Abstract
BACKGROUND
Lipolysis regulates energy homeostasis through the hydrolysis of intracellular triglycerides and the release of fatty acids for use as energy substrates or lipid mediators in cellular processes. Genes encoding proteins that regulate energy homeostasis through lipolysis are thus likely to play an important role in determining susceptibility to metabolic disorders.
METHODS
We sequenced 12 lipolytic-pathway genes in Old Order Amish participants whose fasting serum triglyceride levels were at the extremes of the distribution and identified a novel 19-bp frameshift deletion in exon 9 of LIPE, encoding hormone-sensitive lipase (HSL), a key enzyme for lipolysis. We genotyped the deletion in DNA from 2738 Amish participants and performed association analyses to determine the effects of the deletion on metabolic traits. We also obtained biopsy specimens of abdominal subcutaneous adipose tissue from 2 study participants who were homozygous for the deletion (DD genotype), 10 who were heterozygous (ID genotype), and 7 who were noncarriers (II genotype) for assessment of adipose histologic characteristics, lipolysis, enzyme activity, cytokine release, and messenger RNA (mRNA) and protein levels.
RESULTS
Carriers of the mutation had dyslipidemia, hepatic steatosis, systemic insulin resistance, and diabetes. In adipose tissue from study participants with the DD genotype, the mutation resulted in the absence of HSL protein, small adipocytes, impaired lipolysis, insulin resistance, and inflammation. Transcription factors responsive to peroxisome-proliferator–activated receptor γ (PPAR-γ) and downstream target genes were down-regulated in adipose tissue from participants with the DD genotype, altering the regulation of pathways influencing adipogenesis, insulin sensitivity, and lipid metabolism.
CONCLUSIONS
These findings indicate the physiological significance of HSL in adipocyte function and the regulation of systemic lipid and glucose homeostasis and underscore the severe metabolic consequences of impaired lipolysis. (Funded by the National Institutes of Health and others).
Both excess and insufficient storage of lipids in white adipose tissue are associated with dyslipidemia, insulin resistance, and an increased risk of type 2 diabetes; thus, maintaining normal white-adipose-tissue function is critical for whole-body insulin sensitivity and energy homeostasis.1 Hormone-sensitive lipase (HSL), encoded by LIPE, is an intracellular lipase with many neutral lipid substrates, including triglycerides, diglycerides, monoglycerides, cholesterol esters, and retinyl esters. HSL is ubiquitously expressed and is a key player in lipolysis in adipocytes, steroidogenesis, and spermatogenesis.2
To elucidate the genetic underpinnings of lipid metabolism and white-adipose-tissue function, we sequenced lipolytic-pathway genes in Old Order Amish participants, identified a frameshift mutation in LIPE, and characterized human carriers of this mutation, both systemically and at the level of the adipocyte.
METHODS
We sequenced 12 lipolytic-pathway genes in 24 Old Order Amish study participants whose fasting serum triglyceride levels were at the extremes of the distribution (lower extreme: median, 24 mg per deciliter [range, 14 to 25]; upper extreme: median, 298 mg per deciliter [range, 273 to 608]) and identified a 19-bp deletion in exon 9 of LIPE in one study participant whose triglyceride level was at the upper extreme. We genotyped the deletion in DNA from 2738 study participants enrolled in the Amish Complex Disease Research Program (ACDRP) and carried out tests of association to determine the effect of the deletion on metabolic traits. Biopsy specimens of abdominal subcutaneous white adipose tissue were obtained from 2 study participants who were homozygous for the deletion (DD genotype), 10 who were heterozygous (ID genotype), and 7 who were noncarriers (II genotype) for assessment of adipose histologic characteristics, lipolysis, enzyme activity, cytokine release, and messenger RNA (mRNA) and protein levels. All ACDRP study participants provided written informed consent.
RESULTS
IDENTIFICATION OF THE LIPE MUTATION
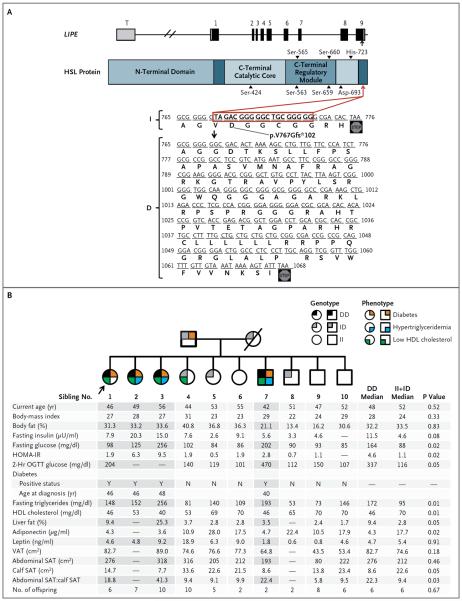
We identified a 19-bp frameshift deletion in exon 9 of LIPE (RefSeq NM_005357: c.2300_2318del; p.V767Gfs⋆102) (Fig. 1A). Of the 2738 participants in the ACDRP study, 140 were heterozygous for the deletion (ID genotype) and 1 was homozygous (DD genotype); 5.1% of Amish persons carry the D allele, as compared with 0.2% of non-Amish persons of European descent. Recruitment of family members of the proband with the DD genotype resulted in the identification of 3 additional DD homozygotes among her 9 siblings (Fig. 1B).
