Abstract
Background
Fetal alcohol spectrum disorder (FASD) is associated with deficits in cerebellar function that can persist through adolescence. Previous studies demonstrated striking inhibition of insulin and insulin-like growth factor (IGF) signaling in ethanol-exposed cerebella.
Objectives
We sought to determine if FASD-induced impairments in motor function were associated with deficits in insulin/IGF signaling in juvenile cerebella. Given the growing evidence that insulin/IGF pathways cross-talk with Notch and Wnt to promote brain development and maturation; we also examined the integrity of canonical Wnt and Notch signaling networks in the brain following chronic prenatal ethanol exposure.
Methods
Pregnant Long Evans rats were fed isocaloric liquid diets containing 0% or 24% ethanol by caloric content from gestation day 6 through delivery. Pups were subjected to rotarod testing on postnatal days (P) 15–16 and sacrificed on P30. Cerebella were used for molecular and biochemical analysis of insulin/IGF-1, canonical Wnt, and Notch signaling mechanisms.
Results
Prenatal ethanol exposures impaired rotarod performance, inhibited signaling through insulin and IGF-1 receptors, IRS-1, and Akt, increased activation of GSK-3β, and broadly suppressed genes mediating the canonical Wnt and Notch networks.
Conclusions
Abnormalities in cerebellar function following chronic prenatal ethanol exposure are associated with inhibition of insulin/IGF, canonical Wnt, and Notch networks that cross-talk via GSK-3β. Effective therapeutic measures for FASD may require multi-pronged support of interrelated signaling networks that regulate brain development.
Keywords: Fetal alcohol spectrum disorder, Insulin, IGF, Wnt, Notch, Cerebellum, Prenatal ethanol exposure, Signal transduction, Multiplex ELISA
Introduction
Alcohol misuse during pregnancy causes neurodevelopmental defects, including microcephaly, cerebellar hypoplasia, motor deficits, and neuro-cognitive impairments ranging from attention deficit hyperactivity disorder to mental retardation. This constellation of abnormalities, together with a number of stereotypical craniofacial defects is termed, ‘fetal alcohol spectrum disorders’ (FASD) [1,2]. A major adverse effect of ethanol on the immature central nervous system (CNS) is to profoundly inhibit insulin and insulin-like growth factor (IGF) signaling [3].
Insulin and IGF regulate neuronal survival and differentiation, myelin formation and maintenance, neuronal migration, plasticity, metabolism, and neurotransmitter supply and responsiveness [4–12]. Ethanol inhibits insulin and IGF-1 receptor tyrosine phosphorylation, receptor tyrosine kinase activation, and insulin receptor substrate (IRS) signaling [13–15]. Importantly, IRS transmits growth, survival, and metabolic signals downstream through phosphotidyl-inositol-3-kinase (PI3K) and Akt, and PI3K/Akt inhibits glycogen synthase kinase 3β (GSK-3β) [3,14–19], which at high levels promotes oxidative stress and apoptosis [12]. Therefore, ethanol’s inhibition of insulin/IGF-1 during development could account for many of the structural and functional phenotypic features of FASD. On the other hand, it is unlikely that this concept reflects the full scope of the problem because many CNS developmental functions are regulated by other signaling networks, including Notch and Wnt, which cross-talk with insulin and IGF pathways [20,21]. Therefore, the adverse effects of prenatal ethanol exposure on CNS development, gene expression, and function could be due to broader impairments of signaling pathways that interconnect with insulin/IGF networks.
Wnt signaling is very complex due to the numerous ligands, receptors, and transcription factors that mediate its physiological functions. The 3 major Wnt signaling pathways are: canonical, Wnt/Calcium, and planar cell polarity [22–24]. In the canonical pathway, Wnt ligands, which comprise a large family of cysteine-rich secreted glycoproteins, bind to Frizzled receptors, inhibiting β-catenin degradation. Accumulation of β-catenin leads to its translocation to the nucleus where it interacts with T cell factor (TCF) transcription factors to regulate gene expression.
The Wnt/Calcium pathway, which is regulated by G coupled phospholipases and proteins, increases cytoplasmic free calcium, and activates protein kinase C, calcium calmodulin mediated kinase II (CK2), and calcineurin (phosphatase) [23,25]. The Wnt planar cell polarity pathway is activated by the RAS homologue gene family member A (RhoA) and RAC1, leading to stimulation of stress kinases such as Jun N-terminal kinase (JNK) and RHO-activated coil-containing protein kinase 1 (ROCK), with attendant remodeling of the cytoskeleton [26–30]. In the CNS, Wnt signaling modulates neuronal proliferation, migration, adhesion, differentiation, and axon outgrowth [31–34]. A potential role for ethanol-mediated impairment of Wnt in FASD was suggested by the recent findings that: 1) both canonical Wnt and insulin/IGF signaling regulate GSK-3β [35,36] and 2) Wnt cross-talks with insulin/IGF networks [20,37–39] through GSK-3β [40]. Mechanistically, ethanol’s activation of GSK-3β could impair Wnt signaling due to increased phosphorylation and attendant degradation of β-catenin [21,24,41–43].
Notch signaling modulates neurogenesis, differentiation, and cell fate during CNS development [44–46]. Notch receptors are located at the cell surface, and Notch signaling is activated by ligand, e.g. Delta, Serrate, Jagged, binding [47]. Subsequent proteolytic cleavage releases Notch’s intracellular domain, which translocates to the nucleus where it binds to transcriptional regulators [47,48] that increase expression of target genes including, Hairy-Enhancer of Split (HES) and HES-related proteins [49,50]. However, Notch signaling is regulated by aspartyl-(asparaginyl)-β-hydroxylase (AAH) [51–53], as Notch (receptor) and Jagged (ligand) have the consensus sequence for AAH hydroxylation [54,55]. AAH’s catalytic activity promotes Notch’s translocation to the nucleus, cell adhesion, and cell motility [56]. AAH is stimulated by insulin/IGF [57,58], and chronic prenatal exposure to ethanol inhibits AAH’s expression and catalytic activity in the developing brain [59].
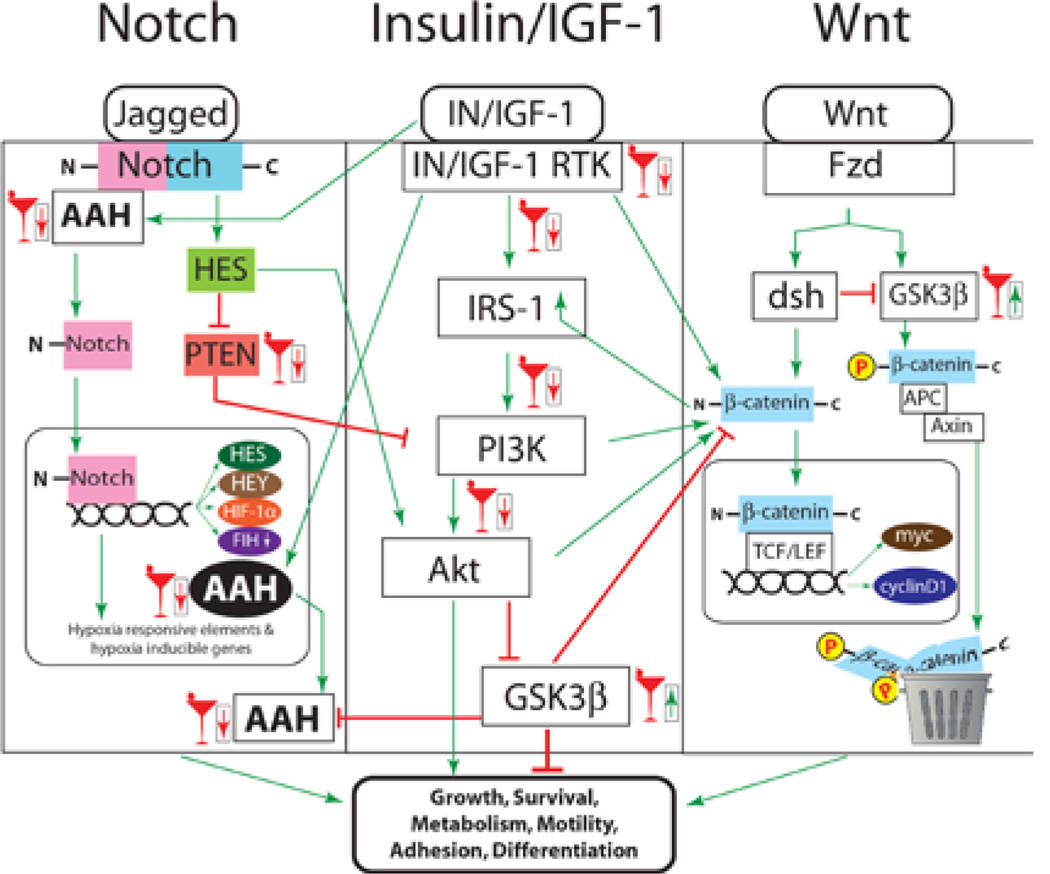
The above discussion highlights critical roles for insulin/IGF, Wnt, and Notch signaling networks in brain development, and provides evidence that chronic prenatal ethanol exposures inhibit insulin/IGF and Notch signaling in immature brains. However, little is known about the effects of ethanol on Wnt signaling, and whether long-term impairments in CNS function caused by prenatal ethanol exposure are due to persistent inhibition of all 3 inter-connecting pathways. Our proposed model for how ethanol disrupts signaling and leads to structural, functional, and molecular abnormalities in the cerebellum is diagramed in figure 1. The present study addresses the overarching hypothesis by examining the integrity of insulin/IGF-1, Wnt, and Notch signaling in relation to the adverse effects of chronic prenatal ethanol exposure on cerebellar function in juvenile rats.
