Abstract
On the strengths of forward genetics and embryology the zebrafish Danio rerio has become an ideal system for the study of early vertebrate development. However, additional tools will be needed to perform more sophisticated analyses and to successfully carry this model into new areas of study such as adult physiology, cancer, and aging. As improved tools make transgenesis more and more efficient, the stage has been set for precise modification of the zebrafish genome such as are done in other model organisms. Genome engineering strategies employing site-specific recombinase (SSR) systems such as Cre/lox and Flp/FRT have become invaluable to the study of gene function in the mouse and Drosophila and are now being exploited in zebrafish as well. My laboratory has begun to use another such SSR, the integrase encoded by the Streptomyces bacteriophage phiC31, for manipulation of the zebrafish genome. The phiC31 integrase promotes recombination between an attachment site in the phage (attP) and another on the bacterial chromosome (attB). Here I describe strategies using the phiC31 integrase to mediate recombination of transgenes containing attP and attB sites in cis to excise elements with spatial and temporal specificity. The feasibility of the intramolecular recombination approach having been established, I discuss prospects for employing phiC31 integrase for intermolecular recombination, i.e. transgene integration at defined sites in the genome.
INTRODUCTION
The zebrafish as a continually expanding model system
In the last twenty years the zebrafish Danio rerio has become well-established as a model organism, particularly for the study of the genetics of early vertebrate development. In the future it is certain that it will become an even more widely used biomedical research model as it reach continues to extend into the such realms as cancer (Amatruda, et al. 2002), infectious disease (Davis, et al. 2002), physiology (Briggs. 2002), behavior (Fetcho, Liu. 1998), and aging (Kishi, et al. 2003). Forward genetic tools already exist and have been usefully exploited for the discovery of new genes and pathways. For example large scale ENU mutagenesis screens first reported 14 years ago produced hundreds of mutants (Driever, et al. 1996; Haffter, et al. 1996), and many of these loci have since been identified at the molecular level. The sequencing of the zebrafish genome is approaching completion (http://www.sanger.ac.uk/Projects/D_rerio/ ) and reverse genetics methods are available (Nasevicius, Ekker. 2000), but additional and more precise genomic tools will be needed to address problems in these new areas.
Transgenic technologies in zebrafish
The ability to generate transgenic lines, by which exogenous DNA can be stably transmitted from generation to generation, is fundamental to the usefulness of the zebrafish as it is for many other model systems. Beyond simple transgenesis is the means to make specific manipulations in the zebrafish genome. In recent years, more efficient methods for obtaining germline integration of foreign genes in zebrafish have been described (Grabher, Wittbrodt. 2008), and increasingly transgenic lines are appearing in the literature (see zfin.org for an up-to-date and searchable listing). For example, using the Tol2 transposon originally isolated from medaka, Kawakami et al (Kawakami, et al. 2004) reported germline transmission rates as high as 50%. Other high efficiency methods include a second transposon, Sleeping Beauty (Davidson, et al. 2003), I-Sce I meganuclease coinjection (Grabher, et al. 2004), and injection of pseudotyped retrovirus into blastula stage embryos (Chen, et al. 2002). Both lines expressing fluorescent markers to label particular cell populations, and to misexpress genes with spatial and temporal precision are needed to realize the full experimental potential of this organism.
Genome modification using site-specific recombinases
Site-specific recombinase approaches extend the usefulness of transgenes by offering a means for their manipulation after they are established in the genome. The Cre/lox and Flp/FRT are most commonly used in mice for the construction of conditional knockout alleles (reviewed in (Branda, Dymecki. 2004)), and for similar gene control and chromosomal rearrangement strategies in Drosophila (reviewed in (Bischof, Basler. 2008)). Both loxP and FRT sites comprise short inverted repeats around a core sequence, and recombination between two loxP sites or two FRT sites results in the exchange of sequences flanking each site but preservation of the loxP/FRT sites themselves, which may be substrates for additional reactions. Therefore, recombination between two loxP/FRT sites flanking a transgene in the presence of Cre/Flp causes the intervening sequence to be excised as a circular molecule. Reintegration is formally possible but thermodynamically highly unlikely. For this reason, Cre and Flp have proven to be very useful for deleting transgenes from chromosomes. While Cre and Flp can catalyze intermolecular recombination, as could occur between a loxP/FRT site on a plasmid and one on a host chromosome, this creates two loxP/FRT sites which can immediately recombine, reversing the integration event (although modifications have been devised to allow Cre and FLP to be used in this way; see Discussion).
The phiC31 bacteriophage uses a distinct integrase to catalyze directed recombination at sequence-specific sites in the Streptomyces genome (Smith, et al. 2010). The integrase of the phiC31 bacteriophage mediates recombination between two different sequences, the attP site in the phage genome and the attB site in the Streptomyces chromosome (Figure 1A). The integrase alone is sufficient for this reaction in the absence of other cofactors from the phage or from Streptomyces and can even catalyze recombination between attB and attP sites in vitro (Thorpe, Smith. 1998). Importantly, unlike Cre/lox and Flp/FRT, the attL and attR sites thus created cannot be acted upon by integrase alone, which requires additional, yet to be identified cofactors to reverse the reaction. Therefore, phiC31 integrase would appear to have great potential as a tool for both intra- and inter-molecular recombination strategies.
Figure 1.
