Abstract
Bla g 4 is a male cockroach specific protein and is one of the major allergens produced by Blattella germanica (German cockroach). This protein belongs to the lipocalin family that comprises a set of proteins that characteristically bind small hydrophobic molecules and play a role in a number of processes such as: retinoid and pheromone transport, prostaglandin synthesis and mammalian immune response. Using NMR and Isothermal Titration Calorimetry we demonstrated that Bla g 4 binds tyramine and octopamine in solution. In addition, crystal structure analysis of the complex revealed details of tyramine binding. As tyramine and octopamine play important roles in invertebrates, and are counterparts to vertebrate adrenergic transmitters, we speculate that these molecules are physiological ligands for Bla g 4. The nature of binding these ligands to Bla g 4 sheds light on the possible biological function of the protein. In addition, we performed a large-scale analysis of Bla g 4 and Per a 4 (an allergen from American cockroach) homologs to get insights into the function of these proteins. This analysis together with a structural comparison of Blag 4 and Per a 4 suggests that these proteins may play different roles and most likely bind different ligands.
Keywords: Cockroach allergen, Bla g 4, Per a 4, tyramine, octopamine, allergy
1. Introduction
According to the National Center for Health Statistics, more than 25 million Americans (8.4% of the US population) had asthma in 2010 and the prevalence of the disease continues to increase. Moreover, asthma is more prevalent among children, women, African-Americans, Native Americans, and those with family incomes below the poverty line. While the disease can be triggered by many different environmental factors, exposure to cockroach allergens has been identified in numerous studies as a prominent risk factor for asthma (Call et al., 1992; Cohn et al., 2006; Eggleston et al., 1998; Gelber et al., 1993; Matsui et al., 2003). Initial studies investigating the role of cockroach allergies in asthma focused on inner city populations, but subsequent evidence suggests that the problem is more widespread. Surveys have estimated that measurable concentrations of cockroach allergens are present in a majority of all US households, and that the rate of sensitization to cockroach allergens among suburban middle-class children with asthma is much larger than previously suspected (Cohn et al., 2006; Matsui et al., 2003).
In 1964, Bernton and Brown, (Bernton and Brown, 1964) were the first to link cockroaches with allergic disease. Since then multiple cockroach-allergen specific proteins have been identified (Arruda, Vailes, Hayden, et al., 1995; Arruda, Vailes, Mann, et al., 1995; Arruda et al., 1997; Helm et al., 1996; Santos et al., 1999). Cockroach allergens are produced mainly by two species: Blattella germanica (German cockroach) and Periplaneta americana (American cockroach). The German cockroach is most commonly found in Europe and the US, while the American cockroach is predominantly found in South America and some regions in Asia. There are 14 cockroach allergens officially registered on the WHO/IUIS list of Allergen Nomenclature (www.allergen.org), eight from B. germanica and six from P. americana. Among all patients sensitive to B. germanica allergens, 95% are sensitized to one (or more) of four allergens: Bla g 1, Bla g 2, Bla g 4, and Bla g 5. Of these allergen proteins, Bla g 1, Bla g 2 and Bla g 4 have been structurally characterized. Bla g 1 is suggested to have a digestive function and represent a novel fold with the capacity to bind lipids (Mueller et al., 2013). Bla g 2 is an atypical aspartic protease (Gustchina et al., 2005), while Bla g 4 is a lipocalin (Arruda, Vailes, Hayden, et al., 1995; Tan et al., 2009).
The lipocalin family comprises a set of proteins that characteristically bind small hydrophobic molecules and play a role in a number of processes, such as retinoid and pheromone transport, prostaglandin synthesis and mammalian immune response. Lipocalins are typified by their ability to bind small molecules; however, no ligands that bind Bla g 4 had been previously identified and the function of the protein is unknown. Bla g 4 is produced in the conglobate gland and apical utricles of the male reproductive system of B. germanica (Fan et al., 2005). Bla g 4 is later transferred along with spermatophore to the female’s genital tract during copulation. The fate of Bla g 4 in females is unknown, but it has been demonstrated that the immunoreactivity of Bla g 4 disappears 24 hours after mating (Fan et al., 2005; Gore and Schal, 2007).
In this paper, we demonstrate that Bla g 4 specifically binds two biogenic amines, tyramine and octopamine. Additionally, we identify the specific binding mode of tyramine with Bla g 4 by X-ray diffraction. Tyramine and octopamine play important roles in invertebrates, and are counterparts to vertebrate adrenergic transmitters. The nature of the binding of Bla g 4 with these ligands sheds light on the possible biological function of the protein.
2. Materials and methods
2.1 Sequence analysis
Sequences were obtained by running PSI-BLAST (Altschul et al., 1997) against the Uniprot database (Uniprot version: 2013_2) (UniProt, 2012) using the sequences of Bla g 4 and Per a 4 (a close homologous protein to Bla g 4 from P. americana) as the queries. As the first step, position-specific scoring matrix (PSSM) profiles were created by performing searches with Bla g 4 and Per a 4 as queries with an expectation value (e-value) of 10−5 for three cycles. As the second step, PSSM profiles were used to perform searches against the same database as for the first step with an e-value of 10−3 until convergence was achieved. Protein structures, homologous to Bla g 4 and Per a 4, obtained for structure analysis were added to the sequence dataset. Identification of a particular sequence PFAM (Punta et al., 2012) membership was achieved by creating a BLAST database from the PFAMs used as source (PF00061 - Lipocalin, PF08212 – Lipocalin-like, PG03973 – Triabin; PFAM database version: 26.0) to create two (AF015 - Lipocalin, AF119 – Triabin family) AllFam (Radauer et al., 2008) allergen families (AllFam database version: 2011-09-12). Sequences obtained from searches against UniProt were subjected to CD-HIT (Fu et al., 2012), where sequences with 80% identity or higher were removed. The created dataset was merged with results from searches against PFAM and pdbaa databases, ultimately returning 1561 non-redundant protein sequences belonging to Lipocalin, Lipocalin-like and Triabin PFAM family and sequences obtained for structural analysis. CLANS (Frickey and Lupas, 2004) was used to create 2D visualization of sequences pairwise similarity by using the Fruchterman-Reingold graph layout algorithm. Clustering was performed with an evalue of 10−6 until convergence was achieved. Allergens found in identified clusters were aligned with MAFFT (Katoh and Standley, 2013) with the L-INS-I option and later adjusted manually in Jalview (Waterhouse et al., 2009) according to the 2D projection of the structural alignment of representative allergens found in the sequence dataset - Bla g 4, Per a 4, Can f 2 and Equ c 1 (PDB IDs: 3EBK, 3EBW, 3L4R and 1EW3) created in Swiss-PdbViewer (Guex and Peitsch, 1997).