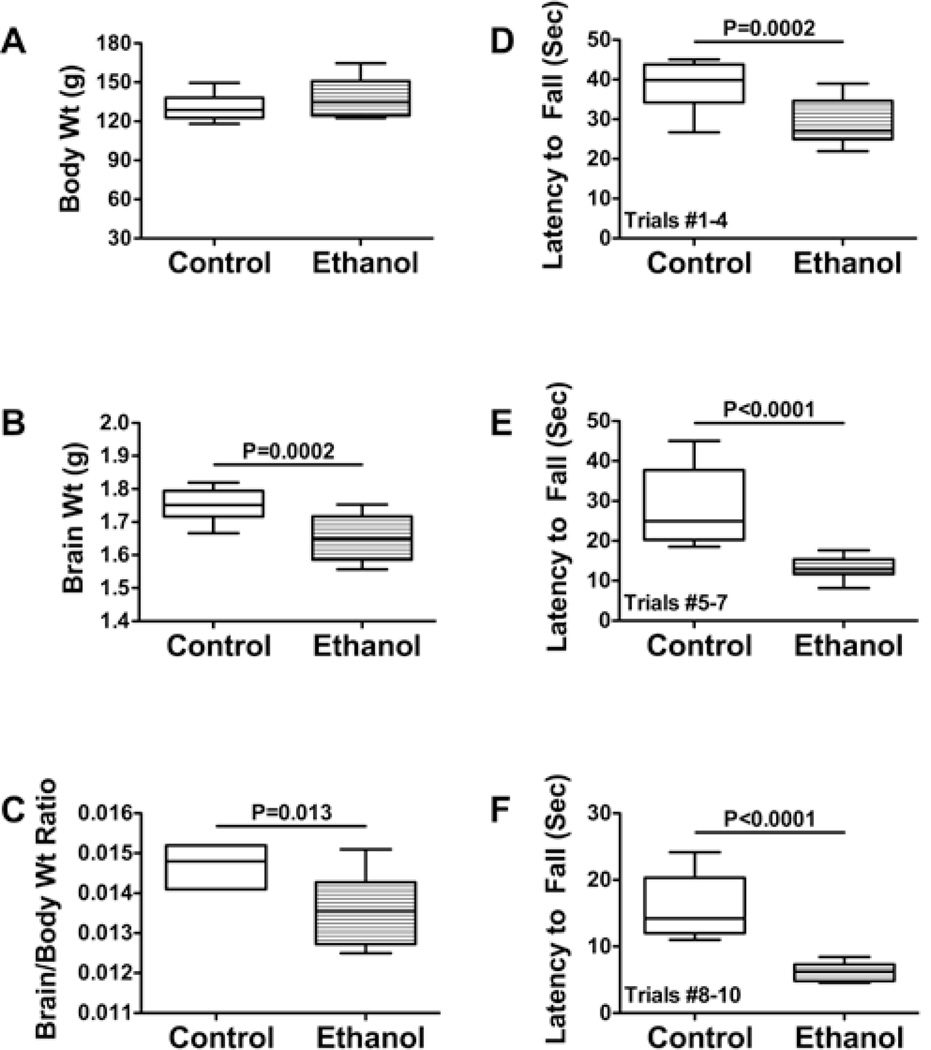
Figure 1.
Effects of prenatal ethanol exposure on body growth, motor performance, and brain weight. Pregnant Long Evans rats were chronically fed with isocaloric liquid diets containing 0% or 24% ethanol. (A) Mean body weights, (B) brain weights, and (C) calculated brain weight/body weight ratios were determined on P30. The Student t-test was used to analyze data in Panels A–C. On P16, the rats were subjected to Rotarod testing with 10 incremental speed trials. Data from Trials (D) 1–4, (E) 5–7, and (F) 8–10 were culled and analyzed using the Mann-Whitney test. Significant inter-group differences are shown above the graphs.
Materials and Methods
Materials
Qiazol reagent, EZ1 RNA universal tissue kit, QuantiTect SYBR Green polymerase chain reaction (PCR) master mix, Wnt array RT2-PCR Profiler and the BIO Robot Z1 were from Qiagen Inc. (Valencia, CA). Monoclonal antibodies to Notch-1, Jagged-1, β-Actin, HES-1, and β-catenin HES-1, and β-catenin were purchased from Abcam Inc. (Cambridge, MA) or Chemicon International (Tecumsula, CA). The A85G6 and A85E6 AAH monoclonal antibodies to AAH and Humbug, respectively, were generated to human recombinant protein [59] and purified over Protein G columns (Healthcare, Piscataway, NJ). Histofix was purchased from Histochoice (Amresco, Solon, OH). The AMV first strand cDNA synthesis kit was obtained from Roche Diagnostics Corporation (Indianapolis, IN). Enzyme-linked immunosorbent assay (ELISA) 96-well plates and the ELISA plate washer were purchased from Nunc (Rochester, NY). Horseradish peroxidase (HRP)-conjugated secondary antibody, Amplex Red soluble fluorophore, and the Akt Pathway Total and Phospho 7-Plex panels were purchased from Invitrogen (Carlsbad, CA). HRP-labeled polymer conjugated secondary antibody used for immunohistochemistry was purchased from Dako Corp (Carpentaria, CA). The SpectraMax M5 microplate reader was purchased from Molecular Devices Corp. (Sunnyvale, CA). Bicinchoninic acid (BCA) reagents were from Pierce Chemical Corp. (Rockford, IL). All other fine chemicals were purchased from CalBiochem (Carlsbad, CA), Pierce (Rockford, IL), or Sigma (St. Louis, MO).
In utero ethanol exposure model
Twenty-four pregnant Long Evans rats were pair-fed with isocaloric liquid diets (BioServ, Frenchtown, NJ) in which ethanol comprised 0% (N=12) or 24% (N=12) of the caloric content. The diets were initiated on gestation day (GD) 6, and continued until parturition [60,61]. GD 0 was the day that sperm was detected in vaginal smears. The 24% ethanol containing diet produced early morning blood alcohol concentrations of 31.3 ± 8.3 µM. Ethanol feeding was begun on GD6 because earlier exposures lead to excessive fetal loss due to impaired placentation [62]. The dams were monitored to ensure equivalent caloric intake and body weight maintenance.
Once the pups were born, the litters were culled to 8 pups per dam, and maternal diets were switched to standard chow. Pups were housed with the dams until they were weaned. The pups were monitored daily and weighed weekly. The offspring were subjected to rotarod testing of motor function on postnatal days (P) 15-P16, and sacrificed on P30. Freshly harvested cerebella were snap frozen in a dry ice-methanol bath and stored at –80°C for RNA and protein studies. The cerebellum was studied because it is a major target of ethanol-induced neurotoxicity [1–3]. The Lifespan-Rhode Island Hospital IACUC committee approved these procedures and the use of rats in experiments.
Rotarod testing
We used rotarod tests to assess effects of chronic pre-natal ethanol exposure on motor function [52,63]. On P15, rats were trained to remain balanced on the rotating Rotamex-5 apparatus (Columbus Instruments) at 1–5 rpm. All tested rats succeeded in the training. On P16, rats (N=12 per group) were administered 10 trials at incremental speeds up to 10 rpm, with 10 minutes rest between trials. The latency to fall was automatically detected and recorded with photocells placed over the rod. However, trials were stopped after 30 seconds to avoid exercise fatigue. Data from trials 1–3 (2–5 rpm), 4–7 (5–7 rpm), and 8–10 (8–10 rpm) were culled and analyzed using the Mann-Whitney test.
Quantitative Reverse Transcriptase Polymerase Chain Reaction (qRT-PCR) analysis
We used qRT-PCR analysis to measure mRNA expression. Tissues were homogenized in Qiazol reagent, and total RNA was isolated using the RNAeasy Mini kit. RNA was reverse transcribed using the AMV First Strand cDNA synthesis kit and random oligodeoxynucleotide primers. The resulting cDNAs were used as templates in probe-based qPCR amplification reactions with gene-specific primer pairs. Primer-probe pairs were designed using ProbeFinder Version 2.45 software (Roche Applied Science, Indianapolis, IN), and mRNA target specificity was verified using NCBI-BLAST (Basic Local Alignment Search Tool). The amplified signals were detected and analyzed using the LightCycler 480 Real-Time PCR System and software (Roche Diagnostics, Indianapolis, IN). Expression levels for genes of interest were normalized to β-actin, which was measured simultaneously in the duplex qPCR reactions. Inter-group statistical comparisons were made using the calculated mRNA/β-actin ratios. In separate reactions, we determined that the mean levels of β-actin mRNA did not differ significantly for control and ethanol-exposed rats. The inclusion of primer-probe pairs to simultaneously measure β-actin enabled us to control for well-to-well differences caused by technical errors in pipetting, and also express levels of gene expression as relative abundance rather than fold differences.