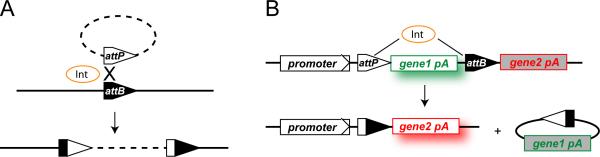
Mechanism of phiC31 integrase action. A) The integrase of the phiC31 Streptomyces bacteriophage (Int) catalyzes recombination between an attachment site in the phage genome (attP) and a site in the bacterial genome (attB). In eukaryotic applications, it has generally been found that recombination is more efficient when the attP site is provided by the host genome rather than by the donor plasmid. B) Generalized strategy for use of phic31 integrase (Int) for transgene excision. PhiC31 integrase (Int) can catalyze recombination between att sites in the same molecule; if these sites are in the same orientation the intervening sequenced will be excised. In this way, the integrase can be be used to inactivate a gene (gene1) and/or activate a second gene (gene2) in a tissue-restricted and/or temporally-regulated manner.
Although its natural environment is prokaryotic it was shown that phiC31 integrase could catalyze intramolecular recombination in human cells in culture (Groth, et al. 2000). Using an intramolecular recombination (excision) assay, minimal attB and attP sites were defined to be 34 basepairs and 39 basepairs, respectively. Integration was demonstrated in human and mouse cells of plasmids bearing attB sites but not attP sites (Thyagarajan, et al. 2001). Directed integration of an attB plasmid into endogenous loci, termed pseudo-attP sites, was observed to occur 10 to 20-fold more efficiently than random integration. In Drosophila, phiC31 integrase has been shown to efficiently recombine attB–bearing plasmids with transgenes containing attP sites (Groth, et al. 2004), and based on this elegant transgenesis strategies have been devised by a number of groups (Bateman, et al. 2006; Bischof, et al. 2007; Fish, et al. 2007; Huang, et al. 2009; Boy, et al. 2010).
Cre and Flp recombinases function in zebrafish
Several reports have now been made of the successful application of the Cre/lox system in zebrafish (Dong, Stuart. 2004; Langenau, et al. 2005; Pan, et al. 2005; Thummel, et al. 2005; Boniface, et al. 2009; Hans, et al. 2009; Hesselson, et al. 2009; Collins, et al. 2010; Jopling, et al. 2010; Kikuchi, et al. 2010; Seok, et al. 2010), and Flp recombinase as well appears to function as expected in the zebrafish embryo (Boniface, et al. 2009). As is beginning to become clear in the mouse and Drosophila, considerable power lies in the ability to combine more than one tool simultaneously (Branda, Dymecki. 2004; Huang, et al. 2009). Because they recognize different target sequences, phiC31 and the other recombinases can potentially be used in parallel to allow independent manipulation of more than one transgene in a single organism. Finally, although there have been reports of chromosomal aberrations resulting from stable expression of phiC31 integrase in human cells (Liu, et al. 2009), the potential genotoxicity of Cre is also well-known (Schmidt, et al. 2000; Loonstra, et al. 2001), and phiC31 may turn out to be a less toxic alternative. For these reasons, the development of phiC31 integrase technology should complement other site-specific recombinases as they are adapted for use in the zebrafish. I focus here on intermolecular recombination (i.e., transgene excision) using the phiC31 integrase; successful intermolecular recombination (transgene integration) has not yet been described in the literature, but practice and experience with the former should facilitate the latter advance.
RATIONALE
Phic31 integrase functions in zebrafish
Although native to Streptomyces bacteria, the integrase encoded by the phiC31 phage can also function in eukaryotic cells, including Drosophila (Groth, et al. 2004), mouse (Belteki, et al. 2003), and frog embryos (Allen, Weeks. 2005) human cells in culture (Groth, et al. 2000), as well as a variety of plant species (Lutz, et al. 2004; Khan, et al. 2005; Rubtsova, et al. 2008; Thomson, et al. 2010). It was therefore not surprising to find that this integrase could also function in zebrafish (Lister. 2010). While the native integrase was active in zebrafish embryos, we found using a plasmid-based reporter assay that a version of the integrase optimized for mouse codon usage (PhiC31o; (Raymond, Soriano. 2007)) gave over twice the frequency of recombination. Others have confirmed the basic utility of this approach (Lu, et al. 2010), but for simplicity I focus below on results from my own laboratory.
To observe recombination in living embryos, we constructed a reporter (XIpGbR) comprising 1) the Xenopus EF1a promoter and rabbit beta-globin intron, 2) a GFP open reading frame and SV40 polyadenylation signal flanked by an attP site and an attB site, and 3) a DsRed-Express open reading frame and polyadenylation signal. To reduce the chance that the att sites might interfere with expression of the GFP and DsRed ORFs, they were shortened to the minimal lengths found to retain activity in a bacterial assay. It was confirmed that this plasmid could be recombined in bacteria and in embryos if and only if phic31 integrase was present. We also confirmed in these transient transgenesis experiments that the GFP and DsRed cassettes were expressed appropriately. (Interestingly, a construct in which the relative position of the attP and attB sites was reversed still expressed GFP, and could be recombined precisely and efficiently, however following excision no expression of DsRed could be observed.)
