Abstract
Purpose
To develop a novel 4-dimensional computed tomography (4D-CT) technique that exploits standard fast helical acquisition, a simultaneous breathing surrogate measurement, deformable image registration, and a breathing motion model to remove sorting artifacts.
Methods and Materials
Ten patients were imaged under free-breathing conditions 25 successive times in alternating directions with a 64-slice CT scanner using a low-dose fast helical protocol. An abdominal bellows was used as a breathing surrogate. Deformable registration was used to register the first image (defined as the reference image) to the subsequent 24 segmented images. Voxel-specific motion model parameters were determined using a breathing motion model. The tissue locations predicted by the motion model in the 25 images were compared against the deformably registered tissue locations, allowing a model prediction error to be evaluated. A low-noise image was created by averaging the 25 images deformed to the first image geometry, reducing statistical image noise by a factor of 5. The motion model was used to deform the low-noise reference image to any user-selected breathing phase. A voxel-specific correction was applied to correct the Hounsfield units for lung parenchyma density as a function of lung air filling.
Results
Images produced using the model at user-selected breathing phases did not suffer from sorting artifacts common to conventional 4D-CT protocols. The mean prediction error across all patients between the breathing motion model predictions and the measured lung tissue positions was determined to be 1.19 ± 0.37 mm.
Conclusions
The proposed technique can be used as a clinical 4D-CT technique. It is robust in the presence of irregular breathing and allows the entire imaging dose to contribute to the resulting image quality, providing sorting artifact–free images at a patient dose similar to or less than current 4D-CT techniques.
Introduction
Respiration-induced motion is a considerable source of uncertainty in radiation therapy treatment planning (1, 2) and has the potential to lead to substantial dose delivery errors if not properly accounted for (3). Respiratory-gated 4-dimensional computed tomography (4D-CT) offers spatial and temporal information for tumor motion and has become indispensable for the highly conformal treatment of lung tumors (2). Despite revolutionizing high-dose conformal lung cancer therapy, conventional 4D-CT methods suffer from image artifacts that can distort structures of interest (4) and inaccurately characterize breathing motion (5, 6) due to their inability to account for irregular breathing.
Current commercial clinical 4D-CT images are typically acquired using either low-pitch helical (7) or ciné acquisition sequences (8, 9). Scans are combined with simultaneous breathing surrogate measurements to retrospectively group acquired projections (helical protocols) or images (cine protocols) according to breathing phase, using either amplitude-based or phase-based sorting (10-13). These techniques perform well under conditions of regular breathing with a consistent breathing depth, but image artifacts arise under conditions of irregular breathing (5), leading to systematic errors in tumor volume and center-of-mass measurements (14).
This paper reports a novel technique for providing quantitative 4D-CT images free of sorting artifacts. A repeated fast helical CT protocol was used, with multiple scans acquired to sample the breathing cycle. A breathing motion model (15) was used to relate tissue motion to tidal volume and airflow as measured with a breathing surrogate. Initial results for 1 patient from this technique were presented previously by Low et al (16). The technique has been developed further, and the results from 10 patients are presented here. The results were evaluated for image quality, presence of artifacts, robustness in the presence of irregular breathing, and spatial accuracy of model-predicted tissue locations.
Methods and Materials
Image acquisition
Figure 1 shows a flow chart describing the workflow of the new technique. Ten patients were enrolled under an institutional review board–approved protocol. Patients were imaged using a Multidetector row CT scanner (Somatom Definition Flash; Siemens Healthcare, Forchhiem, Germany) with a standard fast helical imaging protocol. Images were acquired with a nominal beam collimation (longitudinal field of view) of 64 × 0.6 mm (3.84 cm), a pitch of 1.2 and scanner rotation period of 0.285 second, which resulted in a table movement of 46.08 mm per rotation and a table velocity of 161.7 mm/s. The scans used 120 kV and 40 effective mAs per slice (169 mA for this pitch and rotation time). Images were reconstructed with 1-mm slice thickness, using the Siemens reconstruction kernel B30f. This protocol required approximately 2.5 seconds to scan the entire lung volume from apices to lung bases. In order to adequately sample the breathing motion, the complete protocol involved imaging the patient with 25 individual series (25 separate acquisitions), using alternating scanning directions with approximately 2-second delay between acquisitions. The entire scanning sequence took approximately 140 seconds. The total computed tomography dose index volume (CTDI vol) for the entire scan (all 25 acquisitions) was 4.22 cGy, which is less than a current clinical slow-helical 4D-CT protocol (Siemens Sensation Open, 800mAs, spiral pitch of 0.1, CTDI vol of 6.9 cGy).
Fig. 1.