2.2 Evolutionary analysis
Sequences from AllFam families AF015 and AF119 were mapped on the dataset used for clustering, then were aligned by MAFFT (Katoh and Standley, 2013) with the L-INS-I option to increase accuracy. The obtained sequence alignment was subjected to MEGA5 (Tamura et al., 2011). Phylogeny reconstruction was performed using the maximum-likelihood estimation with WAG (Whelan and Goldman, 2001) amino acid substitution model with gamma-distributed rates among patterns. The bootstrap method with 1000 replications was used to test branch probabilities.
2.3 Structure analysis
Representation of protein structures, homologous to Bla g 4 and Per a 4, were obtained by performing a PSI-BLAST search against the pdbaa. NCBI BLAST database (as of January 2013), was used to create a structural alignment in the VMD (Humphrey et al., 1996) program. Homology between protein structures was measured by using the QH algorithm (O’Donoghue and Luthey-Schulten, 2003). Calculated QH values for given residues (QRES) were then applied to the Bla g 4 structure, instead of B-factor values, and displayed in Pymol (DeLano, 2002).
2.4 Structure determination
Protein production, crystallization, and data collection have been described previously (Tan et al., 2009). Here we present the reinterpretation of diffraction data using a new methodology included in the HKL-3000 package (Minor et al., 2006). During these studies, we reinvestigated data obtained from both Se-Met labeled and native Bla g 4 crystals. The Se-Met and native structures were determined using the Multi-wavelength Anomalous Diffraction (MAD) technique and Molecular Replacement (MR), respectively, by HKL-3000 coupled with SHELXD/C/E (Sheldrick, 2008), MLPHARE (Otwinowski, 1991), DM (Cowtan and Main, 1993), ARP/wARP (Perrakis et al., 1999), MOLREP (Vagin and Teplyakov, 1997), SOLVE/RESOLVE (Terwilliger, 2004), and selected programs from the CCP4 package (Winn et al., 2011). Both the Se-Met derivative and the native crystal structures were re-examined in the P41212 space group. Models were later refined with REFMAC (Murshudov et al., 2011) and COOT (Emsley and Cowtan, 2004), TLS groups were determined with the TLSMD server (Painter and Merritt, 2006), and structure validation was performed using MOLPROBITY (Chen et al., 2010) and ADIT (Yang et al., 2004). Structures and structure factors were deposited to the PDB (Berman et al., 2000) with accession code 4N7D and 4N7C for Se-Met derivative and native Bla g 4 respectively. Refinement statistics are summarized in Table 1.
Table 1.
Refinement and validation statistics. Ramachandran plot was calculated using MOLPROBITY. Numbers in parenthesis refer to the highest resolution shell.
| Protein | Se-Met | Native |
|---|---|---|
| PDB code | 4N7D | 4N7C |
| Resolution (Å) | 50.0–2.1 (2.18–2.10) | 50.0–1.75(1.81–1.75) |
| Unique reflections | 51198 (5140) | 45386 (4520) |
| Completeness (%) | 100.0(100.0) | 99.9(100.0) |
| R (%) | 17.6 | 18.6 |
| Rfree (%) | 21.7 | 21.3 |
| Mean B value (Å2) | 31.6 | 37.4 |
| RMS deviation bond lengths (Å) | 0.018 | 0.024 |
| RMS deviation bond angles (°) | 2.0 | 2.2 |
| Number of amino acid residues | 172 | 174 |
| Number of water molecules | 117 | 127 |
| Ramachandran plot | ||
| Most favored regions (%) | 86.9 | 85.5 |
| Additional allowed regions (%) | 11.9 | 13.2 |
2.5 NMR titration studies
15N-labeled Bla g 4 samples were obtained from bacterial cultures grown in M9 media supplemented with 15N ammonium chloride. 1H-15N HSQC spectra were acquired at 308 K on a Bruker 800 MHz NMR equipped with a cryo-probe. 0.2 mM Bla g 4 samples in 20 mM acetate buffer, pH 4.5 and 10% D2O were used for NMR titration experiments. Titrations with tyramine or octopamine were performed with protein:ligand molar ratios of 1:0, 1:0.5, 1:1, and 1:2. All NMR data was processed using NMRPipe (Delaglio et al., 1995) and analyzed by Sparky (Goddard, T.D. and Kneller, D.G., SPARKY 3, University of California, San Francisco).
2.6 Isothermal titration calorimetry (ITC) experiments
Binding enthalpy was recorded using an iTC200 microcalorimeter (Microcal (GE Healthcare), Piscataway, NJ). Measurements were carried out at 25°C in 20 mM phosphate buffer at pH 8.0. Binding studies were performed with a Bla g 4 concentration of 100 μM, and the titrant, tyramine or octopamine, with an initial concentration of 1 mM. ITC data were processed and fitted using the manufacturer supplied MicroCal Origin software.
2.7 Other computational techniques
Solvent-accessible surface areas were calculated with PDBePISA (Krissinel and Henrick, 2007). Figures were prepared with Pymol and Turbo (Roussel, 1991). Modeling of octopamine binding was performed in COOT, where molecules of the R- and S-octopamine were superposed on the tyramine molecule. DALI (Holm and Rosenstrom, 2010) and FATCAT (Ye and Godzik, 2004) were used to identify similar proteins.