A stable reporter line
A stable transgenic line was generated by co-injecting embryos with XIpGbR, in a Tol2 transposon backbone, along with Tol2 transposase mRNA. From multiple integrations a line (vc2) was selected that has shown strong GFP expression, but no detectable red fluorescence, for several generations. Injection of transgenic embryos with mRNA encoding the phiC31 integrase, however, produces widespread red fluorescence (Figure 2). Sequencing of PCR products from genomic DNA confirms precise excision of the GFP cassette. We have also constructed a red-to-green reporter and are currently attempting to establish lines with this (Figure 3). This reporter was assembled by Gateway cloning, described below.
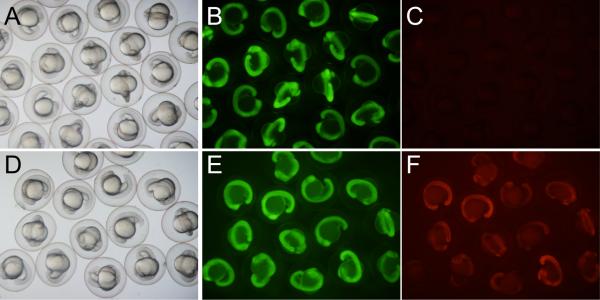
Figure 2.
Recombination in the green-to-red reporter strain, vc2. A)-C) uninjected embryos (shown at 18-somite stage) express GFP strongly (B) but do not express DsRed (C). D)-F) Injection of messenger RNA encoding codon-optimized phiC31 integrase (PhiC31o) induces widespread transgene recombination and expression of DsRed. (Due in part to significant maternal GFP expression, GFP signal is not seen to diminish.)
Figure 3.
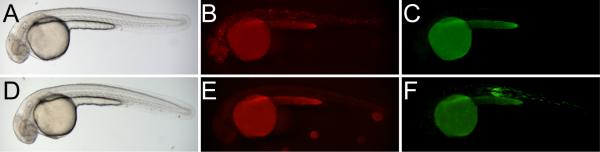
Demonstration of a red-to-green phiC31 integrase reporter generated by Gateway cloning. Wild-type embryos were injected with the plasmid pDestTol2pA2-XIpRbG alone (A-C) or with PhiC31o mRNA (D-F). In the absence of integrase activity mosaic DsRed expression (B) but no GFP expression (C) is observed. With addition of PhiC31o, only mosaic GFP expression (F) is seen.
The complementary arm to the development of transgenes flanked by attachment sites is the isolation of tissue-specific drivers, for expressing the recombinase in restricted patterns. We have begun to use the green-to-red reporter line to test candidate promoters for appropriateness as drivers in transient assays (Figure 4) prior to using them to generate stable lines. The reporter line can then be used to screen prospective founders.
Figure 4.
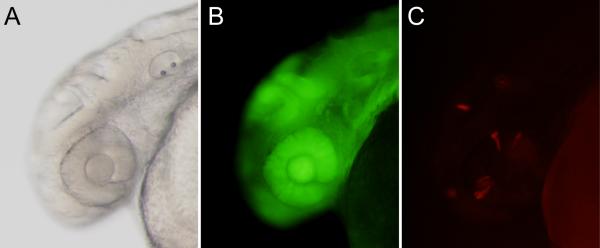
Tissue-specific recombination. A vc2 (green-to-red reporter) embryo injected with the plasmid pDestTol2pA2-mitfa0.9-PhiC31o-pA is shown at approximately 36 hours post-fertilization A) brightfield, B) near ubiquitous GFP expression, C) recombination and expression of DsRed occurs in a small subset of cells.
Conditional activation of phiC31 integrase
While spatial control of recombination may be obtained by tissue-specific expression of integrase, temporal control is another highly desirable feature. Temporal control of recombination in the Cre-lox system has been achieved by fusing the recombinase to a ligand binding domain variant of the estrogen receptor, rendering the chimeric protein inactive unless in the presence of 4-hydroxytamoxifen. It was reported that phiC31 integrase activity can also be regulated by fusion to the ER ligand-binding domain (Sharma, et al. 2008). We tested the inducibility of an analogous PhiC31o-estrogen receptor fusion protein in zebrafish. Embryos from the green-to-red reporter transgenic line were injected with mRNA encoding PhiC31o alone or as an N-terminal fusion to the estrogen receptor variant ERT2. Embryos treated with DMSO or 4-hydroxytamoxifen (4-OHT) beginning 60-90 minutes after fertilization were examined at approximately 30 hours post-fertilization for the presence of DsRed-expressing cells. Recombination was only observed with PhiC31o-ERT2 injection in the presence of 4-OHT (Figure 5), however the fraction of DsRed-positive cells was much lower with PhiC31o-ERT2 than with PhiC31o alone. The precise reasons for the reduced activity of the fusion protein have not yet been determined; however, these results indicate that improvements to this approach may be required to bring its efficiency up to that of constitutively-active integrase. The present version may still be useful for situations where mosaic recombination is desired or sufficient.
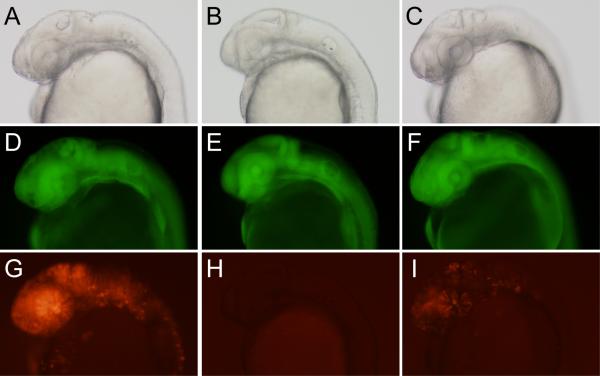
Figure 5.