Figure 1 (facing page). Loss-of-Function Mutation in LIPE with Pedigree Showing Transmission of the Mutation and Metabolic Characteristics.
Hormone-sensitive lipase (HSL) (775 amino acids) is encoded by exons 1 through 9 of LIPE (Panel A). Black shading denotes the exonic coding region, with white representing the 5′- and 3′-untranslated regions. The exon T (in gray) is included only in the testis-specific RNA isoform, which encodes a 1076-amino-acid protein in frame with the adipose-tissue isoform. The arrow indicates the location of the frameshift mutation resulting from a 19-bp deletion in exon 9 (RefSeq NM_005357: c.2300_2318del; p.V767Gfs*102). The mutation is predicted to delete 6 amino acids, alter the sequence of 2 amino acids, and add an additional 90 amino acids to the C-terminal of the protein (I denotes the non mutated allele, and D the mutated allele). The pedigree (Panel B) shows transmission of the HSL mutation in an Amish family. Squares indicate male family members, circles female family members, and the slash a deceased family member. Of the 10 siblings, 4 were identified as homozygous for the mutation. Sibling 1 (arrow) was the proband (the first identified DD homozygote). The table below the pedigree shows individual values for metabolic traits. P values refer to the comparison of the traits among the siblings with the use of the Wilcoxon or Kruskal–Wallis rank-sum test. Sibling 3 is being treated by her physician with glipizide and insulin glargine. Sibling 7 is being treated by his physician with metformin. Sibling 1 and Sibling 2 did not report taking any medications to treat diabetes or dyslipidemia at their most recent visits to the Amish Research Clinic. Sibling 4 has impaired glucose tolerance. Diabetes was defined by a fasting plasma glucose level of at least 126 mg per deciliter (7.0 mmol per liter), a randomly obtained nonfasting plasma glucose level of at least 200 mg per deciliter (11.1 mmol per liter), a plasma glucose level of at least 200 mg per deciliter 2 hours after an oral glucose-tolerance test (OGTT), the use of insulin or prescription oral glucose-lowering agents, or a previous diagnosis of diabetes by a physician. Hypertriglyceridemia was defined as a triglyceride level of more than 150 mg per deciliter (1.7 mmol per liter). A low level of high-density lipoprotein (HDL) cholesterol was defined as less than 55 mg per deciliter (1.4 mmol per liter) for women and less than 45 mg per deciliter (1.2 mmol per liter) for men. Parental genotypes were inferred from the offspring. Phenotype information was unavailable for the parents, with the exception of a diagnosis of diabetes in the father. The mother died of leukemia at 56 years of age. The pedigree was constructed with the use of Fisher Family History.3 The body-mass index is the weight in kilograms divided by the square of the height in meters. The homeostasis model assessment for insulin resistance (HOMA-IR) was used to calculate insulin resistance, according to the following formula: (fasting glucose [in milligrams per deciliter] × fasting insulin [in microunits per milliliter]) ÷ 405. Higher values indicate greater insulin resistance. To convert the values for insulin to picomoles per liter, multiply by 6.945. To convert the values for glucose to millimoles per liter, multiply by 0.05551. To convert the values for triglycerides to millimoles per liter, multiply by 0.01129. To convert the values for cholesterol to millimoles per liter, multiply by 0.02586. SAT denotes subcutaneous adipose tissue, and VAT visceral adipose tissue.