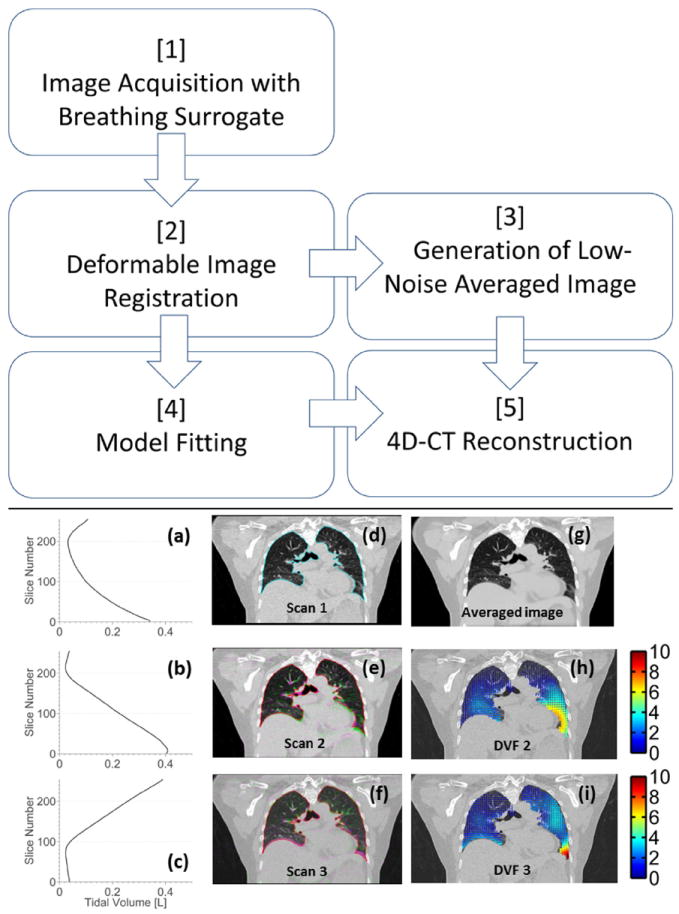
Flow chart describes the technique. (a-c) Tidal volume is plotted against slice number, associated with scans 1-3. (d) Reference scan (scan 1) with lung contour is shown. (e, f) Scans 2 and 3 (pink) are overlaid onto the reference scan (green). (g) Average of 25 images were deformed to the reference image geometry. (h, i) DVF (mm), corresponding to the deformations from each scan to the reference image geometry, are shown. DVF = deformation vector fields. A color version of this figure is available at www.redjournal.org.
Each scan was performed during free breathing. Due to the relative motion of the tissue and the scanner during the acquisition process, the 25 free-breathing images did not correspond to any single breathing phase. As the speed of the helical scan is much greater than the tissue motion, each individual axial slice can be considered to be acquired at a single static respiratory phase and thus contain minimal blurring artifacts. The relative motion between each tissue voxel in the 25 scans was used to determine the parameters of the motion model. An abdominal bellows system (Philips Medical Systems) was used to provide a breathing cycle surrogate. The bellows voltage signal was sampled at 100 Hz, using a PC with analog-to-digital converter controlled using custom LabVIEW software (National Instruments, Austin, TX). Signals were analyzed further using Matlab software (version R2013a; Mathworks, Natick, MA). The bellows signal has been previously shown to be proportional to tidal volume (17, 18). During CT acquisition, the bellows data were synchronized with CT acquisition by monitoring the “x-ray on” signal from the CT scanner. This signal identified when the scanner was acquiring projection data and was synchronized with the DICOM [Digital Imaging and Communication in Medicine] image time tags to correct for the time when projections were not acquired. The bellows voltage signals were smoothed using a central moving average filter, and the rate of signal change was calculated (V/s) to give a surrogate measurement of airflow.
Image registration
The 25 image volumes were each segmented using a semiautomatic region-growing tool to isolate the lungs and mainstem bronchi (MIM version 5.6; MIM Software, Cleveland, OH). Accurate registration of the shear motion between the lungs and chest wall was achieved by conducting 2 separate registrations and modeling procedures for (1) the lungs; and (2) all other tissues, using the lung contour as the boundary between the 2. Deformable image registration was implemented on the segmented lung images, using open source software (Elastix; PaloSanto Solutions, Ecuador) (19) and performed with an Intel i7 PC running at 3.60 GHz. A B-spline registration algorithm (20) was used to register the first segmented image volume to the subsequent 24 segmented image volumes (the target image volume). B-spline interpolation was accelerated with a graphics processor-based parallel algorithm, implemented with an NVidia GeForce GTX 680 GPU (NVIDIA Corporation, Santa Clara, CA). The first image volume was chosen arbitrarily as a reference image. The Mattes mutual information similarity metric (21) with no penalty term was applied. The optimizer was an iterative stochastic gradient descent (22). A 5-level multiresolution approach starting with B-spline grid spacing of 160 mm was used. The registration process was entirely automated, although each registered image volume was visually inspected to ensure correct alignment. The deformation vector fields (DVF) were calculated from the results (19).
