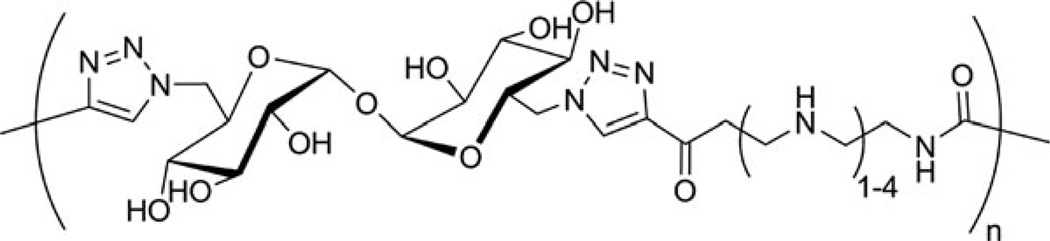
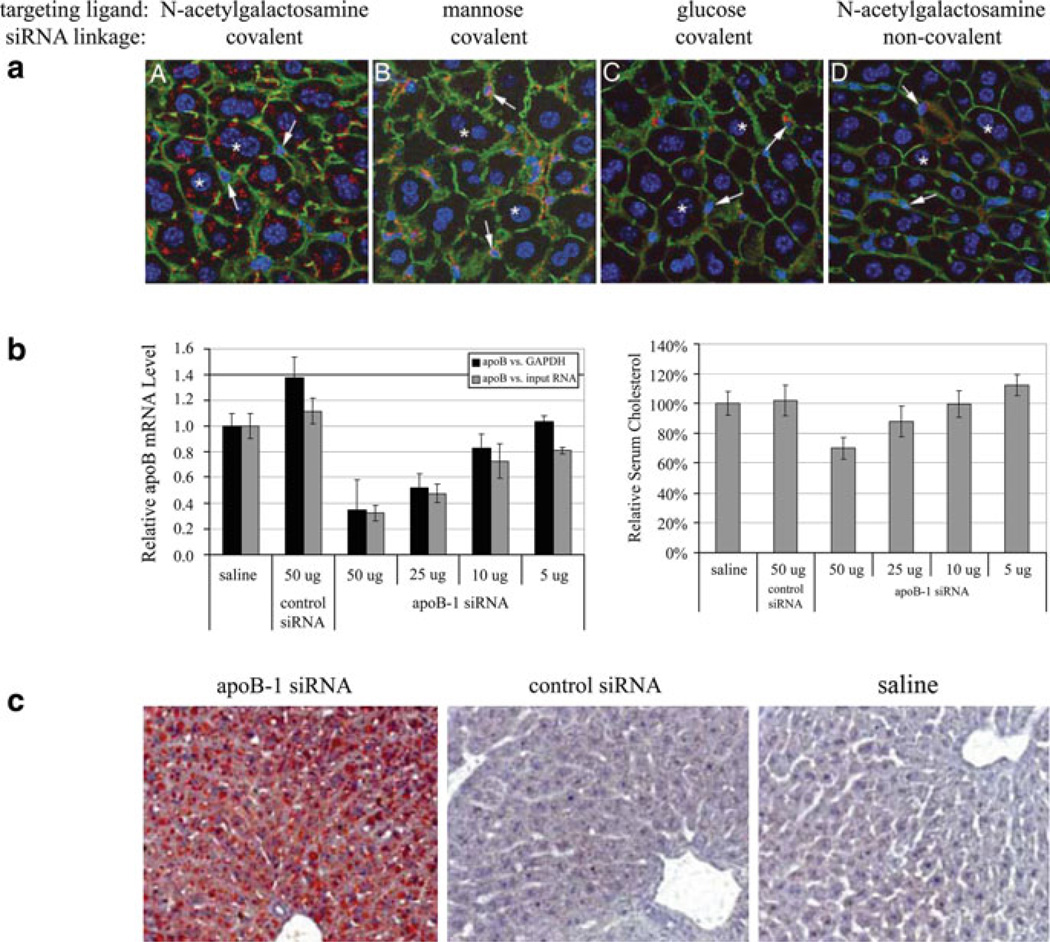
Abstract
Carbohydrates have been investigated and developed as delivery vehicles for shuttling nucleic acids into cells. In this review, we present the state of the art in carbohydrate-based polymeric vehicles for nucleic acid delivery, with the focus on the recent successes in preclinical models, both in vitro and in vivo. Polymeric scaffolds based on the natural polysaccharides chitosan, hyaluronan, pullulan, dextran, and schizophyllan each have unique properties and potential for modification, and these results are discussed with the focus on facile synthetic routes and favorable performance in biological systems. Many of these carbohydrates have been used to develop alternative types of biomaterials for nucleic acid delivery to typical polyplexes, and these novel materials are discussed. Also presented are polymeric vehicles that incorporate copolymerized carbohydrates into polymer backbones based on polyethylenimine and polylysine and their effect on transfection and biocompatibility. Unique scaffolds, such as clusters and polymers based on cyclodextrin (CD), are also discussed, with the focus on recent successes in vivo and in the clinic. These results are presented with the emphasis on the role of carbohydrate and charge on transfection. Use of carbohydrates as molecular recognition ligands for cell-type specific delivery is also briefly reviewed. We contend that carbohydrates have contributed significantly to progress in the field of non-viral DNA delivery, and these new discoveries are impactful for developing new vehicles and materials for treatment of human disease.
Keywords: Carbohydrate, DNA, Nucleic acid delivery, polysaccharide, Transfection
1 Introduction
Nucleic acids have broad potential for use in human therapeutics. The completion of the Human Genome Project has brought the promise of nucleic acid-based drugs to treat a myriad of acquired and inherited human diseases, including HIV, cancer, cystic fibrosis, rheumatoid arthritis, asthma, cardiovascular disease, and neurodegenerative disorders [1–3]. As nucleic acids are large, charged molecules and susceptible to enzymatic degradation, a delivery vehicle is required to condense the polynucleotide into a compact structure which protects it from degradation and facilitates its cellular internalization. Past and present delivery vehicle technology has been centered about the genetically-engineered virus as a means of nucleic acid delivery. Viral vectors have demonstrated successful gene transfer in vivo due to their innate cellular internalization and gene transduction capabilities, and many viral vectors have progressed into the clinic [4, 5]. However, the widespread applicability of viral vehicles is tempered by the potential to elicit unpredictable immune responses and their relative difficulty of manufacture [6, 7]. The potential clinical pitfalls of viral-based nucleic acid delivery have spurred a broad research focus devoted to developing non-viral delivery systems that allows similar gene transduction capacities but have reduced potential for toxicity.
Synthetic materials for nucleic acid condensation can offer marked improvement over viral delivery. Materials can be designed for high nucleic acid loading capacity, cell-specific targeting through chemical conjugation of molecular recognition elements, and biocompatibility, and are better suited to scale up for mass production. These materials are typically cationic, and may contain primary, secondary, and tertiary amines that can be protonated at physiological pH, which is necessary for electrostatic binding with the negatively-charged phosphate groups on the DNA backbone. This cooperative binding event and polycation charge neutralization facilitates compaction of the polymer nucleic acid complexes into small colloidal nanoparticles (termed polyplexes) [8, 9]. Structures such as branched and linear polyethylenimine (PEI) [9–11], poly-l-lysine (PLL) [12–14], spermine [15, 16], and polyamidoamine (PAMAM) [5, 17–19] can bind nucleic acids quite well and have been developed for DNA delivery with varied success. A fine, detailed review of non-viral delivery has been published recently [20]. However, these charge-dense polycations have demonstrated toxicity [4, 8, 21]; thus, design of a nontoxic analog is key to development of a suitable vehicle for human therapy.
Using carbohydrates in nucleic acid delivery is an obvious choice for improving toxicity. Carbohydrates are naturally-available unique scaffolds that have been exploited by synthetic chemists for materials design. Structural features, such as the presence of an anomeric carbon, multiple hydroxyl groups, cyclic structures, and chirality are advantageous for designing biomacromolecules [22–25]. In addition, carbohydrates are readily available, renewable resources; inexpensive materials for introducing hydrophilicity and biocompatibility into polymeric systems. These facets have led to their use in developing novel sustainable materials for biomedical applications [26, 27].
Glycopolymers have broadened the scope of nucleic acid delivery research, as many novel saccharide-based materials have been developed and analyzed for favorable nucleic acid delivery and toxicity profiles. This review provides critical perspective on the progress and favorable results of carbohydrate-based vehicles in nucleic acid delivery. We have focused on glycopolymeric delivery systems, including those derived from pure carbohydrates (chitosan, hyaluronan, pullulan, schizophyllan, dextran, and cyclodextrin) as well as carbohydrate comonomers incorporated into a polymer backbone. Carbohydrates have also been used as molecular recognition elements for targeting receptor-mediated endocytosis and have been conjugated as pendent groups for recognition by cell-surface lectins. Polymers incorporating carbohydrate-mediated targeting will be discussed; however, a full discussion of their use in targeting is beyond the scope of this review.
2 Natural Polysaccharides as Nucleic Acid Delivery Scaffolds
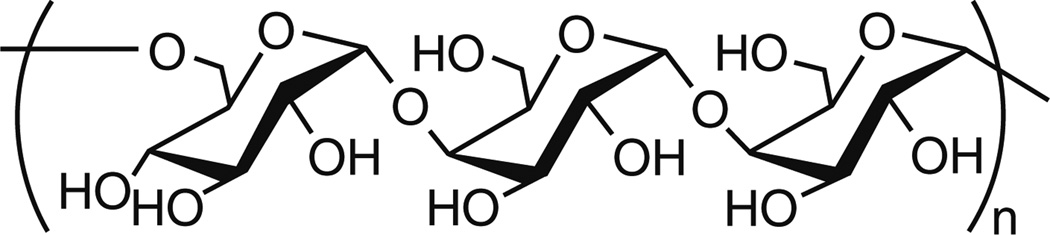
Polysaccharides are complex carbohydrates possessing high structural diversity. They are composed of several monosaccharide units joined together through glycosidic bonds. Typically, polysaccharides are isolated from a natural source, prepared via ring-opening polymerization of anhydro sugars or synthesized by enzymatic polymerization, which provides stereo-control over the polysaccharides synthesized, even at high molecular weight [28]. The natural polysaccharides, such as dextran [29], schizophyllan [30], chitosan [31], hyaluronan [32], and pullulan [33] have all been studied as nucleic acid carriers, and the following section highlights significant recent findings with these polysaccharides.
2.1 Dextran
Dextrans are biodegradable homopolymers of glucose with predominantly α-(1→6) linkages with some branching via α-(1→3) linkage which vary depending on the source of dextran. They are synthesized from sucrose by the action of bacteria, such as Streptococcus mutans or Leuconostoc mesenteroides. The first report of polycation-mediated DNA complexation was published in 1965 on 2-diethyl-aminoethyl (DEAE)-dextran, synthesized from diethyleaminoethyl chloride and dextran [34]. This study was an important milestone in this area because it was the first published example of a non-viral polysaccharide nucleic acid carrier. These vehicles continue to be investigated. The cationic nature of DEAE-dextran enables it to complex effectively with nucleic acids of various types and sizes [29].
About three decades after these first studies, Mack and coworkers [35] used DEAE-dextran to transfect primary cultured human macrophages. Macrophages play a pivotal role in regulating immune response and gene expression and, hence, transfection experiments performed in this study were both interesting and challenging, as macrophages are difficult to transfect. Reproducible luciferase expression was observed with DEAE-dextran, as opposed to with liposome delivery or electroporation. The addition of 100 or 400-µM concentrations of chloroquine also was not seen to enhance gene expression, suggesting that DEAE-dextran particles were not sequestered in endosomes. However, the presence of serum in the transfection medium reduced transgene expression by 60%.
Onishi et al. [36] grafted DEAE-dextran with methyl methacrylate (DDMC), with the hypothesis that methyl methacrylate graft chains could protect the complex from degradation by dextranases present in the cytoplasm, resulting in increased transfection efficiency and decreased cytotoxicity. Transfection experiments were completed in human embryonic kidney (HEK293) cells in serum-containing media. The results showed a fivefold increase in transgene expression with DDMC polyplexes compared with DEAE-dextran polyplexes, supporting the authors’ hypothesis. In addition, the cytotoxicity was shown to be reduced by DDMC grafting during these experiments.
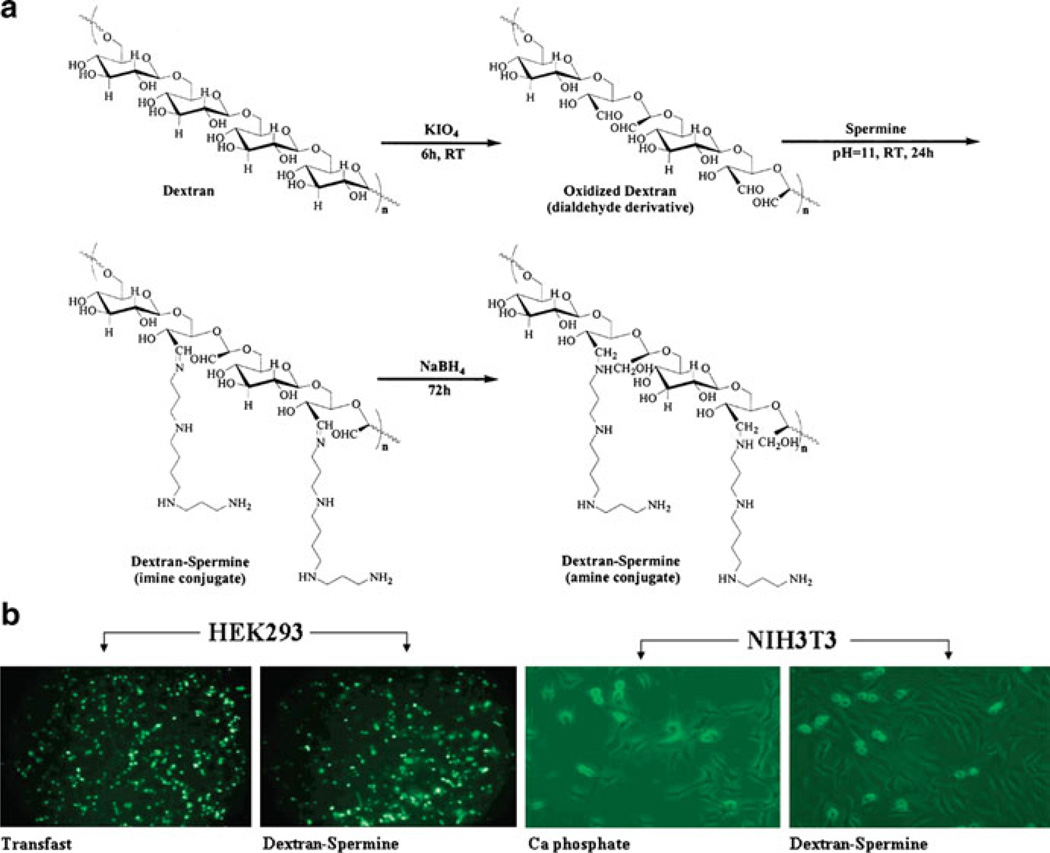
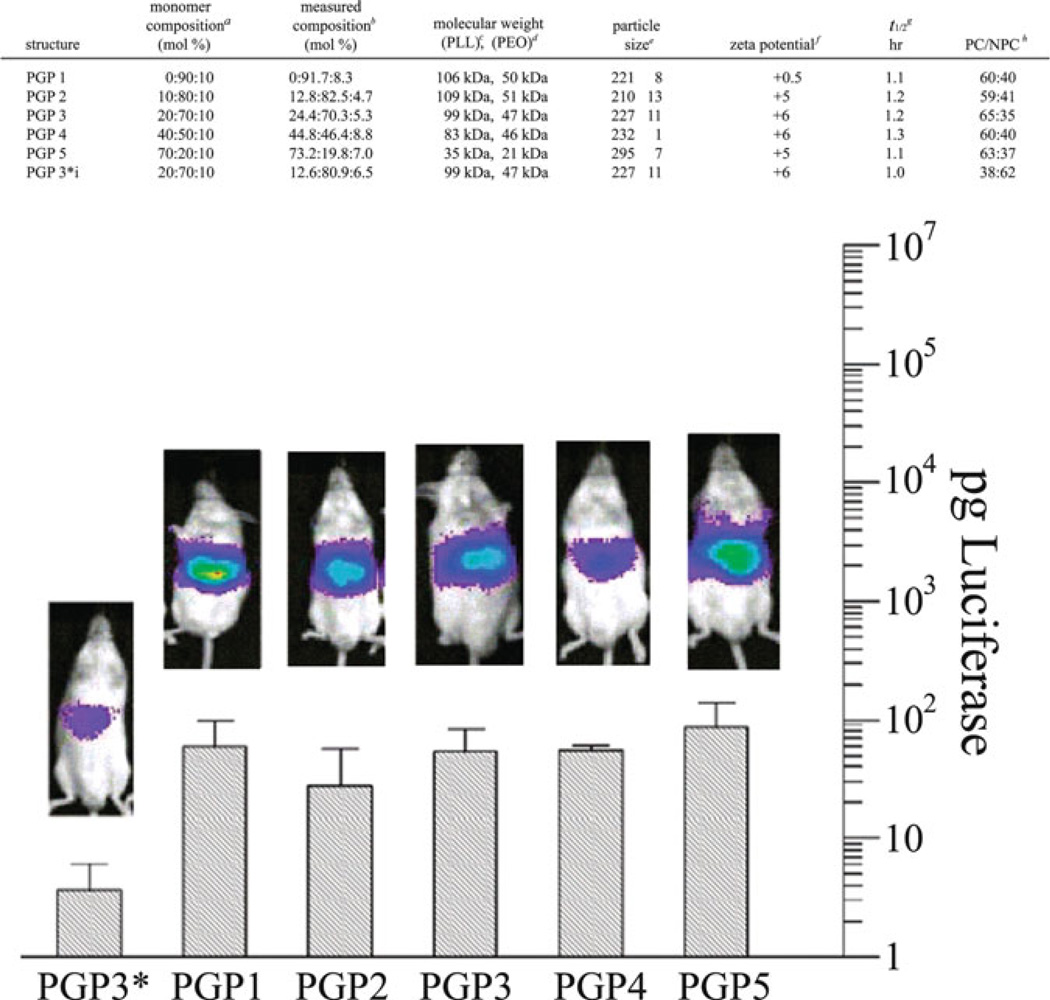
Cationic dextrans (40 kDa) were synthesized by Azzam et al. through conjugation with a variety of oligoamines, including spermine and spermidine [37, 38]. An ethidium bromide exclusion assay (qualitative assay for binding affinity) revealed that the dextran-spermine conjugates bound DNA more strongly than other oligoamine derivatives. Transfection efficiency experiments in NIH3T3 and HEK293 cells showed high gene expression in serum-free media, similar to the positive controls Transfast and DOTAP/Chol (1/1), and much enhanced from calcium phosphate (Fig. 1). The most efficacious vehicles were 6–8 kDa in molecular weight, with a spermine content of 2 µmol/mg and 25–30% branching. Unfortunately, a reduction in gene expression was noted with a similar experiment in serum-containing media. To improve the transfection in serum, Hosseinkhani et al. [39] synthesized a poly(ethylene glycol) (PEG)-containing dextran-spermine conjugate (G7TA141) and three dextran-based spermine conjugates (G7TA103, G7TA107, and G7TA141). When NIH3T3 cells were transfected with these PEGylated vectors in NIH3T3 cells in serum-containing media, they showed higher luciferase expression than their nonPEGylated counterparts, with maximum gene expression observed at a polycation:DNA weight ratio of 5:1. Intramuscular injection of PEGylated G7TA141 in mice at a 5:1 weight ratio (polyplexes dosed at a pDNA dose of 50 µg/mouse) revealed a higher level of transgene (β-galactosidase) expression in mouse muscle than naked pDNA or pDNA complexed with the non-PEGylated analog. Gene expression in the liver was monitored for mice dosed with complexes prepared with dextran-spermine conjugates modified with different levels of PEG conjugation. The results indicated the highest level of β-galactosidase expression in the liver resulted from the 5% PEGylated complexes noted 2 days post-injection. These results demonstrated that delivery in serum-containing media can be improved through PEGylation strategies and elicit favorable results in animal models.
Fig. 1.
(a) Synthesis of dextran-spermine conjugates. (b) Fluorescence micrographs of dextranspermine compared to common transfection reagents in HEK293 and NIH3T3 cells. Adapted with permission from [38]. © 2002 American Chemical Society
Further in vivo studies with dextran-spermine conjugates have been explored recently by Eliyahu et al. [40, 41]. The efficacy of local and systemic delivery in mice was assessed through intramuscular (i.m.) and intranasal (i.n.) injections, respectively. Efficacy, measured by X-gal expression in paraffin-embedded tissue sections, was observed primarily in lung tissue (bronchial epithelial cells, pneumocytes, and alveoli), fibrocytes in the skeletal muscle, and hepatocytes. X-gal expression was higher in each organ when pDNA was delivered by the cationic dextran compared to pDNA only. In comparison, lipoplex (DOTAP/cholesterol lipoplexes) injections resulted in expression only at the site of injection, with distant sites such as liver not being transfected, an observation that did not change with increased lipoplex dosing. Upon histopathological assessment of toxicity, mild inflammation and necrosis were observed in the skeletal muscle, but no toxic effects were seen locally in the lung or liver tissue when pDNA was delivered by dextrans. Systemic toxicity was also low, as injections of polyplex or free polymer showed little effect on organ weight, white blood cell and platelet counts, and serum transaminase levels [41]. PEGylation of spermine-dextran conjugates and decrease in spermine content resulted in lower transgene expression. Systemic transfection of PEGylated dextrans was not dose dependent, since increasing the dose from 6 to 40 µg DNA did not increase transgene expression. This series of initial studies demonstrate the promise of DEAE-dextran as a non-viral delivery vehicle; it remains a promising delivery platform.
Additional recent work has focused on developing dextrans into functional hydrogels for effective delivery of nucleic acids. Singh et al. developed hydrogels containing crosslinked dextran vinyl sulfone and tetra-functional PEG thiols encapsulating siRNA/pDNA-loaded microparticles and dendritic cell chemoattractants for the dual delivery of chemokines and nucleic acids [42]. The chemoattractants were encapsulated in degradable microspheres composed of poly(lactide-co-glycolide) (PLGA); the siRNA is also encapsulated in PLGA and functionalized with PEI before addition of pDNA. Hydrogels were crosslinked in situ via Michael-type addition reactions, and the dextran vinyl sulfone and PEG components were mixed to form hydrogels. The stoichiometry of the components was varied to control the crosslinking density, which can impact the release rate of the microparticles. These materials were nontoxic in multiple cell lines in vitro and exhibited slow release of chemokine from 30% dextran/10% PEG hydrogels. These hydrogels released 70% of encapsulated chemokine after 72 h, indicating that sustained drug release is possible. In primary antigen-presenting cells (APCs), chemokine-induced dendritic cell migration was observed, as well as siRNA-induced knockdown of IL-3. These promising results show that dextran can be used in functional material design for sustained release of drugs and nucleic acids.
Other promising work describing DNA delivery with dextrans has been published recently from the Fréchet laboratory [43]. Acetal-derivatized dextran was solvent evaporated to form dextran nanoparticles which are cleavable under acidic pH [44]. Exploiting the reducing chain ends present on the carbohydrate particles, the authors used alkoxyamine-terminated poly(arginine), commonly referred to as a cell penetrating peptide (CPP) for its purported ability to penetrate cell membranes transiently, to introduce CPP onto the dextran particles through formation of oxime linkages. Using CPP-derivatized dextran particles (containing 20% poly(β-amino ester) polymer) encapsulating a pDNA encoding luciferase, they attained a 60-fold increase in luciferase expression in HeLa cells compared to unmodified particles [43]. These results demonstrate the broad applicability and the future use of dextran in nucleic acid delivery.