EFFECTS OF THE LIPE MUTATION ON METABOLIC TRAITS
Demographic and clinical characteristics of the study participants according to LIPE genotype are shown in Table 1. Carriers of the D allele, as compared with noncarriers, had higher serum triglyceride levels, hepatic fat content, and fasting insulin levels and lower levels of high-density lipoprotein (HDL) cholesterol. In participants without diabetes who completed an oral glucose-tolerance test, the area under the glucose curve and the area under the insulin curve were higher in participants with the ID genotype than in those with the II genotype (Fig. S1 in the Supplementary Appendix, available with the full text of this article at NEJM.org). Heterozygotes had a risk of type 2 diabetes that was 1.8 times as high as the risk among noncarriers (P = 0.02), despite similar body-mass index, and all four participants with the DD genotype received a diagnosis of type 2 diabetes before 50 years of age. In a subgroup of 52 women matched for age and percentage of body fat, assessment of regional fat by means of dual-energy x-ray absorptiometry showed that the 3 women with the DD genotype had a modest decrease in lower-extremity fat as compared with the 49 women with the II or ID genotype (Table S1 in the Supplementary Appendix).
Table 1.
Demographic and Clinical Characteristics of the Study Participants, According to LIPE Deletion Genotype.*
| Characteristic | Genotype | P Value† | ||
|---|---|---|---|---|
| II | ID | DD | ||
| Age | ||||
| No. of participants | 2597 | 140 | 1 | |
| Mean (yr) | 49.9±16.6 | 51.1±16.1 | 39‡ | |
| Sex | ||||
| No. of participants | 2597 | 140 | 1 | |
| Female (%) | 52.8 | 57.9 | 100 | |
| Body-mass index§ | ||||
| No. of participants | 2597 | 140 | 1 | |
| Mean | 27.2±5.1 | 28.0±5.2 | 25.6 | 0.21 |
| Triglycerides¶ | ||||
| No. of participants | 2597 | 140 | 1 | |
| Mean (mg/dl) | 84.6±59.6 | 109.9±71.0 | 145 | 0.003 |
| High-density lipoprotein cholesterol¶ | ||||
| No. of participants | 2597 | 140 | 1 | |
| Mean (mg/dl) | 55.2±15.0 | 49.0±13.3 | 46 | <0.001 |
| Low-density lipoprotein cholesterol¶ | ||||
| No. of participants | 2597 | 140 | 1 | |
| Mean (mg/dl) | 140±43 | 143±44 | 120 | 0.25 |
| Spleen-to-liver fat ratio∥ | ||||
| No. of participants | 837 | 35 | 1 | |
| Mean | 0.80±0.15 | 0.88±0.20 | 1.19 | <0.001 |
| Insulin** | ||||
| No. of participants | 2269 | 116 | 1 | |
| Mean (μU/ml) | 10.6±9.0 | 13.4±7.4 | 17.6 | <0.001 |
| Glucose** | ||||
| No. of participants | 2216 | 124 | 1 | |
| Mean (mg/dl) | 91.3±21.3 | 91.6±23.7 | 84.3 | 0.88 |
| HOMA-IR†† | ||||
| No. of participants | 2114 | 112 | 1 | |
| Mean | 2.5±2.4 | 3.2±2.5 | 3.7 | 0.005 |
| Diabetes | ||||
| No. of participants | 2582 | 140 | 1 | |
| Prevalence (%) | 6.8 | 11.4 | 100 | 0.02‡‡ |
| Systolic blood pressure | ||||
| No. of participants | 2594 | 138 | 1 | |
| Mean (mm Hg) | 121±17 | 123±17 | 113 | 0.14 |
| Diastolic blood pressure | ||||
| No. of participants | 2594 | 138 | 1 | |
| Mean (mm Hg) | 74±10 | 75±11 | 79 | 0.31 |
| Hypertension§§ | ||||
| No. of participants | 2594 | 138 | 1 | |
| Prevalence (%) | 9.8 | 10.1 | 0 | 0.89 |
Plus–minus values are means ±SD. To convert the values for triglycerides to millimoles per liter, multiply by 0.01129. To convert the values for cholesterol to millimoles per liter, multiply by 0.02586. To convert the values for insulin to picomoles per liter, multiply by 6.945. To convert the values for glucose to millimoles per liter, multiply by 0.05551.
P values are for the comparison of mean trait values among the II, ID, and DD genotype groups (additive genetic model) adjusted for age, sex, and family structure. Models that included the interaction between genotype and sex showed no significant sex-specific influence of genotype on any characteristic. P values for comparisons of participants with the II genotype and those with the ID genotype were similar to the additive P values. P = 0.52 for the chi-square test for fit to expectations of Hardy–Weinberg equilibrium.
Shown is the participant's age at the time of the study; the participant is currently 46 years of age.
The body-mass index is the weight in kilograms divided by the square of the height in meters.
Levels of triglycerides, high-density lipoprotein cholesterol, and low-density lipoprotein cholesterol were measured from blood collected after an overnight fast.
