts.
PD Biomarkers of Apoptosis
Cancer cells develop enhanced survival and greater resistance to therapy by evading normal process of programmed cell death or apoptosis by a variety of mechanisms (24, 25). Apoptosis is a complex process and involves multiple pro-survival and pro-apoptotic proteins mainly belonging to a super-family of proteins known as Bcl2 proteins (26). The pro-survival members of Bcl-2 family proteins such as Bcl-2, Bcl-Xl, Mcl-1, Bcl-w, and A1 dynamically engage or sequester pro-apoptotic members of Bcl2 family such as BAX, BAK, BIM, Bad, Puma, Noxa, and Bid to regulate cellular commitment to apoptosis through mitochondrial outer membrane permeabilization (MOMP; 27). It appears that cancer cells dysregulate the delicate balance between pro- and anti-apoptotic proteins by over expression of pro-survival protein members such as Bcl2 in leukemia and colon cancers, by post-translational modification of pro-apoptotic proteins as seen in some breast cancers, and by regulating proteasomal degradation or transcriptional regulation of proteins involved in apoptosis. Several first-in-class drugs are designed and developed with a specific goal of blunting these mechanisms and thus kill cancer cells (24, 28). These drugs selectively target proteins involved in apoptosis pathway and can be roughly grouped into four categories: 1) Bcl2 or BH3 mimetics (29–33); 2) IAP inhibitors or Smac mimetics (34, 35); 3) TRAIL agonist and activating antibodies (36–38); and 4) proteasome inhibitors (39–41). As these newer drugs move to advanced stages of clinical development they present unique challenges for PD monitoring and require effective biomarker implementation strategies. Several members of the Bcl2 family proteins have been explored as potential biomarkers, in preclinical and in a few clinical studies. Overexpression of Bcl-2 (or its family) measured by RT-PCR is the most often studied prognostic biomarker for leukemia, whereas, IHC determination of Ki67 and cleaved caspases are the most explored PD biomarker for drugs targeting apoptosis (42, 43). Additionally, circulating levels of Bcl2 proteins, FASl, TRAIL, surviving, and caspase fragments in blood and immunoblot measurement of Bcl2 family proteins and cleaved caspases in tumor tissues have been studied in some clinical trials. However, none of these biomarkers or their implementation strategies has proven to be effective in determining PD modulation for the drug classes named above (44, 45).
We developed a multiplex biomarker approach, on the Luminex (multiplexed sandwich immunoassay) platform, to study PD modulation by drugs targeting apoptosis (46). This approach involves quantitative measurements of multiple biomarkers covering the intrinsic apoptosis pathway, and not only provides estimates of individual target modulation but also surveys the entire pathway using a single tumor biopsy-thus maximizing the amount of PD information that can be obtained from a single core needle biopsy.
The assays have been developed in collaboration with a commercial provider (RBM Myriad) to assure availability of robust reagents to the wider research community. A unique feature of the multiplex panel TABLE 1 is its precise quantitative measurements of the homo- and hetero-dimers formed in vivo between the various pro-survival and pro-apoptotic proteins, intended to reveal how these oligomers are affected by drug (27, 28, 30). Currently, such assays can only be performed in a qualitative manner by immunoprecipitation techniques (47), which is not feasible in core needle biopsies. The immunoassays we developed utilize well characterized recombinant fusion-proteins as calibrators that mimic the in vivo formed hetero-dimer of Bcl2 proteins. The calibrators also provide a means to compare results from different trials and patients. Finally, the apoptosis process occurs at the interface of the cytosol and mitochondrial membrane where several members of the Bcl2 family are localized. Therefore, central to our strategy for implementation of apoptosis biomarkers was subcellular fractionation of tissue lysates for PD measurements. This strategy also minimizes any in vitro activation of caspases that can occur if mitochondrial contents are mixed with that of cytosol. We implemented a simple protocol to prepare fractionated tissue lysates that can be employed in most clinical laboratories. Although the sample processing and assay method are undergoing thorough validation in a clinical trial to study the PD of an apoptosis modulation, sufficient data has been generated from preclinical studies involving birinapant (Smac mimetic; 46) and selumetinib (MEK inhibitor) (unpublished data) to demonstrate that definitive PD responses can be measured in fractionated extracts. The feasibility of fractionated extractions of 18 g needle biopsies has been demonstrated in biopsies collected from multiple patients (unpublished data), and the protein yields and fractionation’s efficiency conforms to acceptable limits.
Table 1.