2.2 Schizophyllan
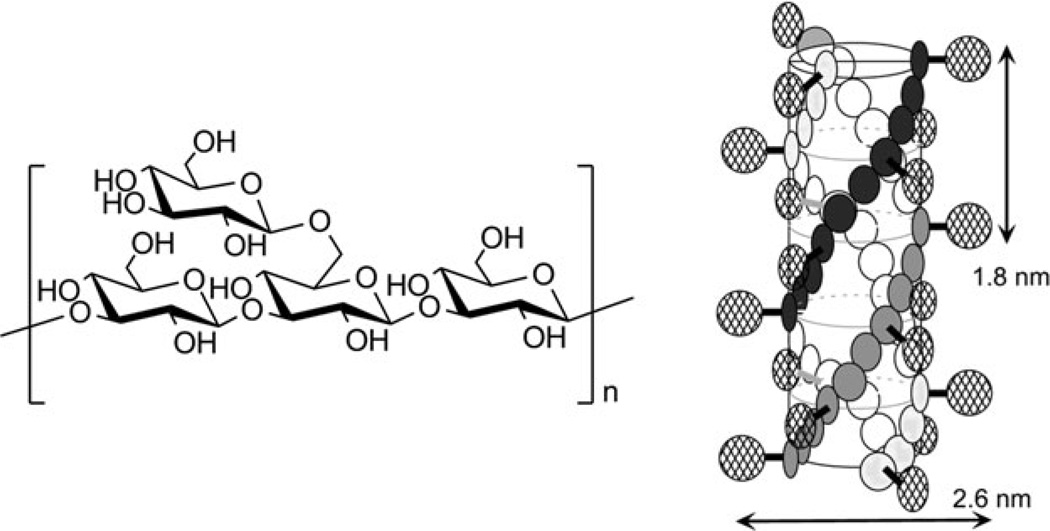
Schizophyllans (SPGs) are naturally-occurring water soluble polysaccharides that are produced by the fungus Schizophyllan commune. SPG belongs to the family of β-(1→3) glucans, and it has one branch through β-(1→6)-d-glucosyl linkage per three glucose units (Fig. 2). The safety of these materials has been well demonstrated, as they have been used as adjuvants for over two decades in drug formulations and in treatment of gynecological cancer [46, 47].
Fig. 2.
Structure of schizophyllan repeat unit and a schematic representation of the triple helix formed in aqueous solution. Figure adapted with permission from [45]. © 2003 Elsevier
In water, SPG exists as a thermodynamically stable triple-helix (Fig. 2) held together by hydrogen bonds. Under special conditions these hydrogen bonds can be broken; for further discussion, it is important to know that, in DMSO, schizophyllan dissociates and exists as a single randomly-coiled chain, but it will re-associate into triple helices upon dilution with water.
The Sakurai lab has reported that SPGcan form complexeswith polycytosine (poly (C)) and polyadenine (poly(A)) [48]. Shortly thereafter, they reported that polydeoxyadenine (poly(dA)) and polydeoxythymine (poly(dT)) can form such complexes with SPG as well [49]. It was also noted that none of these homopolynucleotides form supramolecular structures on their own and, instead, exist in solutions as single chains, thus not self-assembling via hydrogen bond formation. This seems to be a necessary requirement, as other homopolynucleotides (e.g., polyuracil) which form intermolecular and/or intramolecular bonds in solution do not form complexes with SPG. The structure of SPG–polynucleotide complexes is not yet completely clear. Sakurai proposed, based on circular dichroism studies, the SPG–polynucleotide complex is a hetero-triplex similar to the original SPG-triplex, with one of the schizophyllan chains being replaced by one homopolynucleotide chain [49]. Additionally, the complex will form only with single-stranded SPG. Both of these statements were recently challenged [50], as it was shown that SPG–poly(C) complex can be formed with triplex-SPGs that have been previously denatured and renatured, and these complexes have identical melting temperatures to ones that are formed with a single-stranded SPGs. This newer approach allows one to prepare SPG–polynucleotide polyplexes in more physiologically relevant conditions – since no use of DMSO is required – but complex stoichiometry will likely be different.
Since schizophyllan can form complexes only with single stranded homopolynucleotides, this polynucleotide fragment must be incorporated into DNA/RNA that is used for delivery. Nonetheless, it was demonstrated that such polynucleotides can be efficiently complexed with SPG and be protected from degradation by nucleases within the complex [51], as well as act as an antisense inhibitor of a complementary mRNA in cell-free media [52]. It was concluded that, in order to retain its silencing function, the single stranded antisense oligodeoxynucleotide (AS-ODN) must have stronger affinity toward target mRNA than SPG. This effect was confirmed by Koumoto et al. who demonstrated that the presence of the sshomopolynucleotide complementary to the one complexed inside SPG·DNA and SPG·RNA polyplex is sufficient to induce release of the cargo [53].
For the complexation of double-stranded DNA, a more elaborate polynucleotide morphology has been designed via the introduction of homopolynucleotide sequences on the ends for SPG binding [54]. Poly(dA) 80-mer was introduced at both ends of DNA, forming loops which provide protection from degradation by endonucleases, an approach adopted from viruses.
It is possible to avoid incorporation of homopolynucleotide by using ternary complexes between polynucleotide, SPG, and polycation. It was recently demonstrated that such ternary complexes can be prepared with PEI and cationic cellulose (quaternized nitrogen is a charged functional group in cationic cellulose) [55]. However, this kind of approach is a step away from the non-cationic nature of schizophyllan-based delivery systems and will not be discussed here.
Soon after the discovery of the ability of SPG to complex homopolynucleotides, it was demonstrated that schizophyllan complexes with poly(A) or poly(C) will dissociate at pH 4–6 [45]. This property is important as SPG polyplexes could potentially release their polynucleotide cargo in the acidic pH environment of endosomes and/or lysosomes. However, since these polyplexes possess a net negative charge and lack targeting groups, they are inefficient in terms of cellular internalization.
In initial attempts to improve SPG as a nucleic acid delivery vehicle, lactose and mannose were incorporated in schizophyllan by chemical modification of branching glucose units [51, 56]. The chemical modification is chemoselective to the branched glucose units on the main polymer backbone, such that the polymer backbone remains intact. Oxidation by periodate ion requires hydroxyl groups to be vicinal, and that is why the oxidation of schizophyllan main chain is avoided. Aldehydes formed in this step are good handles to be used in further modification, which, as we shall see further, mostly involve reductive amination. Furthermore, these modifications do not interfere with the ability of SPG to form triple-helices and polyplexes. Mannose- and lactose-modified SPGs were demonstrated to protect homopolynucleotides poly(C) and poly(dA) from degradation, and these targeted polyplexes were able to bind to corresponding lectins. It is not clear if similar protection and recognition by receptors can be achieved in the case of AS-ODNs, since data for such experiments was not presented, but in vitro studies demonstrated that lactose-functionalized SPG deliver antisense oligonucleotide to HepG2 cells. Phosphorothioate AS-ODN that would suppress mRNA of c-myb, a proto oncogene that causes cancer when overexpressed, was used. The antisense effect was 10% higher for SPG-mediated delivery and 40% higher for lactose-SPG-mediated delivery compared to naked AS-ODN at 30 µg/mL; it was 10% and 45% higher, respectively, at 60 µg/mL. The delivery efficiency was decreased for lactose-SPG-mediated delivery, but not for SPG-mediated delivery, in the presence of galactose; however, a large concentration of galactose – 20 mM – was used to demonstrate the specificity.
An analogous strategy was used for grafting SPG with folic acid (FA) and, as in the previously-described study, SPG–FA was used for the delivery of phosphorothioate AS-ODN that would suppress c-myb [47]. This time it was noted that after grafting the weight-averaged molecular weight of SPG was decreased from 150 to 90 kDa. The degree of grafting was estimated to be 9% and grafted folic acid could be recognized by folate binding proteins. In vitro experiments showed that SPG–FA complexes can efficiently deliver AS-ODN to KB cells, causing 45% decrease in cell viability, and that delivery efficiency is dependent on the concentration of free folic acid in the media. Importantly, it was demonstrated that scrambled AS-ODN delivered by the same SPG–FA vehicle does not suppress cell growth, proving that cell growth suppression is mediated by the antisense ODN and not a nonspecific effect of the delivery vehicle.
Following initial work on SPGs, Matsumoto et al. [57], in an attempt to improve cellular internalization, modified SPGs with octaarginine (R8), spermine, arginineglycine-aspartic acid tripeptide (RGD), or single amino acids (Arg and Ser). The SPG modification was done using the route described above and was in a 0.5–24.7 mol% range. Antisense delivery experiments were conducted in A375 melanoma cells and the HL-60 leukemia cell line. A 60% decrease in cell growth was observed with AS-ODN complexes formed with R8-SPG, and a 56% decrease in cell growth was revealed with RGD-SPG complexes at a concentration of 12.5 µg/mL in A375 cells. It should be noted that the antisense activity was minimal while using the positive control, Lipofectamine, or SPGs modified with spermine, arginine, or serine. When the same experiment was performed using the sense sequence (S-c-myb), Lipofectamine and spermine-modified SPGs were highly cytotoxic compared to naked S-c-myb, but the rest of the modified SPG-systems showed the level of toxicity similar to naked S-c-myb. An analogous trend was observed when the experiments were performed in HL-60 cells. Thus, the authors demonstrated that octaarginine- or RGD-modified SPGs elicited a more potent antisense effect (likely derived from increased internalization) than Lipofectamine and negligible toxicity comparable to unmodified SPG, thereby providing new insight into schizophyllans as nucleic acid carriers. It is important to note here that, although schizophyllan was modified with cationic (at physiological pH) functionalities, the N/P ratio used for polyplex formulation was less than one. The polyplex ζ-potential was not reported, but authors expect it to be negative based on the N/P ratio and stress that modified SPG is principally different and advantageous compared to polycationic polynucleotide carriers.
Work on the modification of SPG and delivery of AS-ODN to A375 cells was continued with introduction of amino-modified PEG (5 kDa) through previously-described reductive amination [58]. The degree of modification used in this study was 10.1% (The PEGylation strategy was suggested in order to promote fusion of polyplexes with the cell membrane – as opposed to endocytic cellular internalization; this approach is very unusual because PEG is typically used to shield vehicle charge, preventing aggregation in serum and nonspecific associations). Cell culture studies show that inhibition of endocytosis leads to a decreased antisense effect, which suggests that internalization occurs through an endocytic route, rather than by vesicle fusion. Using nigericin, a chemical inhibitor that blocks transport from endosomes to lysosomes, the antisense-mediated decrease in cell growth was preserved for PEG-SPG/AS-ODN but nearly completely abrogated for RGD-SPG/AS-ODN. The authors suggest that this result confirms endosomal escape of PEG-SPA/AS-ODNs prior to transport to the lysosome. Instead, this result could be indicative that the incorporation of PEG on the delivery vehicle affects the internalization route. These polyplexes may also enter the cell through an alternative pathway which does not traffic to lysosomes, in which case nigericin would have no effect. This is evidenced with the RGD-modified polyplex, for which nigericinsensitive trafficking appears to be essential for efficient function. More studies are needed to elucidate the effect of PEG on polyplex trafficking.
PEG modification of SPG was attempted together with galactose modification [59]. Galactose-terminated PEGs of different lengths were introduced into SPG by reductive amination. In vitro studies conducted in serum-containing medium with HepG2 cells demonstrated that, among tested PEG lengths (0.2, 0.6, 2, and 6 kDa) used for SPG modification, the longest one (6 kDa) was the most efficient in reducing cell growth through delivery of AS-ODN. This 10 mol% Gal-PEG-modified SPG delivery vehicle was more efficient in reducing cell growth than 8.7 mol% galactose-modified SPG (Gal8.7-SPG), naked AS-ODN, and SPGs modified by glucose-terminated PEGs. Importantly, there was no difference in the antisense effect in A375 cells (which do not express galactose receptors) when SPGs modified by glucose- and galactose-terminated PEG-SPGs were compared. However, both of these delivery vectors caused a significant (up to 65%) decrease in cell viability; this toxicity is potentially problematic for systemic delivery applications. In addition, this work demonstrates the propensity for internalization of complexes made with galactose-modified PEG-SPGs by cells other than hepatocytes.
Mizu and coworkers [60] utilized the same SPG modification strategy with spermine, R8, RGD, or cholesterol to deliver unmethylated, CpG motif-containing single stranded oligo DNA with (dA)40 tail at the 3′ end into murine macrophage-like cells (J774.A1) to enhance cytokine secretion. Consistent with the study described above, the degree of functionalization was kept low: 0.5 mol% and 6.9 mol% in the cases of R8 and cholesterol, respectively. CpG DNA has been shown to be an effective adjuvant in various vaccines to treat numerous diseases [61]. Previous studies have shown that complexation of phosphorothioate AS-ODNs with modified SPGs reduces their non-specific interactions with proteins and increases their cellular uptake. In this study, the secretion of three different cytokines (IL-6, IL-12, and TNF-α) was assessed. A five- to tenfold increase in cytokine secretion was observed for the modified SPG complexed with CpG DNA over naked CpG DNA. The SPGs modified with octaarginine were found to have the highest efficacy, followed by RGD- and cholesterol-modified SPGs. However, only 20–40% enhancement in cytokine secretion was found when CpG DNA complexed with unmodified SPG was used.
This work with antigen-presenting cells was continued using phosphodiester (PO)-DNA instead of phosphorothioate (PS)-DNA to avoid “unexpected biological responses” [62]. For many antisense oligonucleotides, the phosphodiester bond is replaced by phosphorothioate, which reduces nuclease susceptibility, presumably by introducing chirality [63]. In vitro experiments with mouse primary spleen cells revealed an interleukin (IL) expression trend similar to those described by Mizu et al. [60]. R8-modified SPG elicited the highest expression of IL-6 and IL-12 at both 25 µg/mL and 50 µg/mL DNA concentrations, while spermine-modified SPG yielded a slightly lower response. When PO-DNA was used, four- to sixfold higher DNA concentrations were needed to induce IL secretion comparable to PS-DNA, a finding attributed to lower PO-DNA stability toward nuclease degradation. While further development of SPG vehicles for PO-DNA delivery is needed, the ability to deliver DNA to primary cells by SPG vehicles is a substantial achievement. Followup in vivo experiments in mice showed a significant increase in IL-12 secretion when SPG was used for delivery, as compared to naked DNA. Unfortunately it was not mentioned whether PS-DNA or PO-DNA was used, but the ability to deliver DNA in vivo was demonstrated.
SPG modifications to create cationic vehicles by grafting amines (ethanolamine, spermine, spermidine, and tripropylenetetramine (N4C3)) onto the SPG backbone have also been attempted [46]. In vitro experiments in COS-1 cells revealed that, among amines used for SPG grafting, N4C3-modified SPG was the most efficient in transfection, a result related to its high amine density. Thus 34 kDa SPG containing 41 mol% grafting with N4C3 was fivefold more efficient in transfection (at N/P 10) than PEI (25 kDa); however, it was also the most toxic vehicle – 40% more toxic than PEI. Toxicity followed the same trend as the transfection efficiency, suggesting that the more charge-dense polymers were also the most toxic. The molecular weight of glucan has a role as well, and 80-kDa SPG was the most efficient (among 12-, 34-, 80-, and 150-kDa tested), while 12- and 150-kDa were the least efficient. To reduce toxicity, PEG was introduced via amide linkage with succinimide-activated, carbonyl-terminated PEG. Both 2 and 5-kDa PEG-SPG derivatives showed 100% cell viability, but transfection efficiency was also reduced, becoming similar to PEI. Finally, it was demonstrated that SPG vectors have long-term transfection, with detectable intracellular plasmid DNA over 30 days post-transfection. This sustained DNA residence was speculated to be the result of slow non-enzymatic hydrolysis of the SPG backbone.
Studies utilizing SPG as a nucleic acid carrier have been performed over the last decade and highlight the potential applicability of SPG for nucleic acid delivery applications. The unique ability to form complexes with polynucleotides via hydrogen bonding – without electrostatic interactions – has great promise, since cytotoxicity mediated by positive charge density can thereby be avoided. Moreover, branched glucose units offer attractive and facile modification sites due to selective modification by oxidation or reductive amination; this leaves the main chain intact, preserving the ability to condense polynucleotides. The SPG backbone cannot be cleaved enzymatically in mammals due to the lack of appropriate enzymes, making schizophyllan a good candidate for slow-release delivery vehicle development. Further research on the efficiency and versatility of this polymer will reveal the future of schizophyllan as a non-viral nucleic acid carrier.
2.3 Hyaluronan
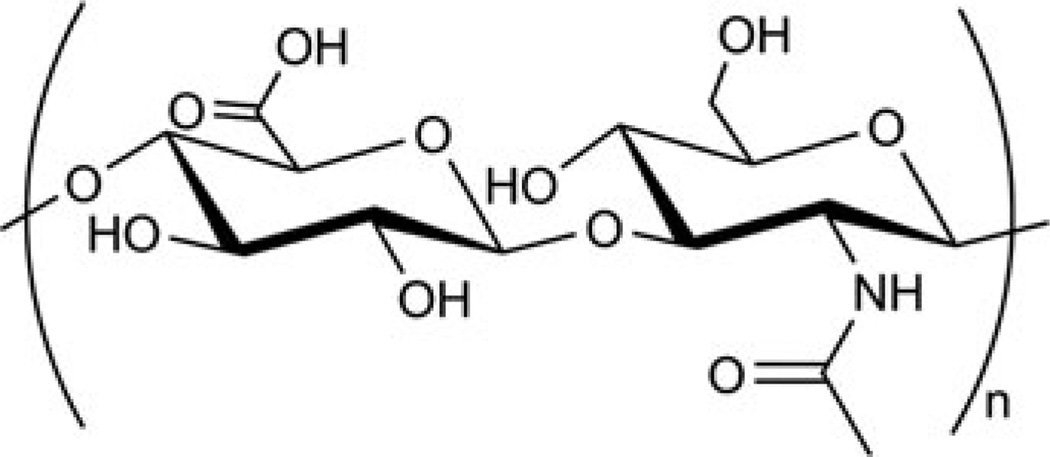
Hyaluronan, also called hyaluronic acid (HA), is a glycosaminoglycan, a major component of the extracellular matrix. It is composed of N-acetyl-d-glucosamine and d-glucuronic acid (Fig. 3) [64]. It has been extensively used in biomedical applications due to its biodegradability as well as lubricating, shock-absorbing, and non-immunogenic properties [65]. As can be seen from the structure, hyaluronic acid has several sites for chemical modifications. Functionalization via the carboxyl group is used most often in gene delivery applications, because there is only one carboxyl group per repeat unit (as opposed to multiple hydroxyl groups). Such modification also removes the negative charge, which is beneficial for the complexation of negatively-charged polynucleotides. As discussed later in this chapter, the carboxyl group can be activated toward nucleophiles in aqueous solution.
Fig. 3.
Structure of hyaluronan
In 2003, Kim and coworkers [65] formulated pDNA encoding platelet-derived growth factor (PDGF) with HA and studied in vitro transfection efficiency in COS-1 cells (Fig. 4). In this study, solutions containing various amounts of pDNA were mixed with HA solution, flash-frozen and lyophilized, yielding DNA-HA matrices. These matrices were then placed in a DMF/H2O solution of adipic dihydrazide and 1-ethyl-3-(3-dimethyl amino)propyl carbodiimide (EDC), a conventional water-soluble, carboxyl-activating agent. An HCl solution was subsequently added to lower the pH. By allowing these mixtures to incubate for various amounts of time, DNA-HA matrices with different degrees of crosslinking could be obtained. It should be pointed out here that the DNA is physically entrapped in a pre-formed, mesh-like HA matrix. While relatively uncommon for nucleic acid delivery, this approach is common for the delivery of small molecule therapeutics.
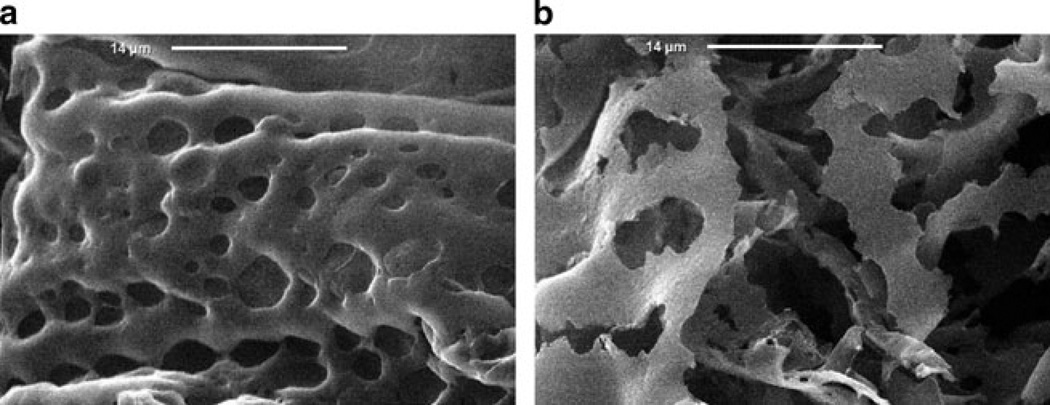
Fig. 4.
SEM images of DNA-HA matrices: (a) before and (b) after incubation in hyaluronidase solution (10 units/ml) for 7 days. Figure adapted with permission from [65]. © 2003 Elsevier
The pDNA release kinetics from the matrices in the presence of the enzyme hyaluronidase (at concentrations that were intended to resemble serum conditions) were shown to be dependent on the pDNA loading and the degree of HA cross-linking. It was observed that pDNA release is faster from the matrices with lesser degrees of crosslinking, and it was suggested that matrices with higher extents of crosslinking could potentially be used for slow release applications.
One such application could be delivery of Has2-pDNA, a plasmid that codes for hyaluronan synthase 2 [66]. This enzyme facilitates the synthesis of larger HA molecules and can prevent post-surgical peritoneal adhesions. In one study, DNA-HA films were prepared using previously-described chemistry; however, lyophilization was replaced with air-drying under sterile conditions and an isopropanol/H2O mixture was used instead of DMF/H2O. The release kinetics of DNA were similar to that from the HA film described previously, but release did not occur until after 7 days. The reason for this delay was not completely clear; the authors suggest that a possible way to overcome the delay is to use a crosslinked DNA-HA film sandwiched between two non-crosslinked DNA-HA films. Non-crosslinked film would be expected to undergo rapid hydrolysis, thus serving as a source of HA during the initial period.
As an extension of the HA film approach, Yun and coworkers [32] synthesized hyaluronan microspheres using the chemistry described above, but the synthesis was completed in emulsion in one step, yielding 5- to 20-µm microspheres. These microspheres were found to be biodegradable and released three times more pDNA when incubated with hyaluronidase in PBS (phosphate buffered saline) solution (vs enzyme-free PBS). As in the case of films, DNA release from the microspheres was dependent on the DNA loading. DNA-HA microspheres were not directly used for transfection; instead, DNA obtained from release experiments was used in transfection of Chinese hamster ovary (CHO) cells using Lipofectamine. The relative levels of transfection over time had the same trend as DNA release from the DNA-HA microspheres and confirmed that released DNA is bioactive.
The transfection capabilities of the HA microspheres were investigated in vivo by injecting the microspheres containing pDNA (encoding β-galactosidase) in rat hind limb muscles [67]. Three weeks post-injection, animals were sacrificed and RT-PCR showed detectable pDNA, indicating that DNA-HA microspheres are suitable for slow DNA release in vivo. In addition, the humanized monoclonal antibody that recognizes E- and P-selectin in modified CHO cells and human umbilical vein endothelial cells (HUVECs) were conjugated to HA microspheres. Antibody-conjugated HA microspheres showed very specific binding to cells expressing E- and P-selectin, demonstrating a great potential for development of site-selective HA delivery vectors [67].
Recently, other approaches and modifications of HA for gene delivery applications have been investigated. Among the most interesting ones are mixed chitosan-hyaluronan based gene delivery systems [68–70] and PEG-HA photocrosslinked hydrogels [71]. HA has also been used to improve the biocompatibility of branched PEI via covalent conjugation [72].
A more sophisticated system was reported recently in which HA is modified with spermine and a lipophilic amine containing a long hydrocarbon chain. This system was shown to be efficient in siRNA complexation, has a very low critical micelle concentration (40–140 mg/L, depending on the length of lipophilic amine chain), and forms cationic micelles with 125–555 nm diameter [73].
2.4 Pullulan
Pullulan is a neutral, water-soluble polysaccharide synthesized from starch by the fungus Aureobasidium pullulans. It is composed of maltotriose units, in which the three glucose units of the maltotriose are linked via α-(1→4) glycosidic linkages and consecutive maltotriose units are linked by a α-(1→6) glycosidic unit (Fig. 5) [74]. The versatility of pullulan has encouraged its usage in a variety of applications, including use as decorative materials in baking, coatings, capsules, and also in soft-chew candies [74, 75]. Perhaps the most significant work has been done with pullulan nanoparticles as drug carriers, where water-insoluble drug molecules [76], vitamins [77], or cholesterol [78] have been encapsulated in the hydrophobic pullulan interior and used in treating a variety of diseases. However, it was not until 2004 that pullulan was used to design biomaterials that could be used to deliver nucleic acid therapeutics.
Fig. 5.