3. Results
3.1 Sequence analysis
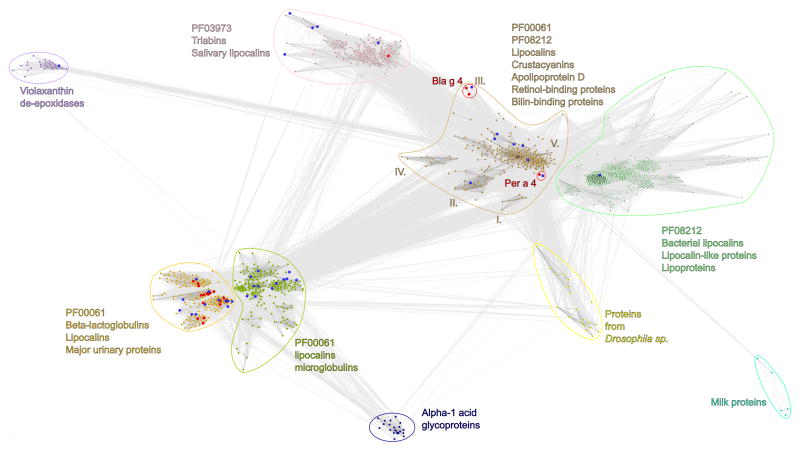
Clustering based on sequence similarity (Fig 1) showed that Bla g 4 and Per a 4, both of which are cockroach allergens, are homologs. The brown cluster indicates where Bla g 4 and Per a 4 allergens belong. Almost all protein sequences in this cluster come from two PFAM families (Lipocalin and Lipocalin-like – PF00061 and PF08212) that are mixed with each other. This cluster contains several subclusters, which are marked with roman numerals. Subcluster marked with an ‘I’ contains chloroplastic lipocalins. Plasma retinol binding protein from various Vertebrata including Canis lupus familiaris, Mus musculus, Rattus norvegicus and Homo sapiens are contained within the cluster marked as ‘II’. Isoforms of the Bla g 4 allergen are found in the cluster marked with ‘III’. As Bla g 4 is part of the Triabin protein family, cluster with this allergen lies between clusters containing proteins from Triabin and Lipocalin protein families. Subcluster ‘IV’, contains uncharacterized proteins from Insecta. The largest subcluster, subcluster ‘V’, is the most diverse of all the other clusters. The proteins within this subcluster with solved structures are: Human apolipoprotein D complexed with progesterone (Protein Data Bank (PDB) codes: 2HZQ, 2HZR), subunit of crustacyanin-A2 (PDB code: 1GKA), apocrustacyanin C2 (PDB code: 1S2P), lipocalin complexed with fluorescein (PDB code: 1N0S), insecticyanin-A (PDB code: 1Z24) and Per a 4 allergen from cockroach (PDB code: 3EBW). Other proteins within this subcluster are apolipoprotein D, chlorophyllide A binding protein, bilin binding protein, as well as bombyrin and biliverdin binding protein. Cluster marked in light green color contains only one protein with known structure, bacterial lipocalin (PDB code: 1QWD). Other proteins inside this cluster are outer membrane lipoproteins Blc, lipocalin-like and lipoprotein-like proteins from the Lipocalin-like PFAM protein family (PF08212) and almost all of those sequences are from various bacterial strains. Cyan cluster contains milk proteins from two species Diploptera punctata and Rhyparobia maderae (cockroaches). Yellow cluster contains various proteins from Drosophila Sp. Navy-blue cluster contains mammalian Alpha-1 acid glycoproteins. Cluster marked in green contains lipocalins, retinoic acid-binding proteins, prostaglandin-D synthetases, siderocalins, prostaglandin isomerases, neutrophil gelatinases and Alpha-1 microglobulins. Several proteins from this cluster have determined structures (PDB codes: 2XST, 3DSZ, 1EPA, 1EPB, 2RD7, 3S26, 2CZT, 2CZU, 2RQ0, 2WWP, 3SAO, 2L5P, 3QKG, 3BX7, 3BX8, 3DTQ). Cluster marked in orange contains β-lactoglobulins, major urinary proteins, salivary lipocalins, and odorant-binding proteins. Furthermore, this cluster contains the largest number of determined protein structures (PDB codes: 1GM6, 2L9C, 1EXS, 2R73, 2R74, 2RA6, 1XKI, 3EYC, 2A2G, 2A2U, 1EW3, 1I04, 1I05, 1I06, 3L4R, 1DZJ, 1DZK, 1DZM, 1DZP, 1E00, 1E02, 1E06, 1HQP, 1YUP, 2HLV). Almost all protein sequences from the green and orange cluster are from the Lipocalin PFAM protein family (PF00061). Small purple cluster in the top left corner of Fig. 1 contains violaxanthin de-epoxidases (PDB codes: 3CQN, 3CQR) from plants and bacteria. Pink cluster contains sequences from the Triabin PFAM protein family (PF03973). This cluster is created from proteins like nitrophorin, thrombin inhibitor or amine binding protein (PDB codes: 1PM1, 2ALL, 2AMM, 1SXX, 1SY2, 1AVG, 1EUO, 1PEE, 1T68, 2A3F, 2ACP, 2AH7, 2AL0, 2HYS, 2GTF, 4GE1, 4GET) and salivary lipocalins. Despite the PFAM classification, which includes Bla g 4 into the Triabin family, this allergen is different from proteins from this PFAM and is more similar to the Lipocalin family.
Fig. 1.
Two-dimensional projection of the CLANS clustering results. Proteins are indicated by dots. Lines indicate sequence similarity detectable with BLAST and are colored by a spectrum of gray shades according to the BLAST p value (black: p value < 10−225, light gray: p value < 10−5). Allergen sequences are marked as red dots. Sequences with known structure are marked as blue dots. Allergen sequences with known structure are marked as red dots with blue boarders. Bla g 4 and Per a 4 allergens protein sequences are in the red circles.
3.2 Evolutionary analysis
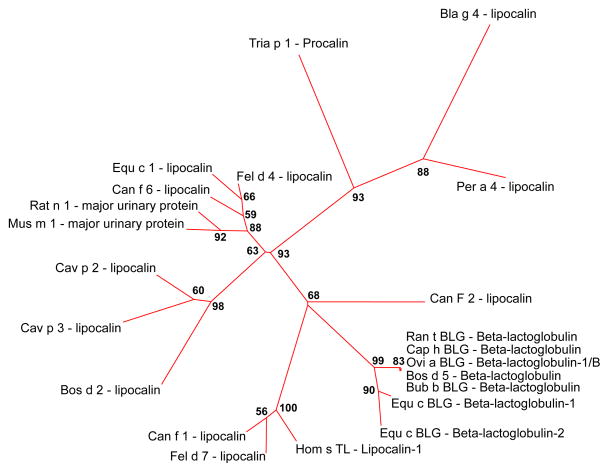
All allergen sequences existing in the dataset, created for sequence analysis, were used to create a Maximum Likelihood phylogenetic tree (Fig. 2). The most closely related allergen group belongs to the beta-lactoglobulin protein family (Bos d 5, Bub b BLG, Ran t BLG, Cap h BLG, Ovi a BLG). These beta-lactoglobulin proteins are so closely related, it is virtually impossible to distinguish between them on this tree, with the exception of horse beta-lactoglobulins (Equ c BLG 1 and Equ c BLG 2). Equally distant from this group is the dog lipocalin allergen (Can f 2) and a small group containing lipocalins from human, dog, and cat (Hum s TL, Can f 1, Fel d 7). Another closely related group is the major urinary proteins from rat (Rat n 1) and mouse (Mus m 1) and lipocalins from dog, cat, and horse (Can f 6, Fel d 4, Equ c 1). The closest group to the one described above contains lipocalin allergens from guinea pig and cattle (Cav p 2, Cav p 3 and Bos d 2). Bla g 4 and Per a 4 are close homologs, however there are several differences between the two. Procalin Tria p 4 from California kissing bug (Triatoma protracta) is the closest homolog to Bla g 4 and Per a 4, however as the sequence alignment of these allergens (Fig. S1) shows not all amino acids involved in tyramine binding are conserved. In Tria p 4 Asp47 and Tyr85 are substituted with Asn45 and Phe89, respectively. Since the data are not sufficient to create a reliable alignment of the area surrounding Tyr122 in Bla g 4 it was omitted in this analysis.
Fig. 2.