Duplex Enzyme Linked Immunosorbent Assay (ELISA)
Cerebellar tissue was homogenized in Nonidet-40 (NP-40) lysis buffer supplemented with protease (1 mM PMSF, 0.1 mM TPCK, 1 mg/ml aprotinin, 1 mg/ml pepstatin A, 0.5 mg/ml leupeptin, 1 mM NaF, 1 mM Na4P2O7) and phosphatase (2 mM Na3VO4) inhibitors using a Retsch Tissue Lyser (Newtown, PA). Samples were centrifuged at 15,000 g for 10 minutes, and the supernatants were used in assays of immunoreactivity. Protein concentrations were measured with the BCA assay. For direct binding ELISAs, 100ng of protein in 50 µl of bicarbonate buffer were adsorbed to the bottoms of MaxiSorp 96-well ELISA plates by overnight incubation at 4°C. After rinsing in Tris buffered saline (TBS; 50 mM Tris-HCl, pH 7.5, 150 mM NaCl), the wells were blocked for 3 hours with 250 µl/well of 2% bovine serum albumin (BSA) in TBS. The samples were then incubated with primary antibody (0.1–0.5 µg/ml) for 1 hour at 37°C. Immunoreactivity was detected with HRP-conjugated secondary antibody (1:10000) and Amplex UltraRed soluble fluorophore. Amplex Red fluorescence was measured (Ex 530/Em 590) in a SpectraMax M5 microplate reader (fluorescence light units; FLU). Subsequently, the samples were incubated with biotin-conjugated antibodies to large ribosomal protein (RPLPO), and immunoreactivity was detected with streptavidin-conjugated alkaline phosphatase (1:1000) and the 4-Methylumbelliferyl phosphate (4-MUP) fluorophore. Fluorescence (Ex360/Em450) intensity was measured in a SpectraMax M5. Non-specific binding was assessed with parallel negative control incubations in which the primary or secondary antibody was omitted. The mean ratios of specific protein/RPLPO fluorescence were used for statistical comparisons.
Multiplex ELISA
We used bead-based multiplex ELISAs to examine the integrity of insulin and IGF-1 signaling networks by measuring immunoreactivity to the insulin receptor (IR), IGF-1 receptor (IGF-1R), IRS-1, Akt, GSK-3β, pYpY1162/1163-IR, pYpY1135/1136-IGF-1R, pS312-IRS-1, pS473-Akt, and pS9-GSK3β according to the manufacturer’s protocol. Samples containing 200 µg of protein were incubated with the beads, and captured antigens were detected with biotinylated secondary antibody and phycoerythrin-conjugated Streptavidin. Plates were read in a Bio-Plex 200 system (Bio-Rad, Hercules, CA). Data are expressed as fluorescence light units corrected for protein concentration.
Statistical analyses
To ensure inclusion of subjects with independent exposures to the control or ethanol-containing liquid diets, data from pupsin different litters (N=12 each) were analyzed in each group. Data corresponding to gene expression or immunoreactivity are depicted in boxplots representing the medians (horizontal bars), 95% confidence intervals (box limits), and range (whiskers) for each group. Planned inter-group comparisons were made using Student t tests. Multivariate analysis of variance (MANOVA) tests were used to identify the significant main effects of ethanol on: 1) upstream mediators of insulin and IGF signaling (trophic factor, receptor and IRS expression and phosphorylation); 2) downstream mediators of insulin/IGF signaling through Akt pathways; 3) Wnt genes and 4) Notch-related signaling genes. This approach was used to consolidate the large number of dependent variable responses in relation to ethanol exposure and correct for repeated measures. Statistical analyses were performed using the GraphPad Prism 5 (San Diego, CA) or NCSS8 (Kaysville, UT) software and significant P-values (<0.05) are indicated within graph panels or tables.
Results
Effects of chronic prenatal ethanol exposure on juvenile cerebellar motor function
In control and ethanol-exposed pups, body weights increased continuously, and at the end of the experiment (P30), the mean body weights were similar for the two groups (Figure 1A). Therefore, chronic prenatal ethanol exposure had no significant effect on postnatal body growth through early adolescence. In contrast, the mean brain weights (Figure 1B) and the calculated mean brain weight/body weight ratios (Figure 1C) were significantly reduced in the ethanol-exposed rats. Rotarod tests of cerebellar function demonstrated significantly shorter mean latencies to fall over all trial blocks, but the largest inter-group difference was observed with the most challenging trials (Figures1D–1F).
Chronic prenatal ethanol exposure causes impairments in insulin/IGF signaling in the brain
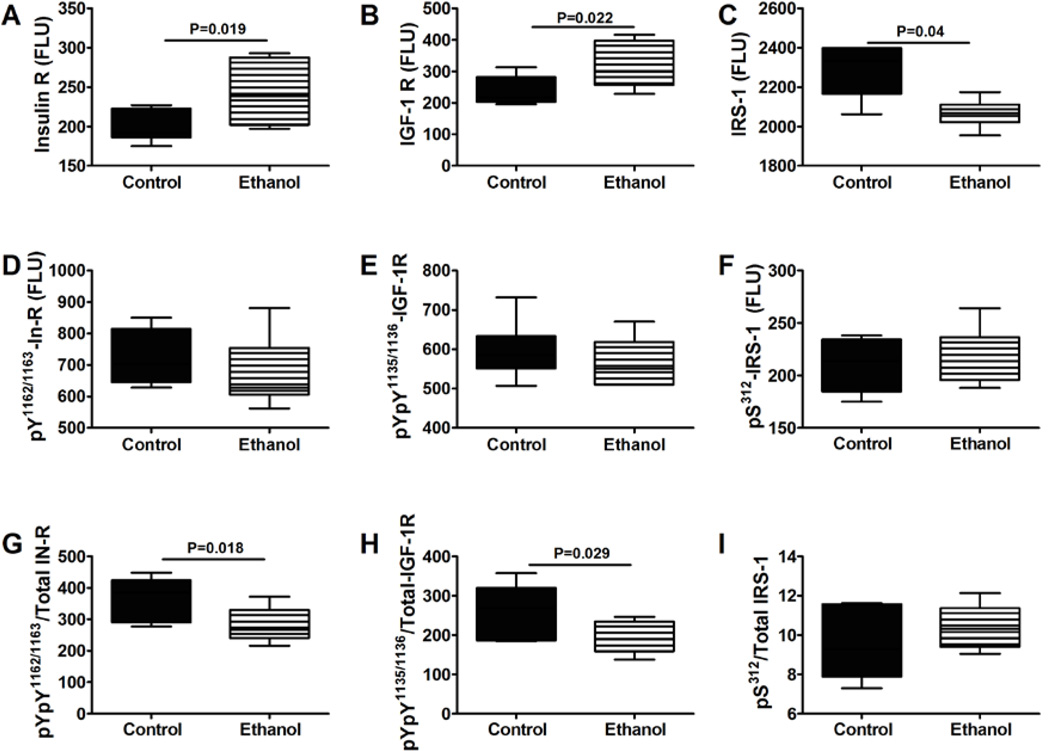
We used multiplex ELISAs to interrogate ethanol’s effects on the integrity of brain insulin/IGF signaling. To examine upstream pathway components, we measured total and phosphorylated levels of insulin receptor, IGF-1 receptor, and IRS-1, and calculated the relative levels of phosphorylation from the ratios of phospho-/total protein (Figure 2). Chronic prenatal ethanol exposure resulted in higher mean levels of insulin and IGF-1 receptor and lower levels of IRS-1 protein (Figures 2A–2C). Although there were no significant inter-group differences in the levels of tyrosine phosphorylated (activated) insulin and IGF-1 receptors (Figures 2D and 2E), the relative levels of tyrosine phosphorylated insulin and IGF-1 receptors were significantly reduced by prenatal ethanol exposure (Figures 2G and 2H). Multivariate ANOVA (MANOVA) testing demonstrated a significant main effects of ethanol on insulin receptor (F=5.53; P=0.04), pYpY1162/1163-IR/total IR (F=5.91; P=0.035), and pYpY1135/1136-IGF-1R/total IGF-1R (F=4.92; P=0.05), and a main effect trend of ethanol on IGF-1 receptor (F=2.96; P=0.10) expression. Therefore, the chronic prenatal ethanol exposures caused insulin and IGF-1 resistance (increased receptor expression vis-à-vis reduced receptor tyrosine phosphorylation) in the brain. Although the mean levels of pS312-IRS-1 and pS312-IRS-1/total IRS-1 were similar in control and ethanol-exposed cerebella (Figures 2F and 2I), the reduced levels of IRS-1 protein vis-a-vis decreased activation of insulin and IGF-1 receptor tyrosine kinases could have further impaired insulin and IGF-1 downstream signaling.
Figure 2.
Cerebellar insulin and IGF-1 resistance following chronic prenatal ethanol exposure. P30 cerebella were used to measure Immunoreactivity to the (A) insulin receptor (IR), (B) IGF-1R, (C) IRS-1, (D) pYpY1162/1163-IR, (E) pYpY1135/1136-IGF-1R, (F) pS312-IRS-1 by multiplex ELISA. Phospho-/total protein ratios for (G) IR, (H) IGF-1R, and (I) IRS-1 were calculated. Inter-group comparisons were made using Student t-tests. Significant differences are indicated within the panels. N=12/group with each sample contributed from a different litter.