Structure of pullulan
In 2002 Hosseinkhani et al. synthesized and evaluated several pullulan derivatives for gene delivery in vivo [79]. This work will not be discussed here in detail, since no in vitro transfection experiments were conducted; however, this study presents an interesting approach to delivery vehicle design. Pullulan was grafted with diethylene triamine pentaacetic acid (using a corresponding anhydride and DMAP(4-(dimethylamino)pyridine)) and with diethylenetriamine and triethylenetetramine (using 1,1’-carbonyldiimidazole (CDI)). After purification, pullulan derivative solutions were mixed with pDNA solutions. This was followed by addition of Zn2+ ions, which were chelated by delivery vectors to allow tighter DNA encapsulation. These complexes showed enhanced gene expression in liver parenchymal cells which lasted for over 14 days.
In their further studies, the Tabata lab have synthesized pullulans grafted with spermine, using the same approach – CDI-mediated coupling [80]. This grafting procedure calls for 15 equivalents of spermine per hydroxyl group (or 69 equivalents of spermine per primary hydroxyl group) of pullulan, yielding pullulan with 12.3% spermine introduction. After the purification, modified pullulan was used for transfection of human bladder cancer (T24) cells. This study revealed that pullulan-g-spermine polyplexes enter the cell through clathrin- and caveolae-mediated endocytosis with involvement of sugar-recognition receptors. The transfection efficiency evaluated by reporter gene expression was tenfold better than Lipofectamine. Moreover, it is mentioned that, according to the authors’ unpublished data, similar enhancement in transfection is observed for Caki-1, ACHN, PC3, LNCaP, HepG2, UMUC-3, EJ, and primary isolated rat bone marrow stromal cells.
Their next study investigated the influence of pullulan molecular weight and the amount of spermine grafting on transfection [81]. Among three tested molecular weights (22.8, 47.3, and 112 kDa), pullulan with an intermediate molecular weight (47.3 kDa) was the most efficient in transfecting HepG2 cells. The optimal amount of grafted spermine for transfection varied for different molecular weight pullulans, and this amount decreased with increasing pullulan molecular weight. However, the optimal molar ratio of polymer to DNA was similar (close to 100) for all three tested molecular weights. Receptor-mediated endocytosis was suggested because transfection inhibition was observed upon pretreatment of cells with asialofetuin, a competitive inhibitor for the asialoglycoprotein receptor on hepatocytes.
Gupta and coworkers have formulated pullulan hydrogel nanoparticles as a pDNA delivery carrier [33]. The in vitro delivery efficacy and cytotoxicity of this approach were determined by β-gal expression and MTT assay, respectively, in HEK293 and COS-7 cells. In this study, water-soluble materials such as pDNA could be encapsulated within the hydrophilic core of these hydrogels and thus transported into the cells. The extent of pDNA protection from nuclease degradation was tested using gel electrophoresis. The results indicated that the pullulan nanoparticles were effective in protecting pDNA against DNase degradation. The cell viability in COS-7 and HEK293 cells determined using an MTT assay indicated that pullulan was relatively nontoxic; however, the cell viability decreased to about 80% (COS-7 cells) and 70% (HEK293 cells) with an increase in dosage to about 20 mg/mL. The cellular uptake mechanism of these nanoparticles was studied using SEM and fluorescent staining of cytoskeleton components in primary human fibroblast (hTERT-bJ1) cells, which revealed that the nanoparticles entered the cells via an endocytic pathway. Following this, transfection was performed in serum-containing media to mimic in vivo conditions in both COS-7 and HEK293 cell lines. The results indicated maximum expression at pullulan concentration of 250 µg/mL. However, the transfection decreased with an increase in polymer concentration, which could be related to the cytotoxicity revealed in the MTT assay. The delivery efficacy was cell-type dependent, with COS-7 cells having higher gene expression than HEK293 cells. The pullulan-containing nanoparticles yielded comparable β-galactosidase expression to Lipofectamine. This was the first study demonstrating the utility of pullulan as a DNA delivery carrier, and these results have been significant in motivating further development of pullulan-based non-viral vectors.
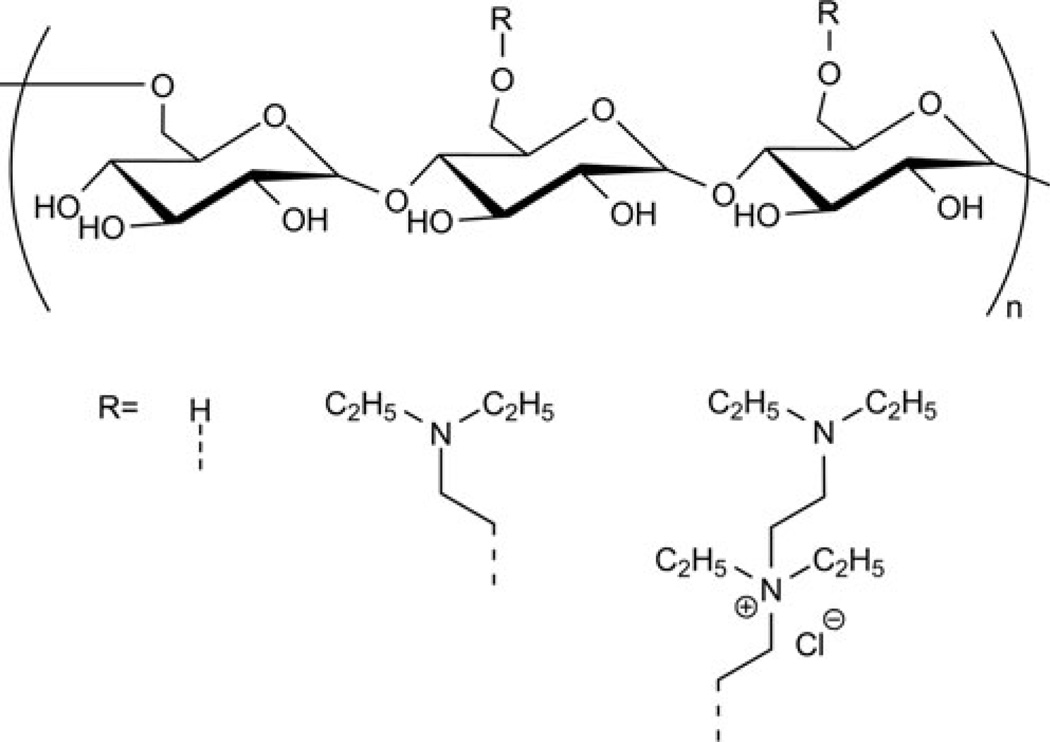
Consequently, San Juan and coworkers chemically cross-linked DEAE-pullulan and synthesized a cationic, 3D pullulan matrix that could be loaded with pDNA (pSEAP) and function as a delivery vehicle (Fig. 6) [82]. In vitro transfection and cytotoxicity experiments were performed in vascular smooth muscle cells (SMCs). In this study, pullulan was first grafted with N,N-diethylamine groups yielding cationic DEAE-pullulan and then chemically crosslinked using POCl3 to form a 3D matrix in the form of a hydrogel disc. All the experiments were performed with both cationic DEAE-pullulan and the crosslinked DEAE-pullulan matrix. The extent of DEAE-pullulan/pDNA binding was studied via dye exclusion assay using PicoGreen. The results indicated that neutral pullulan did not bind pDNA, as no fluorescence quenching was observed. However, fluorescence intensity decreased sharply with DEAE-pullulan, indicating pDNA binding with the cationic molecule (and PicoGreen exclusion). The cell viability determined via the MTT assay indicated no significant toxicity when the SMCs were treated with either pSEAP/pullulan or pSEAP/DEAE–pullulan. The delivery efficacy in media containing serum was 150-fold higher using DEAE-pullulan than for naked pSEAP or a pSEAP/neutral pullulan mixture. The pSEAP extracted from the DEAE–pullulan matrix was found to be protected from nuclease degradation when compared with neutral pullulan, as indicated by gel electrophoresis. Furthermore, the DEAEpullulan matrix has been shown to be nontoxic to SMCs, as revealed via MTT assay. Significant delivery efficacy was noted using DEAE–pullulan matrix at 6 days after initial transfection (when compared to pSEAP only). The nontoxicity and biodegradability of these 3D pullulan hydrogels will be useful towards their development as efficient non-viral DNA carriers for implantable devices to facilitate controlled release from surfaces.
Fig. 6.
Structure of DEAE-pullulan
The research performed by several groups on pullulans has demonstrated their potential as nucleic acid delivery vehicles. Although most of the pullulan-based delivery systems yielded low toxicity, some modifications of the backbone or introduction of substituents resulted in higher toxicity. Such modifications are unavoidable because the parent structure is incapable of efficient delivery and lacks target specificity.
2.5 Chitosan
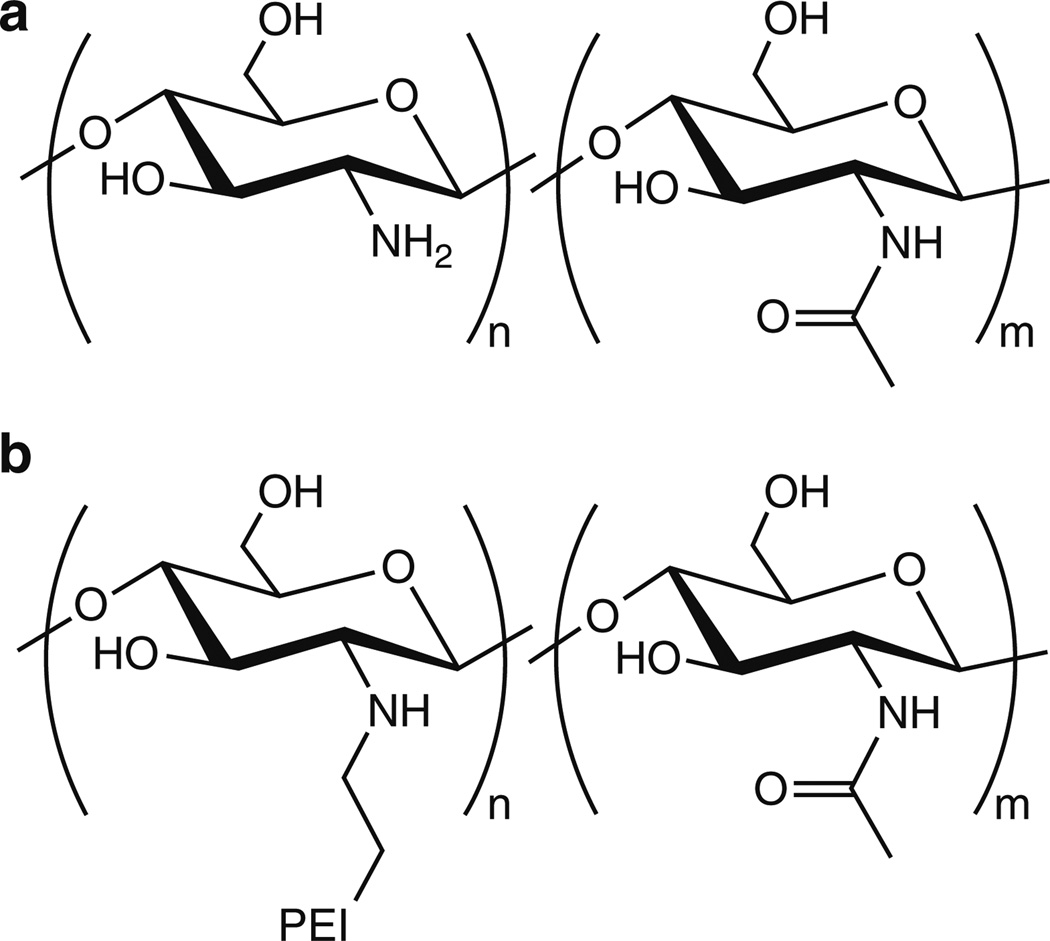
Chitosan is the most widely studied polymeric vehicle for nucleic acid delivery, and the remainder of this section is devoted to its use and development as a delivery vehicle. Chitosan is a polysaccharide composed of glucosamine and N-acetyl glucosamine units bonded via β(1→4) glycosidic bonds (Fig. 7a). The intense study of chitosan stems from its low cost, low toxicity, biodegradability, and the presence of primary and secondary hydroxyls and primary amines – functional groups that can be readily used for modifications via a range of well-established reactions. Amino groups in chitosan have a pKa value of ~6.5, making chitosan positively-charged in neutral and acidic solutions. Chitosan has been found to complex with a variety of polyanions, such as indomethacin [83], sodium hyaluronate [84], pectin, and acacia polysaccharides [85], via electrostatic interactions. This concept has been explored extensively to complex chitosan with therapeutic nucleic acids [31, 86]. The first study describing the formation of chitosan/nucleic acid nanoparticles for gene delivery was reported by Mumper and coworkers in 1995 [87].
Fig. 7.
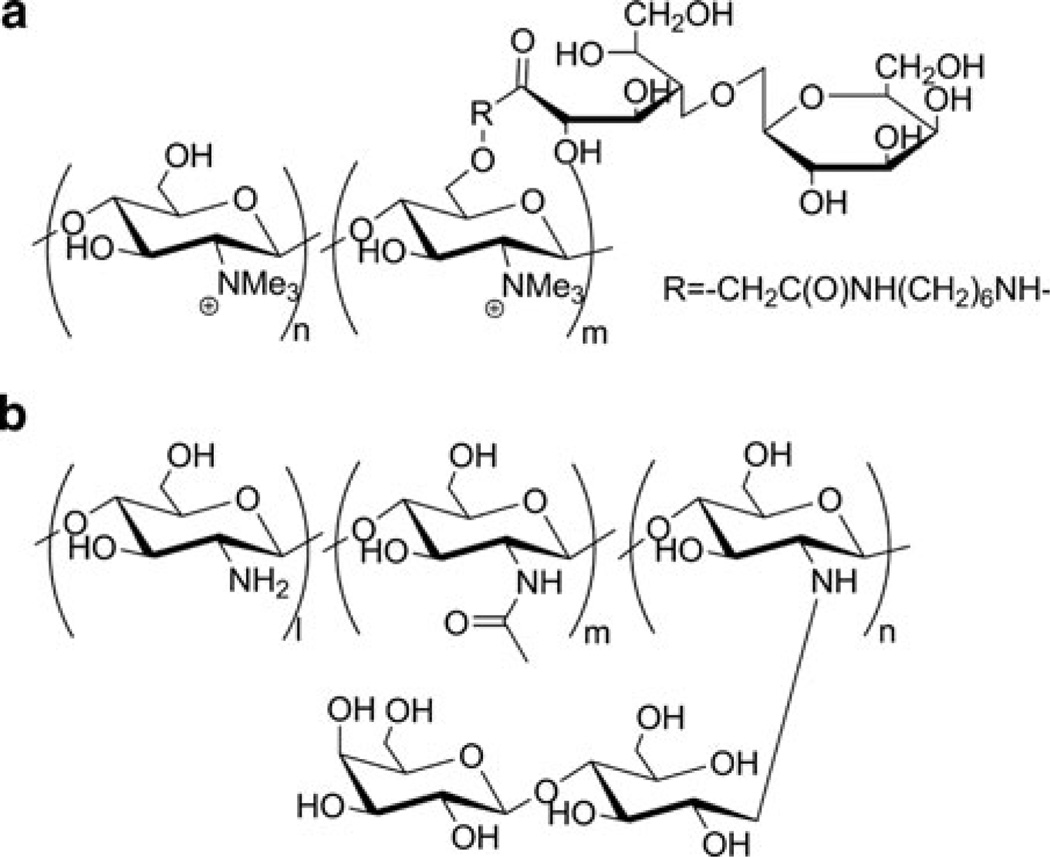
(a) Structure of chitosans. (b) Structure of chitosan-graft-PEI
Chitosan is produced by basic hydrolysis of chitin [88–90]. Chitin is a natural polysaccharide, found widely in fungi and various arthropods, like spiders, insects and crustaceans (shrimp, crabs, lobsters, etc.). In its native state, chitin is a long polymer (molecular weight of 1–10 MDa), insoluble in water and organic solvents. It is made of N-acetyl-2-amino-2-deoxyglucose units linked via β(1→4) glycosidic bonds. Because chitin is semicrystalline and water-insoluble, its hydrolysis is heterogeneous, possibly leading to the formation of localized blocks of N-acetyl-2-amino-2-deoxyglucose units [91]. The influence of these blocks on polyplex formation and transfection is not well documented. In general, the deacetylation degree of commercially-available chitosan is ~80%, but methods affording complete 100% deacetylation have been reported [92]. Notably, by fractionating chitosan via semi-preparative SEC, it was possible to reveal heterogeneity in molecular weights. In the case of the low-molecular-weight fractions of chitosan obtained via degradation with nitrous acid, a deacetylation degree dependence on molecular weight has been shown [93]. Methods for characterization of the deacetylation degree and molecular weight of chitosan and their influence on properties directly related to transfection efficiency, such as biodegradability, mucoadhesion, endothelium permeation enhancement and others, have been extensively reviewed, and the authors suggest using chitosan of ~10 kDa with deacetylation degree ≤80% for gene delivery applications [90].
Chitosan is generally considered nontoxic, with the rare reported toxicity explained by Köping–Höggård et al. as a result of impurities [94]. In their study conducted with ultrapure chitosan, transfection efficiency of 293 cells was shown to be dependent on the polyplex stability, which in turn was dependent on the deacetylation degree of chitosan. A deacetylation degree of at least 65% was found to be required to give efficient transfection. Variations in the molecular weight of chitosan within the range of 31–170 kDa, however, were shown to have no significant effect on polyplex stability and transfection results. In vivo experiments in mice showed that ultrapure chitosan is less efficient in gene delivery to the lung than PEI, but comparable to lipid-based delivery vehicles.
Kiang et al. [95] synthesized chitosans with different deacetylation degrees via acylation of high-deacetylation degree chitosan with acetic anhydride. Cell culture studies with HEK293, HeLa and SW756 cell lines revealed that the transfection efficiency is dependent on both the deacetylation degree and the molecular weight, with chitosan having the highest deacetylation degree being the most efficient. This was attributed to a greater polyplex stability afforded by the high-deacetylation degree chitosan in serum-containing media. However, for intramuscular injection of polyplexes in mice, high-deacetylation degree chitosan was the least efficient, likely due to slower release of the cargo. This study illustrates the potential differences between in vitro and in vivo transfection efficiencies [95].
Similar in vitro results were obtained in a study by Huang et al. [96]. High-deacetylation degree chitosans were also more efficient in protecting DNA from degradation. Transfection of A549 cells with pDNA was made to be more efficient by increasing the deacetylation degree of chitosan. These results showed good correlation between ζ-potential, cellular uptake, and transfection efficiency, suggesting the electrostatic interactions between the nanoparticle and the cell membrane mediate cellular uptake and lead to gene expression [96]. Similar effects were observed with siRNA gene silencing experiments, as polyplex stability and delivery efficiency were generally higher for high-molecular-weight chitosans with higher deacetylation degrees in H1299 human lung carcinoma cells [97].
As discussed in the aforementioned studies, deacetylation degree plays a crucial role in transfection, with the desirable value being in the range of 65–80%. Polyplexes prepared with chitosans having lower amine density, do not protect the cargo from degradation by enzymes, and are not stable in serum. The influence of chitosan molecular weight is less well understood. As previously described, variations in chitosan molecular weight within the range of 31–170 kDa did not affect transfection of 293 cells. However, in an earlier study by MacLaughlin and coworkers [88], it was discovered that a decrease in molecular weight of chitosan from 540 to 7 kDa caused a concomitant decrease in complex size, from 500 to 100 nm. The ability to modify polyplex size can influence the mechanism of endocytosis [98], which likely affects the intracellular fate of the polyplex. In serum-containing media, pDNA delivered with 540-kDa chitosan leads to higher transgene expression than other molecular weight chitosans. In general, it can be speculated that longer chains of high-molecular-weight chitosan are able to form more stable polyplexes with DNA but are less efficient in releasing the cargo than low-molecular-weight chitosans.
The size of the polyplex depends not only on the chemical structure of chitosan but also on the ratio between chitosan and DNA used for polyplex formulation, the concentrations of polymers, and formulation technique. This is commonly described in terms of “N/P ratio,” the ratio of protonatable polymer amines to phosphate groups in the nucleic acid. Increasing N/P ratios typically lead to formation of polyplexes with more positive surface charge, which is evident from ζ-potential measurements. High ζ-potential may seem desirable because it should increase interactions with the negatively-charged cell surface and, hence, lead to higher cellular uptake and transfection. However, high charge density usually results in cytotoxicity, likely caused by the disruption of the cellular membrane [99]. It was shown by Erbacher et al. [100] that N/P ratios greater than 2 are necessary for formation of polyplexes with chitosan (70 kDa was used in this study) having positive ζ-potential. The optimal N/P ratio is specific to each polymer and is usually based on polyplex stability, polyplex ζ-potential, and the ability of the polymer to protect cargo from degradation.
As previously discussed, the protection of pDNA against degrading enzymes is a critical parameter for a non-viral carrier. Such ability is needed for the polyplex to protect the nucleic acid for an extended period of time in the blood while the polyplex circulates and distributes. Research conducted in 1999 by Richardson and coworkers [101] to study the ability of chitosan to protect against DNase degradation revealed that incubation of polyplexes prepared at N/P ratio of 3/1 in the presence of DNase I (8 U, 1 h incubation) protected pDNA from degradation. Other studies of chitosans as gene delivery vehicles confirm that the N/P ratio has to be at least 3/1 to 5/1 in order to provide a sufficient protective effect against DNases.
Shortly after Mumper and coworkers published their original work with chitosan, Murata et al. synthesized quaternary chitosan using MeI in N-methylpyrrolidone (it was also further derivatized by incorporation of galactose) [102, 103]. Since then, quaternization of chitosan has become a primary strategy in the development of chitosan nucleic acid delivery systems. Quaternization introduces pH-independent charges into the polymer backbone and increases the charge density. The efficiency of the quaternization approach was elegantly demonstrated by Thanou and coworkers [104]. They investigated the transfection efficiency and cytotoxicity of quaternized chitosan oligomers (<20 monomer units) in COS-1 and Caco-2 cells. The results demonstrated higher transfection, in both serum-free and serum-containing media, for polyplexes prepared with quaternized chitosan than with unmodified chitosan polyplexes. The increase in transfection was especially significant in COS-1 cells. The efficacy was dependent on the weight ratio of the DNA/chitosan-oligomer polyplexes. The optimal weight ratio for transfection of COS-1 cells in serum-free media was 1:14.MTT assays revealed that the quaternized chitosan remained nontoxic, comparable to unmodified chitosan, in both cell lines.
Kean and coworkers investigated the difference in transfection efficiencies of oligomeric (3–6 kDa) and polymeric (~100 kDa) quaternized chitosans, using monkey kidney fibroblasts (COS-7) and epithelial breast cancer cells (MCF-7) [105]. Both oligomeric and polymeric chitosan at optimized degrees of quaternization (44% for oligomeric chitosan, 57% and 93% for polymeric chitosan) transfected MCF-7 cells more efficiently than PEI as measured by luciferase assay. In the case of COS-7 cells, however, only oligomeric chitosan with 44% quaternization showed transfection comparable to, but not exceeding that of, PEI. Importantly, chitosans showing the highest transfection efficiency showed moderate cytotoxicity, with the polymeric chitosan exhibiting higher toxicity than its oligomeric counterpart.
Attempts to improve transfection of chitosan-based vectors by grafting PEI onto a chitosan backbone have been reported. The goal of such efforts is improvement of the buffering capacity and charge density of chitosan while preserving its low inherent cytotoxicity. One of the first attempts to utilize PEI buffering properties in chitosan based gene delivery systems was made by Kim et al. [106]. PEI was physically added to (not chemically grafted on) chitosan/DNA polyplexes; addition of PEI to water-soluble chitosan/DNA polyplexes increased their transfection efficiency in HeLa, A549, and 293 T cells. Furthermore, a synergistic effect between water-soluble chitosan and PEI was demonstrated in the transfection of 293 T cells. This approach was also efficient in the case of a targeted delivery system – galactosylated chitosan. Addition of moderate amounts (1–2 µg) of PEI to galactosylated chitosan/DNA polyplexes increased the transfection efficiency in HepG2 cells while retaining receptor-mediated cellular internalization. However, addition of a greater amount of PEI (5 µg) decreased cell specificity and, in this case, the transfection efficiency of HepG2 cells by galactosylated chitosan/DNA was not different from that of nongalactosylated water-soluble chitosan/DNA polyplexes.
