SUMMARY
AvGluR1, a glutamate receptor ion channel from the primitive eukaryote Adineta vaga, is activated by alanine, cysteine, methionine and phenylalanine which produce lectin-sensitive desensitizing responses like those to glutamate, aspartate and serine. AvGluR1 LBD crystal structures reveal a novel scheme for binding dissimilar ligands that may be utilized by distantly related odorant/chemosensory receptors. Arginine residues in domain 2 coordinate the γ-carboxyl group of glutamate, while in the alanine, methionine and serine complexes a chloride ion acts as a surrogate ligand, replacing the γ-carboxyl group. Removal of Cl− lowers affinity for these ligands, but not for glutamate, aspartate or for phenylalanine which occludes the anion binding site and binds with low affinity. AvGluR1 LBD crystal structures and sedimentation analysis also provide insights into the evolutionary link between prokaryotic and eukaryotic iGluRs and reveal features unique to both classes, emphasizing the need for additional structure based studies on iGluR-ligand interactions.
INTRODUCTION
Genome sequencing projects provide a rich resource for analysis of ligand recognition and signal transduction mechanisms in diverse families of membrane proteins. Such projects have identified numerous proteins in prokaryotes (Ger et al., 2010), invertebrate eukaryotes (Croset et al., 2010; Srivastava et al., 2008) and plants (Chiu et al., 2002; Lam et al., 1998) which have a modular domain organization characteristic of glutamate receptor ion channels (iGluRs). This modular architecture likely arose from gene fusion events between bacterial periplasmic proteins and primitive ion channels, generating a Venus flytrap receptor ligand binding domain (LBD), interrupted by a pore loop ion channel motif with two membrane spanning segments, as first reported for GluR0 from the cyanobacterium Synechocystis (Chen et al., 1999). AMPA, kainate and NMDA receptors, which are widely expressed in the CNS of vertebrates, together with structurally related proteins in plants, represent a second class of eukaryotic iGluRs. These more complex receptors probably evolved from prokaryotic iGluRs via additional gene fusion events, which attached a 380 residue extracellular amino terminal domain (ATD) and a third transmembrane segment with a cytoplasmic carboxy terminal domain. Despite great significance for understanding the role of iGluRs in organisms that lack complex nervous systems, and for gaining insight into how iGluRs evolved, the majority of iGluR related genes found in prokaryotes, primitive eukaryotes and plants remain virtually uncharacterized.
Recently, a glutamate receptor named AvGluR1 was identified in the freshwater bdelloid rotifer Adineta vaga, and was proposed to be an evolutionary link between prokaryotic and eukaryotic iGluR receptor classes (Janovjak et al., 2011). In common with eukaryotic iGluRs, AvGluR1 has both an amino terminal domain, and three membrane spanning segments with the ‘SYTAN’ motif characteristic of AMPA, kainate and NMDA receptors. On the other hand, like GluR0, the pore loop of AvGluR1 has a K+ channel selectivity filter sequence, TXVGYG, although electrophysiological experiments revealed permeability to both Na+ and K+ (Janovjak et al., 2011). In the present study, we characterized the ligand binding properties, structure and assembly of the AvGluR1 ligand binding domain (LBD). Analysis of AvGluR1 ligand selectivity unexpectedly revealed activation of ion channel gating by the hydrophobic amino acids alanine, cysteine, methionine and phenylalanine, as well as by glutamate, aspartate and serine. To obtain mechanistic insight into how AvGluR1 binds such chemically diverse ligands, we solved six AvGluR1 ligand binding domain crystal structures for complexes with diverse amino acids and discovered a novel binding mechanism in which chloride ions act as a surrogate carboxyl group in the alanine, serine and methionine complexes. A structure based phylogenetic analysis revealed that the AvGluR1 LBD most closely resembles that of prokaryotic iGluRs. However, bound glutamate adopts the same folded conformation as found in NMDA, AMPA and kainate receptors, not the extended conformation found in prokaryote iGluR structures. In AvGluR1 LBD dimers, subunit packing is the same as found in prokaryotic iGluRs, but with a low affinity for dimer assembly characteristic of eukaryote iGluRs.
RESULTS AND DISCUSSION
Ligand binding profile of AvGluR1
Prior studies revealed activation of AvGluR1 by glutamate, aspartate, serine and kainate (Janovjak et al., 2011). To further investigate the selectivity of AvGluR1 we performed radioligand binding assays, using the ligand binding domain of AvGluR1, S1 residues A433-K543 connected via a GT dipeptide linker to S2 residues L656-P788 (Stern-Bach et al., 1994), expressed as a soluble protein in Escherichia coli. The AvGluR1 LBD apo protein exhibited robust binding to [3H] L-glutamate, Kd 203 ± 18 nM (Figure 1A), similar to GluR0 from Synechocystis, Kd 193 nM (Chen et al., 1999), the GluA2 AMPA receptor, Kd 821 nM (Armstrong and Gouaux, 2000), and the GluK1 and GluK2 kainate receptors, Kd 57 nM and 1.4 μM, respectively (Mayer, 2005); by contrast, GluR0 from Nostoc punctiforme binds glutamate with 125-fold lower affinity (Lee et al., 2008). In displacement assays with 100 μM concentrations of twenty genetically coded amino acids, binding of 100 nM [3H] L-glutamate was abolished by glutamate and aspartate, and inhibited by > 50% for glutamine, asparagine, serine, and the hydrophobic amino acids alanine, cysteine, methionine and phenylalanine; histidine, lysine and arginine were inactive (Figure 1B). Concentration displacement curves for the 10 amino acids with highest affinity (Figure 1C and Table S1), and for 14 ligands which have activity at AMPA, kainate or NMDA receptors (Figure 1D and Table S1), permitted quantitative comparisons between different ligands. The sequence of Kd values, Glu 203 nM < Asp 875 nM < Ala 9 μM < Met 15 μM < Ser 24 μM < Gln 37 μM < Cys 46 μM < Asn 81 μM revealed that small hydrophobic amino acids were surprisingly potent compared to glutamate, aspartate and their amides. Binding was stereoselective, and affinity decreased 650-fold for D-Glu (Kd 130 μM), 28-fold for D-Ser (Kd 700 μM) and 14-fold for D-Asp (Kd 12 μM) compared to their L-stereoisomers.
Figure 1.
AvGluR1 ligand binding profile. (A) Saturation binding isotherm for [3H] L-Glu with non-specific binding measured in the presence of 20 mM alanine. (B) Competitive displacement assays with 100 μM concentrations of 20 genetically encoded amino acids; the dashed horizontal line shows the mean binding for 100 nM [3H] L-Glu. (C) Equilibrium dose inhibition curves for displacement of 100 nM [3H] L-Glu by various amino acids. (D) Equilibrium dose inhibition curves for AMPA, kainate and NMDA receptor ligands. Data points are mean ± SEM of three observations for all panels.
Amino acid sequence alignments revealed slightly greater similarity of the AvGluR1 LBD to kainate receptors (22-23% identity) compared to AMPA receptors (18-20 % identity) and NMDA receptors (16-19% identity). Related to this, the kainate receptor preferring agonist 2S,4R-4-methyl glutamate (SYM2081 Kd 49.5 μM) and the GluK1 preferring antagonist UBP-310 (Kd 160 μM) bind with higher affinity than other subtype selective compounds such as NMDA (Kd 9.9 mM), the NMDA receptor antagonist AP5 (Kd 530 μM), and the non-selective antagonist DNQX (Kd 250 μM). Prior measurements of ligand activated ion currents for AvGluR1 showed activation by AMPA and kainate but not NMDA (Janovjak et al., 2011), but displacement assays with [3H] L-glutamate revealed very low affinity for both kainate (Kd 2.7 mM) and NMDA (Kd 9.9 mM), with higher affinity binding of AMPA (Kd 130 μM) and the non-selective iGluR agonist quisqualate (Kd 39 μM).