The accuracy of this deformable registration approach was verified using datasets provided by MD Anderson Cancer Center Deformable Image Registration laboratory (www.dir-lab.com) (23), consisting of thoracic 4D-CT image volumes and a coordinate list of anatomical landmarks that had been manually identified, with expert landmark correspondence used for evaluating DIR spatial accuracy. Registration accuracy was found to be less than 1 voxel for 1500 landmarks across 5 patient data sets.
Motion model fitting
The novel 4D-CT technique described here uses a breathing motion model to relate tissue motion, calculated from the DIR, to tidal volume and airflow, as measured with a breathing surrogate. Low et al (15, 16) have shown that the deformation from an arbitrary reference phase that a piece of lung tissue undergoes during quiet respiration can be described by a linear approximation in tidal volume (v) and airflow (f) as follows
| (1) |
where and relate the deformation of the tissue position to the tidal volume and airflow of the tissue at spatial location , accounting for motion hysteresis. Model parameters are voxel-specific, allowing for spatially heterogeneous lung motion. Time is not explicitly expressed in the model but implicitly considered in the time dependence of the tidal volume and airflow.
The bellows signal associated with each scan represented a surrogate breathing measurement corresponding to each slice of the image. In order to assign each of the deformably registered voxel locations to the correct bellows signal, each of the 25 bellows signals were warped by the z-vector of the corresponding DVF using linear interpolation to produce a 3D set of bellows voltages for each scan, with a single bellows voltage assigned to each voxel of each image.
For each voxel, there exist 25 measured tissue locations calculated from the DIR and 25 related volume and airflow measurements, giving an over-determined system for the 3 vector parameters required to predict the tissue location. The motion model parameters were determined using an orthogonal triangular decomposition to provide the least squares solution to the over-determined system and were solved independently for each individual voxel. The tissue locations predicted by the motion model in the 25 images were compared with the deformably registered tissue locations. This breathing motion model residual was then iteratively minimized to fit the bellows signal for a linear drift with time, and the model parameters were recomputed using the drift-corrected bellows values. The model prediction accuracy was evaluated using a “leave-one-out” procedure, whereby each scan was left out of the final model fitting in turn, and the tissue locations for that scan were predicted using the model generated without that individual scan. This was compared against the deformably registered tissue locations, allowing a model prediction error to be calculated for each voxel of each scan. The mean, standard deviation, and 90th and 95th percentiles of prediction errors for all voxels in the lungs were calculated in each patient, allowing an overall mean value of error to be calculated for the 10 patients in the study.
4D-CT image generation
The motion model parameters as described in equation (1) were used to generate DVFs, which were then used to deform the average image in the reference image geometry to any user-selected breathing phase. Images at end-inhale and end-exhale were generated, and the air content of the lungs (L) was calculated in each phase to allow calibration of the bellows voltage signal (V) to tidal volume (L). 3D linear interpolation was used to warp the averaged reference image. HU value corrections were applied to account for lung parenchyma density changes during breathing. The HU values of each voxel were converted to relative air content by linearly interpolating the HU values between air as measured in the trachea and soft tissue as measured in the liver. At each voxel, the relative air contents of the 25 scan acquisitions were correlated with the bellows voltage signal and the relationships fitted to a line, allowing a HU value correction to be applied per voxel at any breathing phase.
Results
Mean model error across all 10 patients was calculated as 1.21 ± 0.35 mm. Mean value of the 95th percentile of errors was 2.7 ± 0.76 mm. Results from individual patients are shown in Table 1, along with tumor location and size, as contoured by a physician at 25th percentile tidal volume. The maximum tissue motion calculated when each patient inhaled from the 5th to 85th percentile of volume is also shown. Registration accuracy was separately tested using a large set of publicly available landmarks in 4D-CT images and was found to be less than 1 voxel for 1500 landmarks across 5 patient data sets. Registration took less than 15 minutes per patient.
Table 1.