Effects of prenatal ethanol exposure on downstream signaling through Akt and GSK-3β
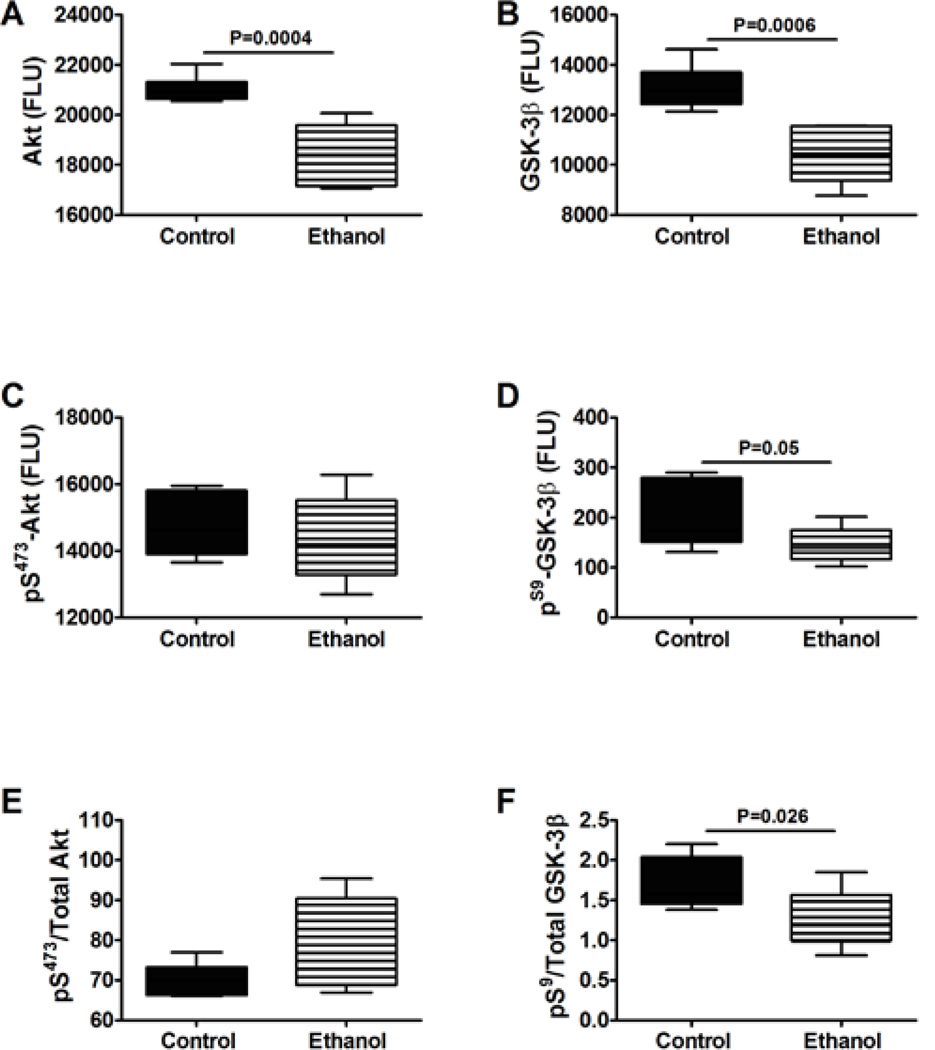
Insulin, IGF-1 and IRS-1 signals downstream to activate Akt and inhibit GSK-3β through phosphorylation of specific Ser residues on these proteins. In addition, signaling through Akt and GSK-3β can be regulated by the levels of protein expression. Multiplex ELISAs demonstrated that chronic prenatal ethanol exposures significantly reduced cerebellar levels of total Akt, but not pS473-Akt or pS473/total Akt (Figures 3A,3C and 3E). Ethanol also significantly reduced the mean levels of GSK-3β, pSer-9-GSK-3β, and the pSer-9-/total GSK-3β ratio (Figures 3B,3D and 3F). MANOVA demonstrated significant main effects of ethanol effects on Akt (F=20.85; P=0.001), GSK-3β (F=12.11; P=0.006), and pSer-9-GSK-3β/total GSK-3β (F=4.99; P=0.05). Since GSK-3β is inactivated by Ser-9 phosphorylation, the net effect of prenatal ethanol exposure was to increase GSK-3βactivity in juvenile cerebella.
Figure 3.
Long-term effects of chronic prenatal ethanol exposure on signaling through Akt and GSK-3β. P30 cerebellar protein homogenates were used to measure (A) Akt, (B) GSK-3β, (C) pS473-Akt, and (D) pS9-GSK3β immunoreactivity by multiplex ELISA. (E,F) Phospho-/total protein ratios were calculated. Inter-group comparisons were made using Student t-tests. Significant differences are indicated within the panels. N=12/group with each sample contributed from a different litter.
Prenatal ethanol exposure impairs Wnt signaling in juvenile brains
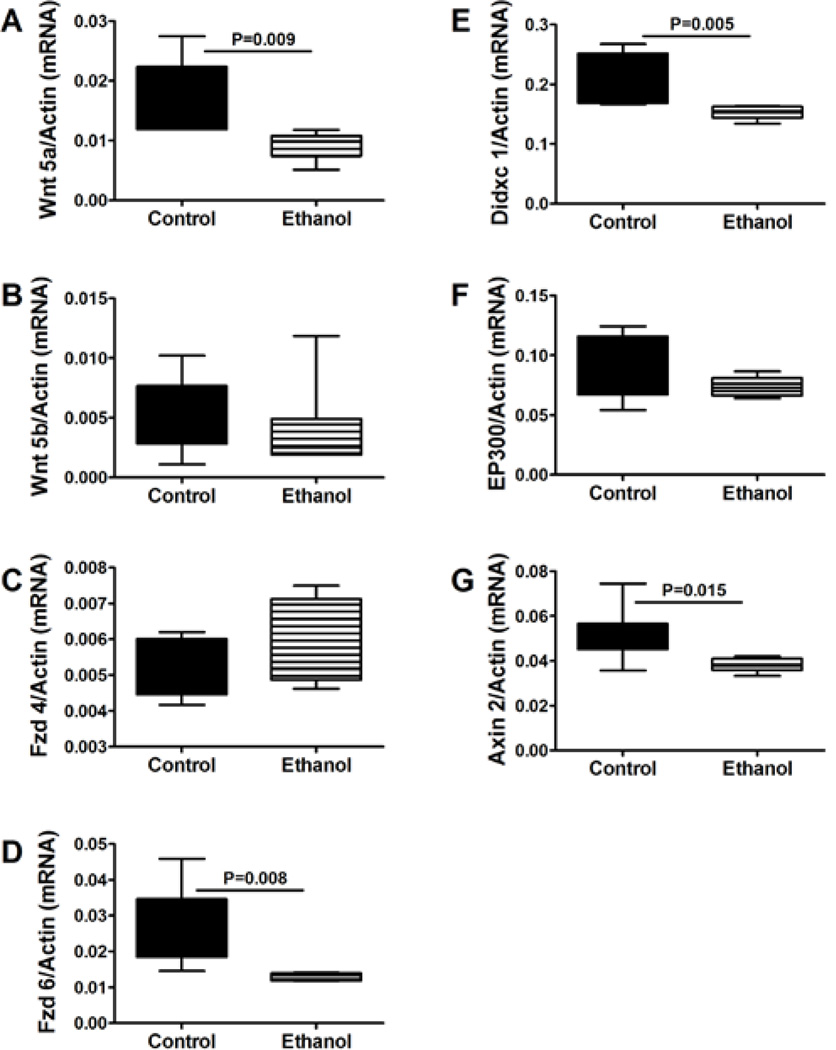
Since insulin/IGF-1 signaling cross-talks with Wnt [20,37–39], we hypothesized that the impaired insulin/IGF-1 signaling in cerebella of chronic prenatal ethanol exposed rats would be associated with down-regulation of Wnt pathway genes (Figure 1). We first performed exploratory qPCR studies in which the expression levels of 84 Wnt pathway and target genes (Supplementary tables 1 and 2) were compared between pooled samples of P20 control and ethanol-exposed rats (N=3 per group with each sample obtained from different litters) using the same model described in the Materials and Methods section (Supplementary Methods). The results suggested that chronic prenatal ethanol exposure inhibits Wnt signaling at all levels of the canonical pathway. From the list of genes in which inter-group differences were 2-fold or greater, or expression levels were relatively high compared with other isoforms of the corresponding ligands or receptors, we focused our further analyses by measuring expression of: Wnt 5a, Wnt 5b, Frizzled (Fzd) 4, Fzd 6, Didxc, EP300, and Axin2 (Figure 4). Chronic prenatal ethanol exposure significantly reduced expression of Wnt5a, Fzd 6, Didxc, and Axin 2, and caused modest but not statistically significant reductions in Wnt 5b and Ep300 in cerebellar tissue. In contrast, Fzd 4 mRNA levels were similar in the control and ethanol-exposed groups. MANOVA testing demonstrated significant main effects of ethanol exposure on Fzd 6 (F=8.45; P=0.015), Didxc (F=9.86; P=0.01), and Axin 2 (F=6.47; P=0.029). Corresponding with the broad inhibition of Wnt pathway genes, ELISA studies demonstrated significantly reduced levels of β-catenin in ethanol-exposed relative to control cerebella (Table 1).