Activation of AvGluR1 by alanine and other hydrophobic amino acids
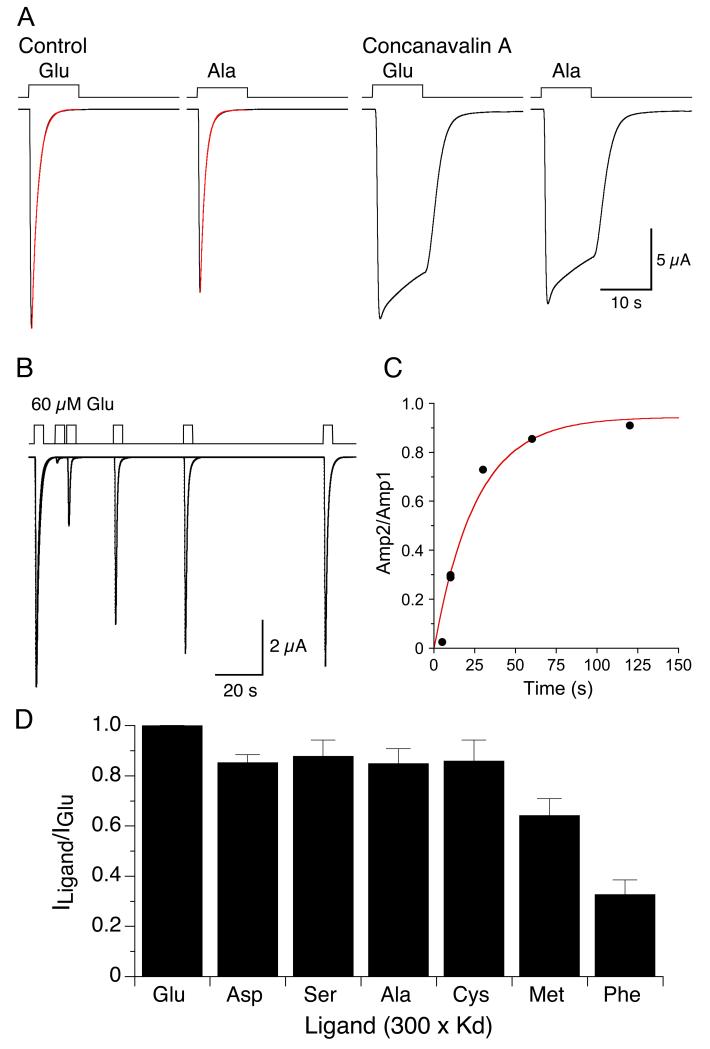
To test whether small hydrophobic amino acids activate ion channel gating we expressed full length AvGluR1 in Xenopus oocytes, and applied ligands at a concentration 300 times the Kd estimated from displacement assays with [3H] L-glutamate. Large inward currents (5.1 ± 1.4 μA, mean ± SD, n = 9) were activated by 60 μM glutamate, with a 10-90% rise time of 240 ± 67 ms, followed by complete desensitization well fit by a single exponential of time constant 626 ± 255 ms (Figure 2A), consistent with prior experiments (Janovjak et al., 2011); the time constant of recovery from desensitization, measured using a twin pulse protocol, was 26 s (Figure 2B and C). Similar responses were recorded for 260 μM aspartate and 7.4 mM serine, 85 ± 3% and 88 ± 6% of the amplitude of those to glutamate. However, AvGluR1 was also activated by hydrophobic amino acids, all of which also evoked complete desensitization (Figure 2A). The amplitude of responses for 2.8 mM alanine and 14 mM cysteine was 85 ± 6%, and 86 ± 8% of those to glutamate, while for 4.5 mM methionine and 63 mM phenylalanine the amplitude was 64 ± 7% and 33 ± 6% (Figure 2D).
Figure 2.
Activation and desensitization of AvGluR1 by hydrophobic amino acids. (A) Responses to 60 μM glutamate and 2.8 mM alanine before and after application of concanavalin A, 0.5 mg/ml 4 min; the onset of desensitization is fit with single exponential functions of time constant 1.25 and 1.0 s for glutamate and alanine respectively. (B) Recovery from desensitization evoked by 60 μM glutamate measured using a twin pulse protocol. (C) The rate of recovery was estimated from a single exponential function fit to the ratio of the test/control pulse amplitude, time constant 25.7 s. (D) Bar plot showing the amplitude of responses to acidic, polar and hydrophobic amino acids applied at 300 times the Kd and normalized to the response to glutamate recorded in the same oocyte; data points show mean ± SEM (n = 6).
In mammalian iGluRs the plant lectin concanavalin A strongly attenuates desensitization for kainate receptors, with only modest effects on AMPA receptors (Partin et al., 1993), most likely by binding to N-linked glycosylated residues that sterically inhibit conformational changes associated with desensitization (Everts et al., 1999; Partin et al., 1993). Of interest, given the greater sequence similarity of AvGluR1 to kainate versus AMPA receptors, and the larger number of predicted N-linked glycosylation for AvGluR1 compared to GluA2, desensitization was strongly attenuated following treatment with 0.5 mg/ml concanavalin A for 4 minutes (Figure 2A).
The structure of AvGluR1 glutamate and aspartate complexes
The results of binding assays and electrophysiological experiments reveal that AvGluR1 ligand selectivity is different from other iGluRs. To elucidate the molecular mechanism we solved AvGluR1 LBD crystal structures for complexes with glutamate, aspartate, serine, alanine, methionine and phenylalanine at resolutions of 1.4 - 1.9 Å (Table 1). The AvGluR1 glutamate and aspartate complex LBD structures were nearly identical, root mean square deviation (RMSD) 0.29 Å for 242 Cα atoms. Omit maps reveal unambiguous electron density for the bound amino acids and seven water molecules (Figure 3), trapped in a roughly pyramidal shaped cavity of volume of 302 ± 1.9 Å3, comparable in size to that for GluK1 (305 ± 6 Å3), but larger than that for GluK2 (255 ± 15 Å3) or GluA2 (218 ± 4 Å3). Similar to GluA2 and GluK2, the cavity for AvGluR1 has an overall positive charge; however, the AvGluR1 cavity is pinched off into a series of smaller vestibules by the side chains of Thr679, Arg676, Arg702 and Asp515, while in AMPA and kainate receptors the cavities have a smoother surface. The α-carboxyl groups of glutamate and aspartate form a bidentate salt-bridge with the guanidinium group of Arg522 in domain 1, and make H-bonds with the main chain amide groups of Thr517 in domain 1 and Ala680 in domain 2. The glutamate and aspartate α-amino groups are bound in a tetrahedral arrangement by the carboxyl group of Asp720 in domain 2, by the Asp515 main chain carbonyl, and by Thr517 hydroxyl group in domain 1. Although this mode of binding is observed in all known eukaryotic and prokaryotic iGluRs, the bound glutamate ligand adopts different conformations in the two receptor classes (Figure S1A). In the Synechocystis and Nostoc punctiforme prokaryotic iGluRs the ligand adopts an extended conformation, (χ2 = 177°), and the γ-carboxyl group interacts with residues in domain 1; by contrast, for eight representative eukaryotic iGluR LBDs (χ2 = −70 ± 6.5°), the ligand has undergone a 107° rotation such that the γ-carboxyl group projects towards and interacts with α-helix F in domain 2; in AvGluR1, glutamate adopts a similar pose (χ2 = −70°). However, a pair of arginine residues in domain 2, that are replaced by hydrophobic or polar residues in other iGluRs (Figure S1B), generates a novel binding mechanism for the terminal carboxyl groups of glutamate and aspartate in AvGluR1. The side chain of Arg676, in a loop preceding α-helix E, forms a salt bridge with the terminal carboxyl groups of both acidic amino acids, while due to the different size and geometry of these ligands, the side chain of Arg702 at the tip of helix F forms a second salt bridge with the bound aspartate but not with glutamate (Figure 3).
Table 1.