Tamoxifen-dependent recombination. Individual vc2 green-to-red reporter embryos injected with mRNA for PhiC31o (A,D,G) or PhiC31o-ERT2 (B,C,E,F,H,I) are shown. With addition of DMSO alone (B,E,H) no DsRed expression is observed, while addition of 300 nM 4-hydroxytamoxifen (C,F,I) induces recombination and red fluorescence (I), although to a much lesser degree than non-chimeric PhiC31o integrase (G).
Gateway-compatible vectors
The use of multisite Gateway cloning technology has facilitated the development of complex vectors for zebrafish transgenesis. To take advantage of this modular approach, we have generated and tested a number of entry vectors for use with the Tol2 kit described by Kwan, et al.(Kwan, et al. 2007). A list of these vectors is given in Table 1. (It is important to note that although there is some shared terminology, the attachment sites and recombinases used in the Gateway system are derived from the lambda phage and are distinct from, and do not cross-react with, those of phiC31.) In addition to middle entry vectors, which can be combined with a promoter of choice to generate tissue-specific integrase “drivers”, we have constructed integrase targets by inserting the EF1α promoter/β-globin intron/attP-attB cassette as a unit into a 5’ entry vector. Versions include flanked GFP-polyA, DsRed-polyA, and polyA alone. These may thus be combined with a middle entry vector of choice (and 3’ entry and destination vectors) in a multi-site Gateway reaction to generate expression clones in which the middle ORF is only expressed after the construct is acted upon by the phiC31 integrase. An example of a red-to-green reporter made in this fashion is shown in Figure 3. (We also tried flanking GFP-polyA with attP and attB sites within a middle entry clone, in combination with a 3’ entry clone containing DsRed with its own start site, but found that the DsRed was only weakly expressed following excision.)
Table 1.
Gateway entry vectors for use with the PhiC31 integrase system
| Name | Type | Description |
|---|---|---|
| p5E-XIpGb | 5′ entry | EF1α promoter w/excisable GFP cassette |
| p5E-XIpRb | 5′ entry | EF1α promoter w/excisable DsRed cassette |
| p5E-XIppAb | 5′ entry | EF1α promoter w/stop cassette |
| pME-PhiC31o | Middle | codon-optimized integrase |
| pME-PhiC31o-ERT2 | Middle | 4-hydroxytamoxifen-inducible integrase |
| pME-nlsPhiC31o | Middle | nuclear-localized integrase |
MATERIALS AND METHODS
pME-PhiC31o was generated by removing the codon-optimized integrase sequence from pPhiC31o (Addgene plasmid 13794; (Raymond, Soriano. 2007)) with EcoRI and XbaI and inserting it into the plasmid pENTR3C-CS, a middle entry vector containing the multiple cloning site of pCS2. p5E-XIpRb was constructed by inserting a BglII-XhoI fragment containing the Xenopus EF1-alpha promoter and rabbit beta-globin intron, attP site, DsRed-Express with poly A signal, and attB site into the BamHI-XhoI sites of p5EMCS (Kwan, et al. 2007). p5E-mitfa0.9 contains 0.9 kilobases of sequence upstream of the translational start site of the mitfa gene , and was made by replacing transferring the mitfa promoter from the plasmid pNP-P+ (Lister, et al. 2001) to the 5’ entry vector p5EMCS (Kwan, et al. 2007) as a SalI-HindIII fragment. pDestTol2pA2-XIpRb-EGFP-pA was generated in a multi-site Gateway LR reaction (Invitrogen) with the entry vectors p5E-XIpRb, pME-EGFP, p3E-polyA, and the destination vector pDestTol2pA2 (Kwan, et al. 2007). pDestTol2pA2-mitfa0.9-PhiC31o-pA was generated in a multi-site Gateway LR reaction with the entry vectors p5E-mitfa0.9, pME-PhiC31o, p3E-polyA, and the destination vector pDestTol2pA2. Additional cloning details for all plasmids are available upon request.
The plasmids pCS2P+PhiC31o and pCS2P+PhiC31o-ERT2 have been previously described, as has generation of the green-to-red reporter transgenic line, Tg(XlEef1a1:attP-GFP-attB-DsRed2)vc2 (Lister. 2010). Messenger RNA for each integrase was synthesized using the SP6 mMessage mMachine kit (Ambion/Applied Biosystems) following plasmid linearization by restriction digest with Not I or BssHII. Microinjections were performed using a pressure injection apparatus from Applied Scientific Instrumentation, on a Nikon stereodissection microscope.
To activate the PhiC31o-ERT2 fusion, 4-hydroxytamoxifen (Sigma cat. no. H7904, made up at a stock concentration 300 uM in DMSO) was added to embryos at 60-90 minutes post-fertilization to a final concentration of 300 nM, with an equal volume of DMSO alone added to the control dish.