Figure 4.
Effects of chronic prenatal ethanol exposure on Wnt signaling in juvenile cerebella. Probe-based qPCR amplification reactions were used to measure (A) Wnt 5a, (B) Wnt 5b, (C) Fzd 4, (D) Fzd 6, (E) Didxc 1A, (F) EP300, and (G) Axin 2. Results were normalized to β-actin, which was measured simultaneously in the duplex qPCR reactions. Inter-group comparisons were made with calculated mRNA/β-actin ratios. N=12/group with each sample contributed from a different litter.
Table 1.
Effects of Chronic Prenatal Ethanol Exposure on Notch and Wnt Pathway Protein Expression in Cerebellum.
| Immunoreactivity | Control | Ethanol | t-Test |
|---|---|---|---|
| Notch 1 | 20002.8 ± 1182.0 | 19892.7 ± 1028.2 | |
| Jagged 1 | 12360.8 ± 590.9 | 9897.8 ± 1031.3 | 0.035 |
| HES-1 | 3883.5 ± 111.2 | 3917.7 ± 159.2 | |
| Humbug | 18863.4 ± 1007.0 | 13628.9 ± 1318.8 | 0.003 |
| AAH | 10340.2 ± 491.6 | 7581.0 ± 710.3 | 0.003 |
| β-Catenin | 4824.5 ± 138.1 | 3828.6 ± 227.2 | <0.001 |
Immunoreactivity was measured in P30 cerebellar tissue by duplex ELISA with results normalized to large ribosomal protein measured in the same wells. Results correspond to mean net fluorescence light units (± S.E.M.) in 50 ng protein. Data were generated from 12 rats per group, with each sample contributed from a different litter. Inter-group comparisons were made with Student T-tests
Chronic prenatal ethanol exposure inhibits Notch pathway genes in juvenile cerebella
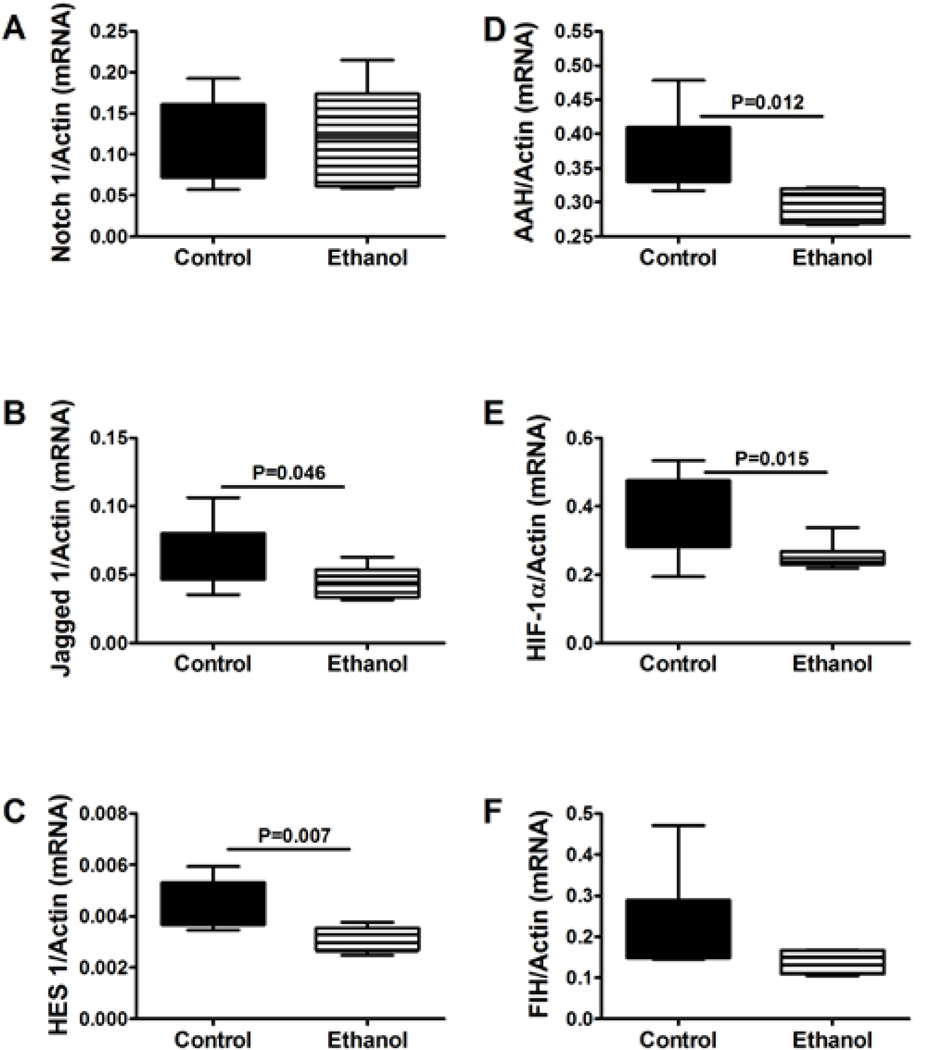
Insulin and IGF-1 signaling regulate AAH and hypoxia-inducible factor-1a (HIF-1α) expression [51], and both AAH and HIF-1α cross-talk with Notch [51]. Activation of insulin/IGF-1, AAH, Notch, and HIF-1α promotes cell motility [51–53,58,64], whereas ethanol inhibition of insulin/IGF-1 signaling and AAH impairs neuronal migration [59,65]. We extended our investigations to determine the degree to which chronic prenatal ethanol exposure impairs Notch signaling in juvenile cerebella. Moreover, we examined expression of other genes and proteins that cross-talk within this network and regulate neuronal motility and/or adhesion, i.e. Humbug (AAH-related molecule that lacks the C-terminal catalytic region) [54], HIF-1α, and factor inducing HIF-1α (FIH) [51]. In these studies, we measured Notch 1, Jagged 1, and HES-1, AAH, HIF-1α, and FIH mRNAs in P30 cerebella. Probe-based qRT-PCR analysis demonstrated significantly lower levels of Jagged 1, HES-1, AAH, and HIF-1α expression in ethanol-exposed relative to control brains. In contrast, no significant inter-group differences were observed with respect to Notch 1 or FIH (Figure 5). MANOVA testing demonstrated significant main effects of ethanol on HIF-1α (F=6.37; P=0.03), AAH (F=19.26; P=0.001), and HES-1 (F=8.95; P=0.014), and main effect trends with respect to Jagged (F=3.47; P=0.09) and FIH (F=3.75; P=0.081). Furthermore, ELISAs demonstrated significantly reduced levels of Jagged 1, Humbug, and AAH, in ethanol-exposed versus control cerebella (Table 1).
Figure 5.
Effects of chronic prenatal ethanol exposure on Notch signaling in juvenilecerebella. Probe-based qPCR amplification reactions were used to measure (A) Notch 1, (B) Jagged 1, (C) HES 1, (D) AAH, (E) HIF-1α, and (F) FIH gene expression with results normalized to β-actin, which was measured simultaneously. Inter-group comparisons were made using the calculated mRNA/β-actin ratios. N=12/group with each sample contributed from a different litter.
Discussion
Chronic prenatal ethanol exposure impairs insulin/IGF-1 signaling in juvenile cerebella
This study demonstrates that motor impairments caused by chronic prenatal ethanol exposure, were associated with persistent insulin/IGF-1 resistance which is characterized by elevated levels of insulin/IGF-1 receptor expression vis-à-vis reduced levels of receptor tyrosine phosphorylation. The associated inhibition of IRS-1 expression could have further impaired insulin/IGF-1 signaling by restricting the substrate needed to transmit signals to downstream pathways. Although it is not readily apparent why brain insulin/IGF-1 resistance persists beyond the period of ethanol exposure, consequences would include ongoing cell loss due to impaired survival mechanisms, increased oxidative stress, and deficits in cholinergic function [61]. The reduced IRS-1 protein levels could have been mediated by insulin/IGF-1 resistance, since insulin and IGF-1 regulate IRS gene expression [66–68].
Insulin/IGF-1 resistance typically reduces Akt [15,18,19,69–71], and increases GSK-3β [19,59,65,72] activity. Paradoxically, in ethanol-exposed cerebella, pS473-Akt levels were not reduced, indicating that the kinase activation mechanisms were intact. On the other hand, Akt protein expression was significantly reduced in ethanol-exposed cerebella; previous studies showed that Akt signaling can be modulated via changes in its protein levels [73,74]. Therefore, the combined effects of insulin/IGF-1 resistance and reduced Akt expression could account for the persistent structural and functional abnormalities observed in ethanol-exposed juvenile cerebella. Moreover, the deleterious effects of insulin/IGF-1 resistance on growth, metabolism, and cell survival were likely exacerbated by activation of GSK-3β (reduced Ser9 phosphorylation), and attendant increased oxidative stress and pro-death signaling in ethanol-exposed cerebella.