Lung volume (85th percentile), tidal volume (85th- to 5th-percentile volume), maximum motion corresponding to the tidal volume, mean and 95th percentile model errors, tumor location, and size (measured at 25th percentile)
| Patient | Lung volume (L) | Tidal volume (L) | Maximum motion (mm) | Mean ± SD (mm) | 95th Percentile (mm) | Tumor location | Tumor size (cm) |
|---|---|---|---|---|---|---|---|
| 1 | 3.9 | 0.3 | 19.3 | 0.84 ± 0.2 | 1.4 | LUL | 3.1 |
| 2 | 2.5 | 0.4 | 24.5 | 0.79 ± 0.2 | 1.2 | RUL | 3.9 |
| 3 | 2.6 | 0.4 | 35.0 | 1.63 ± 0.4 | 2.5 | RUL | 11.6 |
| 4 | 4.1 | 0.3 | 8.0 | 1.23 ± 0.4 | 1.9 | RUL | 4.2 |
| 5 | 6.6 | 0.4 | 28.3 | 1.81 ± 0.5 | 2.6 | RUL | 1.9 |
| 6 | 3.9 | 0.8 | 25.3 | 1.20 ± 0.6 | 2.0 | LUL | 2.3 |
| 7 | 3.6 | 0.4 | 32.8 | 1.29 ± 0.5 | 1.8 | LLL | 1.4 |
| 8 | 4.4 | 0.2 | 22.7 | 0.79 ± 0.2 | 1.7 | RUL | 2.2 |
| 9 | 3.8 | 0.3 | 11.0 | 0.84 ± 0.2 | 1.3 | RUL | 2.5 |
| 10 | 3.9 | 0.3 | 17.4 | 1.48 ± 0.3 | 1.9 | RLL | 3.3 |
| Mean ± SD | 20.4 ± 10.7 | 1.2 ± 0.4 | 2.7 ± 0.8 | 3.6 ± 2.9 |
Abbreviations: LLL = left lower lung; LUL = left upper lung; RUL = right upper lung.
Figure 1 shows an example of images acquired during different breaths for patient 2. The breathing traces (Fig. 1a-c), the overlaid scans (Fig. 1e and f) and the resulting deformation vector fields (Fig. 1h and i) are shown. The free-breathing images are deformed due to the relative motion of the tissue and the scanner during the acquisition process and do not correspond to any single breathing phase. The reference scan was imaged in the foot-to-head direction while the patient was in the process of exhaling; the lungs were approximately mid-to-end inhalation at the start of the scan and deformed to just after end-exhalation by the end of the 2.5-second scan. The effect of the specific reference scan phase on registration accuracy and subsequent model error was investigated by repeating the registrations with reference scans selected to span the range of peak inhalation, mid inhalation, and peak exhalation. No differences were observed in either registration quality or model accuracy.
Due to the low-dose imaging protocol, the pixel-to-pixel statistical variations in the original images were greater than would be achieved with a higher dose (mAs). Pixel-to-pixel noise in Figure 1d in the liver was 46 HU. An average image in the reference image geometry (Fig. 1g) was generated from 25 deformed scans, with the noise characteristics reduced as shown by the visibly reduced mottle in the liver. Mean statistical image noise was reduced to 8 HU, with a noise reduction close to the expected ratio of 5. Residual image registration errors reduced the sharpness of the subsequent image slightly but not to an extent which would reduce clinical usefulness.
Calculation of the patient-specific model parameters on a voxel-by-voxel basis took 6 minutes for a 256- × 512- × 256-voxel image with 1 mm3 resolution. The drift corrections applied to the bellows signals had a mean value of 0.6 ± 0.3 mL/s for the 10 patients. Figure 2 shows an example of the model accuracy across the 25 sampled breathing phases for locations in the upper and lower left lung. Voxel locations are indicated on the left of Figure 2. Measured tissue deformations in the craniocaudal (Fig. 2, CC) direction determined directly from the deformable registration are shown (Fig. 2, crosses) plotted against volume and flow, together with the deformations as predicted by the model (Fig. 2, circles). Amplitudes of motion were up to 2.5 mm and 10.8 mm, for the 2 voxel locations. Figure 2c and g show the tissue deformations as plotted in the CC-versus-anteroposterior (AP) directions, showing increased hysteresis in the upper lung compared with that in the lower lung, as expected (24). Figure 2d and h show the linear fit between HU values and volume used to apply accurate HU values per voxel at user-selected breathing phases. Using the leave-one-out method, mean errors predicting all 25 voxel locations were 0.70 ± 0.30 mm and 0.82 ± 0.32 mm in each voxel, respectively. Mean errors were uncorrelated with scan number in all patients, suggesting the model was stable over the 2.5-minute imaging session and that the surrogate signal was accurately corrected for drift. Figure 3 shows examples from 4 patients, showing images and corresponding error distributions in millimeters.
Fig. 2.