This is the ratio of the attenuation value in the spleen as compared with the attenuation value in the liver, as measured on electron-beam computed tomography. A lower attenuation value indicates more fat in the respective tissue; thus, a higher spleen-to-liver ratio indicates a higher fat content in the liver.
Log-transformed measures of serum insulin and glucose levels after an overnight fast were used for analysis. Untransformed means are presented in the table.
The homeostasis model assessment for insulin resistance (HOMA-IR) was used to calculate insulin resistance, according to the following formula: (fasting glucose [in milligrams per deciliter] × fasting insulin [in microunits per milliliter]) ÷ 405. Higher values indicate greater insulin resistance.
The odds ratio for the risk of type 2 diabetes among participants with the ID genotype as compared with the risk among participants with the II genotype was 1.80 (95% confidence interval, 1.02 to 3.02).
Hypertension was defined as a systolic pressure of 130 mm Hg or more or a diastolic pressure of 85 mm Hg or more, according to the criteria for the metabolic syndrome of the Third Adult Treatment Panel (ATP III) of the National Cholesterol Education Program.
Further evaluation of the proband and her siblings (Fig. 1B) showed that carriers of the D allele (and the homozygotes in particular) had higher triglyceride and insulin levels and lower HDL cholesterol and serum adiponectin levels than did noncarriers, findings that are consistent with population-based data. We observed the expected positive correlation between serum leptin levels and the percentage of body fat in homozygotes for the D allele. Magnetic resonance imaging showed a subtle redistribution of body fat (i.e., decreased lower-extremity fat and increased visceral fat) and — with the exception of the man with the DD genotype, who was lean and very physically active — increased hepatic fat in siblings with the DD genotype as compared with those with the II or ID genotype (Fig. 1B, and Fig. S2 in the Supplementary Appendix).
FUNCTIONAL CHARACTERIZATION OF THE LIPE FRAMESHIFT MUTATION
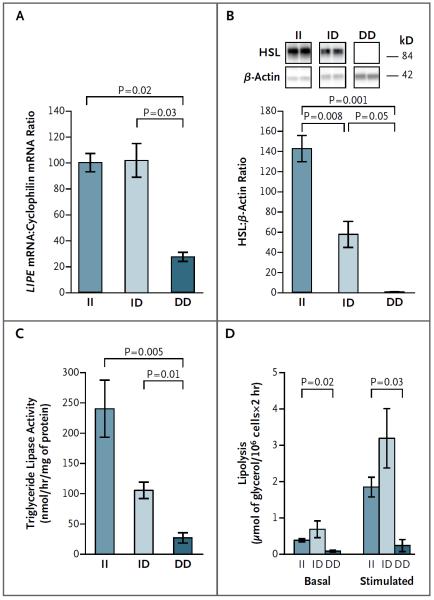
We confirmed the deletion mutation by reverse-transcriptase–polymerase-chain-reaction amplification of LIPE mRNA from abdominal subcutaneous white adipose tissue (Fig. S3 in the Supplementary Appendix). LIPE mRNA levels were lower in tissue samples from participants with the DD genotype than in tissue samples from those with the II genotype; participants with the ID genotype and those with the II genotype had similar LIPE mRNA levels (Fig. 2A). Western blot analysis of white-adipose-tissue extracts showed no detectable HSL protein in participants with the DD genotype and approximately a 50% reduction in HSL protein in participants with the ID genotype, as compared with participants who had the II genotype (Fig. 2B). In vitro overexpression of nonmutated and mutated LIPE showed no significant difference in mRNA expression (Fig. S4A in the Supplementary Appendix) but markedly reduced mutant protein levels (Fig. S4B in the Supplementary Appendix), findings that are consistent with the observed lack of in vivo protein expression and that suggest decreased translation or instability of the mutant HSL protein.
Figure 2. Effect of LIPE Mutation on HSL Protein and Function in Adipose Tissue.
From extracts of abdominal subcutaneous adipose tissue, LIPE messenger RNA (mRNA) levels (Panel A) were measured by means of quantitative real-time polymerase chain reaction and normalized by comparing with cyclophilin mRNA levels. HSL protein levels (Panel B) were measured by means of Western blot analysis with the use of an N-terminal human antibody in which the epitope lies outside the deletion region and were normalized by comparing with β-actin protein levels. In vitro hydrolytic activity against triolein (Panel C) was measured in adipose-tissue homogenates. Phenylisopropyl adenosine–suppressed (basal) and isoproterenol-stimulated lipolysis (Panel D) were measured in isolated adipocytes and expressed per fat-cell weight. Data are means ±SE. A total of 7 study participants had the II genotype, 10 had the ID genotype, and 2 had the DD genotype.