Maximum Likelihood tree created in MEGA5. Numbers at nodes are bootstrap values for given node. Length of branches (red colored) reflects similarity between sequences. Bootstrap values below 50% are not shown.
3.3 Structural analysis of an apo and liganded forms of Bla g 4
As described previously (Tan et al., 2009) Bla g 4 is a compact, globular protein that adopts a typical lipocalin fold (Fig. 3). The core of the protein contains a β-barrel formed by eight anti-parallel strands. Additionally, Bla g 4 has a kinked α-helix that is packed against the β-barrel. Compactness of the molecule is strengthened by two long-range disulfide bonds (Cys10-Cys112 and Cys44-Cys175) that connect the central β-barrel with other structural elements. Both the Se-Met derivative and the native forms of the allergen crystallized in the same space group with very similar unit cell parameters, they represent two different forms of the protein. Namely, the Se-Met derivative is an apo form, while the native version of the protein is a ligand bound form of the protein. Both structures superpose with a Cα rmsd of 0.3 Å and the most significant differences in main chain conformation are observed in the loop regions formed by residues 61–65 and 134–141. Moreover, analysis of the electron density maps for the structure of the native Bla g 4 revealed the presence of a large continuous section of electron density, which does not correspond to any proteinous part of the allergen. The shape of the difference map for this region suggests that tyramine, or a similar small molecule, is present in the structure (Fig. 4). In addition to tyramine corresponding to the electron density, it was also confirmed using NMR titration and ITC experiments that this molecule interacts with the allergen. Bla g 4 interacts with the ligand through both hydrogen bonding and hydrophobic interactions. The amine group of the tyramine participates in three hydrogen bonds, two of which interact independently with the hydroxyl group of Tyr85 and the carboxylate of Asp47. The third hydrogen bond links the amine group to a water molecule, which is also bound to Tyr122. The hydroxyl group of the tyramine does not form any direct hydrogen bonds with the protein, but does interact with the water molecules that fill part of the ligand binding cavity. All water molecules that are in the vicinity of the tyramine’s hydroxyl group form hydrogen bonds with other waters or main chain atoms of the protein. The central part of the ligand is located in a hydrophobic cage formed by the side chains of Ile31, Leu39, Trp45, and Tyr122. Tyr42 and Phe92 form a cap that closes the binding site. In the absence of the ligand, the binding site contains approximately 12 water molecules. Some of the water molecules are displaced after ligand binding, however the position of the water molecules, which act as bridges between the ligand and the protein, do not change appreciably. Furthermore, most of the residues that form the hydrophobic cage do not change conformation upon ligand binding. The only exceptions are Ile31 and Asp47 for which small changes in the conformation of the side chains are observed.
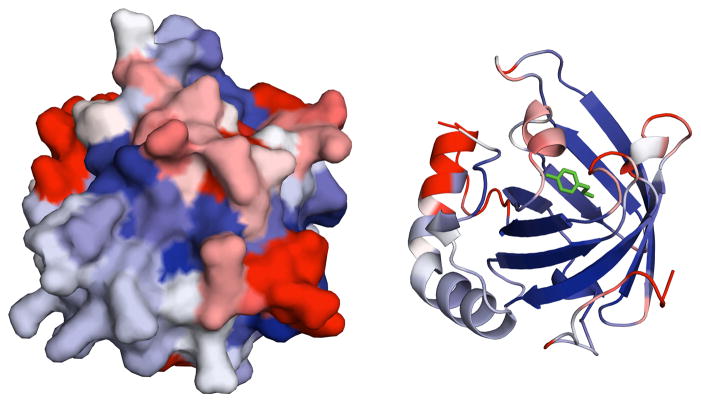
Fig. 3.
Two representations of Bla g 4 (surface on the left and cartoon on the right side) with QRES values displayed in color scale from blue (most structurally conserved) through white to red (not conserved). Tyramine is colored green. QRES values were calculated with VMD 26 from superimposition of close homologs of Bla g 4.
Fig. 4.
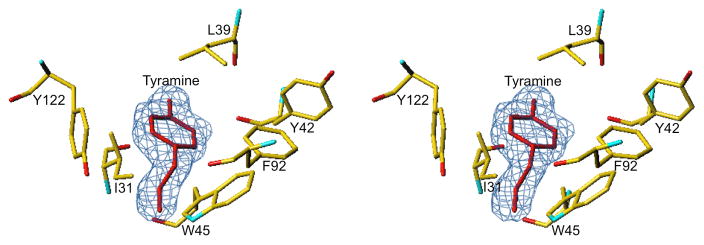
Stereoview of the Bla g 4 ligand-binding site with bound tyramine shown in red. Difference electron density map shown in blue is contoured at 3σ level.
Searches with DALI (Holm and Rosenstrom, 2010) and FATCAT (Ye and Godzik, 2004) identified many other members of the lipocalin family as being structurally similar to Bla g 4, with the structure of Per a 4 being the most similar (~2 Å rmsd). Many of the identified proteins have structures determined in complex with small molecules, like for example biogenic amine-binding protein from saliva of Rhodnius prolixus (PDB code: 4GE1) (Xu et al., 2013), but none of them are bound in the same manner as the tyramine in Bla g 4. Currently (August 2013), there is only one structure (BACE-1; PDB code: 3BRA) of a protein in complex with tyramine in the PDB. BACE-1 is a drug target in Alzheimer’s disease (Kuglstatter et al., 2008) and the mode of tyramine binding is not similar to that observed in the structure of Bla g 4 reported here. Similarly, in the PDB there are only two proteins, human tyrosyl-DNA phosphodiesterase (Tdp1) and human phenylethanolamine N-methyltransfrase (PNMT), for which structures with bound octopamine are reported. While octopamine binding is bridged by vanadate in the Tdp1 structures, its binding in PNMT, which is an adrenaline-synthesizing enzyme, resembles the mode of binding of tyramine in Bla g 4. Namely, the ligand is buried in the protein, and it interacts with the macromolecule through both hydrogen bonding and hydrophobic interactions. Moreover, these interactions also involve bridging water molecules.
A comparison of the Bla g 4 and Per a 4 binding sites revealed partial conservation of the residues that form the hydrophobic cage (Fig. 5). Asp47 and Tyr85, which participate in hydrogen bonding with the amide moiety of tyramine are among those conserved residues. However, some of the residues in Bla g 4 that directly interact with the ligand are not conserved in Per a 4.
Fig. 5.