Taking the idea a step further, Wong and coworkers [107] have grafted low molecular weight PEI (Mn = 0.206 kDa) on water-soluble chitosan (Mn = 3.4 kDa) via cationic polymerization of aziridine, with chitosan amino groups functioning either as initiators or terminators of polymerization (Fig. 7b). It should be noted that such an approach can lead to formation of free, non-bound oligoethyleneamines. However, dialysis performed after polymerization likely removes this side product. The effect of PEI grafting was studied in HeLa, HepG2, and primary hepatocytes. Chitosan-g-PEI complexed pDNA at an N/P ratio of 2.5, but the maximum transfection efficiency in serum-free media was achieved at N/P = 40 for all three tested cell lines. At this N/P ratio, transfection efficiency was similar to PEI (25 kDa) at N/P = 10. The cell viability, assessed via the MTT assay, was reported only for free polymers in HeLa cells and revealed, interestingly, that chitosan-g-PEI had a sevenfold higher LD50 than PEI. Chitosan-g-PEI transfection was investigated in vivo by administration of polyplexes into the common bile duct in rats for liver delivery. Delivery with chitosan-g-PEI at N/P = 10 was 141-fold greater than that of naked DNA, 58-fold greater than PEI (25 kDa), and 3-fold greater than unmodified chitosan.
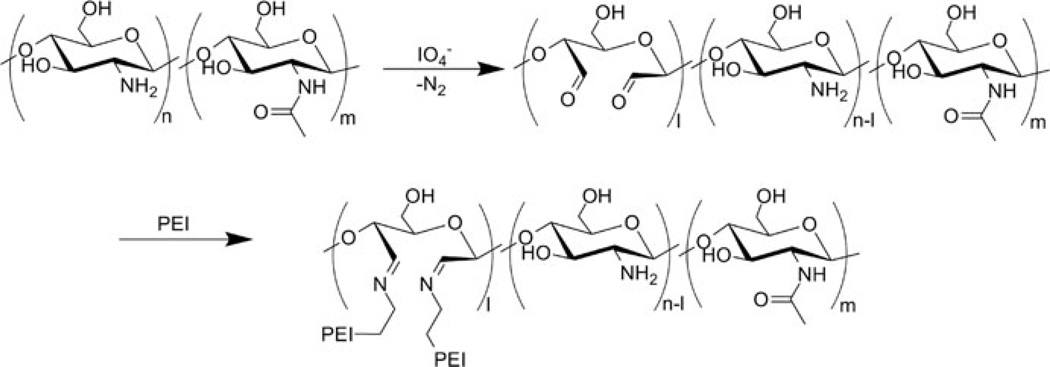
Following their successful tandem use of water soluble chitosan/galactosylated chitosan and PEI for the polyplex formulation, Cho et al. chemically grafted PEI onto chitosan [108, 109]. The grafting was accomplished by partial oxidation of 100 kDa chitosan with periodate followed by reductive amination using 1.8-kDa PEI (Fig. 8). Periodate is widely known to be used in oxidation of vicinal diols, presumably via a five-member intermediate. However, as noted by Vold et al., IO4− oxidative cleavage of 1,2-aminoalcohols is also known [110] and can be successfully used for oxidative cleavage of the C2–C3 bond within the glucosamine unit in chitosan [111]. Thus, it is necessary to exercise careful control of reaction conditions to avoid overoxidation and depolymerization of chitosan.
Fig. 8.
Synthesis of chitosan-graft-PEI. Figure reproduced with permission from [109]. © 2007 Elsevier
Synthesized chitosan-g-PEI was evaluated by transfection in HeLa, HepG2, and 293 T cells. In this case, chitosan-g-PEI was found to complex pDNA stably, with an average polyplex size of 250 nm, and protected pDNA from nuclease degradation. The cytotoxicity of these polymers was much lower than that of PEI (25-kDa) in all three cell lines. Similar to the aforementioned study by Wong et al., the transfection efficiency for this grafted system in serum-free media was higher than unmodified PEI (25-kDa) at N/P = 35 (N/P = 10 was used in the study by Wong et al.). Furthermore, high luciferase expression, similar to that of Lipofectamine, was noted in 293 T cells using chitosan-g-PEI. Interestingly, transgene expression in serum-containing media with chitosan-g-PEI was only slightly decreased in comparison to PEI and Lipofectamine.
To explore further the potential of chitosan-g-PEI vectors, Jiang et al. have synthesized galactosylated chitosan-g-PEI [112] and galactosylated poly(ethylene glycol)-chitosan-g-PEI (Gal-PEG-chitosan-g-PEI) [113], with the latter prepared by linking galactose-terminated PEG carboxylic acid to chitosan-g-PEI via amide bond. Incorporation of PEG is a standard approach to improve the polyplex colloidal stability and prevent undesirable electrostatic interactions with negatively-charged components of plasma and cellular membranes. This approach was successful in this case as well, showing that Gal-PEG-chitosan-g-PEI was less efficient in transfection of HepG2 and HeLa cells than chitosan-g-PEI due to reduced electrostatic interactions. But, more importantly, Gal-PEG-chitosan-g-PEI was better in transfecting HepG2 cells than non-targeted PEG-chitosan-g-PEI, whereas in HeLa cells their transfection efficiency was similar. These results show that cell-specific targeting can be achieved for chitosan-g-PEI vectors.
Recently, new ways to graft PEI on chitosan have emerged. Lu et al. have used a maleic acid anhydride reaction with the amino groups of chitosan, followed by the Michael-type addition of PEI [114]. Lou and coworkers used ethylene glycol diglycidyl ether to link PEI with chitosan through hydroxyl and amino groups of chitosan [115]. Wu et al. have used alkylation of primary hydroxyl groups of chitosan with chloroacetic acid, followed by purification, activation of carboxyl groups with N,N’-dicyclohexylcarbodiimide (DCC) and N-hydroxysuccinimide (NHS), and subsequent coupling with PEI through amide bond formation [116]. It is not clear how self-coupling of chitosan was avoided in this approach, however.
PEI grafting on chitosan is becoming a popular approach for modification of chitosan for gene delivery applications. In recent years several efforts to develop chitosan-g-PEI delivery vectors have been published. These include incorporation of mannose [117] and folic acid [118] derivatives for targeted delivery and application of chitosan-g-PEI for the delivery of siRNA [119].
A means by which the buffering capacity of chitosan delivery vehicles can be improved without significantly increasing their cationic character is to graft the chitosan backbone with imidazole. Imidazole contains a protonatable nitrogen having a pKa of 6.15; thus, imidazole may facilitate endosomal rupture through the proton-sponge mechanism. For this reason, imidazole has been used widely in nucleic acid delivery vectors, and these materials have been discussed elsewhere [120–122].
Kim et al. decorated the chitosan backbone with imidazole groups by coupling of urocanic acid to chitosan through EDC/NHS condensation (Fig. 9) [123]. Urocanic acid-modified water-soluble chitosan (50 kDa) was evaluated for transfection efficiency using 293 T, HeLa, and MCF-7 cells. The modified chitosans were found to bind pDNA and also protected from DNase degradation at charge (N/P) ratios between 5 and 30. The transfection performed in 293 T cells yielded greater transgene expression for urocanic acid-modified chitosan than the unmodified analog, and the efficacy also tended to increase with greater extents of grafting, demonstrating the role of imidazole groups for transfection of 293 T cells. However, no significant enhancement in gene expression was evident when the same experiment was performed in HeLa and MCF-7 cells, thereby indicating that the transfection is highly cell-type dependent. This delivery vehicle was later also used for in vivo studies for aerosol delivery of nucleic acids to the lung, which led to tumor suppression in this model [124, 125].
Fig. 9.
Structure of urocanic acid-modified chitosan
Thiols are typically incorporated into polymers for gene delivery to take advantage of the reducing environment of the cytoplasm. Thiols can be mildly oxidized to produce disulfide (S–S) bonds for delivery vehicle crosslinking, providing protection for the cargo from degradative enzymes and preventing premature nucleic acid release. In the cytosol, the S–S bond can be reduced (primarily due to high concentrations of glutathione), causing changes in polyplex organization that result in release of carried nucleic acid. More information on disulfide usage in polymeric gene delivery systems, disulfide bond synthesis, and mechanism of action can be found in a recent review by Bauhuber et al. [126].
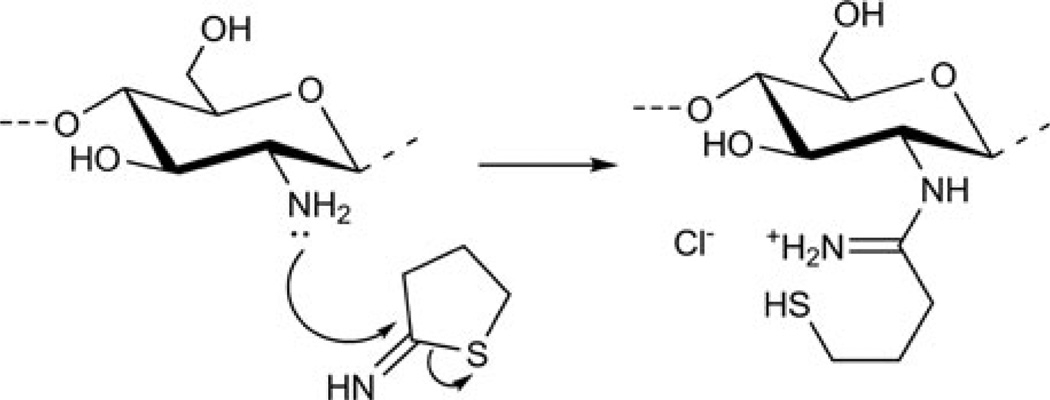
Thiolated chitosans [127] (Fig. 10) have been used for the development of an oral gene delivery vehicle [128]. The synthesis was accomplished in aqueous media using 2-iminothiolane and low-molecular-weight chitosan. Thiolated chitosan complexed pDNA into 125-nm polyplexes with a positive ζ-potential and was able to protect pDNA in artificial intestinal fluids at multiple physiologically-relevant salt concentrations. These complexes were stable at pH = 1.2. Under reducing conditions, thiolated chitosan releases 50% of its pDNA cargo in ~3.5 h, whereas in non-reducing conditions only ~7% of pDNA is released at this time point. Moderate transfection of Caco-2 cells was observed with thiolated chitosan, but it was higher than both controls – naked pDNA and (unmodified) chitosan/pDNA. Based on the stability to artificial intestinal fluids and low pH, as well as low cytotoxicity, it was concluded that thiolated chitosans are candidates to be studied further as oral gene delivery vectors.
Fig. 10.
Synthesis of thiolated chitosan. Figure reproduced with permission from [127]. © 2004 Elsevier
In another study from the same lab [129], thiolated chitosan (12 kDa) was synthesized by conjugation of chitosan with thioglycolic acid. The thiolated chitosan/DNA nanoparticles were more resistant to DNase degradation at pH 4.0 and 5.0 than naked DNA and complexes formed with unmodified chitosan. MTT assays performed at pH 5.0 in Caco-2 cells indicated that the nanoparticles formed using unmodified chitosan and non-crosslinked thiolated chitosan were nontoxic. Cross-linked chitosan at pH 5.0 and both, non-crosslinked and crosslinked thiolated chitosans at pH 4.0, were slightly toxic, with cell viability 80–90% of untreated cells. The transfection experiments performed in Caco-2 cells revealed that both non-crosslinked and crosslinked thiolated chitosan yielded higher transgene expression levels than their unmodified analog in this cell model. In addition, thiolated chitosan/DNA nanoparticles formulated at pH 4.0 exhibited fivefold higher efficiency than unmodified chitosan.
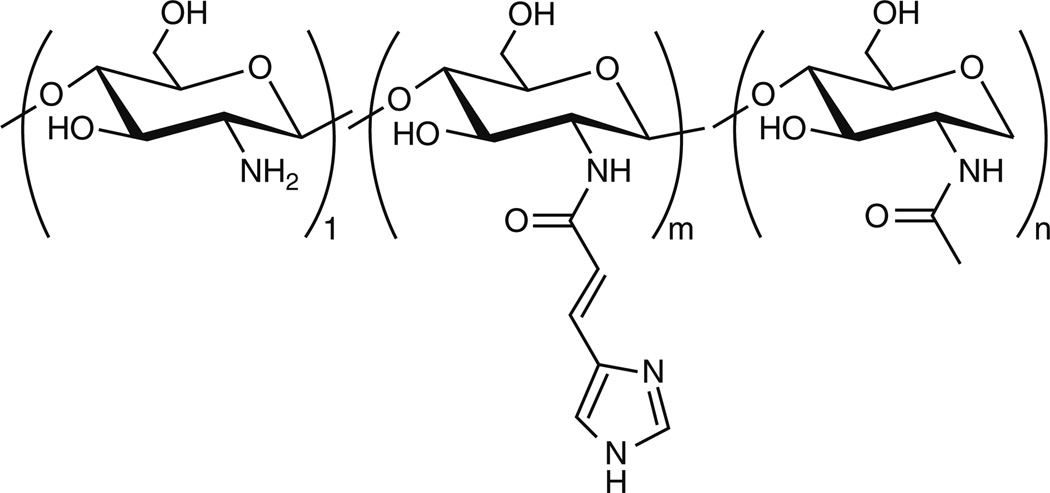
Chitosan has also been grafted with saccharide-based ligands as molecular recognition elements to promote receptor-mediated endocytosis for cell specific delivery in vivo. Extensive research has been completed by various groups in developing chitosan-based delivery vectors that have been conjugated to saccharides for target-specific delivery. The remainder of this section focuses on targeting cell surface lectins with carbohydrates grafted onto a chitosan backbone.
Nearly a decade ago, Murata et al. [102] synthesized quaternary chitosan polymers grafted with galactosyl residues along the side chain (Fig. 11a) and utilized them to transfect HepG2 cells (which express the asialoglycoprotein receptor (ASGPr), for which galactose is a ligand) in serum-free media. The β-galactosidase expression observed in HepG2 cells revealed that the gene expression tended to be higher for galactosylated trimethylchitosan than the non-galactosylated polymer. It was also found that increased amounts of galactose substitution yielded higher transfection efficiency, suggesting a multivalent effect for efficient uptake. Furthermore, the cytotoxic effects of these galactosylated chitosans were similar to those of DEAE-dextran.
Fig. 11.
(a) Structure of galactosylated chitosan. Figure reproduced with permission from [102]. © 1996 Elsevier. (b) Structure of lactose conjugated chitosan. Figure reproduced with permission from [134]. © 2006 American Chemical Society
In a similar study, Park and coworkers [131] synthesized chitosan-based vectors in which the chitosans were conjugated to lactobionic acid, which contains galactose residues. These structures were then grafted with either dextrans (Mn = 5.9 kDa) (GCD), polyethylene glycol (Mn = 5.0 kDa) (GCP), or poly-(vinylpyrrolidone) (GCPVP) to enhance polyplex stability (prevent aggregation). In the first study, galactosyl chitosan (Mn = 4.0 kDa) was conjugated with dextran and examined for delivery efficacy in Chang liver cells (express ASGPr) and HeLa cells (non-ASGPr-expressing). As expected, the dextran conjugation was found to stabilize the polyplexes from aggregation and yielded higher gene expression in Chang liver cells than in HeLa cells. In a related study by the same group, similar results were obtained when galactosyl chitosan (Mn = 7.0 kDa) was conjugated with PEG [132]. Polyplexes formed with this polymer were used to transfect in HeLa, CT-26, and HepG2 cells, and the results were compared to those from the previous study. It was found that these PEGylated analogs had comparable stability to the dextranmediated vectors and were found to protect pDNA from nuclease degradation. The transfection experiments indicated the vehicle had negligible cytotoxicity and that GCP/pDNA yielded transgene (green fluorescent protein) expression in HepG2 cells, but not in HeLa and CT-26 cells, thereby suggesting that the transfection occurs through ASGPr. In a more recent study by the same group, galactosyl chitosan (10 kDa and 50 kDa) were grafted with poly(vinylpyrrolidone) (PVP) using radical polymerization and similarly studied as a hepatocyte-targeting vehicle [130, 133]. PVP has been found to have similar properties to PEG; however, the PVP-modified polyplexes were found to have longer retention time in the blood than the PEG-modified systems.
All of the aforementioned studies demonstrate that chitosan grafted with polycations can be effectively used for hepatocyte-specific delivery applications. In addition, incorporation of flexible hydrophilic groups in the polymer structure provides a steric barrier that prevents aggregation and reduces interactions of the complexes with plasma proteins and phagocytes, thereby increasing the circulation time of these complexes in the plasma and facilitating complexes reaching target cells. However, the syntheses of these grafted systems are slightly more tedious and difficult to manufacture on an industrial scale; in addition, they often result in a highly polydisperse polymer mixture with high batch to batch differences in conjugation efficiency.
Similar results to galactosyl chitosan were seen when lactose-conjugated chitosan (53-kDa) conjugates (lac-chitosan) were synthesized (Fig. 11b) by Hashimoto et al. [134] and used to transfect HepG2 cells. In this study, conjugates were prepared for which the amines along the chitosan backbone were either 8% or 33% functionalized with lactose. Both were found to bind and compact pDNA into ~140-nm polyplexes at N/P ≥ 3 (ζ-potential = +43 mV). Unlike polyplexes prepared with nonlactosylated chitosan, these lac-chitosan-containing polyplexes were stable from aggregation and adsorption after 1 h incubation with bovine serum albumin (polyplexes remained ~150 nm in diameter), thereby indicating that the lactose modification on chitosan results in serum stability. The 8% lac-chitosan/pDNA complexes revealed transfection efficiency in COS-7 cells similar to those made with the unmodified analog, whereas polyplexes formulated with the 33% lacchitosan/DNA had about a twofold lower transfection efficiency than unmodified chitosan/DNA complex. However, in HepG2 cells, a 16-fold enhancement in transgene (luciferase) expression was observed when 8% lac-chitosan/DNA was used for transfection, suggesting receptor-mediated delivery leads to higher gene expression. In both cell lines, Lipofectamine showed much higher gene expression when compared to the conjugated and non-conjugated chitosans.
Inspired by earlier work on galactose-conjugated chitosans, Hashimoto and coworkers [135] synthesized mannose-grafted chitosan (53-kDa) conjugates (man-chitosan) to deliver pDNA in mouse peritoneal macrophages that express the mannose receptor. Here, man-chitosan containing either 5% or 21% modification were synthesized, and mixing with pDNA resulted in formation of ~300-nm polyplexes. Both complexes formed with the man-chitosan derivatives were found to exhibit increased transfection in macrophages compared to pDNA/chitosan polyplexes and yielded comparable transfection to man-PEI/DNA polyplexes in macrophages. When a control experiment was performed in COS-7 cells, the transgene expression of pDNA/5% man-chitosan polyplexes was the same as that of pDNA/chitosan; however, the transfection efficiency of pDNA/chitosan was four times higher than pDNA/21% man-chitosan. The cell viability in experiments in macrophages also revealed negligible cytotoxicity of the man-chitosan polyplexes, which contrasted with the toxicity observed for man-PEI/pDNA complexes. Even though all the above experiments have been promising and have shown effective transfection and target-specificity, particularlywith hepatocytes,most of these in vitro experiments have been performed only in serum-free media. The future of this area depends on performing these experiments in media containing serum, which are a better simulation of in vivo conditions. Also, in vivo data in this field are minimal and more are needed to advance this area toward the clinic. These extensive studies using chitosan have shown that this polysaccharide is indeed very useful for delivering therapeutic DNA into cells and the structure affords nearly limitless potential for chemical modification. However, their transfection efficiencies being lower than other non-viral analogs and viral-vehicles needs to be overcome by chemical and structural modifications. Much further work on this delivery platform is ongoing.
3 Carbohydrate Copolymers
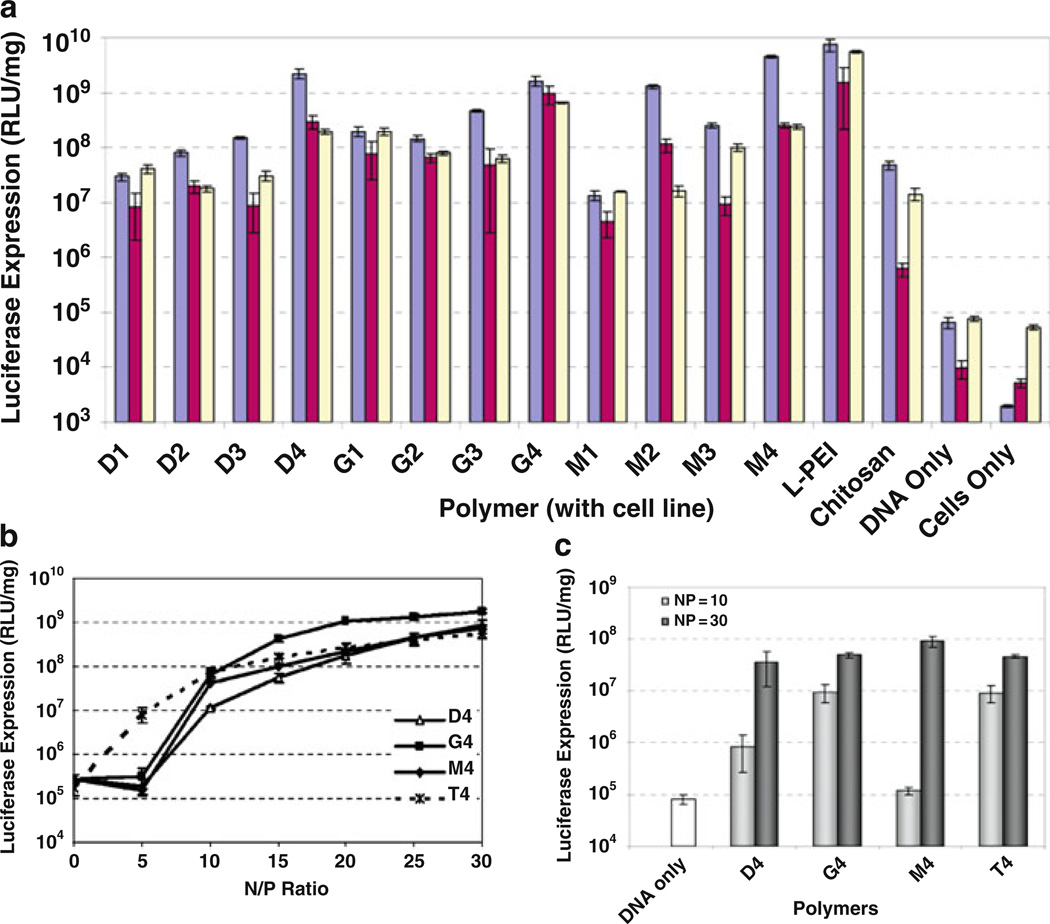
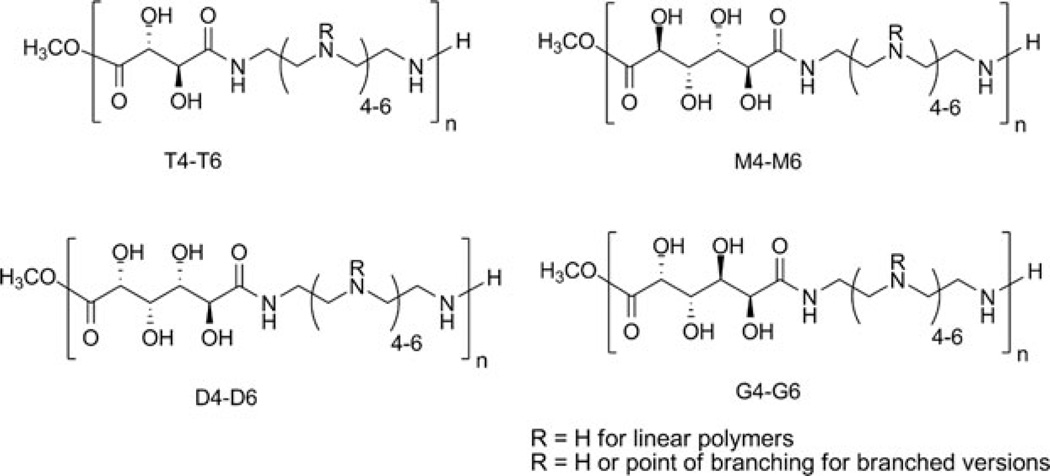
Saccharide copolymers are recently emerging biomaterials with high applicability as nucleic acid delivery vehicles. To date, the structures created can generally be categorized as AABB step-growth type polymers consisting of two different monomers, where one monomer facilitates nucleic acid binding and the other (carbohydrate) monomer imparts biocompatibility. Previous results have shown that saccharide groups contribute to reduction of the cytotoxicity of non-viral vehicles. For example, when relatively toxic polymers, such as PEI, are grafted with carbohydrates, the cytotoxicity is generally decreased (e.g., [136]). For this reason, carbohydrate moieties have been incorporated in the polymer backbone using a variety of synthetic organic reactions, such as polycondensation, cycloaddition, or ring-opening polymerization. The structure of monomers used and synthetic methodologies have been found to influence various parameters, such as solubility, degree of polymerization, branching, and tacticity of the polymers. Furthermore, as with previously-presented systems, the studies in this section also demonstrate that subtle changes in the chemical and structural characteristics of these polycations have a significant effect on the cellular uptake, gene expression, and cell viability. This section describes novel polymers that have been synthesized with a variety of monosaccharides [21, 137], disaccharides [138–140], or cyclic oligosaccharides [141], and their efficacy as non-viral nucleic acid delivery vehicles.