Data collection and refinement statistics
| Data Set | L-Glu | L-Asp | L-Ser | L-Ala | L-Met | L-Phe |
|---|---|---|---|---|---|---|
| DATA COLLECTION | ||||||
| Space group | P21 | P21 | P21 | P21 | P21 | P21 |
|
Unit cell a, b, c (Å) α=γ, β |
55.4, 101.0, 56.7 90, 116.5 |
55.1, 100.4, 56.9 90, 116.4 |
55.4, 100.3,59.9 90, 117.4 |
55.4, 100.7, 56.7 90, 116.4 |
55.0, 100.5, 56.7 90, 116.2 |
55.5, 100.2, 56.8 90, 116.5 |
| Number per a.u. | 2 | 2 | 2 | 2 | 2 | 2 |
| Wavelength (Å) | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| Resolution (Å) a | 30 – 1.37 (1.39) | 40 – 1.66 (1.69) | 40 – 1.94 (1.97) | 40 – 1.72 (1.75) | 40 – 1.60 (1.63) | 40 – 1.92 (1.95) |
| Unique observations | 116416 | 65127 | 43189 | 58814 | 73025 | 42897 |
| Mean redundancy b | 3.8 (3.6) | 3.8 (3.8) | 3.8 (3.8) | 3.9 (3.8) | 3.7 (3.0) | 3.8 (3.8) |
| Completeness (%) b | 97.7 (95.2) | 100 (99.9) | 99.9 (98.7) | 99.0 (98.0) | 99.8 (97.5) | 100 (100) |
| Rmergebc | 0.043 (0.59) | 0.055 (0.58) | 0.050 (0.56) | 0.061 (0.57) | 0.044 (0.50) | 0.065 (0.70) |
| I/σ(I)b | 26.6 (2.0) | 22.3 (2.1) | 27.4 (2.4) | 23.1 (2.5) | 26.2 (2.0) | 21.6 (2.1) |
| REFINEMENT | ||||||
| Resolution (Å) | 29.5 – 1.37 | 35.8 – 1.66 | 35.1 – 1.94 | 29.5 – 1.72 | 29.5 – 1.60 | 29.5 – 1.92 |
| Protein atoms (AC) d | 3988 (310) | 3857 (89) | 3981 (209) | 3883 (115) | 3961 (171) | 3947 (135) |
| Ligand atoms | 20 | 18 | 14 | 12 | 18 | 24 |
| Cl− / glycerol atoms | 4 / 0 | 4 / 0 | 7 / 12 | 6 / 0 | 7 / 0 | 4 / 0 |
| Water atoms | 689 | 435 | 262 | 564 | 516 | 291 |
| Rwork / Rfree (%) e | 13.8 / 16.9 | 15.7 / 18.0 | 14.7 / 18.3 | 15.8 / 19.5 | 15.0 / 17.7 | 15.5 / 18.7 |
| rms deviations | ||||||
| Bond lengths (Å) | 0.014 | 0.010 | 0.012 | 0.011 | 0.010 | 0.011 |
| Bond angles ° | 1.47 | 1.27 | 1.30 | 1.28 | 1.25 | 1.28 |
| Mean B-Values (Å2) | ||||||
| Protein overall | 21.8 | 21.3 | 37.4 | 19.7 | 24.5 | 27.6 |
| MC / SC f | 19.8 / 23.6 | 18.8 / 23.9 | 34.5 / 40.2 | 17.1 / 22.5 | 22.2 / 26.8 | 24.6 / 30.8 |
| Ligand | 13.2 | 13.3 | 26.4 | 9.3 | 16.1 | 21.2 |
| Cl ions / glycerol | 23.3 / − | 22.1 / − | 40.7 / 44.6 | 24.1 / − | 25.1 / − | 25.6 / − |
| Water | 35.6 | 30.8 | 40.9 | 31.2 | 36.5 | 35.2 |
| Ramachandran % g | 98.3 / 0 | 98.3 / 0 | 98.1 / 0 | 98.3 / 0 | 98.6 / 0 | 98.1 / 0 |
| PDB ID Code | 4IO2 | 4IO3 | 4IO4 | 4IO5 | 4IO6 | 4IO7 |
Values in parenthesis indicate the low resolution limit for the highest-resolution shell of data.
Values in parenthesis indicate statistics for the highest-resolution shell of data.
Rmerge = (Σ| II - <II>|) / ΣI |II|, where <II> is the mean L over symmetry–equivalent reflections.
Alternate conformations
Rwork = (Σ ||Fo| − |Fc|| ) / Σ |Fo|, where Fo and Fc denote observed and calculated structure factors, respectively; 5% of the reflections were set aside for the calculation of the Rfree value.
Main chain / Side chain
Preferred / Disallowed conformation
Figure 3.
Mechanism of binding of glutamate and aspartate. (A) Stereoview of an electron density omit map contoured at 5 σ for glutamate and seven water molecules trapped in the AvGluR1 ligand binding cavity. H-bonds anchoring the ligand in the binding site are represented as black dashed lines. Interaction of arginine residues Arg676 and Arg702 are unique among iGluRs. The solvent volume of the ligand binding cavity colored by electrostatic potential is shown as transparent surface, highlighting the positive charge of the cavity. For clarity, domain 1 residues 445-476 and 491-509 have been omitted. This includes Tyr497 that caps the binding site and forms H-bonds with Arg702 and Asp515 side chains. The S1 and S2 segments are colored cyan and gold, respectively. (B) Shows the equivalent view for the aspartate complex for which W4 moves into the position occupied by one of the γ-carboxyl group oxygen atoms in the glutamate complex; the Arg676 side chain was modeled with two conformations.
Water molecules trapped in the ligand binding cavity form a network of hydrogen bonds that link domains 1 and 2, and which mediate additional contacts of the bound glutamate and aspartate ligands to AvGluR1. Water molecules W1 to W6 line up against the base of the pyramidal shaped ligand binding cavity, with W7 occupying the vertex. The side chains of Asp743 in the second interdomain β-strand and Thr517 in domain 1 are connected to the terminal carboxyl groups of glutamate and aspartate via W1, W2 and W3. In a pocket adjacent to α-helix E in domain 2, W5 and W6 play a structural role, linking the main chain amide of Arg676 with the side chain hydroxyl group of Thr679; in the glutamate complex, W4 connects the ligand γ-carboxyl group to this water network, while in the aspartate complex W4 moves 3.4 Å to occupy a position isosteric with one of the ligand γ-carboxyl group oxygen atoms in the glutamate complex. At the vertex of the cavity, W7 links the side chain hydroxyl group of Thr723 with the β- and γ-carboxyl groups of aspartate and glutamate; in the aspartate complex W7 is also connected, via W4, to W8 in a new site created by an alternate conformation of the Arg676 side chain.
To gain further insight into the underlying mechanisms of ligand selectivity for AvGluR1 we performed docking experiments, using the glutamate complex N, C and Cα atoms as a template for least squares superpositions of additional ligands, followed by rotamer selection to obtain the best fit into an omit map. Docking of AMPA, in the conformation found in the GluA2 complex (1FTM), reveals a bad contact of the 5-methyl group with the Asp515 side chain (Figure S2A), accounting for its low affinity (Table S1). This clash is absent for the related agonist quisqualate, the oxadiazolidine ring of which substitutes for W7 and one of the glutamate γ-carboxyl group oxygen atoms. For kainate, which also binds with very low affinity, the 4-propenyl group clashes with Tyr497 and Arg702, while the γ-carboxyl group clashes with Thr679 (Figure S2B). Likewise, the 250-fold lower affinity of the potent kainate receptor agonist 2S,4R-4-methyl glutamate for AvGluR1 can be accounted for by bad contacts of the 4-methyl group with Arg676 and Arg702. Weak binding of NMDA probably originates from a clash between the α-amino N-methyl group and Asp720. By contrast, in NMDA receptors, the aspartate residue equivalent to Asp720 forms a water-mediated contact with the α-amino group, and this water is displaced to accommodate the N-methyl group of NMDA.