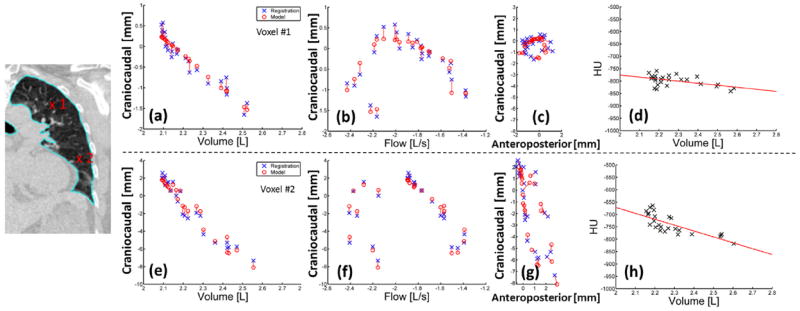
(a-g) Measured deformations from the DIR (x) versus model predicted deformations (o) are shown. CC deformations were plotted against volume (a, e) and flow (b, f) and AP deformations (c, g). (d, h) HU values were plotted against volume with a linear fit (solid line). (a-d) voxel 1 is shown in the upper left lung; (e-h) voxel 2 is shown in the lower left lung (patient 2). AP = anteroposterior; CC = craniocaudal.
Fig. 3.
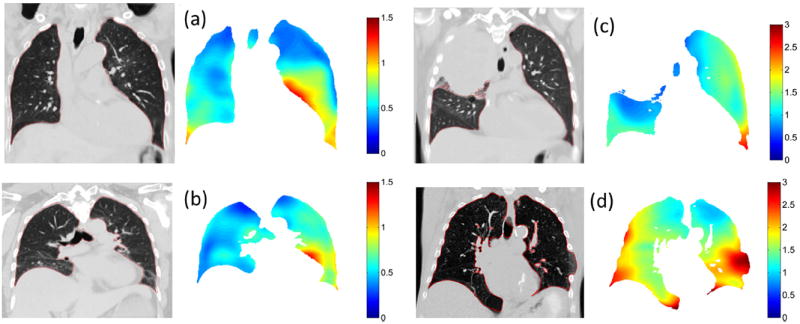
Images at 5th percentile tidal volume and the respective motion model errors (mm) for patients 1 (a), 2 (b), 3 (c), and 5 (d) are shown.
The effect of breathing pattern irregularity on model error was assessed using the method of periodicity analysis based on subspace projections described by Ruan et al (25, 26). No significant correlation was found when model error was compared with tidal volume or variations in either breathing amplitude or period over the 2.5-minute imaging period used to generate model parameters, suggesting the technique is robust in the presence of irregular breathing. The breathing waveforms of the 10 patients in the study were compared with the waveforms from 50 lung cancer patients and found to be representative in terms of irregularity.
Figure 4a and b show a clinical 4D-CT at the 25th percentile tidal volume for patient 4, with image artifacts present throughout the image due to conditions of irregular breathing. Figure 4c and d show images generated with the new technique. Images are low-noise and generated without splitting of the diaphragm or nonphysical distortions of the tumor caused by sorting artifacts shown in Figure 4a and b.
Fig. 4.
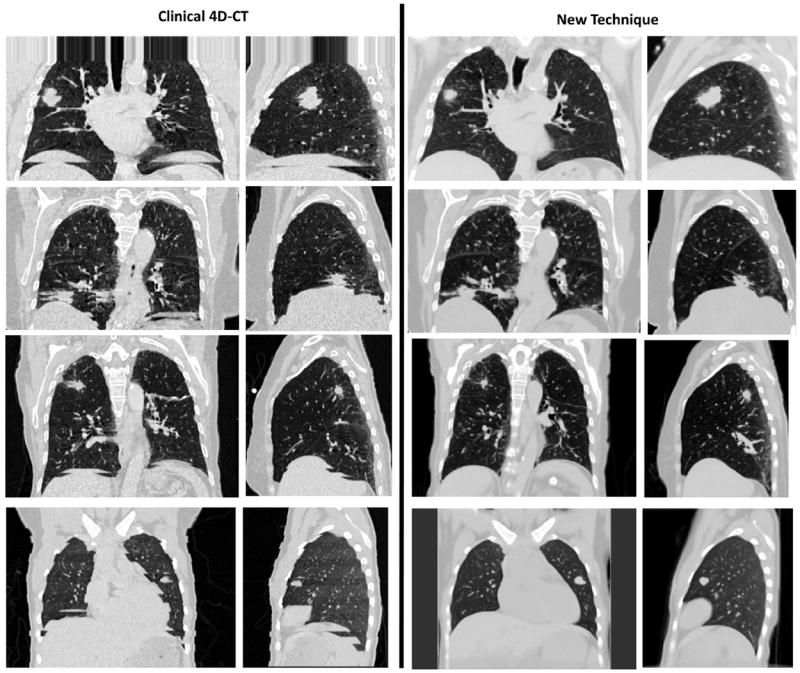
(Left column) Examples of clinical 4D-CT images are shown. (Right column) Sorting artifact–free image was generated by using the new 4D-CT method. Images at the 25th percentile tidal volume, central coronal slice, and right lung sagittal slice are shown in a lung window.