3.1 Monosaccharide-Based Copolymers
The introduction of carbohydrates in polymeric structures could temper the cytotoxicity observed with these vehicles. Two commonly studied polymeric vectors – poly-l-lysine (PLL), a polypeptide consisting of repeating lysine residues, and polyethylenimine (PEI), composed of repeating ethylenediamine units – have shown the ability to deliver DNA for gene expression at a high level in vitro and in vivo [10, 13, 142]. Significant cytotoxicity, likely due to their high charge density and possible membrane-disrupting effects, limits the potential clinical utility of these vehicles. Reduction of the charge density by incorporation of carbohydrates could yield transfection efficiencies greater than those of polysaccharide-based vehicles and afford increased biocompatibility, thus resulting in an improved delivery system. In the first study using this strategy, published by Reineke and coworkers in 2004, dimethyl glucarate was polymerized with diethylenetriamine, triethylenetetramine, tetraethylenepentamine, and pentaethylenehexamine to derive polymers containing 1-4 secondary amines in the polymer repeat unit [137]. These polymers were able to self-assemble with DNA into polyplexes. When transfected into BHK-21 cells, pDNA complexes containing these polymers showed high levels of transgene expression with considerably lower toxicity than PEI. In fact, the analog with four secondary amines, dubbed D4, showed transgene expression comparable to PEI. A similar study was published afterwards by Guan et al. in 2005, using a similar strategy; however, the incorporated charge centers were l-lysines [145]. An acid chloride derivative of galactose was polymerized with oligolysines to develop three polymers with varied amounts of primary amines and spacing between the primary amine and the polymer backbone. These polymers also showed enhanced biocompatibility vs PLL and gene expression was comparable to PLL at low molar concentrations. These polymers did not elicit an immune response when administered to rats via intravenous or subcutaneous injection [145]. These first two studies showed that interrupting the charge density of PEI and PLL is advantageous for achieving low toxicity in vitro and in vivo without sacrificing delivery efficacy.
These favorable results led Reineke and colleagues to expand their initial study using three different carbohydrates to study the effects that hydroxyl number and stereochemistry, as well as amine stoichiometry, have on biological activity. A library of 16 polycations was developed, containing alternating monosaccharides and amine units along the main polymeric backbone, and were termed poly(glycoamidoamine) s (PGAAs) (Fig. 12) [21, 137]. These polymers contain one of four different carbohydrate comonomers: a mixture of dimethyl-d-glucarate, methyl-d-glucarate 1,4-lactone, and methyl-d-glucarate 6,3-lactone (D), dimethyl-mesogalactarate (G), d-mannaro-1,4:6,3-dilactone (M), or dimethyl-l-tartrate (T). These comonomers were polymerized with the same series of oligoamine comonomers as the first study (diethylenetriamine (1), triethylenetetramine (2), tetraethylenepentamine (3), or pentaethylenehexamine (4)) via step-growth polymerization to generate a series of polymers (D1–D4, G1–G4, M1–M4, T1–T4) with degrees of polymerization (n) around 11–14. These initial structures were found to bind and compact pDNA into cationic polyplexes, and increasing the amine number generally led to more efficient DNA binding and smaller polyplex size.
Fig. 12.
Generalized block diagram of PGAA design structure and structures of the 16-polymer library of PGAAs. These polymers allow the direct comparison of changes in amine stoichiometry, as well as hydroxyl number and stereochemistry, on biological properties. Figure adapted with permission from [186]. © 2006 John Wiley & Sons, Inc
This library of cationic glycopolymers was screened for cell viability and transgene expression, and the results were compared to chitosan and PEI to assess the improvements achieved over these vehicles (Fig. 13). To ensure widespread utility, these vehicles were tested in four different cell lines to mimic a wide range of mammalian cell types: BHK-21, HeLa, HepG2, and H9c2(2-1) [21, 144, 146]. High levels of transgene expression was observed in all cell lines; the polymers containing four secondary amines yielded the highest transfection levels (Fig. 13). At the N/P ratio of maximum expression, the luciferase expression efficiency of these vehicles was comparable (within an order of magnitude) to that of PEI and significantly enhanced over chitosan. These results also showed very low toxicity that was comparable to chitosan and significantly lower than PEI. The highest transgene efficiency was observed for G4 and T4, which bind DNA the strongest among the series, and were shown to protect pDNA from degradation by nucleases. Further study in cardiomyoblast (H9c2(2-1)) cells revealed that, despite a significant polyplex size increase in salt and serum, high levels of gene expression were also observed, which were related to high levels of polyplex cellular internalization. Using polyplexes containing FITC-labeled pDNA, the cellular internalization of PGAA polyplexes was assessed in serum-free and serum-containing media. The data show high levels of uptake, with nearly every cell analyzed containing DNA-associated fluorescence under both conditions. These data illustrate the high efficiency by which PGAA polyplexes can enter cells.
Fig. 13.
Transgene expression efficiency of PGAA polymers in multiple mammalian cell types. (a) G, D, and M polyplexes shown high levels of transfection in multiple cell types. PGAAs with four secondary amines/repeat unit in H9c2 cells in (b) serum-free and (c) serum-containing media. Figures adapted with permissions from [21] and [146]. © 2005 and 2006 American Chemical Society
These promising results have led to systematic study on the effects of polymer structure on DNA binding and bioactivity. These further studies probe many aspects of polymer structure to determine the structural elements that lead to efficient delivery. DNA binding of PGAAs occurs through a combination of electrostatics and hydrogen bonding, a direct result of carbohydrate incorporation into the polymer [147]. The close-range hydrogen bonding interactions likely afford greater polyplex stability such that the DNA can remain packaged while inside the cell. Using similar techniques as described above, Lee et al. investigated the effect of increasing the number of secondary amines in the polymer backbone from four to five and six (Fig. 14), as well as the effect of polymer branching, on delivery efficacy and toxicity [148]. The linear and branched polymers yielded polyplexes of generally larger size than those with four secondary amines, but the galactarate and tartrate series with five and six secondary amines did not significantly swell or aggregate in serum-containing media, suggesting the higher secondary amine numbers prevent increasing size in serum. However, these modifications did not result in much enhancement of cellular uptake, as polyplex internalization was mostly unaffected (in some cases it was lower or a little higher and this depended on the cell type). Similar results were observed for transgene expression, as transfection. However, these modifications did have a significant effect on cell viability, as the linear polymers with five and six secondary amines displayed increased toxicity over the series with four secondary amines in the polymer repeat unit. This study confirmed that four secondary amines in the polymer repeat unit yields the highest internalization and transgene expression, and these analogs are the focus of further structure-bioactivity studies.
Fig. 14.
Structures of linear PGAAs
Polymer buffering capacity can have a significant effect on the cellular uptake and gene expression in mammalian cells, as polymer charge likely plays a key role in cell surface binding and either escape or trafficking out of the endosomal/lysosomal path. Liu et al. used titration experiments to calculate the buffering capacity of the d-glucaroamidoamine (D1–D4) and l-tartaroamidoamine (T1–T4), which allows a direct measure of the percentage of amines that can be protonated during endosome acidification [149]. The results of these experiments allow a direct comparison of polymer buffering capacity with cellular internalization and transgene expression. Two new polymers were created by polymerizing the dimethyl-d-glucarate (this is a mixture with lactone derivatives) or dimethyl-l-tartrate with spermine (yielding DS and TS, respectively) to incorporate butylene groups between neighboring secondary amines, thereby increasing the amine spacing. Interestingly, buffering capacity decreased with increasing amine number; however, the delivery efficacy and gene expression increased with increase in amine number. This argues against the proton sponge mechanism of endosomal escape, as this hypothesis states that a higher buffering capacity should promote greater endosomal escape and should lead to increased gene expression. However, when comparing differences in amine spacing, the polymers containing the spermine groups (DS and TS) yielded a substantial decrease in buffering capacity compared to the original PGAAs with ethylene spacing. This suggests that proximal amines (ethylene spacer) have a lower charged state at pH 7.5 due to electrostatic suppression of protonation from neighboring charged amines. However, when comparing the gene expression results, DS and TS had higher gene expression than their PGAA analogs with two ethylene amines (D2 and T2) but the analogs with four amines (D4 and T4) still remained the most efficient delivery systems. Polymers with increased amine spacers also exhibited much higher toxicity, suggesting the charge spacing and charge density plays a significant role in biocompatibility. Buffering capacity also appears to influence the transfection of polymers having the same amine stoichiometry but different carbohydrates, as D4 possesses lower buffering capacity than T4, resulting in higher transgene expression with T4. Increasing cellular uptake was observed with higher amine number, which suggests higher amine density promotes multivalent interactions with the cell surface proteoglycans, facilitating higher uptake. Indeed, cellular uptake was found to be a major contributing factor to efficient transgene expression. This study proves that ethylene spacers between amines lead to more biocompatible gene expression and reveals a complex role of polymer buffering capacity that does not directly correlate with high delivery efficiency and gene expression.
Recent results in the Reineke lab reveal that the PGAAs are biodegradable under physiological conditions, and that removal of the carbohydrate or the oligoethyleneamine groups leads to a polymer that does not degrade under physiological conditions [150]. This points toward a synergistic effect in the presence of both the carbohydrates and ethyleneamine groups in polymer degradation. However, the transgene expression efficiency suffers when non-degradable polymer analogs are used as the delivery vehicle, suggesting that polymer degradation facilitates pDNA release and availability for transcription. This feature can possibly be exploited to develop novel nanodevices for sustained release of nucleic acids. Using dip-coating, the biodegradable polymer T4 and pDNA were deposited in a layer-by-layer assembly on a quartz slide (Fig. 15) [151]. Layer thickness of these devices could be assessed by ellipsometry and monitored by measurement of DNA absorbance. These materials resulted in slow, sustained release of pDNA over time which could be easily delivered into cells for gene expression by another suitable vehicle added separately. The amount of pDNA that was internalized by cells increased with time, consistent with slow release of pDNA. Interestingly, despite more released DNA at the earlier time points, gene expression remained constant, which is likely a function of the pathway of internalization/trafficking of DNA such that only a portion of the internalized DNA reaches the nucleus. This type of study demonstrates the wide utility of these polymers for sustained release. The favorable bioactivity can be further developed for use as a therapeutic delivery vehicle or in novel scaffolds and nanodevices in biomedical applications.
Fig. 15.
Internalization and gene expression of pDNA released from a multilayer assembly. Release of pDNA occurs upon degradation of T4 polyamide. Notable increase in fluorescence intensity over time is observed in the flow histograms. Gene expression (measured by intracellular GFP fluorescence) does not increase at the same rate as DNA uptake. Figure adapted with permission from [151]. © 2009 Elsevier
3.2 Disaccharide-Containing Polymers
Disaccharides can have similar utility to monosaccharides in DNA delivery polymers. Trehalose, a disaccharide composed of two glucose units linked via an α-(1→1) glycosidic bond, has been shown to have cryo- and lyo-protective properties, attributed to an unusually large hydration volume [152]. As a function of these properties, trehalose has been shown to prevent aggregation and fusion of proteins and lipids [153]. Logically, incorporation of these features into a polymer backbone could afford similar characteristics to a DNA delivery system and may prevent aggregation of polyplexes in physiological serum concentrations and ionic strengths. In a pioneering study, Reineke and Davis [138] synthesized a series of polymers via condensation polymerization of amine-functionalized D-trehalose monomers with amidine-based comonomers (AP2 and AP3) (Fig. 16). Based on previous studies [141], six methylene units were used as spacers between the amidine units; however, the distance between the amidine groups and the carbohydrate units was modified to understand further the polymer structure-bioactivity relationships. These polymers self-assemble with pDNA into cationic nanostructures around 80 nm in diameter. The delivery efficiency, in terms of transgene expression and toxicity, in BHK-21 cells in vitro was determined under serum-free conditions to preliminarily assess the bioactivity of these polymers, compared to an analogous structure (AP1) in which the carbohydrate group was replaced with a butylene spacer. Trehalose-based polymers (AP2 and AP3) exhibited improved biocompatibility compared to the non-carbohydrate analog (AP1), which exhibited significant toxicity at low charge (N/P) ratios (including only 20% cell survival at N/P = 5). Toxicity appears to be a function of spacer distance between the amidine and the carbohydrate moieties, with AP2 maintaining ~40% lower toxicity than AP3 at charge ratios greater than 10. The transfection experiments in BHK-21 cells indicated that AP3 yielded an order of magnitude higher luciferase expression at N/P = 5 when compared to AP2. At the same charge ratio, the trehalose containing polymers AP2 and AP3 have two and three orders of magnitude higher luciferase expression, respectively, than AP1. These favorable results confirm the benefit of incorporating trehalose into a polymer structure for gene delivery.
Fig. 16.
Structures of trehalose-containing copolymers. Figure adapted with permission from [138]. © 2003 American Chemical Society
Based on these initial results, new trehalose-based vehicles were developed and their efficacy assessed under physiological conditions, as these previous studies [138] have been performed under serum-free conditions. Previous work has shown that polyplexes can aggregate in vivo and are rapidly cleared from the bloodstream, such that extended circulation times are not achieved [8, 154]. Successful results previously described by Liu et al. [21, 146] and Reineke and Davis [138] prompted Srinivasachari et al. to synthesize a series of trehalose-containing polymers by systematically increasing the amine number (1–4) in the polymer structure, as well as to develop longer polymers to determine whether polymer length plays a role in formation of stable complexes with pDNA [140].
A series of trehalose-based polymers (Tr1–Tr3) with 1,2,3-triazole linkages were synthesized via the “click reaction” of acetylated-diazido trehalose and a series of dialkyne-oligoethyleneamines (1–3) (Fig. 17). Gel electrophoresis revealed that the polymers bind pDNA stably at N/P = 2, with TEM revealing the polyplexes to have either spherical or rod-like morphologies with diameters around 50–125 nm. Dynamic light scattering measurements demonstrated an increase in polyplex size upon incubation in serum-free media (Opti-MEM), suggesting swelling or aggregation of the particles. This effect appeared suppressed in serum-containing media; polyplex resistance to size increases improved with increasing amine stoichiometry. Cellular internalization, transgene expression efficiency, and cell viability were assessed in vitro in HeLa cells under both serum-free and serum-containing experimental conditions [140, 155]. When the HeLa cells were transfected with complexes containing FITC-labeled pDNA in Opti-MEM (N/P = 7), the cellular uptake profile showed that Tr1 and Tr3 were the most effective vehicles, transfecting a higher percentage of cells (>99%) than Tr2 or the positive control, jetPEI. However, in serum-containing media – Dulbecco’s Modified Eagle Medium (DMEM), 99% of cells internalized complexes containing Tr3, more than for Tr1 (76%) or Tr2 (35%) and similar to jetPEI. This could be attributed to the increased stability of Tr3-containing polyplexes from aggregation in the presence of serum proteins. The transfection efficiency results in serum-free and serum-containing media indicated increased bioactivity of Tr3, which yielded higher transgene (luciferase) levels than Tr1 and Tr3. In serum-free media, luciferase expression from Tr2 was comparable to Tr3, whereas in serum-containing media, Tr3 induced one and two orders of magnitude higher gene expression than Tr1 and Tr2, respectively; these results correlated with the enhanced stability of Tr3 in DMEM. The trehalose-based polymers exhibit low (<20%) toxicity in serumcontaining media at N/P = 7, markedly lower than control polymer jetPEI. These results suggest that incorporation of trehalose imparts favorable biological properties. In line with previous polymers, favorable biological activity improves with increasing amine stoichiometry across the polymer series, reaffirming that subtle changes in the polymer structures can measurably influence bioactivity.
Fig. 17.
Structures of trehalose click polymers
To test these theories directly, trehalose-based polymers with four oligoethyleneamine units in the polymer chain were developed. The degree of polymerization of these analogs was varied to assess the influence of polymer length on biological properties [155]. Binding of pDNA and polyplex stability improved as a function of increasing degrees of polymerization in both Opti-MEM and DMEM. The polymers with greater amine stoichiometry have been shown to promote tight DNA binding and favorable polyplex stability due to a combination of electrostatic and hydrogen bonding interactions with the pDNA, with hydrogen bonding likely occurring between secondary amines and/or triazole nitrogens and the guanine/thymine nucleobases (Fig. 18) [156]. In contrast to Tr4, Tr1 did not show evidence of interaction with DNA structure elements, and the interaction appears to be more electrostatic in nature with lower amine stoichiometry. Due to higher amount of secondary amine and triazole groups in the longer Tr4 polymers, binding cooperativity plays a large role in increasing pDNA binding and stability with the increase in degree of polymerization. Indeed, the increase in both the electrostatic and the hydrogen bonding potential of the polymer increases cooperativity.
Fig. 18.
Circular dichroism spectra comparing (a) Tr1 and (b) Tr4. Titration of pDNA with Tr1 results in minimal change in molar ellipticity representative of B-form DNA. Tr4 elicits a shift in ellipticity to a modified B-form, suggesting interaction with DNA base pairs by the polymer. Figure adapted with permission from [156]. © 2008 American Chemical Society
Cytotoxicity was shown to increase with increasing polymer length under serumfree conditions, but the polyplexes were nontoxic in serum in HeLa and H9c2(2-1) cells (Fig. 19). Cellular internalization under both conditions increased with increasing polymer degree of polymerization, as did transgene expression in serum-free media. However, increasing degree of polymerization did not influence transgene expression in serum, as all analogs had similar transgene expression in serum at N/P = 7. Furthermore, all Tr4 analogs were internalized to a higher degree and were less toxic than jetPEI in both serum-free and serum-containing media and, in HeLa cells, higher levels of transgene expression was observed for Tr4. This effect was cell-type dependent, as transgene expression was slightly lower in cardiomyocytes compared to PEI [155]. These results suggest that the stability of these polymers from aggregation in serum-containing media is an important property and may be advantageous in developing these materials towards various in vivo applications.
Fig. 19.
Cellular internalization, transgene expression, and relative cell viability of Tr4. Cellular uptake in (a) serum-free and (b) serum-containing media. Transgene expression and cell viability in (c) HeLa and (d) H9c2(2-1) cells. Figure adapted with permission from [155]. © 2007 American Chemical Society
3.3 Polycationic Cyclodextrin Polymers
Cyclodextrins (CDs) have been popular in developing supramolecular polycations for DNA delivery. Of particular utility is the ability to modify selectively the primary and secondary hydroxyl groups, which has enabled synthesis of a wide variety of biomaterials [157–161]. In contrast to other carbohydrates, CDs contain a hydrophobic interior cavity which has been shown to form inclusion complexes with hydrophobic molecules. CDs have been extensively used in a variety of biomedical applications, and numerous drug molecules have been included in the interior of CDs and utilized towards target-specific delivery [162–167]. Such applications have used CDs as an adjuvant to complex and increase the bioavailability of hydrophobic drugs, an approach used particularly in delivery of inhaled drugs [168]. This hydrophobic “cup” opens up the potential to conjugate functional groups to a hydrophobic molecule for serum stability or cell-specific targeting, such that the hydrophobic molecule is bound within the CD core, and conjugates a pendant molecule for delivery enhancement. This section will describe some notable DNA delivery scaffolds based on CDs, as well as describe recent work with targeted β-CD polymers in clinical and pre-clinical studies for treatment against cancer.
Star polymeric scaffolds based on α-CD have been developed by Yang et al. They were prepared by conjugation of mono-, penta-, nano-, and tetradeca-ethyleneamine units to hydroxyl groups on carbon 6 of glucose moieties using 1,1’-carbonyldiimidazole, such that pendant amine-containing arms stretch from the CD [169]. Since difunctional oligoamines were used, large excess was needed to minimize intra- and intermolecular crosslinking, the purification required precipitation and size exclusion chromatography. The number of oligoamine arms per CD was calculated using 1H-NMR and it varied from 3.4 to 6.8. Unfortunately no attempt was made to explain how the number of oligoamine arms could exceed six. Mw, Mn and polydispersity index of synthesized macromolecules was not reported. These star polymers have been found to compact pDNA stably into spherical cationic nanoparticles ranging between 100 and 200 nm at N/P ≥ 8. These polymers show significant, dose-dependent toxicity in HEK293 and COS-7 cells; this toxicity and resulting transgene expression both increased with increasing oligoethyleneamine content. The transgene expression was comparable in both serum-free and serum-containing media, suggesting that serum does not interfere with polyplex structure. Transgene expression profiles were cell type-dependent, with star polymers exhibiting transgene expression similar to PEI in HEK293 cells but significantly less in COS-7 cells [169]. While the transgene expression profiles in serum are encouraging, high toxicity will likely limit the future utility of these materials.
Cryan and coworkers [170] synthesized a series of polycations by modifying the 6-position of each glucose moiety within the CD structure with a variety of functional groups, such as pyridylamino (AP), alkylimidazole (IM), methoxyethylamino (ME), or primary amine (AM). Ethidium bromide (EtiBr) exclusion experiments indicated that the pDNA binding affinity of these polycationic CDs was wholly dependent on the substituents present in the branching arms. These studies further showed that AP- and AM-modified CDs had higher pDNA binding than ME-CD. These three analogs displayed significantly higher EtiBr exclusion than the IM-CD, a result which could be attributed to increasing hydrophobicity of the IM group. Overall, the N/P ratio required for pDNA binding was quite high (optimal N/P for transfections was 200), necessitating a high concentration of polymer for efficient pDNA compaction. Transgene expression experiments in serum-free conditions revealed high levels of transgene (luciferase) expression compared to uncomplexed pDNA, with AM-substituted CD leading to the highest expression. Addition of chloroquine, used to disrupt endosomal membranes, yielded 10- to 400-fold enhancement in luciferase expression for modified CDs compared to the untreated polymers, suggesting sequestration in endosomal compartments inhibits transfection. In serum, AM-CD showed comparable transgene expression to the cationic lipid, DOTAP. The modified CDs were cytotoxic in a concentration-dependent manner, and were quite toxic at high N/P (the cell viability was around 70% at N/P 200), suggesting limited potential for development as gene delivery agents.
Gonzalez and coworkers synthesized a series of polyamidine-CD polymers via AABB-type condensation of diamino-CD or di(2-aminoethanethio)-CD monomers with difunctionalized-amidine comonomers (six methylene groups between the amidine units). These materials were used to study the effect of spacer length between the charge center and the carbohydrate moiety on pDNA binding and gene expression in BHK-21 and CHO-K1 cells under both serum-free and serumrich conditions [171]. The polymers had a degree of polymerization around 6 that was determined using gel permeation chromatography. Binding studies revealed that a longer spacer between the CDs and the charge center was required for pDNA condensation, likely due to steric constraints with shorter linkers. Using the longer spacers, stable nanoparticles of 150–180 nm at N/P = 10 were attained. Transfection experiments resulted in similar transgene expression as branched PEI (25 kDa) and slightly higher expression than PLL, SuperFect, and Lipofectamine in both cell lines under serum-free conditions. When similar experiments were performed in serum-containing media, a 10% decrease in gene expression was observed for all the polyplexes. The cell viability assay demonstrated that the CD-containing polymers were less toxic than commercial vectors under both conditions.
In a similar study, Hwang et al. [141] studied the effect of spacing between the amidine groups on pDNA binding ability and gene expression by systematic modifications to the number of methylene groups (from 4 to 10) in between the amidine units (Fig. 20). The polymers (β-CDPs) contained four to five repeat units and compacted pDNA into nanoparticles of 12–150 nm in diameter. This study also showed that polymers with four, five, and ten methylene units formed slightly larger particles than the other β-CDPs. The DNase protection ability of these polymers revealed that polymers containing four, six, eight, and ten methylene units protected pDNA against nuclease degradation, while analogs possessing five and seven methylene units offered only partial protection. These results could possibly be due to an effect of odd vs. even methylene spacing on pDNA binding and/or stability. Transfection experiments in BHK-21 cells showed β-CDP6 (6 methylene spacing) yielded higher transgene (luciferase) expression than the other β-CDPs. In addition, nearly 20% reduction in luciferase gene expression was observed when the cells were transfected with β-CDP5 (5 methylene spacing), thereby demonstrating that an optimal spacer length between the amidine units is important for maximal delivery. The cytotoxicity assay at N/P = 50 showed that β-CDP8 and β-CDP7 afforded almost 100% cell viability. However, all the other β-CDPs were found to be toxic, which was also evident via MTT assay. Thus, these experiments demonstrate that an optimal spacing between the amidine units is significant for increased transfection efficiency and decreased cytotoxicity.
Fig. 20.