Chloride ions act as surrogate ligand atoms
Docking experiments also revealed a steric clash with Thr679 for the β-carbon branched amino acids valine, threonine and isoleucine which bind with low affinity (Figure S2), but did not give insight into the mechanism underlying the higher affinity binding of alanine, serine and methionine (Table S1). To address this we solved additional crystal structures for AvGluR1 alanine, serine, methionine and phenylalanine complexes. These revealed similar conformations and extents of domain closure to the acidic amino acid complexes, RMSD 0.19, 0.34, 0.24 and 0.45 Å for 242 Cα atoms superimposed on the glutamate complex, and identical interactions of the ligand α-amino and α-carboxyl groups; likewise, the position of water molecules W1, W2, W5 and W6 was conserved in all six structures (Figure 4 and Figure S3). Strikingly, the volume of the ligand binding pocket and AvGluR1 side chain conformations were essentially identical for all six structures except for the phenylalanine complex. Differences in ligand geometry and chemistry are instead accommodated by the recruitment of Cl− ions as surrogates for the γ-carboxyl group of glutamate, and by rearrangement of solvent structure in the ligand binding pocket. In the alanine complex W4 is displaced and a Cl− ion occupies a position equivalent to one of the ligand γ-carboxyl group oxygen atoms in the glutamate complex; the anion is coordinated by the Arg676 side chain, the main chain amide of Asp720, and by W3 and W7 (Figure 4A). In the serine complex the Cl− ion is displaced by 2.3 Å, and is coordinated by the side chains of both Arg702 and Arg676, and by the main chain amide of Asp720, while the ligand OH group forms H-bonds with W3 and the side chains of Asp720 and Arg676. Water structure in the binding cavity also differs in the alanine and serine complexes, due to movement of W3, which now forms H-bonds linking W2 and W5 (Figure 4B). Surprisingly, side chain conformations, and the location of the Cl− ion in the methionine complex, are essentially identical to those in the serine complex, with the thiomethyl group accommodated by displacement of W3 and W4 (Figure 4C). In the phenylalanine complex the bulky aromatic ring pushes the Arg676 and Arg702 side chains away from the binding site, displacing both the Cl− ion and W3 and W4, increasing the volume of the ligand binding site cavity to 541 Å3 (Figure 4D).
Figure 4.
Anions mediate binding of alanine, serine and methionine. (A) Electron density omit map contoured at 5 σ for alanine, six water molecules, and a Cl− ion trapped in the AvGluR1 ligand binding cavity; coloring and water numbering is the same as for Figure 3. (B) Shows the equivalent view for the serine complex; note the different position of the Cl− ion and displacement of W7. (C) Equivalent view for the methionine complex. (D) Equivalent view for the phenylalanine complex. (E) Equilibrium dose inhibition curves for displacement of 100 nM [3H] L-Glu by glutamate and aspartate in the absence of Cl−; curves for control responses, taken from Figure 1, are plotted as dashed lines. (F) Equilibrium dose inhibition curves for alanine, serine and methionine; note the large rightward shift in in the absence of Cl−.
To test whether Cl− ions play a key role in the binding of neutral and hydrophobic amino acids, as suggested by the crystal structures, the affinity for alanine, serine, methionine, glutamate and aspartate was measured by competitive displacement assays with [3H] L-glutamate, with NaCl in the reaction buffer substituted by an equimolar concentration of 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) titrated to pH 7.4 with NaOH. HEPES was chosen on the basis of pilot experiments that tested protein stability in Cl− free solutions with a range of anion substitutes large enough not to occupy the Cl− binding site. Kd values in Cl− versus HEPES were practically unaffected for glutamate, 0.27 ± 0.02 versus 0.30 ± 0.04 μM, and aspartate, 0.87 ± 0.08 versus 1.41 ± 0.12 μM (Figure 4E). By contrast, the affinity for serine and alanine decreased 48- and 73-fold respectively in the absence of Cl−, from 25 ± 1.9 to 1170 ± 49 μM for serine, and from 9.3 ± 1.2 to 677 ± 96 μM for alanine (Figure 4F). These results suggest that in the alanine and serine complexes the Cl− ion acts as an essential countercharge for the domain 2 binding site arginine residues, the guanidinium groups of which are separated by only 3.5 - 3.6 Å. Surprisingly, the affinity for methionine decreased only 18-fold, from 15.1 ± 2.5 to 270 ± 52 μM in HEPES, although there was strong electron density for a Cl− ion (Figure 4C). Modeling experiments suggest that in the absence of Cl− the bound methionine residue can adopt a different rotamer, positioning the thiomethyl group in the anion binding site, and acting as a barrier between the closely positioned arginine side chains.
Molecular architecture and evolution of the AvGluR1 LBD
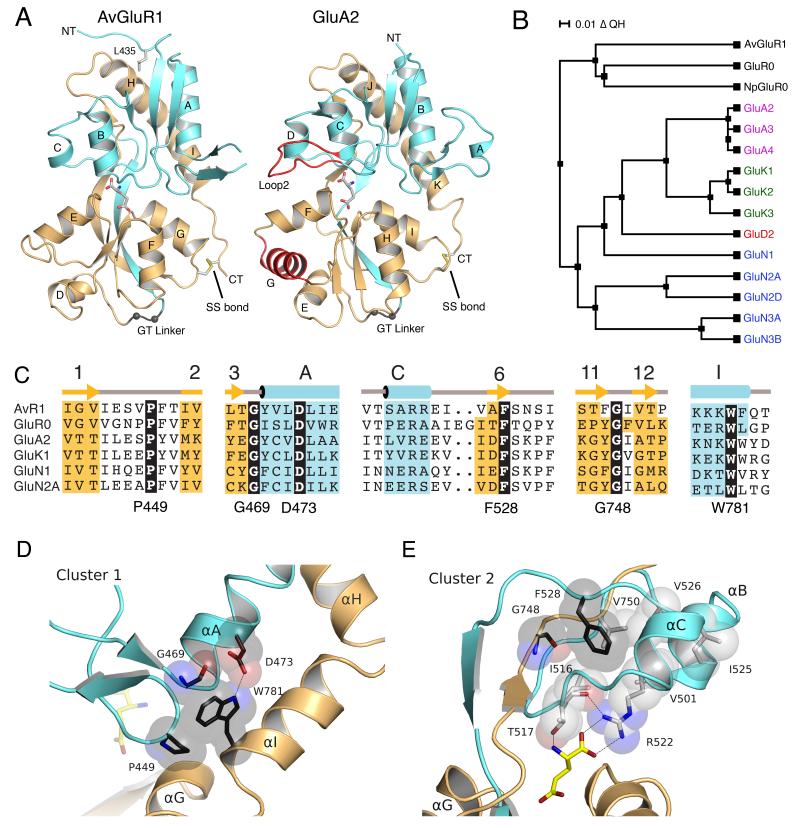
AvGluR1 was proposed to be an evolutionary link between prokaryotic and eukaryotic iGluR receptor classes (Janovjak et al., 2011), and thus we compared its structure to that of these receptor classes. The AvGluR1 LBD crystal structures reveal a two-domain closed-cleft clamshell typical of iGluRs, where the S1 and S2 segments form part of both domain 1 and domain 2 (Figure 5A). A structure based alignment with 14 other iGluR LBD crystal structures from prokaryotic iGluRs, AMPA receptors, kainate receptors and NMDA receptors revealed that 80% of AvGluR1 LBD Cα atoms can be superposed on other iGluR LBD structures with an RMSD of < 2 Å. Similar to bacterial iGluRs, the AvGluR1 LBD has deletions compared to AMPA, kainate and NMDA receptors, and lacks both loop 2, which has been shown to participate in interdomain contacts that stabilize the closed cleft conformation of AMPA and kainate receptor LBDs (Weston et al., 2006a), as well as α-helix G, shown in red for the GluA2 LBD crystal structure (Figure 5A). From this comparison, AvGluR1 appears closer to bacterial receptors than vertebrate iGluRs. However, in common with vertebrate iGluRs, AvGluR1 has a disulfide bond between Cys733 and Cys787, linking the loop following helix G in domain 2 with the C-terminal end of the LBD; both Cys residues are absent in GluR0, suggesting that the presence of this disulfide bond is evolutionarily linked to the addition of a third membrane spanning segment in eukaryotic iGluRs. Despite this, a structure based phylogenetic analysis reveals clustering of AvGluR1 with GluR0 LBDs from Synechocystis and Nostoc punctiforme (Figure 5B).
Figure 5.
Crystal structure of the AvGluR1 LBD reveals prokaryotic iGluR like features. (A) Ribbon diagrams showing a comparison of domain architecture for the glutamate complexes of AvGluR1 and eukaryotic iGluRs represented by GluA2; coloring are the same as in Figure 3, with the GT linker drawn in gray. Secondary structure features conserved in eukaryotic iGluRs but absent in AvGluR1 are colored red in the GluA2 structure. (B) A phylogenetic tree based on additional structural alignments reveals clustering of AvGluR1 with bacterial iGluRs. QH indicates an overall structural similarity score between structures. (C) A structure based sequence alignment for AvGluR1 with representative prokaryotic and eukaryotic iGluRs reveal highly conserved residues widely scattered in linear sequence and not involved in ligand binding (black boxes); cyan and yellow coloring indicates α-helices (and one 310 helix) and β-strands, respectively; α-helices for GluA2 and GluK2 are labeled in italics; gray dots in the AvGluR1 secondary structure schematic indicate six residues in loop 1 for which no main chain electron density was observed; red and green asterisks indicate residues forming direct, or solvent mediated contacts with glutamate, respectively. (D) Structure of the cluster 1 core of conserved residues. (E) Structure of the hydrophobic cluster of core 2 conserved residues.