Multiplex* panels of apoptosis biomarkers
| Panel 1 | Panel 2 | Panel 3 |
|---|---|---|
|
|
|
Multiplexing is achieved on a Luminex platform, which utilizes bead-based technology to multiplex up to 100 assays. The sandwich immunoassay uses a capture antibody covalently linked to beads and a reporter antibody reactive to separate epitope, linked to a fluorescent probe. In the case of heterodimers, the immobilized antibody captures one component and labeled reporter antibody binds to second component of the heterodimers. The validity of each measurement was demonstrated by immunoprecipitation and Western analysis of each biomarker in cancer cell or tumor tissue lysates.
Our experience with PD biomarkers of apoptosis is still evolving and success of this approach will be revealed by the ongoing clinical utility studies. We envision clinical implementation of the assay panel to be drug-specific in at least some cases. For example, the best possible PD biomarker indicators of drug action leading to apoptosis, in a dose and time-dependent manner, can be identified in pre-clinical animal models. Then, that preferred PD biomarker of the apoptosis panel for that drug can then be designated in the clinical protocol as the clinical correlate to be measured in cases where the amount of biopsy material available is limited – a focus that may justify use of just one of the extraction panels for the assay, thereby saving time, money and patient material.
Conclusion
In summary, PD markers are critical for determining whether a given drug is modulating the target in an expected manner. We have recognized in the effort to develop validated PD markers studies that the process requires a meticulous, thorough approach that is unique for each analyte. Not every analyte will be amenable to PD marker development. Some of those developed successfully are included in Text Box 2. Numerous challenges must be faced during the development of PD assays, including baseline specimen heterogeneity in the studied population, intra-tumor heterogeneity, detection methodology, reagent supply and stability, availability of reference materials and generation of standards and controls. We have been surprised to learn that the markers do not always change in the manner expected based on preclinical models, and to learn that the levels in patient samples are often far lower than observed in those preclinical models. These challenges support a dedicated, non-commercial effort to development of assays for the cancer research community.
Text Box 2.
Pharmacodynamic targets
PD assays currently in use in support of DCTD-sponsored clinical trials:
Apoptosis Multiplex IA
Total Topoisomerase IA
PAR IA
HIF1α IA
Total MET IA
pMET IA (pY1234/5)
γH2Ax CTC
p16 CTC
γH2Ax IFA – paraffin sections
Research use assays (RUO), not transferred interlaboratory:
pERK 1/2 (pT202/pY204)
pAKT 1/2 (pS473)
Assays in development, all for paraffin sections:
DNA Repair IFA Multiplex: RAD51, γH2Ax, pNbs1, pATR, ERCC1
Total MET/pMET IFA Multiplex
EMT IFA Multiplex: Vimentin, CTNNb, E-cadherin, N-cadherin, pan-cytokeratin
CTC EMT phenotyping panel
CDK 1/2
Assays with various technical challenges to implementation:
Total AKT and Total ERK – availability of isoform-specific antibodies
Cleaved Caspase 3 (IFA) – low dynamic range of the assay and high sampling variability
Ki-67 – slow rate of biomarker change in tissues, difficult to document an impact in early stage clinical trials with short drug exposures
HIF1α IFA – highly variable tissue distribution pattern for this marker
Footnotes
Note: All procedures involving the use of laboratory animals were provided by Frederick National Laboratory for Cancer Research, which is accredited by AAALAC International and follows the Public Health Service Policy for the Care and Use of Laboratory Animals.
Disclosure of Potential Conflicts of Interest
R. Kinders is a consultant/advisory board member for Trevigen. L. Wang is a consultant/advisory board member for Janssen Diagnostics.