EFFECTS OF THE LIPE MUTATION ON ADIPOSE TISSUE
Dose-dependent reductions in triglyceride lipase activity (Fig. 2C) and cholesterol ester hydrolase activity (Fig. S5A in the Supplementary Appendix) were observed in white adipose tissue from D allele carriers. The content of diglycerides (Fig. S5B in the Supplementary Appendix) and retinyl esters (Fig. S5C in the Supplementary Appendix) in white-adipose-tissue lipid extracts was increased in participants with the DD genotype.
Large adipocytes might be expected in participants with a defect in triglyceride hydrolysis, but adipocyte diameter was significantly smaller in samples of abdominal subcutaneous adipose tissue from participants with the DD genotype than in samples from participants with the II genotype (Fig. S6 in the Supplementary Appendix). The absence of HSL protein was associated with decreased basal lipolysis; after stimulation with isoproterenol, lipolysis in adipocytes increased by a factor of only 2 in participants with the DD genotype, as compared with an increase by a factor of 5 in participants with the ID or II genotype (Fig. 2D). Levels of PNPLA2, encoding adipose triglyceride lipase (ATGL), which catalyzes the first step in the lipolytic pathway, 4 were reduced in adipocytes from participants with the DD genotype as compared with adipocytes from participants with the II genotype (Fig. S7A in the Supplementary Appendix), as were levels of the ATGL protein (Fig. S7B in the Supplementary Appendix).
Sensitivity to the antilipolytic effect of insulin was lower in adipocytes from participants with the DD or ID genotype than in adipocytes from participants with the II genotype; the maximal response to insulin was reduced by approximately 50% in DD homozygotes as compared with II homozygotes (Fig. S8A in the Supplementary Appendix). Protein levels of insulin receptor and insulin receptor substrate 1 were also lower in adipocytes from participants with the DD genotype than in adipocytes from participants with the II genotype (Fig. S8B and S8C in the Supplementary Appendix). In addition, expression of genes known to be up-regulated with insulin resistance (e.g., FOXO1 and FABP5) was increased in white adipose tissue from DD homozygotes (Fig. S10 in the Supplementary Appendix).
The absence of HSL was associated with histologic abnormalities in white adipose tissue and an increase in macrophage infiltration (a hallmark of inflammation) (Fig. S9A in the Supplementary Appendix). Increased mRNA levels of the macrophage chemokine MCP1 (Fig. S9B in the Supplementary Appendix) were consistent with the observed increase in macrophage infiltration. Levels of mRNA encoding interleukin-6 in white adipose tissue (Fig. S9B in the Supplementary Appendix) and levels of interleukin-6 released from white adipose tissue in culture (Fig. S9C in the Supplementary Appendix) were increased in participants with the DD genotype as compared with those with the II genotype.
We observed no significant differences in mRNA levels encoding peroxisome-proliferator–activated receptor γ (PPAR-γ, encoded by PPARG) among the genotype groups; we did, however, observe a comparative decrease in levels of RXRA mRNA, encoding retinoid X receptor α (RXR-α), and in levels of mRNA encoding other key nuclear receptors that regulate lipid and cholesterol metabolism in white adipose tissue from participants with the DD genotype (Fig. S10A in the Supplementary Appendix). As expected, down-regulation of critical transcription regulators of terminal differentiation resulted in marked reduction in the expression of downstream target genes involved in the uptake of free fatty acids, the synthesis of free fatty acids and triglycerides, lipolysis, and glucose transport (Fig. S10B in the Supplementary Appendix).
DISCUSSION
We identified a loss-of-function mutation affecting HSL that results in haploinsufficiency of the protein in heterozygous study participants and the complete absence of the protein in homozygous participants. Our data indicate that HSL is the predominant, if not exclusive, mediator of the hydrolysis of diglycerides, cholesterol esters, and retinyl esters in human white adipose tissue. The mutation was associated with an increased risk of type 2 diabetes in heterozygotes and with the development of diabetes early in adulthood in homozygotes, with accompanying dyslipidemia, hepatic steatosis, and insulin resistance. In addition to defective lipolysis, the absence of HSL resulted in small, inflamed, insulin-resistant adipocytes — evidence of impaired adipogenesis, impaired adipocyte function, or both.