Synthesis of β-CD-based polymer. Reproduced with permission from [141]. © 2001 American Chemical Society
Srinivasachari et al. designed a novel series of macromolecule vehicles using a β-CD core, where the 6-position of the glucose units has been grafted with pendant oligoethyleneamine groups of specified length (where the secondary amine stoichiometry varied from 0 to 4) [172]. To avoid formation of under-substituted impurities, the core and the branching units were conjugated via a 1,2,3-trizole linkage utilizing the high-yielding 1,3-dipolar cycloaddition, termed the “click reaction”. These completely monodisperse β-CD “click clusters” bind and compact pDNA at N/P > 2 into spherical nanoparticles with a diameter around 80–130 nm. Structures with two, three, or four secondary amines in the oligoethyleneamine arms protected pDNA from nuclease degradation when incubated in serum at 37°C for up to 48 h. These macromolecules were able to deliver efficiently Cy5-labeled pDNA into HeLa and H9c2(2-1) cells, and the internalization was comparable to transfection reagents jetPEI and SuperFect with dramatically lower toxicity. Transgene expression in both cell lines increased with increasing secondary amine content, and the analogs with three and four secondary amines showed similar (within an order-of-magnitude) transgene expression as jetPEI and SuperFect.
These promising initial results on the click cluster vehicles were succeeded by analogous studies to incorporate β-CD into a polymeric scaffold for gene delivery [173]. In this case, the β-CD was di-functionalizedwith azide groups and polymerized with dialkyne-functionalized oligoethylenamines to derive linear β-CD polymers containing between one and four secondary amines per repeat unit (Cd1–Cd4; Fig. 21). Based upon earlier favorable results with similar systems [21, 140, 146, 155], several different molecular weights of Cd4 (contains four secondary amines in the repeat unit), were synthesized to examine the effect of variation in the polymer length. Incorporation of β-CD into this polymer showed similar DNA binding and polyplex size and cationic surface profiles as were seen previously for the β-CD click clusters. CD-containing polymers showed significant enhancement in cellular uptake over jetPEI in HeLa cells, with Cd2, Cd3, and Cd4 internalized significantly better that Cd1. With polymer vehicle Cd4, molecular weight did not appear to affect internalization significantly, as similar high levels of polyplex internalization was observed for all lengths. In general, high levels of transgene expressionwere observed with these polymers, and the highest expression was observed with Cd3, slightly higher than Cd4, and both had similar profiles to Jet-PEI. The authors attribute the high delivery of Cd3 to a more flexible, randomly-coiled structure. This assessment is sensible, as lack of polymer rigidity could be important in DNA release, or accessibility of RNA polymerases, depending on how the polymer:DNA polyplex traffics within the cell. Interestingly, despite much more favorable uptake profiles compared to jetPEI, similar transgene expression was observed. This may be due to differences in trafficking mechanisms and rates, which may lead PEI to the nucleus in a more efficient manner. However, if nuclear delivery is not exclusive, this may afford a unique opportunity to deliver siRNA in high levels to the cytosol, an advantageous result for gene knockdown via RNA interference as a therapeutic strategy.
Fig. 21.
Structure of β-CD “click” polymers. Figure reproduced with permission from [173]. © 2009 American Chemical Society
Oligonucleotide delivery with β-CD for targeted cancer therapeutics has been explored extensively by the group of Mark Davis, and the remainder of this section is focused on the groundbreaking in vivo results attained by this laboratory. Previous work in this lab has extensively exploited the hydrophobic interior cavity of β-CD for inclusion of hydrophobic molecules conjugated to targeting groups [175], and this technology will be discussed in greater detail in the next section. In many of these studies, a β-CD polycation end-capped with imidazole groups was used as the delivery vehicle, and the human transferrin protein was used as a targeting ligand due to upregulation of transferrin receptors on tumor cells (Fig. 22). Pun et al. used these targeted β-CD-containing polycations to deliver fluorescently-labeled DNAzymes targeted to the c-myc proto-oncogene – to which tumor cells have been shown to require for proliferation – in tumor-bearing mice [176]. Using whole-body imaging techniques, Cy3-labeled DNAzymes were seen associating with the actin cytoskeleton. The highest levels of sustained fluorescence were observed when polyplexes were administered via intravenous bolus injection (compared to intraperitoneal injection, for which uptake of particles by tumor tissue was low). Unmodified DNAzyme was observed in cryosectioned tumor, liver, and kidneys 8 h post-injection but was not observed after 24 h, suggesting clearance from the body within this time frame. However, DNAzymes delivered in β-CD polyplexes were sustained in the tissues after 24 h, suggesting that the PEG groups afford prolonged circulation time that allows tumor delivery over extended time [176]. These experiments show that targeted delivery of oligonucleotides can be achieved in vivo by β-CD-containing polycations.
Fig. 22.
(a) Structure of β-cyclodextrin-containing polycations (CDP)s and (b) formulation of siRNA-containing targeted nanoparticles formed with CDP. Figure reprinted with permission from [177]. © 2007 National Academy of Sciences, U.S.A
In a mouse model of Ewing’s sarcoma, Hu-Lieskovan et al. used these β-CD-containing vehicles to deliver siRNA against the EWS-FLI1 gene expressed in this disease model [174]. Ewing’s sarcoma (TC71) cells were modified to express stably luciferase and injected into immunocompromised mice to attain a disseminated tumor model, which was verified by bioluminescence imaging and MRI. Mice were dosed twice weekly with transferrin-targeted β-CD polycations complexing siRNA against EWS-FLI1 (siEFBP2), with treatment beginning the same day as injection of tumor cells. Delivery of naked siEFBP2 and targeted polyplexes containing control (non-EWS-FLI1-targeting; siCON1) siRNA did not decrease tumor size compared to control mice, and untargeted siEFBP2 polyplexes showed delayed tumor formation. However, targeted polyplexes containing siEFBP2 reduced tumor growth to 20% of that of control mice, indicating that targeted delivery of siRNA to transferrin-overexpressing tumor cells prevented the tumorigenicity of injected Ewing’s sarcoma cells. Substantial tumor reduction was observed during the course of the experiment (Fig. 23a). Importantly, these repeated treatments did not lead to increased cytokine levels or induce tissue damage, suggesting toxic and immune responses by the animals were minimal. These results show that targeted, systemic delivery of siRNA can treat disseminated cancers with carbohydrate-based non-viral gene delivery in a sequence-specific manner.
Fig. 23.
In vivo performance of targeted cyclodextrin polycations. (a) Growth curves for engrafted tumors. The median integrated tumor bioluminescent signal (photons/s) for each treatment group (n = 8–10) is plotted vs time after cell injection (days). (b) MRI confirmation of tumor engraftment. (c) Dose-dependent effects on cytokine production in non-human primates. Only very high dosage led to significant increases in cytokines. (ab) reprinted with permission from [174], figures 3 and 5. © 2005 American Association for Cancer Research. (c) reprinted with permission from [177]. © 2007 National Academy of Sciences, U.S.A
These promising results have led to further studies in vivo to assess gene knockdown efficacy of vehicles targeted to cancer cells. Heidel et al. used escalating doses of β-CD-siRNA nanoparticles delivered intravenously in monkeys (Fig. 23b). These are fundamental studies, as determination of dose-dependent tolerance levels is crucial for minimization of drug-induced toxicity and/or other untoward side effects. Escalating doses of 3, 9, and 27 mg/kg (with respect to the siRNA component) were administered 3 days apart, followed by a washout period of 11–12 days prior to a final dosing of 3 mg/kg. Serum chemistry and immune effects were measured 6 h post-injection. The lower doses of nanoparticle (3 and 9 mg/kg) were well tolerated and did not lead to significant increases in hematological factors (i.e., coagulation factors) or pro-inflammatory cytokines (Fig. 23c). At the highest dose (27 mg/kg), increases in creatinine and blood urea nitrogen were observed, indicative of acute renal toxicity. Coagulation factors were unaffected by the high dosage, but increases in pro-inflammatory cytokines IL-6 and IFN-γ, indicative of a helper T cell response, were observed shortly after injection of high doses of siRNA. Interestingly, increasing levels of TNF-α were not observed, providing convincing evidence that systemic inflammation is not occurring. Unfortunately, longer term measurements of these cytokines were not reported, so it is unclear whether these effects are transient or chronic. Exposure of the animals to high dosage of siRNA-containing nanoparticles did not lead to high titer antibody formation, suggesting that repeated doses of nanoparticles over time would be well-tolerated by the animals and would not lead to ineffective dosing due to an immune (neutralizing antibody) response. Importantly, high levels of nanoparticles were detected in the blood 5 min post-injection, verifying circulation of nanoparticles and avoiding rapid clearance from the bloodstream [177]. This elegant study confirms the safety of these nanoparticle siRNA delivery systems and suggests that repeating high dosage treatment could be well tolerated in patients. These promising vehicles have shown that high delivery and treatment efficacy can be attained with non-viral nucleic acid delivery, and are among the first carbohydrate-based vehicles to advance to human clinical trials. These preliminary studies have been reviewed recently [178] and provide a bright outlook to the future of the field.
4 Targeted Gene Delivery with Carbohydrates
As described earlier, carbohydrates have been used extensively in their native forms and as components of novel polymeric structures to transfer exogenous nucleic acids into cells. In addition, carbohydrates have also been conjugated to polymeric delivery systems as targeting moieties. While an extensive amount of work has been completed and continues to be published utilizing various carbohydrate structures for targeting, a full review of this area is beyond the scope of this review. As an example of the promise of this extensive research, we briefly review the use of β-d-galactose and β-d-galactosamine cell-specific targeting of polyplexes to hepatocytes, which are commonly studied targeting ligands in this research field. Hepatocytes overexpress the asialoglycoprotein receptor (ASGPr), a lectin that specifically recognizes the β-d-galactose and β-d-galactosamine carbohydrates. Conjugating these sugars as pendant groups on polymeric vehicles has been used extensively to achieve hepatocyte-specific targeting in vitro and in vivo. The following section briefly describes recently-published work to facilitate hepatocyte-selective delivery of DNA.
Early work by Zanta et al. describes galactosylated PEI (lactose was used in a reductive amination with PEI) as a liver-specific delivery system, which maintained tight DNA binding at low N/P ratios [179]. These galactosylated polyplexes were able to transfect NIH 3T3, a non-hepatocyte cell line, indicating incomplete specificity for ASGPr. However, the transgene expression efficiency was less than that observed for unmodified PEI, which can be interpreted as being due to a lack of cell-surface affinity from low levels of ASGPr. By contrast, murine (BNL CL.2) and human (HepG2) hepatocytes showed higher transgene expression from galactosylated PEI than unmodified PEI at low N/P. At high N/P, unmodified PEI demonstrated higher transgene expression, which may be artificially high due to increased toxicity. Incubation of cells with a competitive binding inhibitor to ASGPr, asialofetuin (ASF), led to a decrease in transgene expression, signifying receptor-mediated uptake [179]. A later study by Pun and Davis described the development of a hepatocyte-specific DNA delivery system using β-CD-containing polymers. In this approach, galactosamine was installed at the PEG end of galactose-PEG-tetrapeptide-adamantane via amide bond and this macromolecule was used to create a targeted vehicle by taking advantage of the ability of adamantane to form inclusion complexes with the hydrophobic core of β-CD [175]. Specific delivery to HepG2 cells was partially successful, as inhibition of targeted polyplex internalization was inhibited by ASF. However, as before, significant internalization was still observed for targeted polyplexes inhibited with ASF – this was seen for untargeted polyplexes as well. These studies demonstrate that galactosylation of polymeric vehicles can possibly provide receptor-specific uptake in vitro, but nonspecific delivery is still observed. In addition, transgene expression is used to assess receptor-specific uptake, but this is a downstream event from receptor-mediated internalization. It was clear from these studies that more work was needed to achieve exclusive delivery to hepatocytes and better research tools were needed to assess receptor-mediated internalization.
Further studies have attempted to optimize the amount of galactose conjugation with the specific delivery efficiency. The mole percentage of galactose on PEI was modified by varying the amount of lactose used in reductive amination reaction, to assess the effect of galactose concentration on transfection [180]. Increasing the mol % galactose up to 31.1% did not affect binding of PEI analogs to DNA. However, polyplex size grew with increasing galactose, and this was accompanied by a decrease in zeta potential, which became nearly neutral at high (31.1%) galactose concentration. Increasing galactose has a favorable effect on cell viability – improvement in biocompatibility was directly proportional to percent galactose, which corresponded to lower membrane-damaging effects as evidenced by LDH release. However, receptor-specific delivery was not achieved, as unmodified PEI showed higher levels of luciferase expression than galactosylated analogs. Polyplex size and charge clearly played a role in delivery, as larger, more neutral particles showed lower transgene expression than smaller, cationic polyplexes. A similar study by Ren et al. observed the effect of increasing the number of galactose molecules from one to three on a dendritic structure [26]. Less transgene (luciferase) expression was seen in HepG2 cells than in a non-hepatocyte cell line (BL-6 cells), indicating that hepatocyte specificity is not observed. However, increasing luciferase expression was observed as a function of the number of conjugated galactose molecules, as highest luciferase expression was observed for the tri-galactosylated compounds at low concentration. These results suggest that increasing the number of galactose groups may improve targeting based on a multivalent effect.
A potential problem with assessing receptor-mediated delivery may indeed be use of an in vitro system. HepG2 cells are hepatocellular carcinoma cells, so receptor expression may be more varied than a typical liver cell, and the rapid growth of malignant cells may help to internalize polyplexes non-specifically. Therefore, Nishikawa et al. monitored targeted DNA delivery in vivo to determine pharmacokinetic and biodistribution profiles with poly-l-ornithine-based polymers conjugated with galactose [181]. The conjugation was done using 2-imino-2-methoxyethyl-thio-β-d-galactopyranoside. Conjugation of galactose did not impact the polyplex size, so polyplexes formulated with [32P] DNA were injected intravenously into mice. As these materials are not serum-stabilized in any way, rapid clearance of polyplexes occurred, as the half-life was 8.2 min or less. However, the majority of radioactivity and luciferase expression was observed in the liver (with respect to other organs monitored – lungs, kidneys, spleen, heart), and the delivery to the liver increased with time. More significantly, when parenchymal (hepatocytes) and non-parenchymal (Kupffer, non-hepatocytes) liver cells were separated by collagenase perfusion, much higher radioactivity and luciferase expression was seen in the parenchymal cells than the non-parenchymal cells, suggesting hepatocyte-specific delivery was preferred over non-specific delivery. Some non-specific delivery was still observed – particularly to the lungs – but these are generally promising results that suggest that, in an animal model, hepatocyte specificity can be achieved.
The previously-published work in the fields of hepatocyte-targeted nucleic acid delivery and serum stabilization can be combined to develop smart delivery vehicles that contain structural elements for overcoming cellular barriers. In an elegant study, Chen et al. described the design and synthesis of PEGylated glycopeptides containing a cysteine-terminated triantennary glycopeptide (Fig. 24), PEGylated peptide, and melittin to form polymers through formation of disulfide bonds under oxidative conditions [182].
Fig. 24.
Structure of PEGylated glycopeptide. Figure reproduced with permission from [182]. © 2007 American Chemical Society
These polymers have been designed to achieve serum stability and sustained circulation with the PEGylated peptide, specific targeting to hepatocytes with galactosylated peptide, and increased cellular internalization and endosomal disruption with the fusogenic peptide monomer (melittin). Under the reducing conditions of the cell, the disulfide bonds can be reduced to sulfhydryl groups, degrading the polymer and releasing the pDNA. They developed five PEGylated glycopeptides (PGPs) with increasing melittin concentrations from 0% (PGP1) to 73% (PGP5), as well as a control polymer, PGP3*, that did not contain galactose for targeting. The specific delivery of DNA to hepatocytes was monitored in a murine model (Fig. 25). Polyplexes were administered intravenously by injection into the tail vein, which was followed by hydrodynamic stimulation via saline injection 5 min later to promote high levels of cellular uptake. Polymeric delivery of DNA was retained in the body twice as long as uncomplexed DNA, having a half-life of 1 h. Hepatocyte-specific targeting was assessed by the ratio of DNA in parenchymal vs non-parenchymal liver cells, and the galactosylated polymers led to 50% higher delivery to parenchymal cells than to non-parenchymal cells. A control of nongalactosylated polymer showed 50% higher uptake in non-parenchymal cells vs parenchymal cells. Luciferase expression was measured 24 h post-injection, and significantly higher luciferase expression was observed in the galactosylated analogs (Fig. 25). These results suggest that hepatocyte-favored delivery leading to transgene expression of DNA can be achieved through rational therapeutic design.
Fig. 25.
Characterization and luciferase expression of PGP DNA condensates in vivo. These results show that luciferase expression is dependent on galactose incorporation but independent of amount of melittin. (a) Represents the input mol ratio of Cys-terminated melittin, PEG-peptide, and glycopeptide. (b) Represents the measured mol ratio of Cys-terminated melittin, PEG-peptide, and glycopeptide for each purified PGP. (c) Values are the calculated MW based on polylysine standards. (d) Values are the calculated MW based on PEG standards. (e) The mean particle size determined at a stoichiometry of 0.3 nmol of PGP per mg of DNA. The value represents the mean diameter (nm) based on unimodal analysis. (f) The zeta potential of PGP DNA condensates at a stoichiometry of 0.3 nmol of PGP per mg of DNA. (g) The metabolic half-life of PGP 125I-DNA in triplicate mice. The results are derived from Fig. 6. (h) The PC/NPC ratio of DNA-targeted liver. (i) Represents a control PGP 3 in which galactose has been removed. Figure adapted with permission from [182]. © 2007 American Chemical Society
Targeted delivery of siRNA has also been achieved using rationally-designed polymeric vehicles using galactose as a hepatocyte-targeting group. Rozema et al. describe a polymeric system, dubbed Dynamic PolyConjugates, to attach N-acetylgalactosamine, PEG, and siRNA to a polymeric backbone for hepatocyte-targeting, charge shielding for serum stability, and gene knockdown, respectively [183]. The PEG was conjugated through an acid-labile maleamate linkage for release of PEG in endosomal compartments that can expose polymer amines for endosome disruption, and the siRNA was attached through a disulfide linkage which is cleaved under the reducing conditions of the cytoplasm. Using an siRNA targeted against the mRNA for apolipoprotein B (apoB), a gene expressed by hepatocytes, they were able to achieve 80% knockdown of apoB in mouse primary hepatocytes compared to 60–70% knockdown for a commercially-available transfection reagent, TransIT-siQuest. Mice injected intravenously with N-acetylgalactosylated oligonucleotide polyconjugates showed intracellular delivery of Cy3-oligonucleotide to hepatocytes, compared to no intracellular delivery when the targeting group was mannose or glucose (Fig. 26a). Mice injected with apoB siRNA-containing polyconjugates (Fig. 26b, c) showed dose-dependent knockdown of apoB mRNA two-days post-injection, as well as corresponding decreases in serum cholesterol. Hepatic lipid content was visualized by oil red staining of tissue sections, and mice treated with apoB siRNA showed increased lipid content corresponding in decreasing cholesterol transport from the tissue by apoB (Fig. 26d). These results confirm the liver-specific delivery of siRNA with polymers conjugated to N-acetylgalactosamine.
Fig. 26.
Specific delivery of siRNA to hepatocytes with Dynamic PolyConjugates. (a) Confocal micrographs indicate specific intracellular delivery of oligonucleotides by targeting hepatocytes with N-acetylgalactosamine, as Cy3-labeled oligonucleotide (red) is seen within mouse hepatocytes, compared to when mannose and glucose are used as targeting moieties and Cy3 oligonucleotides are seen in the pericellular regions. (b) RT-qPCR shows dose-dependent decrease in apoB mRNA, corresponding to (c) decreasing serum cholesterol levels. (d) Increased hepatic lipid content (stained with oil red) relative to control siRNA and saline injections confirm knockdown of apoB-mediated cholesterol transport from the liver. Figure adapted with permission from [183]. © 2007 National Academy of Sciences, U.S.A
The results discussed in this section present a brief sampling of the literature focused on hepatocyte-specific delivery with carbohydrate-based targeting ligands. This section focused on galactose for targeting the ASGPr on hepatocytes, but other carbohydrates have also been studied for cell-specific targeting, including lactose for targeting the airway epithelia and mannose for targeting cells of the immune system [143, 184, 185]. While a detailed review of this subfield is beyond the scope of this chapter, work in this area is ongoing and continues to develop polymeric systems for specific delivery of nucleic acids through lectin-mediated targeting.
5 Outlook
More than a decade of intense research has been devoted to developing carbohydrate-based polymers as nucleic acid carriers for the treatment of disease. This review has outlined the progression and successes attained by carbohydrate-based vehicles. As described herein, carbohydrates allow biocompatible transfection of nucleic acids into mammalian cells, enable chemical modification for delivery enhancement, and can be used as targeting moieties for achieving receptor-specific delivery. Indeed, the last few years have yielded substantial progress in the development of carbohydrates as delivery vehicles, including tumor-specific delivery of siRNA for human therapeutics, specific delivery of pDNA to hepatocytes, and development of rationally-designed vehicles designed to promote efficient delivery by systematically bypassing extracellular and intracellular barriers. These promising studies prove that the past years of research have led to valuable information about how DNA delivery vehicles are processed by the cell, and this knowledge is actively applied to develop new, functional materials designed for use in humans. While this prospect has yet to be achieved, the progress in the field is evident and carbohydrate-based nucleic acid delivery vehicles will remain at the forefront of non-viral delivery research. Future studies in this area will continue to develop cutting-edge reagents for delivery of nucleic acids, and progress more promising vehicles towards the clinic for eventual treatment of devastating human disease.
Acknowledgments
We apologize to our many colleagues whose elegant work we were unable to discuss directly in this review. The authors acknowledge NIH New Innovator Award. T.M.R. is a fellow of the Alfred P. Sloan Research Foundation and a recipient of the Camille Dreyfus Teacher-Scholar Award.
Abbreviations
- APC
Antigen-presenting cell
- apoB
Apolipoprotein B
- ASF
Asialofetuin
- ASGP
Asialoglycoprotein receptor
- AS-ODN
Antisense oligodeoxynucleotide
- CD
Cyclodextrin
- CDI
1,1’-Carbonyldiimidazole
- CDP
Cyclodextrin-containing polycations
- CPP
Cell penetrating peptide
- DCC
N,N’-Dicyclohexylcarbodiimide
- DDMC
2-Diethyl-aminoethyl–dextran–methyl methacrylate graft copolymer
- DEAE
2-Diethyl-aminoethyl
- DMEM
Dulbecco’s Modified Eagle Medium
- EDC
1-Ethyl-3-(3-dimethyl amino)propyl carbodiimide
- EtiBr
Ethidium bromide
- HA
Hyaluronic acid
- IL
Interleukin
- N4C3
Tripropylenetetramine
- NHS
N-Hydroxysuccinimide
- NPC
Non-parenchymal
- PAMAM
Polyamidoamine
- PBS
Phosphate buffered saline
- PC
Parenchymal
- PDGF
Platelet-derived growth factor
- PEG
Poly(ethylene glycol)
- PEI
Polyethylenimine
- PGP
PEGylated glycopeptide
- PLGA
Poly(lactide-co-glycolide)
- PLL
Poly-l-lysine
- PO-DNA
Phosphodiester-DNA
- poly(A)
Polyadenine
- poly(C)
Polycytosine
- poly(dA)
Polydeoxyadenine
- poly(dT)
Polydeoxythymine
- PS-DNA
Phosphorothioate-DNA
- PVP
Poly(vinylpyrrolidone)
- R8
Octaarginine
- RGD
Arginine-glycine-aspartic acid
- SMC
Smooth muscle cell
- SPG
Schizophyllan
- TNF
Tumor necrosis factor
Contributor Information
Antons Sizovs, Department of Chemistry, Macromolecules and Interfaces Institute, Virginia Polytechnic Institute and State University, Blacksburg, VA24060, USA.
Patrick M. McLendon, Department of Chemistry, Macromolecules and Interfaces Institute, Virginia Polytechnic Institute and State University, Blacksburg, VA24060, USA; Department of Molecular Cardiovascular Biology, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH 45228, USA.
Sathya Srinivasachari, Department of Chemistry, University of Cincinnati, Cincinnati, OH 45229, USA; Institute for Stem Cell Biology and Regenerative Medicine, Bangalore, India.
Theresa M. Reineke, Email: treineke@vt.edu, Department of Chemistry, Macromolecules and Interfaces Institute, Virginia Polytechnic Institute and State University, Blacksburg, VA24060, USA.