Structure based sequence alignments revealed six amino acids that are conserved in all known bacterial and vertebrate iGluRs but which do not play any direct role in ligand binding. To investigate why these residues, which are scattered in linear sequence (black boxes in Figure 5C and Figure S1B) but conserved in iGluRs with diverse ligand binding properties, we mapped them back to the AvGluR1 crystal structure, and found that they form two distinct clusters which play a key role in organizing domain 1 as a rigid body primed to bind glutamate in a dock and lock mechanism (Abele et al., 2000). The first cluster, composed of Pro449, Gly469, Asp473 and Trp781, links α-helix A with α-helix I (Figure 5D). The second cluster, composed of Phe528 and Gly748, forms part of a conserved hydrophobic core that positions Arg522 in an extended conformation ready to bind glutamate (Figure 5E).
Our initial attempts at crystallization were hindered by inability of thrombin to cleave the N-terminal His tag; this was overcome by extending the native N-terminal sequence by four residues (ARLK) compared to GluA2 and GluK2 LBDs (Armstrong and Gouaux, 2000; Mayer, 2005). In the AvGluR1 LBD structure the side chain of Leu435 in the ARLK sequence is wedged in a conserved hydrophobic pocket located between α-helices A and H and β-strand 1 on the upper surface of domain 1 (Figure 5A). The trapping of Leu435 is of potential significance because this region is involved in the allosteric regulation of iGluR activation and desensitization, and although in the full length GluA2 structure this segment is solvent exposed, the linker was shortened by deletion of six residues. Thus it is possible that in intact iGluRs the ATD-LBD linker adopts a different conformation that buries Leu435, as observed in the AvGluR1 LBD crystal structure.
Sedimentation analysis of AvGluR1 LBD assembly
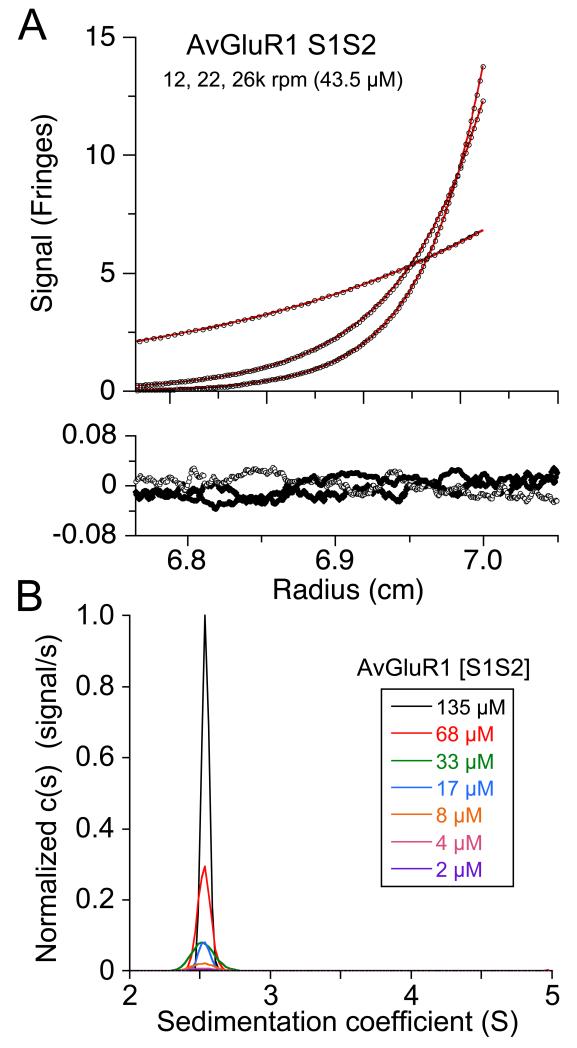
Glutamate receptor ion channels are tetramers in which the extracellular domains assemble as a dimer of dimers (Sobolevsky et al., 2009). A distinguishing feature of eukaryotic versus prokaryotic iGluRs is a large difference in affinity for LBD dimer assembly. In AMPA, kainate and NMDA receptors the LBDs interact very weakly in solution, with a monomer-dimer Kd > 5-10 mM (Furukawa et al., 2005; Sun et al., 2002; Weston et al., 2006b), while the amino terminal domains form dimers at sub micromolar protein concentrations (Jin et al., 2009; Kumar et al., 2011; Rossmann et al., 2011; Zhao et al., 2012); this strong ATD interaction plays a key role in receptor biogenesis. By contrast, for Synechocystis and Nostoc punctiforme iGluRs, which do not have an amino terminal domain, the LBDs interact more strongly, with a monomer-dimer Kd of 0.8 and 5 μM respectively (Lee et al., 2008; Mayer et al., 2001), suggesting that for iGluRs which lack an ATD, LBD interactions are required for efficient receptor assembly. In this context, AvGluR1 stands out because, although like other eukaryotic iGluRs AvGluR1 has a large amino terminal domain (Janovjak et al., 2011), the results of a structure based phylogenetic analysis reveals instead that the LBD of AvGluR1 clusters with bacterial iGluRs (Figure 5B). However, we found using sedimentation analysis that there was no measureable self-association of the AvGluR1 LBD at protein concentrations of up to 135 μM (4 mg/ml), similar to the behavior of other eukaryotic iGluRs. We initially performed a sedimentation equilibrium experiment (Figure 6A), for which a weight-average MW of 27,346 g/mol [95% CI 27,146 −27,546] was determined from a global analysis of nine data sets acquired at 3 speeds (12,000, 22,000 and 26,000 rpm) for 3 loading concentrations (5, 14, and 44 μM), in excellent agreement with the predicted mass of 27,462 g/mol based on the amino acid sequence. Globally fitting the sedimentation equilibrium data to a monomer-dimer association model, χ2 value 6.96, did not improve fit quality compared to a single species model, χ2 value 6.87. To investigate higher protein concentrations we performed a sedimentation velocity experiment with loading concentrations varying from 2 μM to 135 μM (Figure 6B). The isotherm of weighted average sedimentation coefficients did not reveal any concentration dependent shift, with an average value for Sw(S) of 2.53S. Thus, although the AvGluR1 LBD has prokaryotic like structural features, its low affinity oligomerization is like that of other eukaryotic iGluRs.
Figure 6.
Low affinity for dimer assembly by the AvGluR1 LBD. (A) Sedimentation equilibrium scans for the AvGluR1 LBD, initial loading concentration 43.5 μM, at rotor speeds of 12, 22 and 26 krpm, fit with a single species model; the lower panel shows residuals from a global fit of nine data sets: Three loading concentrations, each run at three speeds. (B) Sedimentation velocity profiles for seven loading concentrations varying from 2 μM to 135 μM, reveals only a single species.
AvGluR1 LBD dimer crystal structures
Despite low affinity for self association, AvGluR1 LBD amino acid complexes crystallized as back to back dimers (Figure 7), the canonical arrangement found in the full length GluA2 structure (Sobolevsky et al., 2009). The AvGluR1 glutamate complex dimer has a buried surface of 1305 Å2 per subunit (Figure S4A), with two-fold symmetric contacts between α-helices C and H in the upper lobe forming most of the dimer contact surface, but with no contacts on the dimer axis of symmetry as found in other eukaryotic iGluR LBD dimer assemblies. However, different from AMPA and kainate receptors, but similar to NMDA receptors (Furukawa et al., 2005), α-helix E in domain 2 forms intermolecular contacts with α-helix H in domain 1 of the dimer partner. By comparing the angle between structurally equivalent pairs of α-helices, which form the domain 1 dimer contact surface, we found that subunit orientation in the AvGluR1 dimer differed from that of other eukaryotic iGluRs. For AvGluR1 the angle between α-helix H in the two subunits was 45° (Figure 7A); for GluA2 the two subunits pivot away from each other such that the angle between the equivalent α-helix in the two subunits increases to 61° (Figure 7B). As a result of the more ‘upright’ poise of subunits in the AvGluR1 dimer assembly the distance between threonine Cα atoms in the GT linker that replaces the ion channel segments increases from 40 Å in the GluA2 dimer, to 49 Å in the AvGluR1 dimer. Similar measurements for other AMPA, kainate and NMDA receptor dimer assemblies gave distances of 38, 39, 43, 38 and 36 Å, and angles of 59, 61, 56, 65 and 50° for GluA3, GluA4, GluK1, GluK2 and the GluN1/GluN2A heterodimer. By contrast, GluR0 (49 Å and 35°) also has an upright dimer assembly like that for AvGluR1, indicating that the AvGluR1 dimer assembly retains prokaryotic features consistent with the results of phylogenetic analysis for the LBD monomers (Figure 6B).