DISCUSSION
Use of the Cre/lox system is becoming more widespread in zebrafish (Dong, Stuart. 2004; Langenau, et al. 2005; Pan, et al. 2005; Thummel, et al. 2005; Feng, et al. 2007; Wang, et al. 2008; Collins, et al. 2010; Jopling, et al. 2010; Kikuchi, et al. 2010), and includes conditional approaches (Boniface, et al. 2009; Hans, et al. 2009). Intramolecular recombination in zebrafish mediated by the phiC31 integrase represents another tool now available for the zebrafish researcher (Lister. 2010; Lu, et al. 2010). The XIpGbR green-to-red transgenic line we previously established (Lister. 2010), vc2, is functionally analogous to a G2R Cre reporter that has been reported (Yoshikawa, et al. 2008), and should be useful for the development and screening of integrase-expressing driver lines, as well as for lineage analysis. To facilitate the construction of drivers and reporters/effectors, we have adapted a number of components for use with Gateway cloning (Table 1).
An obvious goal for those working with phiC31 integrase in zebrafish is to demonstrate its utility for targeted transgene integration, and a number of labs including my own are working on this. The issue of exactly where DNA integrates in the genome is important because of the influence of position effects on transgene expression. This will only become more relevant as more investigators seek to study the function of promoters and proteins in vivo at single basepair or single amino acid resolution, and wish to compare variants in the same chromosomal context. While the efficiency of production of transgenic zebrafish continue to improve, at present there is no way to control the locus of transgene integration. In mice, this is typically achieved through homologous recombination in ES cells followed by chimeric embryo generation to establish the alteration in the germline. In zebrafish, ES-like cells have been isolated (Fan, et al. 2004) and homologous recombination has been reported (Fan, et al. 2006), but it has not yet been demonstrated that cells that have undergone the necessary selection regimen retain germline potential when put back into embryos. The lone report of targeted gene insertion in zebrafish has come from a group using Cre recombinase along with mutant loxP sites that once recombined cannot efficiently perform the reverse reaction with each other (Liu, et al. 2007), but only one such event could be verified out of 80,000 injected embryos.
Based on work in other vertebrates, namely mouse and frogs (Belteki, et al. 2003; Allen, Weeks. 2005), intermolecular recombination in zebrafish mediated by phiC31 integrase should be feasible. To make heritable genomic manipulations it is necessary that recombination occur in germ cells. By raising integrase-injected vc2 transgenic embryos to adulthood we determined that phiC31 integrase is active in the zebrafish germline, as it is in mouse (Belteki, et al. 2003). Breeding individual transgenic fish to nontransgenic mates produced a mix of offspring with uniform GFP or DsRed expression almost half the time, regardless of the sex of the transgenic adult (Lister. 2010). This showed that recombination can take place in the germline of either both males and females. Moreover, widespread expression of phiC31 mRNA in early embryos had no obvious deleterious effect on development.
Our initial experiments suggested that codon optimization had a significant effect on the activity of phiC31 integrase in zebrafish cells (Lister. 2010), and additional refinements should improve the likelihood of success of both intra- and intermolecular recombination approaches. When tested side by side with Cre in recombination assays on extrachromosomal and integrated targets in mammalian cells, phiC31 integrase was found to be approximately 50% as active as Cre on plasmids and 10% on integrated transgenes (Andreas, et al. 2002). However, addition of a C-terminal nuclear localization signal increased these to 80% and 50%. Other possibilities for optimizing the efficiency of the system and improving the chances of integration include employing hyperactive phiC31 mutants (Keravala, et al. 2009), as well as the identification and, if possible, specific elimination of interactions with inhibitory proteins (Chen, et al. 2006; Wang, et al. 2010) through directed mutagenesis. The successes observed in zebrafish with the phiC31 integrase within a relatively brief time suggest that the goal of additional uses, notably directed transgenesis through intermolecular recombination (Groth, et al. 2004; Bateman, et al. 2006), will eventually be realized.
SUMMARY
The phiC31 integrase is functional in zebrafish cells and at present offers and alternative, or complement, to existing tools such as Cre and Flp for regulated and precise recombination of transgenes. In the future it may be possible to extend the use of this tool, by exploiting its inherent unidirectionality to direct the integration of transgenes at pre-determined locations in the genome.
ACKNOWLEDGMENTS
I would like to thank AnhThu Nguyen, Katie Lunney, and Leyla Peachy for fish care. This work was supported by grant number R21 HD055311 from the National Institute of Child Health and Human Development.