References
- 1.Doroshow JH, Parchment RE. Oncologic phase 0 trials incorporating clinical pharmacodynamics: from concept to patient. Clin Cancer Res. 2008;14:3658–3663. doi: 10.1158/1078-0432.CCR-07-4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kinders RJ, Hollingshead M, Khin S, Rubinstein L, Tomaszewski JE, Doroshow JH, et al. National Cancer Institute Phase 0 Clinical Trials Team. Preclinical modeling of a phase 0 clinical trial: qualification of a pharmacodynamic assay of poly (ADP-ribose) polymerase in tumor biopsies of mouse xenografts. Clin Cancer Res. 2008;14:6877–6885. doi: 10.1158/1078-0432.CCR-08-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kummar S, Kinders R, Gutierrez ME, Rubinstein L, Parchment RE, Phillips L, et al. Phase 0 clinical trial of the poly(ADP-Ribose) polymerase inhibitor ABT-888 in patients with advanced malignancies. J Clinical Oncol. 2009;27:2705–2711. doi: 10.1200/JCO.2008.19.7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ji J, Kinders RJ, Zhang Y, Rubinstein L, Kummar S, Parchment RE, Tomaszewski JE, Doroshow JH. Modeling pharmacodynamic response to the poly(ADP-ribose) polymerase inhibitor ABT-888 in human peripheral blood mononuclear cells. PLoS One. 2011;6:e26152. doi: 10.1371/journal.pone.0026152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Validated Biomarker Assays. Bethesda, Maryland: The Institute; c2013. [cited 2014 Mar 12]. National Cancer Institute [Internet] Available from: http://dctd.cancer.gov/ResearchResources/ResearchResources-biomarkers.htm. [Google Scholar]
- 6.Pfister TD, Hollingshead M, Kinders R, Zhang Y, Evrard Y, Ji J, et al. Development of an immunoassay for quantification of topoisomerase I in solid tumor tissues. PLoS One. 2012;7:1–13. doi: 10.1371/journal.pone.0050494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pfister TD, Ferry-Galow K, Tariq Mohabbat T, Kinders RJ, Khanna C, Mazcko C, et al. Topoisomerase 1 immunoassay provides proof of target engagement by the indenoisoquinoline class of topoisomerase 1 inhibitors in canine lymphomas [abstract] Mol Cancer Ther. 2013;12(Suppl 11):C278. [Google Scholar]
- 8.Kiehlbauch CC, Aboul-Ela B, Jacobson EL, Ringer DP, Jacobson MK. High resolution fractionation and characterization of ADP-ribose polymers. Anal Biochem. 1993;208:26–34. doi: 10.1006/abio.1993.1004. [DOI] [PubMed] [Google Scholar]
- 9.Heitz F, du Bois A, Rochon J, Scheil-Bertram S, Hils R, Fisseler-Eckhoff A, et al. Requirements to assess feasibility of phase 0 trials during major abdominal surgery: variability of PARP activity. Clin Cancer Res. 2012;18:2632–2637. doi: 10.1158/1078-0432.CCR-12-0021. [DOI] [PubMed] [Google Scholar]
- 10.Antony S, Agama KK, Miao Z, Takagi K, Wright MH, Robles AI, et al. Novel indenoisoquinolines NSC 725776 and NSC 724998 produce Persistent topoisomerase I cleavage complexes and overcome multidrug resistance. Cancer Res. 2007;67:10397–12034. doi: 10.1158/0008-5472.CAN-07-0938. [DOI] [PubMed] [Google Scholar]
- 11.Dickey JS, Redon CE, Nakamura AJ, Baird BJ, Sedelnikova OA, Bonner WM. H2AX: functional roles and potential applications. Chromosoma. 2009;118:683–692. doi: 10.1007/s00412-009-0234-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Redon CE, Nakamura AJ, Sordet O, Dickey JS, Gouliaeva K, Lawrence S, et al. γ-H2AX detection in peripheral blood lymphocytes, splenocytes, bone marrow, xenografts, and skin. Methods Mol Biol. 2011;682:249–270. doi: 10.1007/978-1-60327-409-8_18. [DOI] [PubMed] [Google Scholar]
- 13.Kinders RJ, Hollingshead M, Lawrence S, Ji J, Tabb B, Bonner WM, Pommier Y, et al. Development of a validated immunofluorescence assay for γH2AX as a pharmacodynamic marker of topoisomerase I inhibitor activity. Clin Cancer Res. 2010;16:5447–5457. doi: 10.1158/1078-0432.CCR-09-3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yap TA, Lorente D, Omlin A, Olmos D, de Bono JS. Circulating tumor cells: a multifunctional biomarker. Clin Cancer Res. 2014;20:xx-xx. doi: 10.1158/1078-0432.CCR-13-2664. [DOI] [PubMed] [Google Scholar]