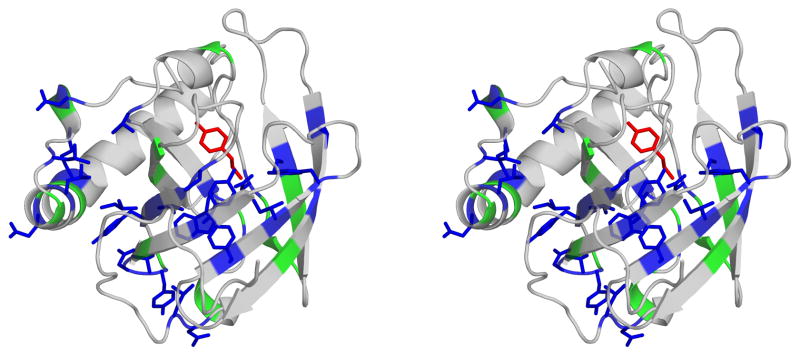
Stereoview of Bla g 4 (shown as ribbon) with bound tyramine (shown as red sticks). Residues that are identical in Bla g 4 and Per a 4 are shown in blue, while residues that are similar are marked in green. For clarity, side chains of similar residues were omitted.
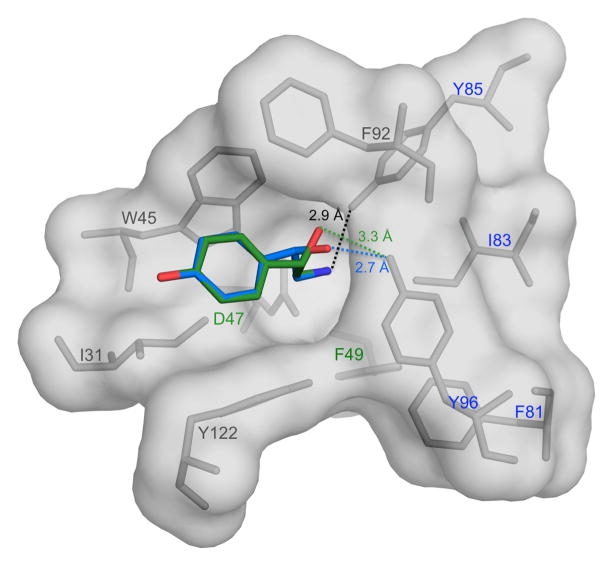
Modeling shown in Fig. 6 suggests that the binding cavity in Bla g 4 is large enough for octopamine to bind. Both NMR and ITC results support this model and indicate that octopamine binds to Bla g 4 in the same manner as tyramine. The only exception is Tyr96, which may form a hydrogen bond with the hydroxyl group of the octopamine. It is apparent that the binding of R-octopamine may also lead to the formation of a stronger hydrogen bond than in the case of S-octopamine because the hydrogen bonding distance from the O7 atom of octopamine to the hydroxyl group of Tyr96 should be significantly shorter (2.7Å vs. 3.3Å respectively, in our model).
Fig. 6.
Octopamine modeled in the Bla g 4 binding site. S-octopamine is shown in green, while R-octopamine is shown in blue. Hydrogen bonding distances from O7 of S-octopamine and R-octopamine to the hydroxyl group of Y96 is 3.3 Å and 2.7 Å respectively. The distance from N8 of S/R-octopamine to the hydroxyl group of Y85 is 2.9 Å. Residues labeled in blue are conserved between Bla g 4 and Per a 4, while residues labeled in green are similar.
Bla g 4 and Per a 4 are cross-reactive despite the fact that their sequence identity is very low (21%). One explanation for this phenomenon assumes that a common IgE epitope exists for both proteins. In order to investigate the existence of such an epitope, structures of both allergens were superposed and continuous surface patches containing identical and similar amino acids were identified (Fig. 7). In addition we used the previously reported studies of Bla g 4 and Per a 4 mutants to determine whether some of the conserved regions may at least partially overlap with IgE epitopes (Tan et al., 2009).
Fig. 7.
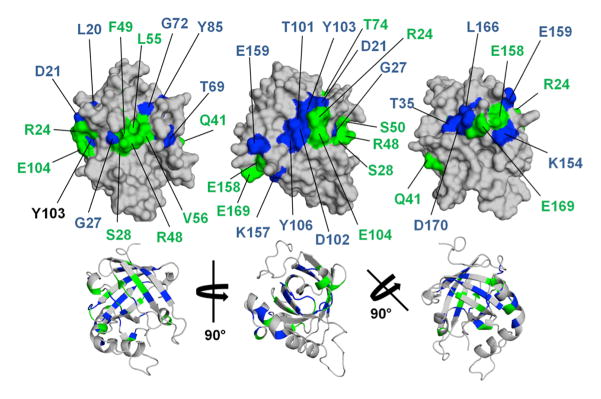
Surface and cartoon representation of Bla g 4. Residues that are identical in Bla g 4 and Per a 4 are highlighted in blue while similar residues are shown in green. The positioning of Bla g 4 is identical in the top panel and bottom panel.
Comparison of the Bla g 4 and Per a 4 structures reveal the presence of three surface patches that contain residues that are conserved in terms of sequence and structure. The first of these patches (P1) is composed of Gly27, Ser28, Arg48, Phe49, Leu55, Val56, and Gly72 (sequence numbering for Bla g 4; identical residues are in bold). The second patch (P2) contains residues Asp21, Arg24, Thr101, Asp102, Tyr103, Glu104, and Tyr106, while the third patch (P3) is composed of Thr35, Lys154, Glu158, Glu159, Leu166, Glu169, and Asp170. Solvent-accessible surface areas for these patches are 270 Å2, 360 Å2, and 530 Å2 respectively.
3.4 Titration of Bla g 4 with tyramine and octopamine
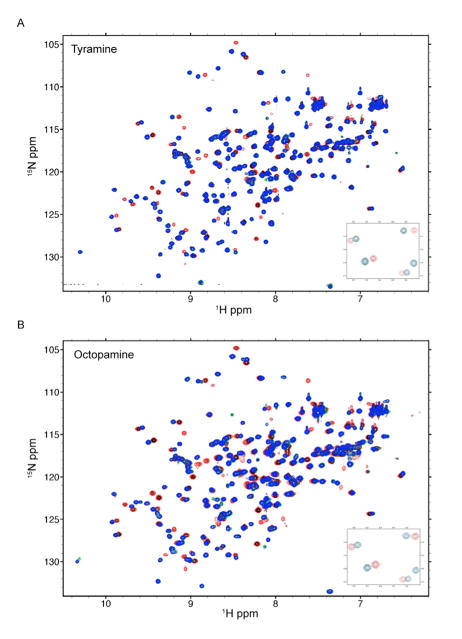
Titration experiments performed using 2D Heteronuclear Single Quantum Correlation (HSQC) NMR showed that Bla g 4 binds to both tyramine and octopamine. Similar patterns of chemical shift perturbation of amide proton cross-peaks were observed for titration with tyramine or octopamine (Figs. 8A and 8B), indicating that both tyramine and octopamine bind to the same binding pocket of Bla g 4. Approximately 40 amide proton cross-peaks were perturbed during the addition of each ligand and the perturbations were saturated with a protein:ligand molar ratio of 1:1, suggesting equimolar binding between Bla g 4 and its ligands. A slow exchange between the apo- and ligand-bound forms of Bla g 4 is indicated by “peak-jumping” instead of “peak-walking” during NMR titration.