REFERENCES
- Allen BG, Weeks DL. Transgenic Xenopus Laevis Embryos can be Generated using phiC31 Integrase. Nat. Methods. 2005;12:975–979. doi: 10.1038/nmeth814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amatruda JF, Shepard JL, Stern HM, Zon LI. Zebrafish as a Cancer Model System. Cancer. Cell. 2002;3:229–231. doi: 10.1016/s1535-6108(02)00052-1. [DOI] [PubMed] [Google Scholar]
- Andreas S, Schwenk F, Kuter-Luks B, Faust N, Kuhn R. Enhanced Efficiency through Nuclear Localization Signal Fusion on Phage PhiC31-Integrase: Activity Comparison with Cre and FLPe Recombinase in Mammalian Cells. Nucleic Acids Res. 2002;11:2299–2306. doi: 10.1093/nar/30.11.2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman JR, Lee AM, Wu CT. Site-Specific Transformation of Drosophila Via phiC31 Integrase-Mediated Cassette Exchange. Genetics. 2006;2:769–777. doi: 10.1534/genetics.106.056945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belteki G, Gertsenstein M, Ow DW, Nagy A. Site-Specific Cassette Exchange and Germline Transmission with Mouse ES Cells Expressing phiC31 Integrase. Nat. Biotechnol. 2003;3:321–324. doi: 10.1038/nbt787. [DOI] [PubMed] [Google Scholar]
- Bischof J, Basler K. Recombinases and their use in Gene Activation, Gene Inactivation, and Transgenesis. Methods Mol. Biol. 2008:175–195. doi: 10.1007/978-1-59745-583-1_10. [DOI] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An Optimized Transgenesis System for Drosophila using Germ-Line-Specific phiC31 Integrases. Proc. Natl. Acad. Sci. U. S. A. 2007;9:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boniface EJ, Lu J, Victoroff T, Zhu M, Chen W. FlEx-Based Transgenic Reporter Lines for Visualization of Cre and Flp Activity in Live Zebrafish. Genesis. 2009;7:484–491. doi: 10.1002/dvg.20526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boy AL, Zhai Z, Habring-Muller A, Kussler-Schneider Y, Kaspar P, Lohmann I. Vectors for Efficient and High-Throughput Construction of Fluorescent Drosophila Reporters using the PhiC31 Site-Specific Integration System. Genesis. 2010;7:452–456. doi: 10.1002/dvg.20637. [DOI] [PubMed] [Google Scholar]
- Branda CS, Dymecki SM. Talking about a Revolution: The Impact of Site-Specific Recombinases on Genetic Analyses in Mice. Dev. Cell. 2004;1:7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
- Briggs J. The Zebrafish: A New Model Organism for Integrative Physiology. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2002;1:R3–9. doi: 10.1152/ajpregu.00589.2001. [DOI] [PubMed] [Google Scholar]
- Chen JZ, Ji CN, Xu GL, Pang RY, Yao JH, Zhu HZ, Xue JL, Jia W. DAXX Interacts with Phage PhiC31 Integrase and Inhibits Recombination. Nucleic Acids Res. 2006;21:6298–6304. doi: 10.1093/nar/gkl890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Burgess S, Golling G, Amsterdam A, Hopkins N. High-Throughput Selection of Retrovirus Producer Cell Lines Leads to Markedly Improved Efficiency of Germ Line-Transmissible Insertions in Zebra Fish. J. Virol. 2002;5:2192–2198. doi: 10.1128/jvi.76.5.2192-2198.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RT, Linker C, Lewis J. MAZe: A Tool for Mosaic Analysis of Gene Function in Zebrafish. Nat. Methods. 2010;3:219–223. doi: 10.1038/nmeth.1423. [DOI] [PubMed] [Google Scholar]
- Davidson AE, Balciunas D, Mohn D, Shaffer J, Hermanson S, Sivasubbu S, Cliff MP, Hackett PB, Ekker SC. Efficient Gene Delivery and Gene Expression in Zebrafish using the Sleeping Beauty Transposon. Dev. Biol. 2003;2:191–202. doi: 10.1016/j.ydbio.2003.07.013. [DOI] [PubMed] [Google Scholar]
- Davis J, Clay H, Lewis J, Ghori N, Herbomel P, Ramakrishnan L. Real-Time Visualization of Mycobacterium-Macrophage Interactions Leading to Initiation of Granuloma Formation in Zebrafish Embryos. Immunity. 2002;6:693–702. doi: 10.1016/s1074-7613(02)00475-2. [DOI] [PubMed] [Google Scholar]
- Dong J, Stuart GW. Transgene Manipulation in Zebrafish by using Recombinases. Methods Cell Biol. 2004:363–379. doi: 10.1016/s0091-679x(04)77020-x. [DOI] [PubMed] [Google Scholar]
- Driever W, Solnica-Krezel L, Schier AF, Neuhauss SC, Malicki J, Stemple DL, Stainier DY, Zwartkruis F, Abdelilah S, Rangini Z, Belak J, Boggs C. A Genetic Screen for Mutations Affecting Embryogenesis in Zebrafish. Development. 1996:37–46. doi: 10.1242/dev.123.1.37. [DOI] [PubMed] [Google Scholar]
- Fan L, Moon J, Crodian J, Collodi P. Homologous Recombination in Zebrafish ES Cells. Transgenic Res. 2006;1:21–30. doi: 10.1007/s11248-005-3225-0. [DOI] [PubMed] [Google Scholar]
- Fan L, Crodian J, Collodi P. Production of Zebrafish Germline Chimeras by using Cultured Embryonic Stem (ES) Cells. Methods Cell Biol. 2004:113–119. doi: 10.1016/s0091-679x(04)77006-5. [DOI] [PubMed] [Google Scholar]