Figure 5 shows a boxplot displaying mean (Fig. 5, o), median (Fig. 5, line), interquartile range (Fig. 5, box), and 5th and 95th percentiles (Fig. 5, whiskers) of model prediction errors for all 10 patients.
Fig. 5.
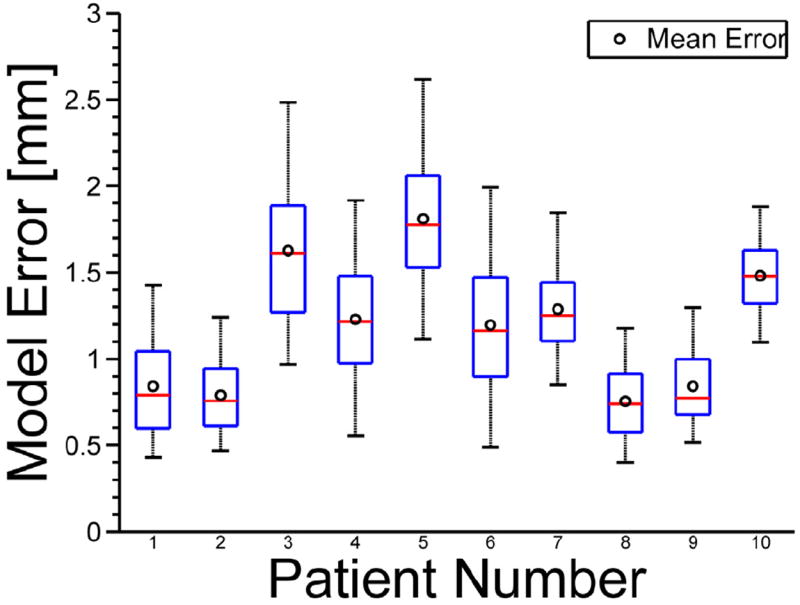
Boxplots show mean (o), median (line), interquartile range (box), and 5th and 95th percentiles (whiskers) of model prediction errors.
Discussion
The results presented here show the novel 4D-CT technique is able to accurately predict lung tissue locations during an arbitrary breath with a mean accuracy of 1.19 ± 0.37 mm across 10 patient data sets. Images are free from sorting artifacts and were generated at a patient dose similar to or less than those used for current 4D-CT techniques. Although techniques to deal with the sorting artifacts present in images created using conventional slow-pitch helical and cine 4D-CT acquisition techniques (2) have been previously proposed using deformable registration (27, 28), the use of fast-helical scans here avoids the resorting of reconstructed images entirely. The technique is robust in the presence of irregular breathing, and the use of a motion model allows images to be generated at any user-specified breathing phase. The acquisition sequence was a set of standard repeated fast-helical CT scans, and the only additional hardware requirement was a data acquisition PC to process the surrogate signal, which could be easily implemented into a clinical workflow. Both the registration and the least-squares fitting procedure could be further optimized using hardware acceleration (GPU) to allow calculation of the motion parameters on a similar timescale as image reconstruction. It is important to note that the 4D-CT technique presented here could be used with any predictive lung motion model (eg, finite element or biomechanical models) (29), not only the heuristic linear motion model used for this study. To our knowledge, we are the first to propose a 4D-CT technique based on registration and deformation of fast helical scans acquired during free breathing.
The motion model accuracy is patient-specific and varies throughout the lung. Regions with large amounts of hysteresis in the upper lungs are accurately modeled in all patients. Regions in the lower lung with increased motion correspond to increased absolute error. Regions at the posterior of the lung in all patients have been observed to suffer from reduced registration accuracy, and correspondingly increased model error, which could be improved with registration parameter optimization. Central regions of the lungs near the mediastinum are accurately modeled, which is important for treatment planning due to the increased risk of radiation therapy–induced bronchial injury in these regions (30, 31). Regions close to the heart show an increase in error due to uncompensated cardiac motion (Fig. 3a and b) which may become significant for radiation therapy treatment planning in cases where tumors are attached to the aorta (32, 33). Initial results using an additional modeling term synchronized to a patient’s electrocardiogram suggest this motion can be successfully accounted for (34). Patient 3 had decreased right lung function due to a collapsed upper right lung (note large motion and tumor volume in Table 1), and an increased relative motion in the left lung, leading to increased motion modeling errors overall (Fig. 3c). Patient 5 was barrel chested (note large lung volume in Table 1) and had a missing rib in the lower left lung, causing the lung to expand outside of the ribcage, corresponding to an increased model error in this region (Fig. 3d). The model assumes motion is linear with tidal volume and airflow, which would not be applicable in this case. Addition of a nonlinear airflow term may better model tissue motion around the ribs and chest wall and is the subject of further work.