Fig. 8.
HSQC spectra of Bla g 4 titrated with (A) tyramine and (B) octopamine. The contours of the amide proton cross-peaks are colored according to molar ratio of Bla g 4:ligand at 1:0 (red), 1:0.5 (black), 1:1 (green) and 1:2 (blue). The inserts show magnified portions of the HSQC spectra indicating “peak-jumping” with increasing concentration of ligand.
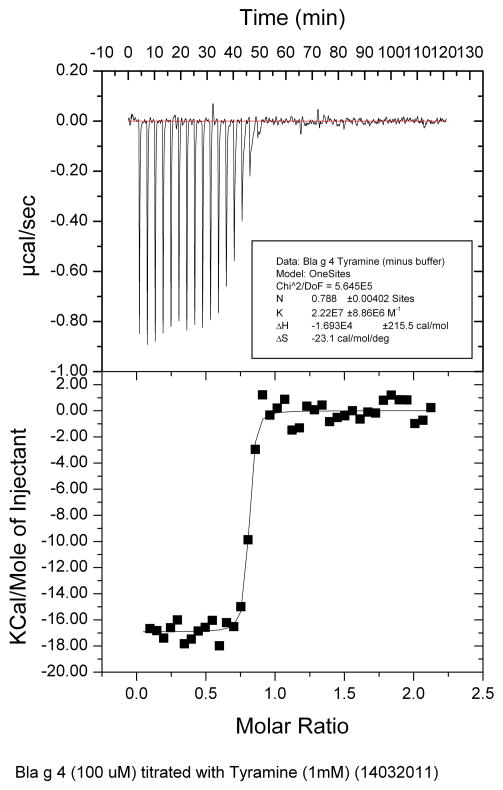
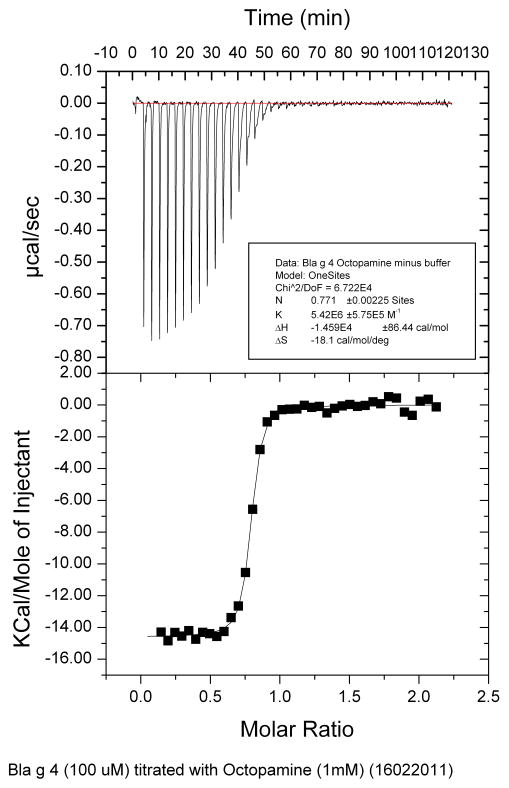
ITC results showed that the tyramine has a slightly higher affinity (Ka) than octopamine for Bla g 4. The enthalpy changes (ΔH) are similar for both tyramine and octopamine for Bla g 4 and the number of binding sites for both ligands is determined to be close to 1 (Table 2, Figs. 9–10), agreeing with NMR titration data.
Table 2.
ITC titration binding parameters results. Raw data shown in supplemental figures 1–2.
| Ka, (M−1 × 106) | ΔH (kcal/mol) | Binding sites, n | |
|---|---|---|---|
| Tyramine | 22.2 ± 8.9 | −16.93 ± 0.22 | 0.788 ± 0.004 |
| Octopamine | 5.4 ± 0.6 | −14.59 ± 0.09 | 0.771 ± 0.002 |
Fig. 9.
Results of ITC binding studies of Bla g 4 and tyramine.
Fig. 10.
Results of ITC binding studies of Bla g 4 and octopamine.
4. Discussion
Bla g 4 is a member of a lipocalin family of proteins, and binds small molecule ligands much like other proteins from this family. Re-examination of the Se-Met and native structures of Bla g 4 resulted in new insight into biochemical/biological function of the allergen. Most likely, different protein production protocols caused the native protein to crystallize with an endogenous ligand, while the Se-Met variant of the protein crystallized without the ligand. Examination of the electron density indicated that tyramine was the ligand bound to Bla g 4. This was then verified through both NMR titration and isothermal titration calorimetry experiments. In addition, both these techniques indicated that tyramine and octopamine are bound by the allergen in a similar manner.
The comparison of Bla g 4 and Per a 4 suggest that these proteins most likely bind different ligands or have different modes for tyramine/octopamine binding. Bla g 4 residues Asp47 and Tyr85, which form hydrogen bonds with the amine group of tyramine, are also conserved in Per a 4. It suggests that the physiological ligand for Per a 4 also contains an amine group and it may be at least partially similar to the tyramine. In addition, Tyr96, which may participate in octopamine binding, is also conserved in Per a 4.
It is highly likely that these molecules, or one of them, are/is physiological ligand(s) for Bla g 4. This finding is consistent with several studies of the role of tyramine/octopamine in reproduction of insects (Lange, 2009). For example, it was found that tyramine is present in oviducts (Donini and Lange, 2004) and spermatheca of locust (da Silva and Lange, 2008; Lange, 2009). It was also found (Hirashima et al., 2007) that there is post-mating suppression of pheromone production by tyramine in B. mori, while octopamine had no effect on this process.
Our analysis revealed the presence of three conserved patches on the surface of Bla g 4 and Per a 4 molecules. These regions are potential candidates for being conformational IgE epitopes that are common for both allergens. Patch P2 seems to be especially suitable for the binding of a cross-reactive antibody or it may partially overlap with an IgE epitope. A significant part of this patch is composed of residues that are identical in Bla g 4 and Per a 4, and its area is large enough to be considered a potential antibody binding site, or part of an epitope. In addition, sequence polymorphism of Bla g 4 in regions corresponding to patches P1 (Ser28) and P2 (Arg24) results in some cases of increased similarity between Bla g 4 and Per a 4 (Jeong et al., 2008; Jeong et al., 2009). Results of IgE binding studies using Bla g 4 derived overlapping peptides (Shin et al., 2009) identified the existence of three protein regions that contain the antibody binding sites. Two of these regions correspond to patches P1 and P2. However, the major IgE epitope identified by Shin et al. (residues 118–152) does not correspond to the region that is similar for both Bla g 4 and Per a 4. Analysis of IgE binding to Bla g 4 and Per a 4 mutants also suggests that patch P2 may be in some cases responsible for cross-reactivity. However, there are no experimental data that analyze role of residues forming patch P3 in cross-reactivity between these two cockroach allergens.