Phenotypic characterization of carriers of the LIPE deletion revealed that HSL plays a previously unsuspected role in metabolic health. Lower lipolytic rates would be expected to be associated with increased adipose stores5; however, we observed reduced white-adipose-tissue storage. In addition, we observed increased white-adipose-tissue inflammation. Although the sequential contribution of white-adipose-tissue dysfunction and white-adipose-tissue inflammation to metabolic disease has been difficult to establish,6 our data indicate that inflammation is secondary to the primary metabolic defect in white adipose tissue, a phenomenon evident in the genetic disruption of other genes involved in adipocyte lipid-droplet storage (e.g., PLIN1 and CIDEC).7,8
Given our observation of dysmorphic and dysfunctional white adipose tissue in study participants who were homozygous for the LIPE deletion, we hypothesized that HSL deficiency might result in decreased activation of PPAR-γ–regulated pathways in white adipose tissue. We observed no significant difference in PPARG mRNA levels among the genotype groups, but in participants with the DD genotype we observed a marked reduction in the expression of RXRA (RXR-α is an obligate heterodimer partner of PPAR-γ); other critical transcription regulators of terminal differentiation; and genes involved in lipid and glucose metabolic pathways, including the uptake of free fatty acids, the synthesis of free fatty acids and triglycerides, lipolysis, and glucose transport. ATGL is an established PPAR-γ target,9 and its decreased expression in LIPE-null adipocytes probably contributes to the global lipolytic defect but is unlikely to contribute to insulin resistance and type 2 diabetes, because human PNPLA2 mutations that affect ATGL function are not associated with obesity and type 2 diabetes.10 The distinctive gene-expression profile of white adipose tissue in participants with the DD genotype supports our finding that the absence of HSL leads, through altered transcriptional regulation, to impaired adipogenesis, the inability to maintain mature adipocyte functions, or both.
Much of our understanding of the biologic functions of HSL is based on HSL-knockout mouse models. Like the participants with the DD genotype whom we describe here, HSL-knockout mice have normal weight; adipocytes that vary in size, with an accumulation of diglycerides, cholesterol esters, and retinyl esters; increased necrotic cell death; and macrophage infiltration. However, the knockout mice have few or no systemic abnormalities in lipid or glucose homeostasis.11–14 In contrast, haploinsufficiency or absence of HSL in our study participants was characterized by a strong systemic phenotype, with ectopic lipid storage, dyslipidemia, insulin resistance, and diabetes. Impaired lipid storage in human adipocytes probably contributes to ectopic triglyceride accumulation in the liver, which may then contribute to dyslipidemia and systemic insulin resistance. The absence of ectopic lipid storage in HSL-knockout mice suggests an alternative, yet uncharacterized, compensatory mechanism for handling excess lipids. Serum lipid levels also differ markedly between mice and humans: HSL-knockout mice have lower fasting triglyceride and higher HDL cholesterol levels than mice without the deletion, whereas the study participants who were homozygous for the deletion mutation had higher fasting triglyceride levels and lower HDL cholesterol levels than noncarriers. Established interspecies differences in lipoprotein metabolism may contribute to these reversed lipid-level findings. Such phenotypic differences support a key role for HSL in the regulation of lipid and glucose metabolism in humans that is absent or different in mouse models. These differences are consistent with the findings for other lipid-storage regulatory genes, such as PPARG, PLIN1, and CIDEC.7,8,15–18
Despite the relatively subtle metabolic phenotype in HSL-knockout mice, gene-expression profiles in these mice support a regulatory role for HSL in adipogenesis and adipocyte function.11–14,19,20 One study showed normalization of serum biochemical values, white-adipose-tissue storage, and gene expression after treatment of HSL-knockout mice with a PPAR-γ agonist.21 Another study showed restoration of retinyl ester hydrolase activity, partial restoration of white-adipose-tissue stores, and normalization of gene expression after treatment with dietary retinoic acid,22 findings that suggest that HSL may be responsible for the release of endogenous ligands for activation of PPAR-γ, RXR-α, or both. Furthermore, ATGL-mediated lipid-droplet hydrolysis of cellular triglycerides was shown to be important in generating essential lipid mediators for PPAR-α and PPAR-δ activation in nonadipose tissues in ATGL-knockout mice.23 It seems that both ATGL and HSL mediate the delivery of specific PPAR and RXR ligands in adipose and nonadipose tissues, despite differences in substrate affinities and specificities.
Although we observed systemic traits such as hepatic steatosis, hypertriglyceridemia, and insulin resistance in study participants who were heterozygous for the LIPE deletion, we did not observe features of marked adipose-tissue dysfunction; these findings suggest that in humans, HSL may have key metabolic functions in nonadipose tissues (e.g., skeletal muscle, the liver, and beta cells) that have not yet been identified. We also note that male HSL-knockout mice are sterile, whereas the male study participant who was homozygous for the LIPE deletion had two children. Further evaluation is needed to determine whether there are differential effects of this mutation on spermatogenesis in male mice and in men.