References
- 1.Pickler RH, Munro CL. Gene therapy for inherited disorders. J Pediatr Nurs. 1995;10:40–47. doi: 10.1016/S0882-5963(05)80097-1. [DOI] [PubMed] [Google Scholar]
- 2.Anderson WF. Human gene therapy. Science. 1992;256:803–813. [Google Scholar]
- 3.Mulligan RC. The basic science of gene therapy. Science. 1993;260:926–932. doi: 10.1126/science.8493530. [DOI] [PubMed] [Google Scholar]
- 4.Wiethoff CM, Middaugh CR. Barriers to nonviral gene delivery. J Pharm Sci. 2003;92(2):203–217. doi: 10.1002/jps.10286. [DOI] [PubMed] [Google Scholar]
- 5.Braun CS, Vetro JA, Tomalia DA, et al. Structure/function relationships of polyamidoamine/ DNA dendrimers as gene delivery vehicles. J Pharm Sci. 2005;94(2):423–436. doi: 10.1002/jps.20251. [DOI] [PubMed] [Google Scholar]
- 6.Behr J-P. Synthetic gene-transfer vectors. Acc Chem Res. 1993;26:274–279. doi: 10.1021/ar200213g. [DOI] [PubMed] [Google Scholar]
- 7.Luo D, Saltzman WM. Synthetic DNA delivery systems. Nat Biotechnol. 2000;18:33–37. doi: 10.1038/71889. [DOI] [PubMed] [Google Scholar]
- 8.Davis ME. Non-viral gene delivery systems. Curr Opin Biotechnol. 2002;13(2):128–131. doi: 10.1016/s0958-1669(02)00294-x. [DOI] [PubMed] [Google Scholar]
- 9.Kircheis R, Wagner E. Polycation/DNA complex for in vivo gene delivery. Gene Ther Regul. 2000;1:95–114. [Google Scholar]
- 10.Godbey WT, Wu KK, Mikos AG. Poly(ethylenimine) and its role in gene delivery. J Control Release. 1999;60(2-3):149–160. doi: 10.1016/s0168-3659(99)00090-5. [DOI] [PubMed] [Google Scholar]
- 11.Godbey WT, Wu KK, Mikos AG. Size matters: molecular weight affects the efficiency of poly(ethylenimine) as a gene delivery vehicle. J Biomed Mater Res B. 1999;45(3):268–275. doi: 10.1002/(sici)1097-4636(19990605)45:3<268::aid-jbm15>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 12.Mannisto M, Vanderkerken S, Toncheva V, et al. Structure-activity relationships of poly(L-lysines): effects of pegylation and molecular shape on physicochemical and biological properties in gene delivery. J Control Release. 2002;83(1):169–182. doi: 10.1016/s0168-3659(02)00178-5. [DOI] [PubMed] [Google Scholar]
- 13.Kim SW. Polylysine copolymers for gene delivery. Gene Transfer. 2007:461–471. [Google Scholar]
- 14.Kwoh DY, Coffin CC, Lollo CP, et al. Stabilization of poly-L-lysine/DNA polyplexes for in vivo gene delivery to the liver. Biochim Biophys Acta. 1999;1444(2):171–190. doi: 10.1016/s0167-4781(98)00274-7. [DOI] [PubMed] [Google Scholar]
- 15.Smith DK. Dendritic supermolecules–towards controllable nanomaterials. Chem Commun. 2006;1:34–44. doi: 10.1039/b507416a. [DOI] [PubMed] [Google Scholar]
- 16.Eliyahu H, Siani S, Azzam T, et al. Relationships between chemical composition, physical properties and transfection efficiency of polysaccharide–spermine conjugates. Biomaterials. 2006;27:1646–1655. doi: 10.1016/j.biomaterials.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 17.Hong S, Bielinska AU, Mecke A, et al. Interaction of poly(amidoamine) dendrimers with supported lipid bilayers and cells: hole formation and the relation to transport. Bioconjug Chem. 2004;15(4):774–782. doi: 10.1021/bc049962b. [DOI] [PubMed] [Google Scholar]
- 18.Kraemer M. Dendritic polyamines: simple access to new materials with defined treelike structures for application in nonviral gene delivery. ChemBioChem. 2004;5(8):1081. doi: 10.1002/cbic.200300905. [DOI] [PubMed] [Google Scholar]
- 19.Svenson S, Tomalia DA. Dendrimers in biomedical applications – reflections on the field. Adv Drug Deliv Rev. 2005;57(15):2106–2129. doi: 10.1016/j.addr.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 20.Mintzer MA, Simanek EE. Nonviral vectors for gene delivery. Chem Rev. 2009;109:259–303. doi: 10.1021/cr800409e. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, Reineke TM. Hydroxyl stereochemistry and amine number within poly(glycoamidoamine) s affect intracellular DNA delivery. J Am Chem Soc. 2005;127(9):3004–3015. doi: 10.1021/ja0436446. [DOI] [PubMed] [Google Scholar]
- 22.Velter I, La Ferla B, Nicotra F. Carbohydrate-based molecular scaffolding. J Carbohydr Chem. 2006;25:97–138. [Google Scholar]
- 23.Varma AJ, Kennedy JF, Galgali P. Synthetic polymers functionalized by carbohydrates: a review. Carbohydr Polym. 2004;56:429–445. [Google Scholar]
- 24.Yudovin-Farber I, Domb AJ. Cationic polysaccharides for gene delivery. Mat Sci Eng. 2007;27:595–598. [Google Scholar]
- 25.Miyata T, Nakamae K. Polymers with pendant saccharides – ‘glycopolymers’. Trends Polym Sci. 1997;5(6):198–205. [Google Scholar]
- 26.Ren T, Zhang G, Liu D. Synthesis of galactosyl compounds for targeted gene delivery. Bioorg Med Chem. 2001;9(11):2969–2978. doi: 10.1016/s0968-0896(01)00203-6. [DOI] [PubMed] [Google Scholar]
- 27.Lundquist JJ, Toone EJ. The cluster glycoside effect. Chem Rev. 2002;102(2):555–578. doi: 10.1021/cr000418f. [DOI] [PubMed] [Google Scholar]
- 28.Ladmiral V, Melia E, Haddleton DM. Synthetic glycopolymers: an overview. Eur Polym J. 2004;40:431–449. [Google Scholar]
- 29.Mehvar R. Dextrans for targeted and sustained delivery of therapeutic and imaging agents. J Control Release. 2000;69:1–25. doi: 10.1016/s0168-3659(00)00302-3. [DOI] [PubMed] [Google Scholar]
- 30.Sakurai K, Uezu K, Numata M, et al. b-1,3-Glucan polysaccharides as novel onedimensional hosts for DNA/RNA, conjugated polymers and nanoparticles. Chem Commun. 2005;35:4383–4398. doi: 10.1039/b506673p. [DOI] [PubMed] [Google Scholar]
- 31.Borchard G. Chitosans for gene delivery. Adv Drug Deliv Rev. 2001;52(2):145–150. doi: 10.1016/s0169-409x(01)00198-3. [DOI] [PubMed] [Google Scholar]
- 32.Yun YH, Goetz DJ, Yellen P, et al. Hyaluronan microspheres for sustained gene delivery and site-specific targeting. Biomaterials. 2004;25(1):147–157. doi: 10.1016/s0142-9612(03)00467-8. [DOI] [PubMed] [Google Scholar]
- 33.Gupta M, Gupta AK. Hydrogel pullulan nanoparticles encapsulating pBUDLacZ plasmid as an efficient gene delivery carrier. J Control Release. 2004;99(1):157–166. doi: 10.1016/j.jconrel.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 34.Vaheri A, Pagano JS. Infectious poliovirus RNA: a sensitive method of assay. Virology. 1965;27(3):434–436. doi: 10.1016/0042-6822(65)90126-1. [DOI] [PubMed] [Google Scholar]
- 35.Mack KD, Wei R, Elbagarri A, et al. A novel method for DEAE-dextran mediated transfection of adherent primary cultured human macrophages. J Immunol Methods. 1998;211:79–86. doi: 10.1016/s0022-1759(97)00194-4. [DOI] [PubMed] [Google Scholar]
- 36.Onishi Y, Eshita Y, Murashita A, et al. Synthesis and characterization of 2-diethylaminoethyl- dextran-methyl methacrylate graft copolymer for nonviral gene delivery vector. J Appl Polym Sci. 2005;98:9–14. [Google Scholar]
- 37.Azzam T, Eliyahu H, Makocitzki A, et al. Dextran-spermine conjugate: an efficient vector for gene delivery. Macromol Symp. 2003;195:247–261. [Google Scholar]
- 38.Azzam T, Raskin A, Makovitzki A, et al. Cationic polysaccharides for gene delivery. Macromolecules. 2002;35(27):9947–9953. [Google Scholar]
- 39.Hosseinkhani H, Azzam T, Tabata Y, et al. Dextran–spermine polycation: an efficient nonviral vector for in vitro and in vivo gene transfection. Gene Ther. 2004;11:194–203. doi: 10.1038/sj.gt.3302159. [DOI] [PubMed] [Google Scholar]
- 40.Eliyahu H, Joseph A, Schillemans JP, et al. Characterization and in vivo performance of dextran-spermine polyplexes and DOTAP/cholesterol lipoplexes administered locally and systemically. Biomaterials. 2007;28(14):2339–2349. doi: 10.1016/j.biomaterials.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 41.Eliyahu H, Joseph A, Azzam T, et al. Dextran-spermine-based polyplexes – evaluation of transgene expression and of local and systemic toxicity in mice. Biomaterials. 2006;27(8):1636–1645. doi: 10.1016/j.biomaterials.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 42.Singh A, Suri S, Roy K. In-situ crosslinking hydrogels for combinatorial delivery of chemokines and siRNA-DNA carrying microparticles to dendritic cells. Biomaterials. 2009;30(28):5187–5200. doi: 10.1016/j.biomaterials.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beaudette TT, Cohen JA, Bachelder EM, et al. Chemoselective ligation in the functionalization of polysaccharide-based particles. J Am Chem Soc. 2009;131(30):10360–10361. doi: 10.1021/ja903984s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bachelder EM, Beaudette TT, Broaders KE, et al. Acetal-derivatized dextran: an acidresponsive biodegradable material for therapeutic applications. J Am Chem Soc. 2008;130(32):10494–10495. doi: 10.1021/ja803947s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sakurai K, Iguchi R, Mizu M, et al. Polysaccharide-polynucleotide complexes. Part 7. Hydrogen-ion and salt concentration dependence of complexation between schizophyllan and single-stranded homo RNAs. Bioorg Chem. 2003;31(3):216–226. doi: 10.1016/s0045-2068(03)00034-8. [DOI] [PubMed] [Google Scholar]
- 46.Nagasaki T, Hojo M, Uno A, et al. Long-term expression with a cationic polymer derived from a natural polysaccharide: schizophyllan. Bioconjug Chem. 2004;15:249–259. doi: 10.1021/bc034178x. [DOI] [PubMed] [Google Scholar]
- 47.Hasegawa T, Fujisawa T, Haraguchi S, et al. Schizophyllan–folate conjugate as a new non-cytotoxic and cancer-targeted antisense carrier. Bioorg Med Chem Lett. 2005;15(2):327–330. doi: 10.1016/j.bmcl.2004.10.071. [DOI] [PubMed] [Google Scholar]
- 48.Sakurai K, Shinkai S. Molecular recognition of adenine, cytosine, and uracil in a singlestrand RNA by a natural polysaccharide: schizophyllan. J Am Chem Soc. 2000;122:4520–4521. [Google Scholar]
- 49.Sakurai K, Mizu M, Shinkai S. Polysaccharide-polynucleotide complexes. 2. Complementary polynucleotide mimic behavior of a natural polysaccharide: schizophyllan in the macromolecular comple with a single strand RNA: poly(C) Biomacromolecules. 2001;2:641–650. doi: 10.1021/bm000121r. [DOI] [PubMed] [Google Scholar]
- 50.Sletmoen M, Naess SN, Stokke BT. Structure and stability of polynucleotide- (1, 3)-[beta]-d-glucan complexes. Carbohydr Polym. 2009;76(3):389–399. [Google Scholar]
- 51.Hasegawa T, Umeda M, Matsumoto T, et al. Lactose-appended schizophyllan is a potential candidate as a hepatocyte-targeted antisense carrier. Chem Commun. 2004;4:382–383. doi: 10.1039/b313426a. [DOI] [PubMed] [Google Scholar]
- 52.Mizu M, Koumoto K, Anada T, et al. Antisense oligonucleotides bound in the polysaccharide complex and the enhanced antisense effect due to the low hydrolysis. Biomaterials. 2004;25:3117–3123. doi: 10.1016/j.biomaterials.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 53.Koumoto K, Mizu M, Sakurai K, et al. Polysaccharide/polynucleotide complexes. Part 6. Chem Biodivers. 2004;1(3):520–529. doi: 10.1002/cbdv.200490045. [DOI] [PubMed] [Google Scholar]
- 54.Anada T, Karinaga R, Koumoto K, et al. Linear double-stranded DNA that mimics an infective tail of virus genome to enhance transfection. J Control Release. 2005;108(2-3):529–539. doi: 10.1016/j.jconrel.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 55.Takeda Y, Shimada N, Kaneko K, et al. Ternary complex consisting of DNA, polycation, and a natural polysaccharide of schizophyllan to induce cellular uptake by antigen presenting cells. Biomacromolecules. 2007;8(4):1178–1186. doi: 10.1021/bm0611937. [DOI] [PubMed] [Google Scholar]
- 56.Hasegawa T, Fujisawa T, Numata M, et al. Schizophyllans carrying oligosaccharide appendages as potential candidates for cell-targeted antisense carrier. Org Biomol Chem. 2004;2(21):3091–3098. doi: 10.1039/B412124B. [DOI] [PubMed] [Google Scholar]
- 57.Matsumoto T, Numata M, Anada T, et al. Chemically modified polysaccharide schizophyllan for antisense oligonucleotides delivery to enhance the cellular uptake efficiency. Biochem Biophys Acta. 2004;1670:91–104. doi: 10.1016/j.bbagen.2003.10.019. [DOI] [PubMed] [Google Scholar]
- 58.Karinaga R, Koumoto K, Mizu M, et al. PEG-appended beta-(1–>3)-d-glucan schizophyllan to deliver antisense-oligonucleotides with avoiding lysosomal degradation. Biomaterials. 2005;26(23):4866–4873. doi: 10.1016/j.biomaterials.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 59.Karinaga R, Anada T, Minari J, et al. Galactose-PEG dual conjugation of beta-(1–>3)- d-glucan schizophyllan for antisense oligonucleotides delivery to enhance the cellular uptake. Biomaterials. 2006;27(8):1626–1635. doi: 10.1016/j.biomaterials.2005.08.023. [DOI] [PubMed] [Google Scholar]
- 60.Mizu M, Koumoto K, Anada T, et al. A polysaccharide carrier for immunostimulatory CpG DNA to enhance cytokine secretion. J Am Chem Soc. 2004;126(27):8372–8373. doi: 10.1021/ja031978+. [DOI] [PubMed] [Google Scholar]
- 61.Klinman DM, Klaschik S, Sato T, et al. CpG oligonucleotides as adjuvants for vaccines targeting infectious diseases. Adv Drug Deliv Rev. 2009;61(3):248–255. doi: 10.1016/j.addr.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 62.Shimada N, Coban C, Takeda Y, et al. Apolysaccharide carrier to effectively deliver native phosphodiester CpG DNA to antigen-presenting cells. Bioconjug Chem. 2007;18(4):1280–1286. doi: 10.1021/bc0700178. [DOI] [PubMed] [Google Scholar]
- 63.Dias N, Stein CA. Antisense oligonucleotides: basic concepts and mechanisms. Mol Cancer Ther. 2002;1(5):347–355. [PubMed] [Google Scholar]
- 64.Pouyani T, Prestwich GD. Functionalized derivatives of hyaluronic acid oligosaccharides: drug carriers and novel biomaterials. Bioconjug Chem. 1994;5(4):339–347. doi: 10.1021/bc00028a010. [DOI] [PubMed] [Google Scholar]
- 65.Kim A, Checkla DM, Dehazya P, et al. Characterization of DNA-hyaluronan matrix for sustained gene transfer. J Control Release. 2003;90:81–95. doi: 10.1016/s0168-3659(03)00175-5. [DOI] [PubMed] [Google Scholar]
- 66.Kim AP, Yellen P, Yun YH, et al. Delivery of a vector encoding mouse hyaluronan synthase 2 via a crosslinked hyaluronan film. Biomaterials. 2005;26:1585–1593. doi: 10.1016/j.biomaterials.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 67.Yun YH, Chen W. Microspheres formulated from native hyaluronan for applications in gene therapy. In: Mansoor MA, editor. Polymeric gene delivery: principles and applications. CRC Press LLC, USA; 2005. pp. 475–486. [Google Scholar]
- 68.de la Fuente M, Seijo B, Alonso MJ. Bioadhesive hyaluronan-chitosan nanoparticles can transport genes across the ocular mucosa and transfect ocular tissue. Gene Ther. 2008;15(9):668–676. doi: 10.1038/gt.2008.16. [DOI] [PubMed] [Google Scholar]
- 69.de la Fuente M, Seijo B, Alonso MJ. Design of novel polysaccharidic nanostructures for gene delivery. Nanotechnology. 2008;19(7):075105/1–075105/9. doi: 10.1088/0957-4484/19/7/075105. [DOI] [PubMed] [Google Scholar]
- 70.Duceppe N, Tabrizian M. Factors influencing the transfection efficiency of ultra low molecular weight chitosan/hyaluronic acid nanoparticles. Biomaterials. 2009;30(13):2625–2631. doi: 10.1016/j.biomaterials.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 71.Wieland JA, Houchin-Ray TL, Shea LD. Non-viral vector delivery from poly(ethylene glycol)-hyaluronic acid hydrogels. J Control Release. 2007;120(3):233–241. doi: 10.1016/j.jconrel.2007.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saraf A, Hacker MC, Sitharaman B, et al. Synthesis and conformational evaluation of a novel gene delivery vector for human mesenchymal stem cells. Biomacromolecules. 2008;9(3):818–827. doi: 10.1021/bm701146f. [DOI] [PubMed] [Google Scholar]
- 73.Shen Y, Li Q, Tu J, et al. Synthesis and characterization of low molecular weight hyaluronic acid-based cationic micelles for efficient siRNA delivery. Carbohydr Polym. 2009;77(1):95–104. [Google Scholar]
- 74.Bruneel D, Schacht E. End group modification of pullulan. Polymer. 1995;36:169–172. [Google Scholar]
- 75.Kaneo Y, Tanaka T, Nakano T, et al. Evidence for receptor-mediated hepatic uptake of pullulan in rats. J Control Release. 2001;70:365–373. doi: 10.1016/s0168-3659(00)00368-0. [DOI] [PubMed] [Google Scholar]
- 76.Nabi IR, Le PU. Caveolae/raft-dependent endocytosis. J Cell Biol. 2003;161(4):673–677. doi: 10.1083/jcb.200302028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Na K, Lee ES, Bae YH. Self-assembled nanoparticles of hydrophobically-modified polysaccharide bearing vitamin H as a targeted anti-cancer drug delivery system. Eur J Pharm Biopharm. 2003;18(2):165–173. doi: 10.1016/s0928-0987(02)00257-9. [DOI] [PubMed] [Google Scholar]
- 78.Akiyoshi K, Kobayashi S, Shichibe S, et al. Self-assembled hydrogel nanoparticle of cholesterol-bearing pullulan as a carrier of protein drugs: complexation and stabilization of insulin. J Control Release. 1998;54(3):313–320. doi: 10.1016/s0168-3659(98)00017-0. [DOI] [PubMed] [Google Scholar]
- 79.Hosseinkhani H, Aoyama T, Ogawa O, et al. Liver targeting of plasmid DNA by pullulan conjugation based on metal coordination. J Control Release. 2002;83(2):287–302. doi: 10.1016/s0168-3659(02)00201-8. [DOI] [PubMed] [Google Scholar]
- 80.Kanatani I, Ikai T, Okazaki A, et al. Efficient gene transfer by pullulan-spermine occurs through both clathrin- and raft/caveolae-dependent mechanisms. J Control Release. 2006;116(1):75–82. doi: 10.1016/j.jconrel.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 81.Jo J, Ikai T, Okazaki A, Nagane K, Yamamoto M, Hirano Y, Tabata Y. Expression profile of plasmid DNA obtained using spermine derivatives of pullulan with different molecular weights. J Biomater Sci Polym Ed. 2007;18(7):883–899. doi: 10.1163/156856207781367756. [DOI] [PubMed] [Google Scholar]
- 82.San Juan A, Hlawaty H, Chaubet F, et al. Cationized pullulan 3D matrices as new materials for gene transfer. J Biomed Mater Res A. 2007;82A(2):354–362. doi: 10.1002/jbm.a.31062. [DOI] [PubMed] [Google Scholar]
- 83.Imai T, Shiraishi S, Saito H, et al. Interaction of indomethacin with low molecular weight chitosan, and improvements of some pharmacuetical properties of indomethacin by low molecular weight chitosans. Int J Pharm. 1991;67:11–20. [Google Scholar]
- 84.Takayama K, Hirata M, Machida Y, et al. Effect of interpolymer complex formation on bioadhesive property and drug release phenomenon of compressed tablet consisting of chitosan and sodium hyaluronate. Chem Pharm Bull. 1990;38:1993–1997. doi: 10.1248/cpb.38.1993. [DOI] [PubMed] [Google Scholar]
- 85.Meshali MM, Gabr KE. Effect of interpolymer complex formation of chitosan with pectin or acacia on the release behavior of chlorpromazine HCl. Int J Pharm. 1993;89:177–181. [Google Scholar]
- 86.Mao H-Q. Chitosan-DNA nanoparticles as gene carriers: synthesis, characterization and transfection efficiency. J Control Release. 2001;70(3):399–421. doi: 10.1016/s0168-3659(00)00361-8. [DOI] [PubMed] [Google Scholar]
- 87.Mumper R, Wang JJ, Claspell JM, et al. Novel polymeric condensing carriers for gene delivery. Proc Intl Sym Control Release Bioact Mater. 1995;22:178–179. [Google Scholar]
- 88.MacLaughlin FC, Mumper RJ, Wang J, et al. Chitosan and depolymerized chitosan oligomers as condensing carriers for in vivo plasmid delivery. J Control Release. 1998;56:259–272. doi: 10.1016/s0168-3659(98)00097-2. [DOI] [PubMed] [Google Scholar]
- 89.Dodane V, Vilivalam VD. Pharmaceutical applications of chitosan. Pharm Sci Technol Today. 1998;1(6):246–253. [Google Scholar]
- 90.Aranaz I, Mengibar M, Harris R, et al. Functional characterization of chitin and chitosan. Curr Chem Biol. 2009;3:203–230. [Google Scholar]
- 91.Aiba S-i. Studies on chitosan: 3. Evidence for the presence of random and block copolymer structures in partially N-acetylated chitosans. Int J Biol Macromol. 1991;13(1):40–44. doi: 10.1016/0141-8130(91)90008-i. [DOI] [PubMed] [Google Scholar]
- 92.Mima S, Miya M, Iwamoto R, et al. Highly deacetylated chitosan and its properties. J Appl Polym Sci. 1983;28(6):1909–1917. [Google Scholar]
- 93.Nguyen S, Hisiger S, Jolicoeur M, et al. Fractionation and characterization of chitosan by analytical SEC and 1H NMR after semi-preparative SEC. Carbohydr Polym. 2009;75(4):636–645. [Google Scholar]
- 94.Köping-Höggård M, Tubulekas I, Guan H, Edwards K, Nilsson M, Vårum KM, Artursson P. Chitosan as a nonviral gene delivery system. Structure–property relationships and characteristics compared with polyethylenimine in vitro and after lung administration in vivo . Gene Ther. 2001;8:1108–1121. doi: 10.1038/sj.gt.3301492. [DOI] [PubMed] [Google Scholar]
- 95.Kiang T, Wen J, Lim HW, et al. The effect of the degree of chitosan deacetylation on the efficiency of gene transfection. Biomaterials. 2004;25:5293–5301. doi: 10.1016/j.biomaterials.2003.12.036. [DOI] [PubMed] [Google Scholar]