Figure 7.
Crystal structure and functional analysis of the AvGluR1 LBD dimer assembly. (A and B) The AvGluR1 dimer has an ‘upright’ orientation compared to the GluA2 dimer; α-Helix H and its symmetry mate in AvGluR1, and α-helix J and its symmetry mate in GluA2, are drawn as transparent cylinders; the angle between vectors running through the center of these helices is 45° in AvGluR1 and 61° in GluA2. (C) Stereoview of the AvGluR1 LBD dimer assembly viewed parallel to the dimer interface formed by α-helices C and H, with a ribbon diagram for subunits colored gold and rose; side chains mediating salt bridges and polar interactions are drawn as sticks connected by dashed lines; chloride ions are drawn as spheres; Leu766 is located in the lower third of α-helix H. (D) Desensitization is abolished by the S520C/L766C mutation; the left panel shows a control response to 100 μM glutamate recorded from wild type AvGluR1, with the onset of desensitization fit with a single exponential of time constant 460 ms; the next three panels show responses to 100 μM glutamate recorded from one oocyte for the AvGluR1 S520C/L766C mutant before, during, and 20 min after application of 10 mM DTT for 5 min.
The AvGluR1 dimer interface is formed primarily by polar interactions between the back faces of α-helices C and H along with contributions from α-helices E, I, β-strands 6 and 12 (Figure 7C). Inter molecular contacts are generated by salt bridges between Arg523 and Asp762, Glu524 and Lys759, Asp743 and Arg769; by hydrogen bonds between the side chains of Ser520 and Glu770, Arg687 and the main chain carbonyl of Glu770, and between Thr746 and Arg769; hydrophobic side chain interactions are sparse and limited to contacts between Val518 and Leu766. The upright arrangement of the AvGluR1 dimer assembly creates a solvent filled tunnel with an electropositive surface potential (Figure S4B) that penetrates the length of α-helix H and which contains four Cl− ions coordinated by the Arg523 and Arg769 side chains. In kainate receptor GluK1 and GluK2 LBD dimer assemblies a single Cl− ion also binds at the dimer interface, but is buried in a small cavity instead of a solvent filled tunnel (Chaudhry et al., 2009; Plested and Mayer, 2007). Its is probable that as for GluK1 and GluK2, the binding of Cl− ions helps to stabilize AvGluR1 dimer assembly by acting as a counter charge to arginine side chains present in the dimer interface.
In AMPA and kainate receptors, consistent with models that predict rupture of the LBD dimer assembly during desensitization (Sun et al., 2002), introducing Cys mutations to form disulfide crosslinks between α-helices D and J in adjacent subunit pairs abolishes desensitization (Weston et al., 2006b). At the structurally equivalent positions in AvGluR1, Ser520 and Leu766, we modeled Cys residues and found that these predicted formation of disulfide bonds with good geometry; consistent with this, desensitization in response to 100 μM glutamate was completely abolished for the AvGluR1 S520C/L766C mutant (Figure 7D). Prolonged applications of the reducing agent DTT reversibly inhibited responses to glutamate for the S520C/L766C mutant but did not induce desensitization like that for wild type AvGluR1; after removal of DTT responses to glutamate recovered without application of oxidizing reagents. The smaller amplitude of responses for the S520C/L766C mutant under reducing conditions, without measurable desensitization, suggests that the Cys mutations might also perturb gating, perhaps via steric effects, as seen for the equivalent GluA2 mutant (Weston et al., 2006b).
In conclusion the ligand binding properties of AvGluR1 are distinct, and resemble neither those in eukaryotic glutamate receptors, nor their previously characterized prokaryotic precursors. It is widely believed that the two domain Venus flytrap structure of iGluR LBDs evolved from the large family of bacterial periplasmic binding proteins whose members utilize the same scaffold to selectively bind a wide variety of ligands, including small oxyanions, mono- and oligo-saccharides, amino acids, oligopeptides, polyamines and vitamins. AvGluR1 represents a new example where the same protein molecule is able to bind to chemically diverse amino acids, using ions as substitute ligands. This raises the question as to why glutamate was selected as a neurotransmitter amongst a wide spectrum of candidate amino acids and small molecules, and if fine tuning was subsequently required during evolution to remove sensitivity to other amino acids, as a necessary step for high fidelity information processing at synapses. Binding of the glutamate γ-carboxyl group by arginine residues in AvGluR1 has precedent from the mechanism found in mGluR GPCRs, for which Arg and Lys residues play a similar role; however in mGluRs these residues are located in domain 1 not in domain 2 (Kunishima et al., 2000). Intriguingly the binding of AP4 to mGluR4 requires Cl− ions (Kuang and Hampson, 2006), while for mGluR1 a putative anion binding site remote from the glutamate γ-carboxyl group has been proposed to create a pocket for binding of subtype selective ligands (Acher et al., 2011), but structural information on the anion binding site for mGluRs is lacking. Consistent with its proposed role as an evolutionary intermediate (Janovjak et al., 2011), structure based phylogenetic analysis revealed that the AvGluR1 LBD most closely resembles prokaryotic iGluRs, although the bound glutamate ligand adopts the same twisted conformation found in AMPA, kainate and NMDA receptors, instead of the extended conformation found in prokaryotic iGluRs. An analysis of dimer structures also reveals AvGluR1 LBD packing like that in prokaryotic iGluRs, while analytical ultracentrifugation experiments revealed instead a low affinity for dimer assembly like that for eukaryotic iGluRs.
EXPERIMENTAL PROCEDURES
Construct Design and protein expression
Design of the AvGluR1 LBD construct was based on domain boundaries demarcated previously for AMPA and kainate receptor LBDs, but with a 4 residue extension at the N-terminus, and included S1 residues A433-K543 and S2 residues L656-P788 joined by a GT linker. These were isolated from the full length cDNA by overlap PCR, cloned into pET22b with an N-terminal MH8SSGLVPRGS affinity tag and thrombin cleavage site, sequenced, and expressed in E. coli Origami B(DE3) induced with 30 μM IPTG for 15 hours at 18 °C. The soluble fraction from bacterial cell lysates was purified using Ni-NTA chromatography, followed by thrombin cleavage, and ion exchange chromatography using SP sepharose. Final yields were typically 8-10 mg from 12 liter cultures.
Ligand binding assays
Reactions were carried out in a ligand binding buffer (LBB) containing 150 mM NaCl, 2 mM EDTA, 10 mM HEPES pH 7.0 and 10% glycerol. Apo protein was generated by extensive dialysis against LBB, with 8 changes over 5 days, for a total volume exchange of 1020. Reactions were set up on ice in 96-well plates with 250 nM apo protein and 10 μg/ml BSA in 200 μl LBB per well with [3H] L-Glu (50.6 Ci/mmol) diluted 1:20 with [1H] L-Glu; non specific binding was measured in the presence of 20 mM alanine. Competing ligands were added either at 100 μM, or as a concentration series, and incubated for 120 min prior to filtering through Millipore multiscreen IP filter plates pre-wet with 50% ethanol, and washed twice with ice cold LBB both before and after filtration of the binding reactions. Plates were dried thoroughly, sealed with clear adhesive plastic tape, incubated with 50 μl/well Optiphase Supermix (Perkin Elmer) scintillation fluid and counted on a liquid scintillation counter (Wallac Trilux 1450 microbeta).