5. Conclusions
Bla g 4 is a male cockroach specific protein that is a member of the lipocalin family of proteins, and binds small molecule ligands. It binds both tyramine and octopamine, and most likely the function of the protein is to deliver a neurotransmitter to a female. The structural analysis shows that tyramine binding does not cause any significant changes in the ligand binding site. We speculate that the delivery of tyramine causes post-mating inactivation of pheromone production in a female. However, it cannot be excluded that Blag 4 works as a carrier for both tyramine and octopamine, which is consistent with the NMR and ITC results indicating that octopamine binds to Bla g 4 in the same manner as tyramine. Comparison of Bla g 4 and Per a 4 structures and sequences suggests that these proteins bind different ligands, and therefore have different physiological roles. Currently lack of experimental data does not allow for the identification of Bla g 4 and Per a 4 fragments that are responsible for cross-reactivity.
Supplementary Material
Highlights.
Bla g 4 is a major cockroach allergen that belongs to the lipocalin family.
It was determined that Bla g 4 binds tyramine and octopamine.
Structural analysis provides details for tyramine binding.
Most likely Bla g 4 and Per a 4 bind different ligands.
Acknowledgments
The work described in this paper was supported partially by GM53163 grant.
Abbreviations
- Se-Met
selenomethionine
- PDB
Protein Data Bank
- rmsd
root mean square derivative
- HSQC NMR
heteronuclear single quantum correlation nuclear magnetic resonance
- Tdp1
human tyrosyl-DNA phosphodiesterase
- PNMT
human phenylethanolamine N-methyltransfrase
- ITC
isothermal titration calorimetry
- IgE
immunoglobulin E
- PSSM
position-specific scoring matrix
- e-value
expectation value
- MAD
multi-wavelength anomalous diffraction
- MR
molecular replacement
Appendix A. Supplementary Data
Supplementary material associated with this article can be found, in the online version, at http://dx.doi.org/XXXXXXX.
Footnotes
Accession numbers: The atomic coordinates and the structure factors have been deposited to the Protein Data Band under accession codes: 4N7C for native Bla g 4 and 4N7D for the Se-Met Bla g 4 structure.
Conflicts of Interest
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic acids research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arruda LK, Vailes LD, Hayden ML, Benjamin DC, Chapman MD. Cloning of cockroach allergen, Bla g 4, identifies ligand binding proteins (or calycins) as a cause of IgE antibody responses. The Journal of biological chemistry. 1995;270:31196–31201. doi: 10.1074/jbc.270.52.31196. [DOI] [PubMed] [Google Scholar]
- Arruda LK, Vailes LD, Mann BJ, Shannon J, Fox JW, Vedvick TS, Hayden ML, Chapman MD. Molecular cloning of a major cockroach (Blattella germanica) allergen, Bla g 2. Sequence homology to the aspartic proteases. The Journal of biological chemistry. 1995;270:19563–19568. doi: 10.1074/jbc.270.33.19563. [DOI] [PubMed] [Google Scholar]
- Arruda LK, Vailes LD, Platts-Mills TA, Hayden ML, Chapman MD. Induction of IgE antibody responses by glutathione S-transferase from the German cockroach (Blattella germanica) The Journal of biological chemistry. 1997;272:20907–20912. doi: 10.1074/jbc.272.33.20907. [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank, Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernton HS, Brown H. Insect Allergy--Preliminary Studies of the Cockroach. The Journal of allergy. 1964;35:506–513. doi: 10.1016/0021-8707(64)90082-6. [DOI] [PubMed] [Google Scholar]
- Call RS, Smith TF, Morris E, Chapman MD, Platts-Mills TA. Risk factors for asthma in inner city children. The Journal of pediatrics. 1992;121:862–866. doi: 10.1016/s0022-3476(05)80329-4. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. 2010;D66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn RD, Arbes SJ, Jr, Jaramillo R, Reid LH, Zeldin DC. National prevalence and exposure risk for cockroach allergen in U.S. households. Environmental health perspectives. 2006;114:522–526. doi: 10.1289/ehp.8561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowtan KD, Main P. Improvement of macromolecular electron-density maps by the simultaneous application of real and reciprocal space constraints. Acta crystallographica. 1993;49:148–157. doi: 10.1107/S0907444992007698. [DOI] [PubMed] [Google Scholar]
- da Silva R, Lange AB. Tyramine as a possible neurotransmitter/neuromodulator at the spermatheca of the African migratory locust, Locusta migratoria. Journal of insect physiology. 2008;54:1306–1313. doi: 10.1016/j.jinsphys.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. Journal of biomolecular NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. 2002. [Google Scholar]
- Donini A, Lange AB. Evidence for a possible neurotransmitter/neuromodulator role of tyramine on the locust oviducts. Journal of insect physiology. 2004;50:351–361. doi: 10.1016/j.jinsphys.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Eggleston PA, Rosenstreich D, Lynn H, Gergen P, Baker D, Kattan M, Mortimer KM, Mitchell H, Ownby D, Slavin R, Malveaux F. Relationship of indoor allergen exposure to skin test sensitivity in inner-city children with asthma. The Journal of allergy and clinical immunology. 1998;102:563–570. doi: 10.1016/s0091-6749(98)70272-6. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta crystallographica. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fan Y, Gore JC, Redding KO, Vailes LD, Chapman MD, Schal C. Tissue localization and regulation by juvenile hormone of human allergen Bla g 4 from the German cockroach, Blattella germanica (L) Insect molecular biology. 2005;14:45–53. doi: 10.1111/j.1365-2583.2004.00530.x. [DOI] [PubMed] [Google Scholar]
- Frickey T, Lupas A. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics (Oxford, England) 2004;20:3702–3704. doi: 10.1093/bioinformatics/bth444. [DOI] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics (Oxford, England) 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelber LE, Seltzer LH, Bouzoukis JK, Pollart SM, Chapman MD, Platts-Mills TA. Sensitization and exposure to indoor allergens as risk factors for asthma among patients presenting to hospital. The American review of respiratory disease. 1993;147:573–578. doi: 10.1164/ajrccm/147.3.573. [DOI] [PubMed] [Google Scholar]
- Gore JC, Schal C. Cockroach allergen biology and mitigation in the indoor environment. Annual review of entomology. 2007;52:439–463. doi: 10.1146/annurev.ento.52.110405.091313. [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Gustchina A, Li M, Wunschmann S, Chapman MD, Pomes A, Wlodawer A. Crystal structure of cockroach allergen Bla g 2, an unusual zinc binding aspartic protease with a novel mode of self-inhibition. Journal of molecular biology. 2005;348:433–444. doi: 10.1016/j.jmb.2005.02.062. [DOI] [PubMed] [Google Scholar]