Prenatal ethanol exposure impairs Wnt signaling in juvenile brains
Wnt signaling helps mediate brain morphogenesis, cell proliferation, migration, differentiation, and axonal outgrowth, and it cross-talks with insulin/IGF pathways. However, the available data about ethanol’s effects on Wnt signaling in the developing brain are limited. Previous studies showed that chronic prenatal followed by postnatal ethanol exposures lead to persistently high brain levels of Wnt and β-catenin protein, which would impair neuronal maturation [75,76]. Our study showed that the long-term effects of just chronic prenatal ethanol exposures include inhibition of genes at multiple levels of the canonical Wnt signaling pathway, i.e. Wnt ligands (Wnt 5a), receptors (Frizzled 6), and components of the destruction complex (Didxc and Axin 2), and decreased β-catenin immune reactivity. Since insulin/IGF-1 networks cross-talk with Wnt via GSK-3β [21,35,39,77], increased GSK-3β activity caused by insulin/IGF-1 resistance could have further impaired Wnt signaling in juvenile cerebella. Mechanistically, activated GSK-3β phosphorylates β-catenin, causing it to be targeted for degradation via the ubiquitin-proteasome pathway [24,43,77]. Correspondingly, β-catenin protein levels were significantly reduced in chronic ethanol-exposed brains. Therefore, the long-term adverse effects of chronic prenatal ethanol exposures on cerebellar development and function are likely mediated in part by inhibition of gene expression and disruption of insulin/IGF cross-talk with the canonical Wnt pathway.
Effects of chronic prenatal ethanol exposure on Notch pathway genes
Notch signaling mediates neuronal migration and plasticity [44–46,78]. Therefore, inhibition of Notch could result in significant structural and functional abnormalities in the developing brain. Notch signaling is activated by AAH [53,56,58,64,79], as both Notch and Jagged proteins contain the consensus sequence for AAH’s catalytic activity [54,56]. AAH promotes cell motility, and its expression and catalytic activity are regulated by insulin/IGF-1 [59]. Correspondingly, ethanol inhibition of insulin/IGF-1 signaling decreases AAH expression and catalytic activity, and impairs cerebellar neuron migration [59], but the effects on Notch have not yet been determined. AAH and Notch are also regulated by HIF-1α which, in addition hypoxia, is regulated by insulin/IGF-1 [51]. Therefore, Notch, Jagged, AAH, and HIF-1α signaling networks are inter-related through insulin/IGF-1(Figure 1) [51,52].
The studies herein demonstrate that chronic prenatal ethanol exposure causes inhibition of Notch (reduced Jagged and HES-1 expression), HIF-1α, AAH and insulin/IGF-1 signaling in juvenile cerebella. Reduced AAH and HIF-1α were likely mediated by insulin/IGF-1 resistance, whereas reduced Jagged-1 expression was probably mediated by inhibition of AAH [51]. Jagged-1 is a ligand for Notch receptor, and impaired expression of Notch ligands inhibits Notch signaling [80]. Decreased HES-1 mRNA corresponds with ethanol’s inhibitory effects on Notch signaling, as HES-1 is a downstream target gene of Notch [49,50].
These results demonstrate that the functional cerebellar abnormalities in prenatal ethanol-exposed juvenile rats are associated with inhibition of signaling through three major inter-related signaling pathways: insulin/IGF-1, canonical Wnt and Notch. This suggests that the pathogenic basis for the neurodevelopmental defects in FASD is complex and mediated by disruption of several major inter-connected signaling networks that cross-talk through insulin/IGF-1 and GSK-3β. Our proposed model of how these three pathways cross-talk and how ethanol disrupts their signaling is diagramed in figure 6. The limitations of the study design are: 1) we did not validate the results by treating the rats with inhibitors or activators of insulin, Wnt, or Notch signaling; 2) it is not possible from the results to determine whether the impairments in Wnt and Notch signaling are caused by insulin/IGF resistance, or vice versa; and 3) it is not possible to determine the degree to which specific impairments in insulin, Wnt, or Notch signaling mediate the phenotypic abnormalities in our model. The availability of specific inhibitors and/or activators of insulin, Wnt, and Notch signaling will enable these questions to be addressed mechanistically. Studies currently in progress are designed to determine the degree to which restoration of insulin/IGF signaling with insulin sensitizer agents abrogates impairments in Notch and Wnt signaling, together with the structural and functional abnormalities in the cerebellum. However, due to the complex nature of the cross-talk among several major signal transduction networks, we anticipate that multi-pronged therapeutic approaches will be required to prevent or reduce the severity of deficits caused by prenatal ethanol exposure.
Figure 6.
Proposed effects of ethanol on cross-talk signaling of insulin/IGF-1 with Notch and Wnt pathways in the developing brain. Ethanol inhibits insulin/IGF-1 signaling by impairing ligand-receptor binding, activation of receptor tyrosine kinases, tyrosine phosphorylation of IRS-1, and transmission of signals downstream through Akt pathways. In addition, ethanol activates phosphatases (PTP-1b and PTEN) that negatively regulate signaling through insulin/IGF-1 receptors and PI3K. Ethanol inhibits insulin/IGF-1 regulated genes that mediate Notch signaling, e.g. AAH. The net effect is inhibition of Notch target genes such as HES and HEY, and reduced cross-talk with the insulin/IGF-1 pathways via inhibition of PTEN and activation Akt. Ethanol disrupts Notch signaling at other levels in the cascade, including via ligand and receptor gene expression. Canonical Wnt signaling is regulated by GSK-3β, which phosphorylates β-catenin, targeting it for proteolytic destruction. Ethanol may impair Wnt signaling by inhibiting insulin/IGF-1 suppression of GSK-3β or by directly activating GSK-3β. The resulting high levels of GSK-3β activity could dampen positive cross-talk with Wnt-activated cellular functions.
Acknowledgements
Research was supported by AA-11431, AA-12908, and AA-16126 from the National Institutes of Health.
References
- 1.Riley EP, Infante MA, Warren KR. Fetal alcohol spectrum disorders: an overview. Neuropsychol Rev. 2011;21:73–80. doi: 10.1007/s11065-011-9166-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mattson SN, Crocker N, Nguyen TT. Fetal alcohol spectrum disorders: neuropsychological and behavioral features. Neuropsychol Rev. 2011;21:81–101. doi: 10.1007/s11065-011-9167-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de la Monte SM, Wands JR. Role of central nervous system insulin resistance in fetal alcohol spectrum disorders. J Popul Ther Clin Pharmacol. 2010;17:e390–e404. [PMC free article] [PubMed] [Google Scholar]
- 4.de la Monte SM, Wands JR. Review of insulin and insulin-like growth factor expression, signaling, and malfunction in the central nervous system: relevance to Alzheimer’s disease. J Alzheimers Dis. 2005;7:45–61. doi: 10.3233/jad-2005-7106. [DOI] [PubMed] [Google Scholar]
- 5.Chesik D, De Keyser J, Wilczak N. Insulin-like growth factor system regulates oligodendroglial cell behavior: therapeutic potential in CNS. J Mol Neurosci. 2008;35:81–90. doi: 10.1007/s12031-008-9041-2. [DOI] [PubMed] [Google Scholar]
- 6.Gong X, Xie Z, Zuo H. Invivo insulin deficiency as a potential etiology for demyelinating disease. Med Hypotheses. 2008;71:399–403. doi: 10.1016/j.mehy.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 7.Liang G, Cline GW, Macica CM. IGF-1 stimulates de novo fatty acid biosynthesis by Schwann cells during myelination. Glia. 2007;55:632–641. doi: 10.1002/glia.20496. [DOI] [PubMed] [Google Scholar]
- 8.Ye P, Kollias G, D’Ercole AJ. Insulin-like growth factor-I ameliorates demyelination induced by tumor necrosis factor-alpha in transgenic mice. J Neurosci Res. 2007;85:712–722. doi: 10.1002/jnr.21181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laviola L, Natalicchio A, Perrini S, Giorgino F. Abnormalities of IGF-I signaling in the pathogenesis of diseases of the bone, brain, and fetoplacental unit in humans. Am J Physiol Endocrinol Metab. 2008;295:E991–E999. doi: 10.1152/ajpendo.90452.2008. [DOI] [PubMed] [Google Scholar]
- 10.Broughton SK, Chen H, Riddle A, Kuhn SE, Nagalla S, et al. Large-scale generation of highly enriched neural stem-cell-derived oligodendroglial cultures: maturation-dependent differences in insulin-like growth factor-mediated signal transduction. J Neurochem. 2007;100:628–638. doi: 10.1111/j.1471-4159.2006.04171.x. [DOI] [PubMed] [Google Scholar]