Electrophysiological analysis
Two electrode voltage clamp recordings were performed using stage 5-6 Xenopus laevis oocytes injected with AvGluR1 cRNA, agarose cushion electrodes filled with 3 M KCl (Schreibmayer et al., 1994), and a custom made recording chamber to allow rapid solution exchange, with ligands applied using computer controlled valves as described previously (Panchenko et al., 1999). The extracellular solution contained (in mM) 100 NaCl, 1 KCl, 2 CaCl2, 1 MgCl2 and 5 HEPES titrated to pH 7.4 with NaOH; in some experiments the divalent ion concentration was changed to 10 μM CaCl2 and 2 mM MgCl2.
Crystallization and structure determination
Crystallization experiments used protein dialyzed against 50 mM NaCl, 10 mM TrisCl pH 8.0, 1 mM EDTA and either 2 mM L-Glu or 10 mM of either L-Asp, L-Ala, L-Ser or L-Met, or 50 mM L-Phe. Trays were setup in hanging drop format at 20 °C using a protein concentration of 5 to 10 mg/ml at a 1:1 or 1:2 protein:reservoir volume ratio, with a reservoir containing 0.1 M BisTris propane pH 6.5, 50 or 100 mM NaCitrate and 17.5 to 20.0% PEG 3350. Diffraction quality crystals were obtained by streak seeding, cryoprotected by serial transfers to glycerol, final concentration 10-15%, and flash frozen in liquid nitrogen. Data were collected at APS beamline ID22, using 1 Å radiation and a MAR300 CCD detector. Diffraction data were indexed, scaled and merged using HKL2000 (Otwinowski and Minor, 1997). The AvGluR1 LBD glutamate complex was solved by molecular replacement with Phaser (McCoy et al., 2007) using a GluK2 monomer (1S50) as a search probe after deletion of residues 18-34 (loop 1), 63-72 (loop 2) and 254-259 (C terminus), with mutation of all side chains to Ser; two molecules were located in the asymmetric unit with rotation and translation Z scores of 6.3, 5.7 for protomer 1 and 5.6, 10.7 for protomer 2. The solution was successfully built using Phenix Autobuild (Adams et al., 2010). The aspartate, alanine, methionine and phenylalanine complexes were solved by Fourier difference techniques, using the refined coordinates for the AvGluR1 LBD glutamate complex stripped of alternate conformations and heteroatoms as a starting model; for the serine complex molecular replacement was required due to a difference in cell dimensions (Table 1), with rotation and translation Z scores of 8.7, 8.1 for protomer 1 and 10.8, 17.9 for protomer 2. Iterative cycles of refinement and model building were carried out using Phenix (Adams et al., 2010) and Coot (Emsley et al., 2010) with either 4 TLS groups or for the L-Glu complex individual anisotropic B-factors. For the glutamate and alanine complexes the location of Cl− ions was confirmed by calculation of anomalous difference Fourier maps using data collected at 1.5418 Å in the home lab. Models were validated with Molprobity (Chen et al., 2010), with the following scores, where 100% is the best amongst structures of comparable resolution: glutamate 0.95 (99%); aspartate 0.96 (100%); serine 1.01 (100%); alanine 1.12 (99%); methionine (0.94) 100% phenylalanine (0.98) 100%; Cavity calculations were performed using VOIDOO with a probe radius of 1.4 Å on a 1.0 Å grid (Kleywegt, 1994); LSQMAN was used for superpositions (Kleywegt, 1996). Figures were generated using PyMol (Schrödinger, LLC).
Structural analysis
The pairwise structural alignment algorithm DALI was used for the identification of AvGluR1 LBD homologs with known structure (Holm and Rosenstrom, 2010). Structure based sequence alignments were generated based on a progressive pairwise heuristic algorithm as implemented in MUSTANG (Konagurthu et al., 2006). Structure based phylogeny was calculated based on the structural similarity score (QH) using the MultiSeq module of VMD (Humphrey et al., 1996). The value of QH indicates an overall structural similarity score between two structures and is calculated using the equation QH = ℵ−1 [qaln + qgap], where ℵ is the normalization that accounts the contribution from both contacts between the aligned regions as well as between the residue present in the aligned position and the gap region, qaln represents the fraction of Cα-Cα distances that are similar between the two aligned structures, and qgap introduces a penalty term to account for the presence of insertions with the QH value decreasing with larger perturbations (O’Donoghue and Luthey-Schulten, 2005).
Analytical Ultracentrifugation
Sedimentation velocity (SV) and equilibrium (SE) experiments were performed in a ProteomeLab XL-I analytical ultracentrifuge (Beckman Coulter, Indianapolis IN). Samples were prepared by dilution of a concentrated protein stock using reference buffer (20 mM phosphate buffer pH 7.5, 150 mM NaCl, 1 mM EDTA and 2 mM L-glutamate) and loaded into cell housings with either 3, or 12 mm double-sector charcoal-filled epon centerpieces and sapphire windows. For SV, the evolution of the concentration gradient was recorded using absorbance optics at wavelengths of 250 and 280 nm and interference detection at a rotor speed of 50 krpm and temperature of 20 °C. Data were analyzed in SEDFIT using a c(s) distribution model with maximum entropy regularization (P = 0.68) and systematic noise decomposition leaving the meniscus position and weight-average frictional ratio as fitting parameters (Schuck, 2003). The buffer density (1.006995 g/mL) and viscosity (1.0373 cP) were measured using a DMA500M density meter or an AmVn microviscometer from Anton Paar (Graz, Austria). For SE, the equilibrium concentration profiles were recorded using interference detection at rotor speeds of 12, 22, and 26 krpm and a temperature of 10 °C. Data were analyzed in SEDPHAT using a single species model or a monomer-dimer equilibrium association model with mass conservation constraints (Vistica et al., 2004). The buffer density (1.009043 g/mL) and viscosity (1.3316 cP) were measured using a DMA500M density meter or an AmVn microviscometer from Anton Paar (Graz, Austria), respectively.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. E. Isacoff for the AvGluR1 cDNA; the Swartz lab, NINDS for oocytes; Andi Balbo and Carla Glasser for technical assistance. Synchrotron diffraction data was collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source (APS), Argonne National Laboratory. Use of APS was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. This work was supported by the intramural research program of NICHD, NIH, DHHS.
Footnotes
ACCESSION CODES
Atomic coordinates and structure factors have been deposited in the PDB with accession codes of 4IO2, 4IO3, 4IO4, 4IO5, 4IO6 and 4IO7 (Table 1).