- Helm R, Cockrell G, Stanley JS, Brenner RJ, Burks W, Bannon GA. Isolation and characterization of a clone encoding a major allergen (Bla g Bd90K) involved in IgE-mediated cockroach hypersensitivity. The Journal of allergy and clinical immunology. 1996;98:172–180. doi: 10.1016/s0091-6749(96)70240-3. [DOI] [PubMed] [Google Scholar]
- Hirashima A, Yamaji H, Yoshizawa T, Kuwano E, Eto M. Effect of tyramine and stress on sex-pheromone production in the pre- and post-mating silkworm moth, Bombyx mori. Journal of insect physiology. 2007;53:1242–1249. doi: 10.1016/j.jinsphys.2007.06.018. [DOI] [PubMed] [Google Scholar]
- Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic acids research. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. Journal of molecular graphics. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Jeong KY, Lee H, Shin KH, Yi MH, Jeong KJ, Hong CS, Yong TS. Sequence polymorphisms of major German cockroach allergens Bla g 1, Bla g 2, Bla g 4, and Bla g 5. International archives of allergy and immunology. 2008;145:1–8. doi: 10.1159/000107460. [DOI] [PubMed] [Google Scholar]
- Jeong KY, Yi MH, Jeong KJ, Lee H, Hong CS, Yong TS. Sequence diversity of the Bla g 4 cockroach allergen, homologous to lipocalins, from Blattella germanica. International archives of allergy and immunology. 2009;148:339–345. doi: 10.1159/000170388. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Molecular biology and evolution. 2013 doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. Journal of molecular biology. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Kuglstatter A, Stahl M, Peters JU, Huber W, Stihle M, Schlatter D, Benz J, Ruf A, Roth D, Enderle T, Hennig M. Tyramine fragment binding to BACE-1. Bioorganic & medicinal chemistry letters. 2008;18:1304–1307. doi: 10.1016/j.bmcl.2008.01.032. [DOI] [PubMed] [Google Scholar]
- Lange AB. Tyramine: from octopamine precursor to neuroactive chemical in insects. General and comparative endocrinology. 2009;162:18–26. doi: 10.1016/j.ygcen.2008.05.021. [DOI] [PubMed] [Google Scholar]
- Matsui EC, Wood RA, Rand C, Kanchanaraksa S, Swartz L, Curtin-Brosnan J, Eggleston PA. Cockroach allergen exposure and sensitization in suburban middle-class children with asthma. The Journal of allergy and clinical immunology. 2003;112:87–92. doi: 10.1067/mai.2003.1588. [DOI] [PubMed] [Google Scholar]
- Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta crystallographica. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- Mueller GA, Pedersen LC, Lih FB, Glesner J, Moon AF, Chapman MD, Tomer KB, London RE, Pomes A. The novel structure of the cockroach allergen Bla g 1 has implications for allergenicity and exposure assessment. The Journal of allergy and clinical immunology. 2013 doi: 10.1016/j.jaci.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta crystallographica. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donoghue P, Luthey-Schulten Z. On the evolution of structure in aminoacyl-tRNA synthetases. Microbiology and molecular biology reviews : MMBR. 2003;67:550–573. doi: 10.1128/MMBR.67.4.550-573.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z. In: Isomorphous replacement and anomalous scattering. Wolf W, Evans PR, Leslie AGW, editors. Warrington, UK: Daresbury Laboratory; 1991. [Google Scholar]
- Painter J, Merritt EA. TLSMD web server for the generation of multi-group TLS models. Journal of Applied Crystallography. 2006;39:109–111. [Google Scholar]
- Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nature structural biology. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD. The Pfam protein families database. Nucleic acids research. 2012;40:D290–301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radauer C, Bublin M, Wagner S, Mari A, Breiteneder H. Allergens are distributed into few protein families and possess a restricted number of biochemical functions. The Journal of allergy and clinical immunology. 2008;121:847–852. e847. doi: 10.1016/j.jaci.2008.01.025. [DOI] [PubMed] [Google Scholar]
- Roussel ACC. “Turbo Frodo” Silicon Graphics Geometry Partners Directory. Silicon Graphics; 86, Mountain View, CA: 1991. [Google Scholar]
- Santos AB, Chapman MD, Aalberse RC, Vailes LD, Ferriani VP, Oliver C, Rizzo MC, Naspitz CK, Arruda LK. Cockroach allergens and asthma in Brazil: identification of tropomyosin as a major allergen with potential cross-reactivity with mite and shrimp allergens. The Journal of allergy and clinical immunology. 1999;104:329–337. doi: 10.1016/s0091-6749(99)70375-1. [DOI] [PubMed] [Google Scholar]
- Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- Shin KH, Jeong KY, Hong CS, Yong TS. IgE binding reactivity of peptide fragments of Bla g 4, a major German cockroach allergen. The Korean journal of parasitology. 2009;47:31–36. doi: 10.3347/kjp.2009.47.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular biology and evolution. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan YW, Chan SL, Ong TC, Yit le Y, Tiong YS, Chew FT, Sivaraman J, Mok YK. Structures of two major allergens, Bla g 4 and Per a 4, from cockroaches and their IgE binding epitopes. The Journal of biological chemistry. 2009;284:3148–3157. doi: 10.1074/jbc.M807209200. [DOI] [PubMed] [Google Scholar]
- Terwilliger T. SOLVE and RESOLVE: automated structure solution, density modification and model building. Journal of synchrotron radiation. 2004;11:49–52. doi: 10.1107/s0909049503023938. [DOI] [PubMed] [Google Scholar]
- UniProt C. Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic acids research. 2012;40:D71–75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. Journal of Applied Crystallography. 1997;30:1022–1025. [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics (Oxford, England) 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Molecular biology and evolution. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta crystallographica. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Chang BW, Mans BJ, Ribeiro JM, Andersen JF. Structure and ligand-binding properties of the biogenic amine-binding protein from the saliva of a blood-feeding insect vector of Trypanosoma cruzi. Acta crystallographica. 2013;69:105–113. doi: 10.1107/S0907444912043326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Guranovic V, Dutta S, Feng Z, Berman HM, Westbrook JD. Automated and accurate deposition of structures solved by X-ray diffraction to the Protein Data Bank. Acta Crystallographica Section D. 2004;60:1833–1839. doi: 10.1107/S0907444904019419. [DOI] [PubMed] [Google Scholar]
- Ye Y, Godzik A. FATCAT: a web server for flexible structure comparison and structure similarity searching. Nucleic acids research. 2004;32:W582–585. doi: 10.1093/nar/gkh430. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.