In summary, we identified a deletion in LIPE, which, when homozygous, results in the complete absence of the HSL protein. Although a defect in an enzyme that is critical for lipolysis would be expected to result in excess lipid storage, we observed the opposite, providing evidence that HSL plays a key role in maintaining adipogenesis and adipocyte function. Furthermore, our results showed marked down-regulation of PPAR-γ–dependent genes that are responsible for adipogenesis and the maintenance of adipocyte function and also showed secondary inflammation, insulin resistance, and impaired glucose and lipid metabolism. These findings are consistent with the hypothesis that HSL deficiency in humans results in decreased production of endogenous ligands for PPAR-γ, RXR-α, or both. Although PPAR-γ agonists are effective agents for the treatment of type 2 diabetes, they can have untoward side effects.24 Our results suggest that activation of HSL with consequent generation of endogenous ligands for PPAR-γ, RXR-α, or both may serve as an alternative means of reversing dyslipidemia and glucose intolerance in the metabolic syndrome and type 2 diabetes. Overall, our findings reveal the physiological significance of HSL in adipocyte function and the regulation of systemic lipid and glucose homeostasis and underscore the severe metabolic consequences of impaired lipolysis.
Supplementary Material
Acknowledgments
Supported by grants from the National Institutes of Health (R01DK075017, R01DK74828, U01HL072515, R01DK54261, U01HL84756, U01GM074518, R01HL104193, and R01HL088119), the American Heart Association (11PRE7610008, to Dr. Albert), the University of Maryland Multidisciplinary Clinical Research Career Development Program (NIH K12RR023250, to Dr. Damcott), the Mid-Atlantic Nutrition and Obesity Research Center (NIH P30DK072488), the Baltimore Diabetes Research Center (NIH P60DK079637), the American Diabetes Association, the U.S. Department of Agriculture National Institute of Food and Agriculture (National Research Initiative Competitive Grant 2007-35205-17883), and the Geriatric Research Education and Clinical Center, Baltimore Veterans Affairs (VA) Medical Center, and by a VA Research Career Scientist Award (to Dr. Ryan).
We thank our Amish liaisons and Amish Research Clinic staff, as well as the Amish community for cooperation and support; Cecilia Holm for the hormone-sensitive lipase antibody; Michael Quon and Xiao Jian Sun for the insulin receptor and insulin receptor substrate 1 antibodies; Richa Agarwala and Alejandro Schäffer for assistance in pedigree construction; and Kathy Ryan for data management.
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
REFERENCES
- 1.Frayn KN. Adipose tissue as a buffer for daily lipid flux. Diabetologia. 2002;45:1201–10. doi: 10.1007/s00125-002-0873-y. [DOI] [PubMed] [Google Scholar]
- 2.Haemmerle G, Zimmermann R, Zechner R. Letting lipids go: hormone-sensitive lipase. Curr Opin Lipidol. 2003;14:289–97. doi: 10.1097/00041433-200306000-00009. [DOI] [PubMed] [Google Scholar]
- 3.Beiler K. Fisher family history: descendants and history of Christian Fisher (1757–1838) Eby's Quality Printing; Ronks, PA: 1988. [Google Scholar]
- 4.Zimmermann R, Strauss JG, Haemmerle G, et al. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 2004;306:1383–6. doi: 10.1126/science.1100747. [DOI] [PubMed] [Google Scholar]
- 5.Kolditz CI, Langin D. Adipose tissue lipolysis. Curr Opin Clin Nutr Metab Care. 2010;13:377–81. doi: 10.1097/MCO.0b013e32833bed6a. [DOI] [PubMed] [Google Scholar]
- 6.Berg AH, Scherer PE. Adipose tissue, inflammation, and cardiovascular disease. Circ Res. 2005;96:939–49. doi: 10.1161/01.RES.0000163635.62927.34. [DOI] [PubMed] [Google Scholar]