REFERENCES
- Abele R, Keinanen K, Madden DR. Agonist-induced isomerization in a glutamate receptor ligand-binding domain. A kinetic and mutagenetic analysis. J Biol Chem. 2000;275:21355–21363. doi: 10.1074/jbc.M909883199. [DOI] [PubMed] [Google Scholar]
- Acher FC, Selvam C, Pin JP, Goudet C, Bertrand HO. A critical pocket close to the glutamate binding site of mGlu receptors opens new possibilities for agonist design. Neuropharmacology. 2011;60:102–107. doi: 10.1016/j.neuropharm.2010.07.002. [DOI] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica. Section D, Biological crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong N, Gouaux E. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: Crystal structures of the GluR2 ligand binding core. Neuron. 2000;28:165–181. doi: 10.1016/s0896-6273(00)00094-5. [DOI] [PubMed] [Google Scholar]
- Chaudhry C, Plested AJ, Schuck P, Mayer ML. Energetics of glutamate receptor ligand binding domain dimer assembly are modulated by allosteric ions. Proc Natl Acad Sci U S A. 2009;106:12329–12334. doi: 10.1073/pnas.0904175106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GQ, Cui C, Mayer ML, Gouaux E. Functional characterization of a potassium-selective prokaryotic glutamate receptor. Nature. 1999;402:817–821. doi: 10.1038/45568. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta crystallographica. Section D, Biological crystallography. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu JC, Brenner ED, DeSalle R, Nitabach MN, Holmes TC, Coruzzi GM. Phylogenetic and expression analysis of the glutamate-receptor-like gene family in Arabidopsis thaliana. Molecular biology and evolution. 2002;19:1066–1082. doi: 10.1093/oxfordjournals.molbev.a004165. [DOI] [PubMed] [Google Scholar]
- Croset V, Rytz R, Cummins SF, Budd A, Brawand D, Kaessmann H, Gibson TJ, Benton R. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS genetics. 2010;6:e1001064. doi: 10.1371/journal.pgen.1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica. Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everts I, Petroski R, Kizelsztein P, Teichberg VI, Heinemann SF, Hollmann M. Lectin-induced inhibition of desensitization of the kainate receptor GluR6 depends on the activation state and can be mediated by a single native or ectopic N-linked carbohydrate side chain. J Neurosci. 1999;19:916–927. doi: 10.1523/JNEUROSCI.19-03-00916.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa H, Singh SK, Mancusso R, Gouaux E. Subunit arrangement and function in NMDA receptors. Nature. 2005;438:185–192. doi: 10.1038/nature04089. [DOI] [PubMed] [Google Scholar]
- Ger MF, Rendon G, Tilson JL, Jakobsson E. Domain-based identification and analysis of glutamate receptor ion channels and their relatives in prokaryotes. PloS one. 2010;5:e12827. doi: 10.1371/journal.pone.0012827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. Journal of Molecular Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Janovjak H, Sandoz G, Isacoff EY. A modern ionotropic glutamate receptor with a K(+) selectivity signature sequence. Nat Commun. 2011;2:232. doi: 10.1038/ncomms1231. [DOI] [PubMed] [Google Scholar]
- Jin R, Singh SK, Gu S, Furukawa H, Sobolevsky AI, Zhou J, Jin Y, Gouaux E. Crystal structure and association behaviour of the GluR2 amino-terminal domain. EMBO J. 2009;28:1812–1823. doi: 10.1038/emboj.2009.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleywegt GJ. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta crystallographica. Section D, Biological crystallography. 1994;50:178–185. doi: 10.1107/S0907444993011333. [DOI] [PubMed] [Google Scholar]
- Kleywegt GJ. Use of non-crystallographic symmetry in protein structure refinement. Acta crystallographica. Section D, Biological crystallography. 1996;52:842–857. doi: 10.1107/S0907444995016477. [DOI] [PubMed] [Google Scholar]
- Konagurthu AS, Whisstock JC, Stuckey PJ, Lesk AM. MUSTANG: a multiple structural alignment algorithm. Proteins. 2006;64:559–574. doi: 10.1002/prot.20921. [DOI] [PubMed] [Google Scholar]
- Kuang D, Hampson DR. Ion dependence of ligand binding to metabotropic glutamate receptors. Biochemical and biophysical research communications. 2006;345:1–6. doi: 10.1016/j.bbrc.2006.04.064. [DOI] [PubMed] [Google Scholar]
- Kumar J, Schuck P, Mayer ML. Structure and assembly mechanism for heteromeric kainate receptors. Neuron. 2011;71:319–331. doi: 10.1016/j.neuron.2011.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunishima N, Shimada Y, Tsuji Y, Sato T, Yamamoto M, Kumasaka T, Nakanishi S, Jingami H, Morikawa K. Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor. Nature. 2000;407:971–977. doi: 10.1038/35039564. [DOI] [PubMed] [Google Scholar]
- Lam HM, Chiu J, Hsieh MH, Meisel L, Oliveira IC, Shin M, Coruzzi G. Glutamate-receptor genes in plants. Nature. 1998;396:125–126. doi: 10.1038/24066. [DOI] [PubMed] [Google Scholar]
- Lee JH, Kang GB, Lim HH, Jin KS, Kim SH, Ree M, Park CS, Kim SJ, Eom SH. Crystal structure of the GluR0 ligand-binding core from Nostoc punctiforme in complex with L-glutamate: structural dissection of the ligand interaction and subunit interface. J Mol Biol. 2008;376:308–316. doi: 10.1016/j.jmb.2007.10.081. [DOI] [PubMed] [Google Scholar]
- Mayer ML. Crystal Structures of the GluR5 and GluR6 Ligand Binding Cores: Molecular Mechanisms Underlying Kainate Receptor Selectivity. Neuron. 2005;45:539–552. doi: 10.1016/j.neuron.2005.01.031. [DOI] [PubMed] [Google Scholar]
- Mayer ML, Olson R, Gouaux E. Mechanisms for ligand binding to GluR0 ion channels: crystal structures of the glutamate and serine complexes and a closed apo state. J Mol Biol. 2001;311:815–836. doi: 10.1006/jmbi.2001.4884. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donoghue P, Luthey-Schulten Z. Evolutionary profiles derived from the QR factorization of multiple structural alignments gives an economy of information. J Mol Biol. 2005;346:875–894. doi: 10.1016/j.jmb.2004.11.053. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–344. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Panchenko VA, Glasser CR, Partin KM, Mayer ML. Amino acid substitutions in the pore of rat glutamate receptors at sites influencing block by polyamines. J Physiol (Lond) 1999;520:337–357. doi: 10.1111/j.1469-7793.1999.t01-1-00337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partin KM, Patneau DK, Winters CA, Mayer ML, Buonanno A. Selective modulation of desensitization at AMPA versus kainate receptors by cyclothiazide and concanavlin A. Neuron. 1993;11:1069–1082. doi: 10.1016/0896-6273(93)90220-l. [DOI] [PubMed] [Google Scholar]
- Plested AJ, Mayer ML. Structure and mechanism of kainate receptor modulation by anions. Neuron. 2007;53:829–841. doi: 10.1016/j.neuron.2007.02.025. [DOI] [PubMed] [Google Scholar]
- Rossmann M, Sukumaran M, Penn AC, Veprintsev DB, Babu MM, Greger IH. Subunit-selective N-terminal domain associations organize the formation of AMPA receptor heteromers. Embo J. 2011;30:959–971. doi: 10.1038/emboj.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreibmayer W, Lester HA, Dascal N. Voltage clamping of Xenopus laevis oocytes utilizing agarose-cushion electrodes. Pflügers Arch. 1994;426:453–458. doi: 10.1007/BF00388310. [DOI] [PubMed] [Google Scholar]
- Schuck P. On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal Biochem. 2003;320:104–124. doi: 10.1016/s0003-2697(03)00289-6. [DOI] [PubMed] [Google Scholar]
- Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Begovic E, Chapman J, Putnam NH, Hellsten U, Kawashima T, Kuo A, Mitros T, Salamov A, Carpenter ML, et al. The Trichoplax genome and the nature of placozoans. Nature. 2008;454:955–960. doi: 10.1038/nature07191. [DOI] [PubMed] [Google Scholar]
- Stern-Bach Y, Bettler B, Hartley M, Sheppard PO, O’Hara PJ, Heinemann SF. Agonist-selectivity of glutamate receptors is specified by two domains structurally related to bacterial amino acid binding proteins. Neuron. 1994;13:1345–1357. doi: 10.1016/0896-6273(94)90420-0. [DOI] [PubMed] [Google Scholar]
- Sun Y, Olson R, Horning M, Armstrong N, Mayer M, Gouaux E. Mechanism of glutamate receptor desensitization. Nature. 2002;417:245–253. doi: 10.1038/417245a. [DOI] [PubMed] [Google Scholar]
- Vistica J, Dam J, Balbo A, Yikilmaz E, Mariuzza RA, Rouault TA, Schuck P. Sedimentation equilibrium analysis of protein interactions with global implicit mass conservation constraints and systematic noise decomposition. Anal Biochem. 2004;326:234–256. doi: 10.1016/j.ab.2003.12.014. [DOI] [PubMed] [Google Scholar]
- Weston MC, Gertler C, Mayer ML, Rosenmund C. Interdomain interactions in AMPA and kainate receptors regulate affinity for glutamate. J Neurosci. 2006a;26:7650–7658. doi: 10.1523/JNEUROSCI.1519-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston MC, Schuck P, Ghosal A, Rosenmund C, Mayer ML. Conformational restriction blocks glutamate receptor desensitization. Nature Structural and Molecular Biology. 2006b;13:1120–1127. doi: 10.1038/nsmb1178. [DOI] [PubMed] [Google Scholar]
- Zhao H, Berger AJ, Brown PH, Kumar J, Balbo A, May CA, Casillas E, Jr., Laue TM, Patterson GH, Mayer ML, Schuck P. Analysis of high-affinity assembly for AMPA receptor amino-terminal domains. J Gen Physiol. 2012;139:371–388. doi: 10.1085/jgp.201210770. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.