- 15.Wang L, Pfister T, Parchment R, Kummar S, Rubinstein L, Evrard Y, et al. Monitoring drug-induced gH2AX as a pharmacodynamic marker in individual circulating tumor cells. Clin Cancer Res. 2010;16:1073–1084. doi: 10.1158/1078-0432.CCR-09-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park SR, Speranza G, Piekarz R, Wright JJ, Kinders RJ, Wang LH, et al. A multi-histology trial of fostamatinib in patients with advanced colorectal, non-small cell lung, head and neck, thyroid, and renal cell carcinomas, and pheochromocytomas. Cancer Chemother Pharmacol. 2013;71:981–990. doi: 10.1007/s00280-013-2091-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kummar S, Ji J, Morgan R, Lenz HJ, Puhalla SL, Belani S, et al. A phase 1 study of veliparib in combination with metronomic cyclophosphamide in adults with refractory lymphomas. Clin Cancer Res. 2012;18:1726–1734. doi: 10.1158/1078-0432.CCR-11-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meng S, Tripathy D, Frenkel EP, Shete S, Naftalis EZ, Huth JF, et al. Circulating tumor cells in patients with breast cancer dormancy. Clin Cancer Res. 2004;10:8152–8162. doi: 10.1158/1078-0432.CCR-04-1110. [DOI] [PubMed] [Google Scholar]
- 19.Wyllie A, Morris R, Smith A, Dunlop D. Chromatin cleavage in apoptosis: association with condensed chromatin morphology and dependence on macromolecular synthesis. J Pathol. 1984;142:67–74. doi: 10.1002/path.1711420112. [DOI] [PubMed] [Google Scholar]
- 20.Mbebi C, Sée V, Mercken L, Pradier L, Muller U, Loeffler JP. Amyloid precursor protein family-induced neuronal death is mediated by impairment of the neuroprotective calcium/calmodulin protein kinase IV-dependent signaling pathway. J Biol Chem. 2002;277:20979–20990. doi: 10.1074/jbc.M107948200. [DOI] [PubMed] [Google Scholar]
- 21.Zoli W, Ulivi P, Tesei A, Fabbri F, Rosetti M, Maltoni R, et al. Addition of 5-fluorouracil to doxorubicin-paclitaxel sequence increases caspase-dependent apoptosis in breast cancer cell lines. Breast Cancer Res. 2005;7:R681–R689. doi: 10.1186/bcr1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Avondoglio D, Scott T, Kil WJ, Sproull M, Tofilon PJ, Camphausen K. High throughput evaluation of gamma-H2AX. Rad Oncol. 2009;4:31–35. doi: 10.1186/1748-717X-4-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Parchment RE, Tomaszewski JE, Doroshow JH, Ji J, et al. Quantitative gamma-H2AX immunoassay for pharmacodynamic monitoring of DNA damage by chemotherapeutic agents or radiation [abstract] Cancer Res. 2010;70(Suppl 8):A1763. [Google Scholar]
- 24.Cotter TG. Apoptosis and cancer: the genesis of a research field. Nature Rev Cancer. 2009;9:501–507. doi: 10.1038/nrc2663. [DOI] [PubMed] [Google Scholar]
- 25.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 26.Cory S, Adams JM. The Bcl2 family: regulators of the cellular life-or-death switch. Nat Rev Cancer. 2002;2:647–656. doi: 10.1038/nrc883. [DOI] [PubMed] [Google Scholar]
- 27.Chipuk JE, Green DR. How do BCL-2 proteins induce mitochondrial outer membrane permeabilization? Trends Cell Biol. 2008;18:157–164. doi: 10.1016/j.tcb.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davids MS, Letai A. Targeting the B-cell lymphoma/leukemia 2 family in cancer. J Clin Oncol. 2012;30:3127–3135. doi: 10.1200/JCO.2011.37.0981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Touzeau C, Dousset C, Le Gouill S, Sampath D, Leverson JD, Souers AJ, et al. The Bcl-2 specific BH3 mimetic ABT-199: a promising targeted therapy for t(11;14) multiple myeloma. Leukemia. 2013;28:210–212. doi: 10.1038/leu.2013.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 31.Chiappori AA, Schreeder MT, Moezi MM, Stephenson JJ, Blakely J, et al. A phase I trial of pan-Bcl-2 antagonist obatoclax administered as a 3-h or a 24-h infusion in combination with carboplatin and etoposide in patients with extensive-stage small cell lung cancer. British J Cancer. 2012;106:839–845. doi: 10.1038/bjc.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yazbeck VV, Li C, Grandis JR, Zang Y, Johnson DE. Single-agent obatoclax (GX15-070) potently induces apoptosis and pro-survival autophagy in head and neck squamous cell carcinoma cells. Oral Oncol. 2014;50:120–127. doi: 10.1016/j.oraloncology.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Souers AJ, Leverson JD, Boghaert ER, Ackler SL, Catron ND, Chen J, et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med. 2013;19:202–208. doi: 10.1038/nm.3048. [DOI] [PubMed] [Google Scholar]