- 11.Joseph D’Ercole A, Ye P. Expanding the mind: insulin-like growth factor I and brain development. Endocrinology. 2008;149:5958–5962. doi: 10.1210/en.2008-0920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dudek H, Datta SR, Franke TF, Birnbaum MJ, Yao R, et al. Regulation of neuronal survival by the serine-threonine protein kinase Akt. Science. 1997;275:661–665. doi: 10.1126/science.275.5300.661. [DOI] [PubMed] [Google Scholar]
- 13.Banerjee K, Mohr L, Wands JR, de la Monte SM. Ethanol inhibition of insulin signaling in hepatocellular carcinoma cells. Alcohol Clin Exp Res. 1998;22:2093–2101. [PubMed] [Google Scholar]
- 14.Mohr L, Tanaka S, Wands JR. Ethanol inhibits hepatocyte proliferation in insulin receptor substrate 1 transgenic mice. Gastroenterology. 1998;115:1558–1565. doi: 10.1016/s0016-5085(98)70036-8. [DOI] [PubMed] [Google Scholar]
- 15.Xu J, Yeon JE, Chang H, Tison G, Chen GJ, et al. Ethanol impairs insulin-stimulated neuronal survival in the developing brain: role of PTEN phosphatase. J Biol Chem. 2003;278:26929–26937. doi: 10.1074/jbc.M300401200. [DOI] [PubMed] [Google Scholar]
- 16.de la Monte SM, Ganju N, Banerjee K, Brown NV, Luong T, et al. Partial rescue of ethanol-induced neuronal apoptosis by growth factor activation of phosphoinositol-3-kinase. Alcohol Clin Exp Res. 2000;24:716–726. [PubMed] [Google Scholar]
- 17.He J, de la Monte S, Wands JR. Acute ethanol exposure inhibits insulin signaling in the liver. Hepatology. 2007;46:1791–1800. doi: 10.1002/hep.21904. [DOI] [PubMed] [Google Scholar]
- 18.Yeon JE, Califano S, Xu J, Wands JR, De La Monte SM. Potential role of PTEN phosphatase in ethanol-impaired survival signaling in the liver. Hepatology. 2003;38:703–714. doi: 10.1053/jhep.2003.50368. [DOI] [PubMed] [Google Scholar]
- 19.de la Monte SM, Wands JR. Chronic gestational exposure to ethanol impairs insulin-stimulated survival and mitochondrial function in cerebellar neurons. Cell Mol Life Sci. 2002;59:882–893. doi: 10.1007/s00018-002-8475-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yi F, Sun J, Lim GE, Fantus IG, Brubaker PL, et al. Cross talk between the insulin and Wnt signaling pathways: evidence from intestinal endocrine L cells. Endocrinology. 2008;149:2341–2351. doi: 10.1210/en.2007-1142. [DOI] [PubMed] [Google Scholar]
- 21.Beildeck ME, Gelmann EP, Byers SW. Cross-regulation of signaling pathways: an example of nuclear hormone receptors and the canonical Wnt pathway. Exp Cell Res. 2010;316:1763–1772. doi: 10.1016/j.yexcr.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cadigan KM, Peifer M. Wnt signaling from development to disease: insights from model systems. Cold Spring Harb Perspect Biol. 2009;1:a002881. doi: 10.1101/cshperspect.a002881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sugimura R, Li L. Noncanonical Wnt signaling in vertebrate development, stem cells, and diseases. Birth defects research. Part C: Embryo Today: Reviews. 2010;90:243–256. doi: 10.1002/bdrc.20195. [DOI] [PubMed] [Google Scholar]
- 24.Rao TP, Kühl M. An updated overview on Wnt signaling pathways: a prelude for more. Circ Res. 2010;106:1798–1806. doi: 10.1161/CIRCRESAHA.110.219840. [DOI] [PubMed] [Google Scholar]
- 25.Miller JR, Hocking AM, Brown JD, Moon RT. Mechanism and function of signal transduction by the Wnt/beta-catenin and Wnt/Ca2+ pathways. Oncogene. 1999;18:7860–7872. doi: 10.1038/sj.onc.1203245. [DOI] [PubMed] [Google Scholar]
- 26.Shulman JM, Perrimon N, Axelrod JD. Frizzled signaling and the developmental control of cell polarity. Trends Genet. 1998;14:452–458. doi: 10.1016/s0168-9525(98)01584-4. [DOI] [PubMed] [Google Scholar]
- 27.Wansleeben C, Meijlink F. The planar cell polarity pathway in vertebrate development. Dev Dyn. 2011;240:616–626. doi: 10.1002/dvdy.22564. [DOI] [PubMed] [Google Scholar]
- 28.LaMonica K, Bass M, Grabel L. The planar cell polarity pathway directs parietal endoderm migration. Dev Biol. 2009;330:44–53. doi: 10.1016/j.ydbio.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.James RG, Conrad WH, Moon RT. Beta-catenin-independent Wnt pathways: signals, core proteins, and effectors. Methods Mol Biol. 2008;468:131–144. doi: 10.1007/978-1-59745-249-6_10. [DOI] [PubMed] [Google Scholar]
- 30.Tanegashima K, Zhao H, Dawid IB. WGEF activates Rho in the Wnt-PCP pathway and controls convergent extension in Xenopus gastrulation. EMBO J. 2008;27:606–617. doi: 10.1038/emboj.2008.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patapoutian A, Reichardt LF. Roles of Wnt proteins in neural development and maintenance. Curr Opin Neurobiol. 2000;10:392–399. doi: 10.1016/s0959-4388(00)00100-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galceran J, Miyashita-Lin EM, Devaney E, Rubenstein JL, Grosschedl R. Hippocampus development and generation of dentate gyrus granule cells is regulated by LEF1. Development. 2000;127:469–482. doi: 10.1242/dev.127.3.469. [DOI] [PubMed] [Google Scholar]
- 33.Ikeya M, Lee SM, Johnson JE, McMahon AP, Takada S. Wnt signalling required for expansion of neural crest and CNS progenitors. Nature. 1997;389:966–970. doi: 10.1038/40146. [DOI] [PubMed] [Google Scholar]
- 34.Salinas PC, Fletcher C, Copeland NG, Jenkins NA, Nusse R. Maintenance of Wnt-3 expression in Purkinje cells of the mouse cerebellum depends on interactions with granule cells. Development. 1994;120:1277–1286. doi: 10.1242/dev.120.5.1277. [DOI] [PubMed] [Google Scholar]
- 35.Ding VW, Chen RH, McCormick F. Differential regulation of glycogen synthase kinase 3beta by insulin and Wnt signaling. J Biol Chem. 2000;275:32475–32481. doi: 10.1074/jbc.M005342200. [DOI] [PubMed] [Google Scholar]
- 36.Pearl LH, Barford D. Regulation of protein kinases in insulin, growth factor and Wnt signalling. Curr Opin Struct Biol. 2002;12:761–767. doi: 10.1016/s0959-440x(02)00386-x. [DOI] [PubMed] [Google Scholar]
- 37.Liu H, Fergusson MM, Wu JJ, Rovira II, Liu J, et al. Wnt signaling regulates hepatic metabolism. Sci Signal. 2011;4:ra6. doi: 10.1126/scisignal.2001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoon JC, Ng A, Kim BH, Bianco A, Xavier RJ, et al. Wnt signaling regulates mitochondrial physiology and insulin sensitivity. Genes Dev. 2010;24:1507–1518. doi: 10.1101/gad.1924910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bommer GT, Feng Y, Iura A, Giordano TJ, Kuick R, et al. IRS1 regulation by Wnt/beta-catenin signaling and varied contribution of IRS1 to the neoplastic phenotype. J Biol Chem. 2010;285:1928–1938. doi: 10.1074/jbc.M109.060319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamazaki H, Yanagawa Si. Axin and the Axin/Arrow-binding protein DCAP mediate glucose-glycogen metabolism. Biochem Biophys Res Commun. 2003;304:229–235. doi: 10.1016/s0006-291x(03)00582-5. [DOI] [PubMed] [Google Scholar]
- 41.Tanneberger K, Pfister AS, Kriz V, Bryja V, Schambony A, et al. Structural and functional characterization of the Wnt inhibitor APC membrane recruitment 1 (Amer1) J Biol Chem. 2011;286:19204–19214. doi: 10.1074/jbc.M111.224881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roberts DM, Pronobis MI, Poulton JS, Waldmann JD, Stephenson EM, et al. Deconstructing the βcatenin destruction complex: mechanistic roles for the tumor suppressor APC in regulating Wnt signaling. Mol Biol Cell. 2011;22:1845–1863. doi: 10.1091/mbc.E10-11-0871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pierfelice T, Alberi L, Gaiano N. Notch in the vertebrate nervous system: an old dog with new tricks. Neuron. 2011;69:840–855. doi: 10.1016/j.neuron.2011.02.031. [DOI] [PubMed] [Google Scholar]
- 45.Cornell RA, Eisen JS. Notch in the pathway: the roles of Notch signaling in neural crest development. Semin Cell Dev Biol. 2005;16:663–672. doi: 10.1016/j.semcdb.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 46.Lasky JL, Wu H. Notch signaling, brain development, and human disease. Pediatr Res. 2005;57:104R–109R. doi: 10.1203/01.PDR.0000159632.70510.3D. [DOI] [PubMed] [Google Scholar]
- 47.Bray S, Bernard F. Notch targets and their regulation. Curr Top Dev Biol. 2010;92:253–275. doi: 10.1016/S0070-2153(10)92008-5. [DOI] [PubMed] [Google Scholar]