- 7.Rubio-Cabezas O, Puri V, Murano I, et al. Partial lipodystrophy and insulin resistant diabetes in a patient with a homozygous nonsense mutation in CIDEC. EMBO Mol Med. 2009;1:280–7. doi: 10.1002/emmm.200900037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gandotra S, Le Dour C, Bottomley W, et al. Perilipin deficiency and autosomal dominant partial lipodystrophy. N Engl J Med. 2011;364:740–8. doi: 10.1056/NEJMoa1007487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kershaw EE, Schupp M, Guan HP, Gardner NP, Lazar MA, Flier JS. PPAR-gamma regulates adipose triglyceride lipase in adipocytes in vitro and in vivo. Am J Physiol Endocrinol Metab. 2007;293(6):E1736–E1745. doi: 10.1152/ajpendo.00122.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischer J, Lefèvre C, Morava E, et al. The gene encoding adipose triglyceride lipase (PNPLA2) is mutated in neutral lipid storage disease with myopathy. Nat Genet. 2007;39:28–30. doi: 10.1038/ng1951. [DOI] [PubMed] [Google Scholar]
- 11.Osuga J, Ishibashi S, Oka T, et al. Targeted disruption of hormone-sensitive lipase results in male sterility and adipocyte hypertrophy, but not in obesity. Proc Natl Acad Sci U S A. 2000;97:787–92. doi: 10.1073/pnas.97.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang SP, Laurin N, Himms-Hagen J, et al. The adipose tissue phenotype of hormone-sensitive lipase deficiency in mice. Obes Res. 2001;9:119–28. doi: 10.1038/oby.2001.15. [DOI] [PubMed] [Google Scholar]
- 13.Haemmerle G, Zimmermann R, Hayn M, et al. Hormone-sensitive lipase deficiency in mice causes diglyceride accumulation in adipose tissue, muscle, and testis. J Biol Chem. 2002;277:4806–15. doi: 10.1074/jbc.M110355200. [DOI] [PubMed] [Google Scholar]
- 14.Grober J, Lucas S, Sörhede-Winzell M, et al. Hormone-sensitive lipase is a cholesterol esterase of the intestinal mucosa. J Biol Chem. 2003;278:6510–5. doi: 10.1074/jbc.M208513200. [DOI] [PubMed] [Google Scholar]
- 15.Toh SY, Gong J, Du G, et al. Up-regulation of mitochondrial activity and acquirement of brown adipose tissue-like property in the white adipose tissue of fsp27 deficient mice. PLoS One. 2008;3(8):e2890. doi: 10.1371/journal.pone.0002890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tansey JT, Sztalryd C, Gruia-Gray J, et al. Perilipin ablation results in a lean mouse with aberrant adipocyte lipolysis, enhanced leptin production, and resistance to diet-induced obesity. Proc Natl Acad Sci U S A. 2001;98:6494–9. doi: 10.1073/pnas.101042998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Semple RK, Chatterjee VK, O'Rahilly S. PPAR gamma and human metabolic disease. J Clin Invest. 2006;116:581–9. doi: 10.1172/JCI28003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gray SL, Dalla Nora E, Vidal-Puig AJ. Mouse models of PPAR-gamma deficiency: dissecting PPAR-gamma's role in metabolic homoeostasis. Biochem Soc Trans. 2005;33:1053–8. doi: 10.1042/BST0331053. [DOI] [PubMed] [Google Scholar]
- 19.Haemmerle G, Zimmermann R, Strauss JG, et al. Hormone-sensitive lipase deficiency in mice changes the plasma lipid profile by affecting the tissue-specific expression pattern of lipoprotein lipase in adipose tissue and muscle. J Biol Chem. 2002;277:12946–52. doi: 10.1074/jbc.M108640200. [DOI] [PubMed] [Google Scholar]
- 20.Harada K, Shen WJ, Patel S, et al. Resistance to high-fat diet-induced obesity and altered expression of adipose-specific genes in HSL-deficient mice. Am J Physiol Endocrinol Metab. 2003;285(6):E1182–E1195. doi: 10.1152/ajpendo.00259.2003. [DOI] [PubMed] [Google Scholar]
- 21.Shen WJ, Yu Z, Patel S, Jue D, Liu LF, Kraemer FB. Hormone-sensitive lipase modulates adipose metabolism through PPARγ. Biochim Biophys Acta. 2011;1811:9–16. doi: 10.1016/j.bbalip.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ström K, Gundersen TE, Hansson O, et al. Hormone-sensitive lipase (HSL) is also a retinyl ester hydrolase: evidence from mice lacking HSL. FASEB J. 2009;23:2307–16. doi: 10.1096/fj.08-120923. [DOI] [PubMed] [Google Scholar]
- 23.Haemmerle G, Moustafa T, Woelkart G, et al. ATGL-mediated fat catabolism regulates cardiac mitochondrial function via PPAR-α and PGC-1. Nat Med. 2011;17:1076–85. doi: 10.1038/nm.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rizos CV, Elisaf MS, Mikhailidis DP, Liberopoulos EN. How safe is the use of thiazolidinediones in clinical practice? Expert Opin Drug Saf. 2009;8:15–32. doi: 10.1517/14740330802597821. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.