- 34.Fulda S, Vucic D. Targeting IAP proteins for therapeutic intervention in cancer. Nat Rev Drug Disc. 2012;11:109–124. doi: 10.1038/nrd3627. [DOI] [PubMed] [Google Scholar]
- 35.Krepler C, Chunduru SK, Halloran MB, He X, Xiao M, Vultur A, et al. The novel SMAC mimetic birinapant exhibits potent activity against human melanoma cells. Clin Cancer Res. 2013;19:1784–1794. doi: 10.1158/1078-0432.CCR-12-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng W, Yoshida A, Ueda T. YM155 induces caspase-8 dependent apoptosis through downregulation of survivin and Mcl-1 in human leukemia cells. Biochem Biophys Res Comm. 2013;435:52–57. doi: 10.1016/j.bbrc.2013.04.036. [DOI] [PubMed] [Google Scholar]
- 37.Sayers TJ. Targeting the extrinsic apoptosis signaling pathway for cancer therapy. Cancer Immunol Immunother. 2011;60:1173–1180. doi: 10.1007/s00262-011-1008-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vogler M, Walczak H, Stadel D, Haas TL, Genze F, Jovanovic M, et al. Targeting XIAP bypasses Bcl-2-mediated resistance to TRAIL and cooperates with TRAIL to suppress pancreatic cancer growth in vitro and in vivo. Cancer Res. 2008;68:7956–7965. doi: 10.1158/0008-5472.CAN-08-1296. [DOI] [PubMed] [Google Scholar]
- 39.Lesinski GB, Raig ET, Guenterberg K, Brown L, Go MR, Shah NN, et al. IFN-alpha and bortezomib overcome Bcl-2 and Mcl-1 overexpression in melanoma cells by stimulating the extrinsic pathway of apoptosis. Cancer Res. 2008;68:8351–8360. doi: 10.1158/0008-5472.CAN-08-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lucas CD, Allen KC, Dorward DA, Hoodless LJ, Melrose LA, Marwick JA, et al. Flavones induce neutrophil apoptosis by down-regulation of Mcl-1 via a proteasomal-dependent pathway. FASEB J. 2013;27:1084–1094. doi: 10.1096/fj.12-218990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Podar K, Gouill SL, Zhang J, Opferman KT, Zorn E, Tai YT, et al. A pivotal role for Mcl-1 in Bortezomib-induced apoptosis. Oncogene. 2008;27:721–731. doi: 10.1038/sj.onc.1210679. [DOI] [PubMed] [Google Scholar]
- 42.Hector S, Prehn JH. Apoptosis signaling proteins as prognostic biomarkers in colorectal cancer: a review. Biochim Biophys Acta. 2009;1795:117–129. doi: 10.1016/j.bbcan.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 43.Stearns V, Jacobs LK, Fackler M, Tsangaris TN, Rudek MA, Higgins M, et al. Biomarker modulation following short-term vorinostat in women with newly diagnosed primary breast cancer. Clin Cancer Res. 2013;19:4008–4016. doi: 10.1158/1078-0432.CCR-13-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codony J, Servat X, Garcia-Albeniz C, Pericay V, Alonso P, Escudero C, et al. Soluble FAS in the prediction of benefit from cetuximab and irinotecan for patients with advanced colorectal cancer. Med Oncol. 201330:428. doi: 10.1007/s12032-012-0428-0. [DOI] [PubMed] [Google Scholar]
- 45.Yalcin AD, Gumuslu S, Parlak GE, Bisgin A. Soluble trail as a marker of efficacy of allergen-specific immunotherapy in patients with allergic rhinoconjunctivitis. Med Sci Monit. 2012;18:CR617–CR621. doi: 10.12659/MSM.883488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Srivastava AK, Jaganathan S, Stephen LL, Hollingshead MG, Scull J, Kinders RJ, et al. Progress on development of multiplex panel of 15 biomarkers to support development of anticancer drugs targeting apoptosis [abstract] Cancer Res. 2013;73(Suppl 8):3366. [Google Scholar]
- 47.Rahmani M, Aust MM, Attkisson E, Williams DC, Jr, Ferreira-Gonzalez A, Grant S. Inhibition of Bcl-2 antiapoptotic members by obatoclax potently enhances sorafenib-induced apoptosis in human myeloid leukemia cells through a Bim-dependent process. Blood. 2012;119:6089–6098. doi: 10.1182/blood-2011-09-378141. [DOI] [PMC free article] [PubMed] [Google Scholar]