- 48.D’Souza B, Meloty-Kapella L, Weinmaster G. Canonical and non-canonical Notch ligands. Curr Top Dev Biol. 2010;92:73–129. doi: 10.1016/S0070-2153(10)92003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kageyama R, Ohtsuka T, Hatakeyama J, Ohsawa R. Roles of bHLH genes in neural stem cell differentiation. Exp Cell Res. 2005;306:343–348. doi: 10.1016/j.yexcr.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 50.Kageyama R, Ohtsuka T, Tomita K. The bHLH gene Hes1 regulates differentiation of multiple cell types. Mol Cells. 2000;10:1–7. doi: 10.1007/s10059-000-0001-0. [DOI] [PubMed] [Google Scholar]
- 51.Lawton M, Tong M, Gundogan F, Wands JR, de la Monte SM. Aspartyl-(asparaginyl) beta-hydroxylase, hypoxia-inducible factor-alpha and Notch cross-talk in regulating neuronal motility. Oxid Med Cell Longev. 2010;3:347–356. doi: 10.4161/oxim.3.5.13296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Silbermann E, Moskal P, Bowling N, Tong M, de la Monte SM. Role of aspartyl-(asparaginyl)-β-hydroxylase mediated notch signaling in cerebellar development and function. Behav Brain Funct. 2010;6:68. doi: 10.1186/1744-9081-6-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cantarini MC, de la Monte SM, Pang M, Tong M, D’Errico A, et al. Aspartyl-asparagyl beta hydroxylase over-expression in human hepatoma is linked to activation of insulin-like growth factor and notch signaling mechanisms. Hepatology. 2006;44:446–457. doi: 10.1002/hep.21272. [DOI] [PubMed] [Google Scholar]
- 54.Dinchuk JE, Focht RJ, Kelley JA, Henderson NL, Zolotarjova NI, et al. Absence of post-translational aspartyl beta-hydroxylation of epidermal growth factor domains in mice leads to developmental defects and an increased incidence of intestinal neoplasia. J Biol Chem. 2002;277:12970–12977. doi: 10.1074/jbc.M110389200. [DOI] [PubMed] [Google Scholar]
- 55.Dahlbäck B, Hildebrand B, Linse S. Novel type of very high affinity calcium-binding sites in beta-hydroxyasparagine-containing epidermal growth factor-like domains in vitamin K-dependent protein S. J Biol Chem. 1990;265:18481–18489. [PubMed] [Google Scholar]
- 56.Lavaissiere L, Jia S, Nishiyama M, de la Monte S, Stern AM, et al. Overexpression of human aspartyl(asparaginyl)beta-hydroxylase in hepatocellular carcinoma and cholangiocarcinoma. J Clin Invest. 1996;98:1313–1323. doi: 10.1172/JCI118918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de la Monte SM, Tamaki S, Cantarini MC, Ince N, Wiedmann M, et al. Aspartyl-(asparaginyl)-beta-hydroxylase regulates hepatocellular carcinoma invasiveness. J Hepatol. 2006;44:971–983. doi: 10.1016/j.jhep.2006.01.038. [DOI] [PubMed] [Google Scholar]
- 58.Lahousse SA, Carter JJ, Xu XJ, Wands JR, de la Monte SM. Differential growth factor regulation of aspartyl-(asparaginyl)-beta-hydroxylase family genes in SH-Sy5y human neuroblastoma cells. BMC Cell Biol. 2006;7:41. doi: 10.1186/1471-2121-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.de la Monte SM, Tong M, Carlson RI, Carter JJ, Longato L, et al. Ethanol inhibition of aspartyl-asparaginyl-beta-hydroxylase in fetal alcohol spectrum disorder: potential link to the impairments in central nervous system neuronal migration. Alcohol. 2009;43:225–240. doi: 10.1016/j.alcohol.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gundogan F, Elwood G, Longato L, Tong M, Feijoo A, et al. Impaired placentation in fetal alcohol syndrome. Placenta. 2008;29:148–157. doi: 10.1016/j.placenta.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Soscia SJ, Tong M, Xu XJ, Cohen AC, Chu J, et al. Chronic gestational exposure to ethanol causes insulin and IGF resistance and impairs acetylcholine homeostasis in the brain. Cell Mol Life Sci. 2006;63:2039–2056. doi: 10.1007/s00018-006-6208-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gundogan F, Elwood G, Greco D, Rubin LP, Pinar H, et al. Role of aspartyl-(asparaginyl) beta-hydroxylase in placental implantation: Relevance to early pregnancy loss. Hum Pathol. 2007;38:50–59. doi: 10.1016/j.humpath.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 63.de la Monte SM, Tong M, Bowling N, Moskal P. si-RNA inhibition of brain insulin or insulin-like growth factor receptors causes developmental cerebellar abnormalities: relevance to fetal alcohol spectrum disorder. Mol Brain. 2011;4:13. doi: 10.1186/1756-6606-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sepe PS, Lahousse SA, Gemelli B, Chang H, Maeda T, et al. Role of the aspartyl-asparaginyl-beta-hydroxylase gene in neuroblastoma cell motility. Lab Invest. 2002;82:881–891. doi: 10.1097/01.lab.0000020406.91689.7f. [DOI] [PubMed] [Google Scholar]
- 65.Carter JJ, Tong M, Silbermann E, Lahousse SA, Ding FF, et al. Ethanol impaired neuronal migration is associated with reduced aspartyl-asparaginyl-beta-hydroxylase expression. Acta Neuropathol. 2008;116:303–315. doi: 10.1007/s00401-008-0377-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang X, Vaag A, Hansson M, Groop L. Down-regulation of insulin receptor substrates (IRS)-1 and IRS-2 and Src homologous and collagen-like protein Shc gene expression by insulin in skeletal muscle is not associated with insulin resistance or type 2 diabetes. J Clin Endocrinol Metab. 2002;87:255–259. doi: 10.1210/jcem.87.1.8144. [DOI] [PubMed] [Google Scholar]
- 67.Hoene M, Lehmann R, Hennige AM, Pohl AK, Häring HU, et al. Acute regulation of metabolic genes and insulin receptor substrates in the liver of mice by one single bout of treadmill exercise. J Physiol. 2009;587:241–252. doi: 10.1113/jphysiol.2008.160275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hirashima Y, Tsuruzoe K, Kodama S, Igata M, Toyonaga T, et al. Insulin down-regulates insulin receptor substrate-2 expression through the phosphatidylinositol 3-kinase/Akt pathway. J Endocrinol. 2003;179:253–266. doi: 10.1677/joe.0.1790253. [DOI] [PubMed] [Google Scholar]
- 69.Pang M, de la Monte SM, Longato L, Tong M, He J, et al. PPARdelta agonist attenuates alcohol-induced hepatic insulin resistance and improves liver injury and repair. J Hepatol. 2009;50:1192–1201. doi: 10.1016/j.jhep.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chakraborty AK, Liang K, DiGiovanna MP. Co-targeting insulin-like growth factor I receptor and HER2: dramatic effects of HER2 inhibitors on nonoverexpressing breast cancer. Cancer Res. 2008;68:1538–1545. doi: 10.1158/0008-5472.CAN-07-5935. [DOI] [PubMed] [Google Scholar]
- 71.Gao L, Zhang X, Wang FR, Cao MF, Zhang XJ, et al. Chronic ethanol consumption up-regulates protein-tyrosine phosphatase-1B (PTP1B) expression in rat skeletal muscle. Acta Pharmacol Sin. 2010;31:1576–1582. doi: 10.1038/aps.2010.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Luo J. Lithium-mediated protection against ethanol neurotoxicity. Front Neurosci. 2010;4:41. doi: 10.3389/fnins.2010.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Roy HK, Olusola BF, Clemens DL, Karolski WJ, Ratashak A, et al. AKT proto-oncogene overexpression is an early event during sporadic colon carcinogenesis. Carcinogenesis. 2002;23:201–205. doi: 10.1093/carcin/23.1.201. [DOI] [PubMed] [Google Scholar]
- 74.Kumar S, Bryant CS, Chamala S, Qazi A, Seward S, et al. Ritonavir blocks AKT signaling, activates apoptosis and inhibits migration and invasion in ovarian cancer cells. Mol Cancer. 2009;8:26. doi: 10.1186/1476-4598-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Singh AK, Gupta S, Jiang Y, Younus M, Ramzan M. In vitro neurogenesis from neural progenitor cells isolated from the hippocampus region of the brain of adult rats exposed to ethanol during early development through their alcohol-drinking mothers. Alcohol Alcohol. 2009;44:185–198. doi: 10.1093/alcalc/agn109. [DOI] [PubMed] [Google Scholar]
- 76.Karaçay B, Li S, Bonthius DJ. Maturation-dependent alcohol resistance in the developing mouse: cerebellar neuronal loss and gene expression during alcohol-vulnerable and -resistant periods. Alcohol Clin Exp Res. 2008;32:1439–1450. doi: 10.1111/j.1530-0277.2008.00720.x. [DOI] [PubMed] [Google Scholar]
- 77.Doble BW, Woodgett JR. GSK-3: tricks of the trade for a multi-tasking kinase. J Cell Sci. 2003;116:1175–1186. doi: 10.1242/jcs.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.De Ferrari GV, Inestrosa NC. Wnt signaling function in Alzheimer’s disease. Brain Res Brain Res Rev. 2000;33:1–12. doi: 10.1016/s0165-0173(00)00021-7. [DOI] [PubMed] [Google Scholar]
- 79.Ince N, de la Monte SM, Wands JR. Overexpression of human aspartyl (asparaginyl) beta-hydroxylase is associated with malignant transformation. Cancer Res. 2000;60:1261–1266. [PubMed] [Google Scholar]
- 80.de La Coste A, Freitas AA. Notch signaling: distinct ligands induce specific signals during lymphocyte development and maturation. Immunol Lett. 2006;102:1–9. doi: 10.1016/j.imlet.2005.06.014. [DOI] [PubMed] [Google Scholar]