- Feng H, Langenau DM, Madge JA, Quinkertz A, Gutierrez A, Neuberg DS, Kanki JP, Look AT. Heat-Shock Induction of T-Cell lymphoma/leukaemia in Conditional Cre/lox-Regulated Transgenic Zebrafish. Br. J. Haematol. 2007;2:169–175. doi: 10.1111/j.1365-2141.2007.06625.x. [DOI] [PubMed] [Google Scholar]
- Fetcho JR, Liu KS. Zebrafish as a Model System for Studying Neuronal Circuits and Behavior. Ann. N. Y. Acad. Sci. 1998:333–345. doi: 10.1111/j.1749-6632.1998.tb09060.x. [DOI] [PubMed] [Google Scholar]
- Fish MP, Groth AC, Calos MP, Nusse R. Creating Transgenic Drosophila by Microinjecting the Site-Specific phiC31 Integrase mRNA and a Transgene-Containing Donor Plasmid. Nat. Protoc. 2007;10:2325–2331. doi: 10.1038/nprot.2007.328. [DOI] [PubMed] [Google Scholar]
- Grabher C, Wittbrodt J. Recent Advances in Meganuclease-and Transposon-Mediated Transgenesis of Medaka and Zebrafish. Methods Mol. Biol. 2008:521–539. doi: 10.1007/978-1-60327-483-8_36. [DOI] [PubMed] [Google Scholar]
- Grabher C, Joly JS, Wittbrodt J. Highly Efficient Zebrafish Transgenesis Mediated by the Meganuclease I-SceI. Methods Cell Biol. 2004:381–401. doi: 10.1016/s0091-679x(04)77021-1. [DOI] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP. Construction of Transgenic Drosophila by using the Site-Specific Integrase from Phage phiC31. Genetics. 2004;4:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth AC, Olivares EC, Thyagarajan B, Calos MP. A Phage Integrase Directs Efficient Site-Specific Integration in Human Cells. Proc. Natl. Acad. Sci. U. S. A. 2000;11:5995–6000. doi: 10.1073/pnas.090527097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, van Eeden FJ, Jiang YJ, Heisenberg CP, Kelsh RN, Furutani-Seiki M, Vogelsang E, Beuchle D, Schach U, Fabian C, Nusslein-Volhard C. The Identification of Genes with Unique and Essential Functions in the Development of the Zebrafish, Danio Rerio. Development. 1996:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- Hans S, Kaslin J, Freudenreich D, Brand M. Temporally-Controlled Site-Specific Recombination in Zebrafish. PLoS ONE. 2009;2:e4640. doi: 10.1371/journal.pone.0004640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselson D, Anderson RM, Beinat M, Stainier DY. Distinct Populations of Quiescent and Proliferative Pancreatic Beta-Cells Identified by HOTcre Mediated Labeling. Proc. Natl. Acad. Sci. U. S. A. 2009;35:14896–14901. doi: 10.1073/pnas.0906348106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Zhou W, Dong W, Watson AM, Hong Y. From the Cover: Directed, Efficient, and Versatile Modifications of the Drosophila Genome by Genomic Engineering. Proc. Natl. Acad. Sci. U. S. A. 2009;20:8284–8289. doi: 10.1073/pnas.0900641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling C, Sleep E, Raya M, Marti M, Raya A, Belmonte JC. Zebrafish Heart Regeneration Occurs by Cardiomyocyte Dedifferentiation and Proliferation. Nature. 2010;7288:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M. A Transposon-Mediated Gene Trap Approach Identifies Developmentally Regulated Genes in Zebrafish. Dev. Cell. 2004;1:133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Keravala A, Lee S, Thyagarajan B, Olivares EC, Gabrovsky VE, Woodard LE, Calos MP. Mutational Derivatives of PhiC31 Integrase with Increased Efficiency and Specificity. Mol. Ther. 2009;1:112–120. doi: 10.1038/mt.2008.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MS, Khalid AM, Malik KA. Phage phiC31 Integrase: A New Tool in Plastid Genome Engineering. Trends Plant Sci. 2005;1:1–3. doi: 10.1016/j.tplants.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, Evans T, Macrae CA, Stainier DY, Poss KD. Primary Contribution to Zebrafish Heart Regeneration by gata4(+) Cardiomyocytes. Nature. 2010;7288:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi S, Uchiyama J, Baughman AM, Goto T, Lin MC, Tsai SB. The Zebrafish as a Vertebrate Model of Functional Aging and very Gradual Senescence. Exp. Gerontol. 2003;7:777–786. doi: 10.1016/s0531-5565(03)00108-6. [DOI] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien CB. The Tol2kit: A Multisite Gateway-Based Construction Kit for Tol2 Transposon Transgenesis Constructs. Dev. Dyn. 2007;11:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- Langenau DM, Feng H, Berghmans S, Kanki JP, Kutok JL, Look AT. Cre/lox-Regulated Transgenic Zebrafish Model with Conditional Myc-Induced T Cell Acute Lymphoblastic Leukemia. Proc. Natl. Acad. Sci. U. S. A. 2005;17:6068–6073. doi: 10.1073/pnas.0408708102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister JA. Transgene Excision in Zebrafish using the phiC31 Integrase. Genesis. 2010;2:137–143. doi: 10.1002/dvg.20595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister JA, Close J, Raible DW. Duplicate Mitf Genes in Zebrafish: Complementary Expression and Conservation of Melanogenic Potential. Dev. Biol. 2001;2:333–344. doi: 10.1006/dbio.2001.0379. [DOI] [PubMed] [Google Scholar]