- 96.Huang M, Fong C-W, Khor E, et al. Transfection efficiency of chitosan vectors: effect of polymer molecular weight and degree of deacetylation. J Control Release. 2005;106(3):391–406. doi: 10.1016/j.jconrel.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 97.Liu X, Howard KA, Dong M, et al. The influence of polymeric properties on chitosan/ siRNA nanoparticle formulation and gene silencing. Biomaterials. 2007;28(6):1280–1288. doi: 10.1016/j.biomaterials.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 98.Rejman J, Oberle V, Zuhorn IS, et al. Size-dependent internalization of particles via the pathways of clathrin- and caveolae-mediated endocytosis. Biochem J. 2004;377(1):159–169. doi: 10.1042/BJ20031253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Choksakulnimitr S, Masuda S, Tokuda H, et al. In vitro cytotoxicity of macromolecules in different cell culture systems. J Control Release. 1995;34(3):233–241. [Google Scholar]
- 100.Erbacher P, Zou S, Bettinger T, et al. Chitosan-based vector/DNA complexes for gene delivery: biophysical characteristics and transfection ability. Pharm Res. 1998;15(9):1332–1339. doi: 10.1023/a:1011981000671. [DOI] [PubMed] [Google Scholar]
- 101.Richardson SCW, Kolbe HVJ, Duncan R. Potential of low molecular mass chitosan as a DNA delivery system: biocompatibility, body distribution and ability to complex and protect DNA. Int J Pharm. 1999;178(2):231–243. doi: 10.1016/s0378-5173(98)00378-0. [DOI] [PubMed] [Google Scholar]
- 102.Murata J, Ohya Y, Ouchi T. Possibility of application of quaternary chitosan having pendant galactose residues as gene delivery tool. Carbohydr Polym. 1996;29(1):69–74. [Google Scholar]
- 103.Murata J, Ohya Y, Ouchi T. Design of quaternary chitosan conjugate having antennary galactose residues as a gene delivery tool. Carbohydr Polym. 1997;32(2):105–109. [Google Scholar]
- 104.Thanou M, Florea BI, Geldof M, et al. Quaternized chitosan oligomers as novel gene delivery vectors in epithelial cell lines. Biomaterials. 2002;23:153–159. doi: 10.1016/s0142-9612(01)00090-4. [DOI] [PubMed] [Google Scholar]
- 105.Kean T, Roth S, Thanou M. Trimethylated chitosans as non-viral gene delivery vectors: cytotoxicity and transfection efficiency. J Control Release. 2005;103:643–653. doi: 10.1016/j.jconrel.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 106.Kim TH, Kim SI, Akaike T, et al. Synergistic effect of poly(ethylenimine) on the transfection efficiency of galactosylated chitosan/DNA complexes. J Control Release. 2005;105(3):354–366. doi: 10.1016/j.jconrel.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 107.Wong K, Sun G, Zhang X, et al. PEI-g-chitosan, a novel gene delivery system with transfection efficiency comparable to polyethylenimine in vitro and after liver administration in vivo . Bioconjug Chem. 2006;17:152–158. doi: 10.1021/bc0501597. [DOI] [PubMed] [Google Scholar]
- 108.Jiang H-L, Kim Y-K, Arote R, et al. Chitosan-graft-polyethylenimine as a gene carrier. J Control Release. 2007;117:273–280. doi: 10.1016/j.jconrel.2006.10.025. [DOI] [PubMed] [Google Scholar]
- 109.Kim T-H, Jiang H-L, Jere D, et al. Chemical modification of chitosan as a gene carrier in vitro and in vivo . Prog Polym Sci. 2007;32(7):726–753. [Google Scholar]
- 110.Nicolet BH, Shinn LA. The action of periodic acid on a-amino alcohols. J Am Chem Soc. 1939;61(6):1615. [Google Scholar]
- 111.Vold IMN, Christensen BE. Periodate oxidation of chitosans with different chemical compositions. Carbohydr Res. 2005;340(4):679–684. doi: 10.1016/j.carres.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 112.Jiang H, Kwon J, Kim Y, et al. Galactosylated chitosan-graft-polyethylenimine as a gene carrier for hepatocyte targeting. Gene Ther. 2007;14(19):1389–1398. doi: 10.1038/sj.gt.3302997. [DOI] [PubMed] [Google Scholar]
- 113.Jiang H-L, Kwon J-T, Kim E-M, et al. Galactosylated poly(ethylene glycol)-chitosangraft- polyethylenimine as a gene carrier for hepatocyte-targeting. J Control Release. 2008;131(2):150–157. doi: 10.1016/j.jconrel.2008.07.029. [DOI] [PubMed] [Google Scholar]
- 114.Lu B, Xu X-D, Zhang X-Z, et al. Low molecular weight polyethylenimine grafted N-maleated chitosan for gene delivery: properties and in vitro transfection studies. Biomacromolecules. 2008;9(10):2594–2600. doi: 10.1021/bm8004676. [DOI] [PubMed] [Google Scholar]
- 115.Lou Y-L, Peng Y-S, Chen B-H, et al. Poly(ethylene imine)-g-chitosan using EX-810 as a spacer for nonviral gene delivery vectors. J Biomed Mater Res A. 2009;88A(4):1058–1068. doi: 10.1002/jbm.a.31961. [DOI] [PubMed] [Google Scholar]
- 116.Wu Y, Liu C, Zhao X, et al. A new biodegradable polymer: PEGylated chitosan-g-PEI possessing a hydroxyl group at the PEG end. J Polym Res. 2008;15(3):181–185. [Google Scholar]
- 117.Jiang H-L, Kim Y-K, Arote R, et al. Mannosylated chitosan-graft-polyethylenimine as a gene carrier for Raw 264.7 cell targeting. Int J Pharm. 2009;375(1-2):133–139. doi: 10.1016/j.ijpharm.2009.03.033. [DOI] [PubMed] [Google Scholar]
- 118.Jiang H-L, Xu C-X, Kim Y-K, et al. The suppression of lung tumorigenesis by aerosoldelivered folate-chitosan-graft-polyethylenimine/Akt1 shRNA complexes through the Akt signaling pathway. Biomaterials. 2009;30(29):5844–5852. doi: 10.1016/j.biomaterials.2009.07.017. [DOI] [PubMed] [Google Scholar]
- 119.Jere D, Jiang H-L, Kim Y-K, et al. Chitosan-graft-polyethylenimine for Akt1 siRNA delivery to lung cancer cells. Int J Pharm. 2009;378(1-2):194–200. doi: 10.1016/j.ijpharm.2009.05.046. [DOI] [PubMed] [Google Scholar]
- 120.Pack DW, Putnam D, Langer R. Design of imidazole-containing endosomolytic biopolymers for gene delivery. Biotechnol Bioeng. 2000;67(2):217–223. [PubMed] [Google Scholar]
- 121.Muzzarelli RAA, Mattioli-Belmonte M, Tietz C, et al. Stimulatory effect on bone formation exerted by a modified chitosan. Biomaterials. 1994;15(13):1075–1081. doi: 10.1016/0142-9612(94)90093-0. [DOI] [PubMed] [Google Scholar]
- 122.Midoux P, Monsigny M. Efficient gene transfer by histidylated polylysine/pDNA complexes. Bioconjug Chem. 1999;10(3):406–411. doi: 10.1021/bc9801070. [DOI] [PubMed] [Google Scholar]
- 123.Kim TH, Ihm JE, Choi YJ, et al. Efficient gene delivery by urocanic acid-modified chitosan. J Control Release. 2003;93(3):389–402. doi: 10.1016/j.jconrel.2003.08.017. [DOI] [PubMed] [Google Scholar]
- 124.Jin H, Kim T, Hwang S, et al. Aerosol delivery of urocanic acid–modified chitosan/ programmed cell death 4 complex regulated apoptosis, cell cycle, and angiogenesis in lungs of K-ras null mice. Mol Cancer Ther. 2006;5(4):1041. doi: 10.1158/1535-7163.MCT-05-0433. [DOI] [PubMed] [Google Scholar]
- 125.Jin H, Xu CX, Kim HW, et al. Urocanic acid-modified chitosan-mediated PTEN delivery via aerosol suppressed lung tumorigenesis in K-rasLA1 mice. Cancer Gene Ther. 2008;15(5):275–283. doi: 10.1038/sj.cgt.7701116. [DOI] [PubMed] [Google Scholar]
- 126.Bauhuber S, Hozsa C, Breunig M, et al. Delivery of nucleic acids via disulfide-based carrier systems. Adv Mater. 2009;21(32-33):3286–3306. doi: 10.1002/adma.200802453. [DOI] [PubMed] [Google Scholar]
- 127.Bernkop-Schnürch A, Hornof M, Guggi D. Thiolated chitosans. Eur J Pharm Biopharm. 2004;57(1):9–17. doi: 10.1016/s0939-6411(03)00147-4. [DOI] [PubMed] [Google Scholar]
- 128.Schmitz T, Bravo-Osuna I, Vauthier C, et al. Development and in vitro evaluation of a thiomer-based nanoparticulate gene delivery system. Biomaterials. 2007;28(3):524–531. doi: 10.1016/j.biomaterials.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 129.Martien R, Loretz B, Thaler M, et al. Chitosan–thioglycolic acid conjugate: an alternative carrier for oral nonviral gene delivery? J Biomed Mater Res A. 2007;82A(1):1–9. doi: 10.1002/jbm.a.31135. [DOI] [PubMed] [Google Scholar]
- 130.Park IK, Ihm JE, Park YH, et al. Galactosylated chitosan (GC)-graft-poly(vinyl pyrrolidone) (PVP) as hepatocyte-targeting DNA carrier. Preparation and physicochemical characterization of GC-graft-PVP/DNA complex (1) J Control Release. 2003;86:349–359. doi: 10.1016/s0168-3659(02)00365-6. [DOI] [PubMed] [Google Scholar]
- 131.Park YK, Park YH, Shin BA, et al. Galactosylated chitosan-graft-dextran as hepatocyte- targeting DNA carrier. J Control Release. 2000;69(1):97–108. doi: 10.1016/s0168-3659(00)00298-4. [DOI] [PubMed] [Google Scholar]
- 132.Park IK, Kim TH, Park YH, et al. Galactosylated chitosan-graft-poly(ethylene glycol) as hepatocyte-targeting DNA carrier. J Control Release. 2001;76:349–362. doi: 10.1016/s0168-3659(01)00448-5. [DOI] [PubMed] [Google Scholar]
- 133.Park IK, Ihm JE, Park YH, et al. Galactosylated chitosan (GC)-graft-poly(vinyl pyrrolidone) (PVP) as hepatocyte-targeting DNA carrier: In vivo transfection. Arch Pharm Res. 2003;27(12):1284–1289. doi: 10.1007/BF02975895. [DOI] [PubMed] [Google Scholar]
- 134.Hashimoto M, Morimoto M, Saimoto H, et al. Lactosylated chitosan for DNA delivery into hepatocytes: the effect of lactosylation on the physicochemical properties and intracellular trafficking of pDNA/chitosan complexes. Bioconjug Chem. 2006;17(2):309–316. doi: 10.1021/bc050228h. [DOI] [PubMed] [Google Scholar]
- 135.Hashimoto M, Morimoto M, Saimoto H, et al. Gene transfer by DNA/mannosylated chitosan complexes into mouse peritoneal macrophages. Biotechnol Lett. 2006;28(11):815–821. doi: 10.1007/s10529-006-9006-x. [DOI] [PubMed] [Google Scholar]
- 136.Forrest ML, Gabrielson N, Pack DW. Cyclodextrin-polyethylenimine conjugates for targeted in vitro gene delivery. Biotechnol Bioeng. 2005;89(4):416–423. doi: 10.1002/bit.20356. [DOI] [PubMed] [Google Scholar]
- 137.Liu Y, Wenning L, Lynch M, et al. New poly(D-glucaramidoamine)s induce DNA nanoparticle formation and efficient gene delivery into mammalian cells. J Am Chem Soc. 2004;126(24):7422–7423. doi: 10.1021/ja049831l. [DOI] [PubMed] [Google Scholar]
- 138.Reineke TM, Davis ME. Structural effects of carbohydrate-containing polycations on gene delivery. 1. Carbohydrate size and its distance from charge centers. Bioconjug Chem. 2003;14(1):247–254. doi: 10.1021/bc025592k. [DOI] [PubMed] [Google Scholar]
- 139.Reineke TM, Davis ME. Structural effects of carbohydrate-containing polycations on gene delivery. 2. Charge center type. Bioconjug Chem. 2003;14(1):255–261. doi: 10.1021/bc025593c. [DOI] [PubMed] [Google Scholar]
- 140.Srinivasachari S, Liu Y, Zhang G, et al. Trehalose click polymers inhibit nanoparticle aggregation and promote pDNA delivery in serum. J Am Chem Soc. 2006;128(25):8176–8184. doi: 10.1021/ja0585580. [DOI] [PubMed] [Google Scholar]
- 141.Hwang SJ, Bellocq NC, Davis ME. Effects of structure of beta-cyclodextrin-containing polymers on gene delivery. Bioconjug Chem. 2001;12(2):280–290. doi: 10.1021/bc0001084. [DOI] [PubMed] [Google Scholar]
- 142.Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. A versatile vector for gene and oligonuceotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci USA. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Grosse S, Aron Y, Honore I, Thevenot G, Danel C, Roche A-C, Monsigny M, Fajac I. Lactosylated polyethylenimine for gene transfer into airway epithelial cells: role of the sugar moiety in cell delivery and intracellular trafficking of the complexes. J Gene Med. 2004;2004(6):345–356. doi: 10.1002/jgm.515. [DOI] [PubMed] [Google Scholar]
- 144.Liu Y, Wenning L, Lynch M, et al. Gene delivery with novel poly(L-tartaramidoamine) In: Svenson S, editor. Polymeric drug delivery, volume I: particulate drug carriers. Washington, DC: American Chemical Society; 2006. pp. 217–227. [Google Scholar]
- 145.Metzke M, O’Connor N, Maiti S, et al. Saccharide–peptide hybrid copolymers as biomaterials. Angew Chem Int Ed. 2005;44:6529–6533. doi: 10.1002/anie.200501944. [DOI] [PubMed] [Google Scholar]
- 146.Liu Y, Reineke TM. Poly(glycoamidoamine)s for gene delivery: stability of polyplexes and efficacy with cardiomyoblast cells. Bioconjug Chem. 2006;17(1):101–108. doi: 10.1021/bc050275+. [DOI] [PubMed] [Google Scholar]
- 147.Prevette LE, Kodger TE, Reineke TM, et al. Deciphering the role of hydrogen bonding in enhancing pDNA-polycation interactions. Langmuir. 2007;23(19):9773–9784. doi: 10.1021/la7009995. [DOI] [PubMed] [Google Scholar]
- 148.Lee C-C, Liu Y, Reineke TM. General structure-activity relationship for poly(glycoamidoamine) s: the effect of amine density on cytotoxicity and DNA delivery efficiency. Bioconjug Chem. 2008;19(2):428–440. doi: 10.1021/bc7001659. [DOI] [PubMed] [Google Scholar]
- 149.Liu Y, Reineke TM. Poly(glycoamidoamine)s for gene delivery. structural effects on cellular internalization, buffering capacity, and gene expression. Bioconjug Chem. 2007;18(1):19–30. doi: 10.1021/bc060029d. [DOI] [PubMed] [Google Scholar]
- 150.Liu Y, Reineke TM. Degradation of poly(glycoamidoamine) DNA delivery vehicles: polyamide hydrolysis at physiological conditions promotes DNA release. Biomacromolecules. 2010;11(2):316–325. doi: 10.1021/bm9008233. [DOI] [PubMed] [Google Scholar]
- 151.Taori VP, Liu Y, Reineke TM. DNA delivery in vitro via surface release from multilayer assemblies with poly(glycoamidoamine)s. Acta Biomater. 2009;5(3):925–933. doi: 10.1016/j.actbio.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 152.Paiva C, Panek A. Biotechnological applications of the disaccharide trehalose. Biotechnol Annu Rev. 1996;2:293. doi: 10.1016/s1387-2656(08)70015-2. [DOI] [PubMed] [Google Scholar]
- 153.Lins RD, Pereira CS, Hünenberger PH. Trehalose-protein interaction in aqueous solution. Proteins. 2004;55(1):177–186. doi: 10.1002/prot.10632. [DOI] [PubMed] [Google Scholar]
- 154.Wagner E. Pharm Res. 2004;21:8–14. doi: 10.1023/b:pham.0000012146.04068.56. [DOI] [PubMed] [Google Scholar]
- 155.Srinivasachari S, Liu Y, Prevette LE, et al. Effects of trehalose click polymer length on pDNA complex stability and delivery efficacy. Biomaterials. 2007;28:2885–2898. doi: 10.1016/j.biomaterials.2007.01.037. [DOI] [PubMed] [Google Scholar]
- 156.Prevette LE, Lynch ML, Kizjakina K, et al. Correlation of amine number and pDNA binding mechanism for trehalose-based polycations. Langmuir. 2008;24(15):8090–8101. doi: 10.1021/la800120q. [DOI] [PubMed] [Google Scholar]
- 157.Ortiz-Mellet C, Benito JM, Garcia Fernandez JM, et al. Cyclodextrin-scaffolded glycoclusters. Chem Eur J. 1998;4(12):2523–2531. [Google Scholar]
- 158.Perez-Balderas F, Ortega-Munoz M, Morales-Sanfrutos J, et al. Multivalent neoglycoconjugates by regiospecific cycloaddition of alkynes and azides using organic-soluble copper catalysts. Org Lett. 2003;5(11):1951–1954. doi: 10.1021/ol034534r. [DOI] [PubMed] [Google Scholar]
- 159.Garcia-Lopez JJ, Hernandez-Mateo F, Isac-Garcia J, et al. Synthesis of per-glycosylated beta-cyclodextrins having enhanced lectin binding affinity. J Org Chem. 1999;64(2):522–531. [Google Scholar]
- 160.Gadella A, Defaye J. Selective halogenation at primary postions of cyclomaltooligosaccharides and a synthesis of per-3,6-anhydro cyclomaltooligosaccharides. Angew Chem Int Ed. 1991;30:78–80. [Google Scholar]
- 161.Andre S, Kaltner H, Furuike T, et al. Persubstituted cyclodextrin-based glycoclusters as inhibitors of protein-carbohydrate recognition using purified plant and mammalian lectins and wild-type and lectin-gene-transfected tumor cells as targets. Bioconjug Chem. 2004;15(1):87–98. doi: 10.1021/bc0340666. [DOI] [PubMed] [Google Scholar]
- 162.Bellocq NC, Pun SH, Jensen GS, et al. Transferrin-containing, cyclodextrin polymerbased particles for tumor-targeted gene delivery. Bioconjug Chem. 2003;14(6):1122–1132. doi: 10.1021/bc034125f. [DOI] [PubMed] [Google Scholar]
- 163.Davis ME, Bellocq NC. Cyclodextrin-containing polymers for gene delivery. J Incl Phenom Macro Chem. 2001;44:17–22. doi: 10.1021/bc0001084. [DOI] [PubMed] [Google Scholar]
- 164.Croyle MA, Roessler BJ, Hsu C-P, et al. Beta-cyclodextrins enhance adenoviralmediated gene delivery to the intestine. Pharm Res. 1998;15(9):1349–1355. doi: 10.1023/a:1011985101580. [DOI] [PubMed] [Google Scholar]
- 165.Boger J, Corcoran RJ, Lehn J-M. Cyclodextrin chemistry. Selective modification of all primary hydroxyl groups of alpha- and beta-cyclodextrins. Helv Chim Acta. 1978;61:2190–2218. [Google Scholar]
- 166.Mocanu G, Vizitiu D, Carpov A. Cyclodextrin polymers. J Bioact Compat Polym. 2001;16:315–328. [Google Scholar]
- 167.Zhao Q, Temsamani J, Agrawal S. Use of cyclodextrin and its derivatives as carriers for oligonucleotide delivery. Antisense Res Dev. 1995;5(3):185–192. doi: 10.1089/ard.1995.5.185. [DOI] [PubMed] [Google Scholar]
- 168.Salem LB, Bosquillon C, Dailey LA, et al. Sparing methylation of [beta]-cyclodextrin mitigates cytotoxicity and permeability induction in respiratory epithelial cell layers in vitro . J Control Release. 2009;136(2):110–116. doi: 10.1016/j.jconrel.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 169.Yang C, Li H, Goh SH, et al. Cationic star polymers consisting of [alpha]-cyclodextrin core and oligoethylenimine arms as nonviral gene delivery vectors. Biomaterials. 2007;28(21):3245–3254. doi: 10.1016/j.biomaterials.2007.03.033. [DOI] [PubMed] [Google Scholar]
- 170.Cryan S-A. Cell transfection with polycationic cyclodextrin vectors. Eur J Pharm Sci. 2004;21(5):625–633. doi: 10.1016/j.ejps.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 171.Gonzalez H, Hwang SJ, Davis ME. New class of polymers for the delivery of macromolecular therapeutics. Bioconjug Chem. 1999;10(6):1068–1074. doi: 10.1021/bc990072j. [DOI] [PubMed] [Google Scholar]
- 172.Srinivasachari S, Fichter KM, Reineke TM. Polycationic b-cyclodextrin “click clusters”: monodisperse and versatile scaffolds for nucleic acid delivery. J Am Chem Soc. 2008;130:4618–4627. doi: 10.1021/ja074597v. [DOI] [PubMed] [Google Scholar]
- 173.Srinivasachari S, Reineke TM. Versatile supramolecular pDNA vehicles via “click polymerization” of [beta]-cyclodextrin with oligoethyleneamines. Biomaterials. 2009;30(5):928–938. doi: 10.1016/j.biomaterials.2008.09.067. [DOI] [PubMed] [Google Scholar]
- 174.Hu-Lieskovan S, Heidel JD, Bartlett DW, Davis ME, Triche TJ. Sequencespecific knockdown of EWS-EFI1 by targeted, nonviral delivery of small interfering RNA inhibits tumor growth in a murine model of metastatic Ewing’s sarcoma. Cancer Res. 2005;65:8984–8992. doi: 10.1158/0008-5472.CAN-05-0565. [DOI] [PubMed] [Google Scholar]
- 175.Pun SH, Davis ME. Development of a nonviral gene delivery vehicle for systemic application. Bioconjug Chem. 2002;13:630–639. doi: 10.1021/bc0155768. [DOI] [PubMed] [Google Scholar]
- 176.Pun SH, Tack F, Bellocq NC, Cheng J, Grubbs BH, Jensen GS, Davis ME, Brewster M, Janicot M, Janssens B, Floren W, Bakker A. Targeted delivery of RNA-cleaving DNA enzyme (DNAzyme) to tumor tissue by transferrin-modifed, cyclodextrin-based particles. Cancer Biol Ther. 2004;3(7):641–650. doi: 10.4161/cbt.3.7.918. [DOI] [PubMed] [Google Scholar]
- 177.Heidel JD, Yu ZP, Liu JYC, et al. Administration in non-human primates of escalating intravenous doses of targeted nanoparticles containing ribonucleotide reductase subunit M2 siRNA. Proc Natl Acad Sci USA. 2007;104(14):5715–5721. doi: 10.1073/pnas.0701458104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Davis ME. The first targeted delivery of siRNA in humans via a self-assembling, cyclodextrin polymer-based nanoparticle: from concept to clinic. Mol Pharm. 2009;6(3):659–668. doi: 10.1021/mp900015y. [DOI] [PubMed] [Google Scholar]
- 179.Zanta M-A, Boussif O, Adib A, Behr J-P. In vitro gene delivery to hepatocytes with galactosylated polyethylenimine. Bioconjug Chem. 1997;8:839–844. doi: 10.1021/bc970098f. [DOI] [PubMed] [Google Scholar]
- 180.Kunath K, von Harpe A, Fischer D, et al. Galactose-PEI-DNA complexes for targeted gene delivery: degree of substitution affects complex size and transfection efficiency. J Control Rel. 2003;88(1):159–172. doi: 10.1016/s0168-3659(02)00458-3. [DOI] [PubMed] [Google Scholar]
- 181.Nishikawa M, Yamauchi M, Morimoto K, et al. Hepatocyte-targeted in vivo gene expression by intravenous injection of plasmid DNA complexed with synthetic multifunctional gene delivery system. Gene Ther. 2000;7(7):548–555. doi: 10.1038/sj.gt.3301140. [DOI] [PubMed] [Google Scholar]
- 182.Chen C-p, Kim J-s, Liu D, et al. Synthetic PEGylated glycoproteins and their utility in gene delivery. Bioconjug Chem. 2007;18(2):371–378. doi: 10.1021/bc060229p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Rozema DB, Lewis DL, Wakefield DH, et al. Dynamic polyconjugates for targeted in vivo delivery of siRNA to hepatocytes. Proc Natl Acad Sci USA. 2007;104(32):12982–12987. doi: 10.1073/pnas.0703778104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Weiss SI, Sieverling N, Niclasen M, et al. Uronic acids functionalized polyethyleneimine (PEI)-polyethyleneglycol (PEG)-graft-copolymers as novel synthetic gene carriers. Biomaterials. 2006;27(10):2302–2312. doi: 10.1016/j.biomaterials.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 185.Diebold SS, Kursa M, Wagner E, Cotten M, Zenke M. Mannose polyethylenimine conjugates for targeted DNA delivery into dendritic cells. J Biol Chem. 1999;274:19087–19094. doi: 10.1074/jbc.274.27.19087. [DOI] [PubMed] [Google Scholar]
- 186.Reineke TM. Poly(glycoamidoamine)s: cationic glycopolymers for DNA delivery. J Polym Sci A Polym Chem. 2006;44(24):6895–6908. [Google Scholar]