Conclusions
The technique proposed here can be used with equipment currently available in the clinic, although further validation is required before clinical use. It is robust in the presence of irregular breathing and allows the entire imaging dose to contribute to the resulting image quality, providing nearly motion artifactefree images at a patient dose similar to or less than that used in current 4D-CT techniques.
Summary.
A novel 4-dimensional computed tomography (CT) technique was developed using standard fast helical CT scanning, deformable image registration (DIR), and a breathing motion model to generate low-noise CT images at user-selected breathing phases. Images do not contain artifacts that arise from sorting of images present in amplitude- or phase-based sorting techniques. Results from 10 patients are presented. The technique was shown to have a motion error on the order of 1 mm and to be robust in the presence of irregular breathing.
Footnotes
Conflict of interest: none.
References
- 1.Kyriakou E, McKenzie DR. Changes in lung tumor shape during respiration. Phys Med Biol. 2012;57:919–935. doi: 10.1088/0031-9155/57/4/919. [DOI] [PubMed] [Google Scholar]
- 2.Keall PJ, Mageras GS, Balter JM, et al. The management of respiratory motion in radiation oncology report of AAPM task group 76. Med Phys. 2006;33:3874–3900. doi: 10.1118/1.2349696. [DOI] [PubMed] [Google Scholar]
- 3.Bortfeld T, Jiang SB, Rietzel E. Effects of motion on the total dose distribution. Semin Radiat Oncol. 2004;14:41–51. doi: 10.1053/j.semradonc.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Bao Y, Zhang L, et al. Assessment of respiration-induced motion and its impact on treatment outcome for lung cancer. Biomed Res Int. 2013;2013:872739. doi: 10.1155/2013/872739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen GTY, Kung JH, Beaudette KP. Artifacts in computed tomography scanning of moving objects. Semin Radiat Oncol. 2004;14:19–26. doi: 10.1053/j.semradonc.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 6.Watkins WT, Li R, Lewis J, et al. Patient-specific motion artifacts in 4dct. Med Phys. 2010;37:2855–2861. doi: 10.1118/1.3432615. [DOI] [PubMed] [Google Scholar]
- 7.Ford EC, Mageras GS, Yorke E, et al. Respiration-correlated spiral CT: A method of measuring respiratory-induced anatomic motion for radiation treatment planning. Med Phys. 2003;30:88–97. doi: 10.1118/1.1531177. [DOI] [PubMed] [Google Scholar]
- 8.Pan T. Comparison of helical and cine acquisitions for 4D-CT imaging with multislice CT. Med Phys. 2005;32:627–634. doi: 10.1118/1.1855013. [DOI] [PubMed] [Google Scholar]
- 9.Langner UW, Keall PJ. Prospective displacement and velocity-based cine 4D CT. Med Phys. 2008;35:4501–4512. doi: 10.1118/1.2977539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu W, Parikh PJ, Hubenschmidt JP, et al. A comparison between amplitude sorting and phase-angle sorting using external respiratory measurement for 4D CT. Med Phys. 2006;33:2964–2974. doi: 10.1118/1.2219772. [DOI] [PubMed] [Google Scholar]
- 11.Abdelnour AF, Nehmeh SA, Pan T, et al. Phase and amplitude binning for 4D-CT imaging. Phys Med Biol. 2007;52:3515–3529. doi: 10.1088/0031-9155/52/12/012. [DOI] [PubMed] [Google Scholar]
- 12.Santoro JP, Yorke E, Goodman KA, et al. From phase-based to displacement-based gating: A software tool to facilitate respiration-gated radiation treatment. J Appl Clin Med Phys. 2009;10:2982. doi: 10.1120/jacmp.v10i4.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wink NM, Panknin C, Solberg TD. Phase versus amplitude sorting of 4D-CT data. J Appl Clin Med Phys. 2006;7:77–85. doi: 10.1120/jacmp.v7i1.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarker J, Chu A, Mui K, et al. Variations in tumor size and position due to irregular breathing in 4D-CT: A simulation study. Med Phys. 2010;37:1254–1260. doi: 10.1118/1.3298007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Low DA, Parikh PJ, Lu W, et al. Novel breathing motion model for radiotherapy. Int J Radiat Oncol Biol Phys. 2005;63:921–929. doi: 10.1016/j.ijrobp.2005.03.070. [DOI] [PubMed] [Google Scholar]