- Liu J, Skjorringe T, Gjetting T, Jensen TG. PhiC31 Integrase Induces a DNA Damage Response and Chromosomal Rearrangements in Human Adult Fibroblasts. BMC Biotechnol. 2009;31 doi: 10.1186/1472-6750-9-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WY, Wang Y, Qin Y, Wang YP, Zhu ZY. Site-Directed Gene Integration in Transgenic Zebrafish Mediated by Cre Recombinase using a Combination of Mutant Lox Sites. Mar. Biotechnol. (NY) 2007;4:420–428. doi: 10.1007/s10126-007-9000-x. [DOI] [PubMed] [Google Scholar]
- Loonstra A, Vooijs M, Beverloo HB, Allak BA, van Drunen E, Kanaar R, Berns A, Jonkers J. Growth Inhibition and DNA Damage Induced by Cre Recombinase in Mammalian Cells. Proc. Natl. Acad. Sci. U. S. A. 2001;16:9209–9214. doi: 10.1073/pnas.161269798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Maddison LA, Chen W. PhiC31 Integrase Induces Efficient Site-Specific Excision in Zebrafish. Transgenic Res. . 2010 doi: 10.1007/s11248-010-9394-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz KA, Corneille S, Azhagiri AK, Svab Z, Maliga P. A Novel Approach to Plastid Transformation Utilizes the phiC31 Phage Integrase. Plant J. 2004;6:906–913. doi: 10.1111/j.1365-313x.2004.02015.x. [DOI] [PubMed] [Google Scholar]
- Nasevicius A, Ekker S. Effective Targeted Gene ‘Knockdown’ in Zebrafish. Nat. Genet. 2000;220472325:216–20. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- Pan X, Wan H, Chia W, Tong Y, Gong Z. Demonstration of Site-Directed Recombination in Transgenic Zebrafish using the Cre/loxP System. Transgenic Res. 2005;2:217–223. doi: 10.1007/s11248-004-5790-z. [DOI] [PubMed] [Google Scholar]
- Raymond CS, Soriano P. High-Efficiency FLP and PhiC31 Site-Specific Recombination in Mammalian Cells. PLoS ONE. 2007;1:e162. doi: 10.1371/journal.pone.0000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsova M, Kempe K, Gils A, Ismagul A, Weyen J, Gils M. Expression of Active Streptomyces Phage phiC31 Integrase in Transgenic Wheat Plants. Plant Cell Rep. 2008;12:1821–1831. doi: 10.1007/s00299-008-0604-z. [DOI] [PubMed] [Google Scholar]
- Schmidt EE, Taylor DS, Prigge JR, Barnett S, Capecchi MR. Illegitimate Cre-Dependent Chromosome Rearrangements in Transgenic Mouse Spermatids. Proc. Natl. Acad. Sci. U. S. A. 2000;25:13702–13707. doi: 10.1073/pnas.240471297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seok SH, Na YR, Han JH, Kim TH, Jung H, Lee BH, Emelyanov A, Parinov S, Park JH. Cre/loxP-Regulated Transgenic Zebrafish Model for Neural Progenitor-Specific Oncogenic Kras Expression. Cancer. Sci. 2010;1:149–154. doi: 10.1111/j.1349-7006.2009.01393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N, Moldt B, Dalsgaard T, Jensen TG, Mikkelsen JG. Regulated Gene Insertion by Steroid-Induced PhiC31 Integrase. Nucleic Acids Res. 2008;11:e67. doi: 10.1093/nar/gkn298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MC, Brown WR, McEwan AR, Rowley PA. Site-Specific Recombination by phiC31 Integrase and Other Large Serine Recombinases. Biochem. Soc. Trans. 2010;2:388–394. doi: 10.1042/BST0380388. [DOI] [PubMed] [Google Scholar]
- Thomson JG, Chan R, Thilmony R, Yau YY, Ow DW. PhiC31 Recombination System Demonstrates Heritable Germinal Transmission of Site-Specific Excision from the Arabidopsis Genome. BMC Biotechnol. 2010;17 doi: 10.1186/1472-6750-10-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe HM, Smith MC. In Vitro Site-Specific Integration of Bacteriophage DNA Catalyzed by a Recombinase of the resolvase/invertase Family. Proc. Natl. Acad. Sci. U. S. A. 1998;10:5505–5510. doi: 10.1073/pnas.95.10.5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel R, Burket CT, Brewer JL, Sarras MP, Jr, Li L, Perry M, McDermott JP, Sauer B, Hyde DR, Godwin AR. Cre-Mediated Site-Specific Recombination in Zebrafish Embryos. Dev. Dyn. 2005;4:1366–1377. doi: 10.1002/dvdy.20475. [DOI] [PubMed] [Google Scholar]
- Thyagarajan B, Olivares EC, Hollis RP, Ginsburg DS, Calos MP. Site-Specific Genomic Integration in Mammalian Cells Mediated by Phage phiC31 Integrase. Mol. Cell. Biol. 2001;12:3926–3934. doi: 10.1128/MCB.21.12.3926-3934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BY, Xu GL, Zhou CH, Tian L, Xue JL, Chen JZ, Jia W. PhiC31 Integrase Interacts with TTRAP and Inhibits NFkappaB Activation. Mol. Biol. Rep. 2010;6:2809–2816. doi: 10.1007/s11033-009-9829-3. [DOI] [PubMed] [Google Scholar]
- Wang L, Zhang Y, Zhou T, Fu YF, Du TT, Jin Y, Chen Y, Ren CG, Peng XL, Deng M, Liu TX. Functional Characterization of Lmo2-Cre Transgenic Zebrafish. Dev. Dyn. 2008;8:2139–2146. doi: 10.1002/dvdy.21630. [DOI] [PubMed] [Google Scholar]
- Yoshikawa S, Kawakami K, Zhao XC. G2R Cre Reporter Transgenic Zebrafish. Dev. Dyn. 2008;9:2460–2465. doi: 10.1002/dvdy.21673. [DOI] [PMC free article] [PubMed] [Google Scholar]