- 16.Low DA, White BM, Lee PP, et al. A novel CT acquisition and analysis technique for breathing motion modeling. Phys Med Biol. 2013;58:L31–L36. doi: 10.1088/0031-9155/58/11/L31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Werner R, White B, Handels H, et al. Technical note: Development of a tidal volume surrogate that replaces spirometry for physiological breathing monitoring in 4D CT. Med Phys. 2010;37:615–619. doi: 10.1118/1.3284282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu W, Low DA, Parikh PJ, et al. Comparison of spirometry and abdominal height as four-dimensional computed tomography metrics in lung. Med Phys. 2005;32:2351–2357. doi: 10.1118/1.1935776. [DOI] [PubMed] [Google Scholar]
- 19.Klein S, Staring M, Murphy K, et al. Elastix: A toolbox for intensity-based medical image registration. IEEE Trans Med Imaging. 2010;29:196–205. doi: 10.1109/TMI.2009.2035616. [DOI] [PubMed] [Google Scholar]
- 20.Staring M, Klein S, Pluim JP. A rigidity penalty term for nonrigid registration. Med Phys. 2007;34:4098–4108. doi: 10.1118/1.2776236. [DOI] [PubMed] [Google Scholar]
- 21.Mattes D, Haynor DR, Vesselle H, et al. Nonrigid multimodality image registration. Proc SPIE, Medical Imaging 2001: Image Processing. 2001 Jul 3;4322:1609–1620. [Google Scholar]
- 22.Thevenaz P, Unser M. Optimization of mutual information for multiresolution image registration. IEEE Trans Image Process. 2000;9:2083–2099. doi: 10.1109/83.887976. [DOI] [PubMed] [Google Scholar]
- 23.Castillo R, Castillo E, Guerra R, et al. A framework for evaluation of deformable image registration spatial accuracy using large landmark point sets. Phys Med Biol. 2009;54:1849–1870. doi: 10.1088/0031-9155/54/7/001. [DOI] [PubMed] [Google Scholar]
- 24.Zhao T, Lu W, Yang D, et al. Characterization of free breathing patterns with 5D lung motion model. Med Phys. 2009;36:5183–5189. doi: 10.1118/1.3246348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruan D, Fessler JA, Balter JM, et al. Exploring breathing pattern irregularity with projection-based method. Med Phys. 2006;33:2491–2499. doi: 10.1118/1.2207253. [DOI] [PubMed] [Google Scholar]
- 26.Ruan D, Fessler JA, Balter JM, et al. Real-time profiling of respiratory motion: Baseline drift, frequency variation and fundamental pattern change. Phys Med Biol. 2009;54:4777–4792. doi: 10.1088/0031-9155/54/15/009. [DOI] [PubMed] [Google Scholar]
- 27.Wolthaus JWH, Sonke J-J, van Herk M, et al. Reconstruction of a time-averaged midposition ct scan for radiotherapy planning of lung cancer patients using deformable registrationa. Med Phys. 2008;35:3998–4011. doi: 10.1118/1.2966347. [DOI] [PubMed] [Google Scholar]
- 28.Wu G, Lian J, Shen D. Improving image-guided radiation therapy of lung cancer by reconstructing 4D-CT from a single free-breathing 3D-CT on the treatment day. Med Phys. 2012;39:7694–7709. doi: 10.1118/1.4768226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McClelland JR, Hawkes DJ, Schaeffter T, et al. Respiratory motion models: A review. Med Image Anal. 2013;17:19–42. doi: 10.1016/j.media.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 30.Miller KL, Shafman TD, Anscher MS, et al. Bronchial stenosis: An underreported complication of high-dose external beam radiotherapy for lung cancer? Int J Radiat Oncol Biol Phys. 2005;61:64–69. doi: 10.1016/j.ijrobp.2004.02.066. [DOI] [PubMed] [Google Scholar]
- 31.Timmerman R, McGarry R, Yiannoutsos C, et al. Excessive toxicity when treating central tumors in a phase II study of stereotactic body radiation therapy for medically inoperable early-stage lung cancer. J Clin Oncol. 2006;24:4833–4839. doi: 10.1200/JCO.2006.07.5937. [DOI] [PubMed] [Google Scholar]
- 32.Ekberg L, Holmberg O, Wittgren L, et al. What margins should be added to the clinical target volume in radiotherapy treatment planning for lung cancer? Radiother Oncol. 1998;48:71–77. doi: 10.1016/s0167-8140(98)00046-2. [DOI] [PubMed] [Google Scholar]
- 33.Seppenwoolde Y, Shirato H, Kitamura K, et al. Precise and real-time measurement of 3D tumor motion in lung due to breathing and heartbeat, measured during radiotherapy. Int J Radiat Oncol Biol Phys. 2002;53:822–834. doi: 10.1016/s0360-3016(02)02803-1. [DOI] [PubMed] [Google Scholar]
- 34.White B, Thomas D, Lamb J, et al. We-a-134-08: Modeling cardiac induced lung tissue motion for a quantitative breathing motion model. Med Phys. 2013;40:470. doi: 10.1118/1.4866888. [DOI] [PMC free article] [PubMed] [Google Scholar]


