Abstract
Primate gastrointestinal microbial communities are becoming increasingly appreciated for their relevance to comparative medicine and conservation, but the factors that structure primate “microbiomes” remain controversial. This study examined a community of primates in Kibale National Park, Uganda, to assess the relative importance of host species and location in structuring gastrointestinal microbiomes. Fecal samples were collected from primates in intact forest and from primates in highly disturbed forest fragments. People and livestock living nearby were also included, as was a geographically distant population of related red colobus in Kenya. A culture-free microbial community fingerprinting technique was used to analyze fecal microbiomes from 124 individual red colobus (Procolobus rufomitratus), 100 individual black-and-white colobus (Colobus guereza), 111 individual red-tailed guenons (Cercopithecus ascanius), 578 human volunteers, and 364 domestic animals, including cattle (Bos indicus and B. indicus × B. taurus crosses), goats (Caprus hircus), sheep (Ovis aries), and pigs (Sus scrofa). Microbiomes sorted strongly by host species, and forest fragmentation did not alter this pattern. Microbiomes of Kenyan red colobus sorted distinctly from microbiomes of Ugandan red colobus, but microbiomes from these two red colobus populations clustered more closely with each other than with any other species. Microbiomes from red colobus and black-and-white colobus were more differentiated than would be predicted by the phylogenetic relatedness of these two species, perhaps reflecting heretofore underappreciated differences in digestive physiology between the species. Within Kibale, social group membership influenced intra-specific variation among microbiomes. However, intra-specific variation was higher among primates in forest fragments than among primates in intact forest, perhaps reflecting the physical separation of fragments. These results suggest that, in this system, species-specific processes such as gastrointestinal physiology strongly structure microbial communities, and that primate microbiomes are relatively resistant to perturbation, even across large geographic distances or in the face of habitat disturbance.
Keywords: microbiome, non-human primate, Uganda, Colobinae, Cercopithecinae, forest fragmentation
INTRODUCTION
Advances in molecular techniques have enabled detailed studies of microbial communities and the roles of “microbiomes” in human and animal health [Dominguez-Bello et al., 2011; Shendure & Ji, 2008; Turnbaugh et al., 2007]. The microbiome concept considers the community of fungi, bacteria, bacteriophages, viruses, and protozoa that inhabit a host and play important roles in digestion, development [McFall-Ngai, 2002], metabolic disorders [Turnbaugh et al., 2009], behavior [Ezenwa et al., 2012], and immune function [Round & Mazmanian, 2009]. Furthermore, a robust microbiome may prevent colonization of the host by disease-causing microorganisms [Adams et al., 2007; Croswell et al., 2009; Dillon et al., 2005; Koch & Schmid-Hempel, 2011]. For these reasons, understanding the factors that structure microbiomes has emerged as a research priority [Caporaso et al., 2011; Robinson et al., 2010; Spor et al., 2011; Ursell et al., 2012; Yeoman et al., 2011].
Non-human primates (hereafter, “primates”) have long been recognized as important model systems for understanding human health; thus, there is growing interest in the primate microbiome [Degnan et al., 2012; Frey et al., 2006; McKenna et al., 2008; Ochman et al., 2010; Szekely et al., 2010; Uenishi et al., 2007; Xu et al., 2010; Yildirim et al., 2010]. Studies to date have shown that host phylogeny and environmental factors such as diet play strong roles in structuring primate microbiomes. For example, a study by Ochman et al. [2010] of fecal microbiomes of chimpanzees (Pan troglodytes), bonobos (Pan paniscus), and gorillas (Gorilla gorilla) revealed that patterns of microbial community structure recapitulated host phylogeny, supporting earlier studies across a range of species indicating broad patterns of host-microbiome co-evolution [Ley et al., 2008a,b; McFall-Ngai, 2002; Moran et al., 2008; Ochman et al., 2010; Yeoman et al., 2011]. Work by Frey et al. [2006] on wild gorillas and Degnan et al. [2012] on wild chimpanzees revealed evidence of temporal stability in primate microbiomes, lending support to a growing body of research suggesting that gastrointestinal microbiomes may be acquired early in life, quickly reaching a stable-state climax community that is resistant to perturbation [Degnan et al., 2012; Koenig et al., 2011; Turnbaugh et al., 2009]. Degnan et al. [2012] also provided evidence of microbial partitioning by social group, suggesting socio-demographic influences on microbial community structure. Recent work by Amato et al. [2013] demonstrates plasticity of microbiomes of black howler monkeys (Alouatta pigra) in response to habitat degradation [Amato et al., 2013].
The “heirloom vs. souvenir” dichotomy was originally developed as a conceptual framework for parasite–host dynamics, but it has also been used to describe the processes by which microbial communities may be structured [Banks et al., 2009; Kliks, 1990]. Under the “heirloom” model, maternal inoculation of offspring with a Specific collection of microbes at birth creates a shared microbial community among closely related individuals. In this case, microbiomes are inherited and stable. Alternatively, under the “souvenir” model, microbial communities frequently shift in response to extrinsic factors such as host stress, reproductive status, nutritional circumstances, or health state [Caporaso et al., 2011; Costello et al., 2009]. In this case, dynamic colonization or extinction events constantly re-shape microbial communities such that individuals with a history of common environmental exposures develop similar microbiomes [Dethlefsen et al., 2007]. Applied to wild primates, the “heirloom” model would be reflected as phylogenetic clustering of microbiomes irrespective of geography, whereas the “souvenir” model would be reflected as geographic clustering of microbiomes irrespective of phylogeny. Partitioning the effects of phylogeny and geography would require a system in which multiple phylogenetically divergent species inhabit the same environment. Such systems have been used to assess kin-associated bacterial communities of multiple frog species living in shared ponds, revealing strong evidence of species-specific partitioning [McKenzie et al., 2012]. However, we are aware of no such studies that have addressed these questions in mammals or primates.
The diverse and abundant primate community of Kibale National Park (KNP), Uganda, contains species with different digestive physiologies and phylogenetic relationships that occupy the same habitats. By comparing the fecal microbiomes of these primate species across habitat types, we examine the degree to which primate microbiomes are structured as “heirlooms” versus “souvenirs.” To build upon previous work on cross-species microbial transmission in shared habitats, we also examine the fecal microbiomes of people and livestock living close to this primate community. To extend our analyses spatially, we include fecal microbiomes from geographically distant red colobus in Kenya. Our results reveal how fecal microbiomes differ among hosts and across environments, and how primate gastrointestinal microbiomes are structured in this setting.
METHODS
Study Site and Sample Collection
Kibale National Park is a moist evergreen forest near the foothills of the Rwenzori Mountains in Uganda (795 km2, 0°13′–0°41′ N, 30°19′–30°32′E) and is surrounded by a mosaic landscape of densely populated human settlements, farmlands, and remnant forest fragments. Fecal samples were collected between January 2008 and December 2010 from two social groups of each species of primate living within the intact forest in KNP (N = 65) and from primates in 20 highly disturbed forest fragments outside the park (N = 270), as previously described [Goldberg et al., 2008]. Because these fragments are typically close in size or smaller than the home range of a primate social group, only one social group inhabits each fragment. The forest fragments are relatively recently isolated (since approximately the 1950s) and close to KNP, with fragments having been connected to KNP as recently as 1999. For comparison, samples from human volunteers (N = 578) and their domestic animals (N = 364), including cattle (Bos indicus and B. indicus × B. taurus crosses), goats (Caprus hircus), sheep (Ovis aries), and pigs (Sus scrofa), living near the forest fragments were also collected as previously described [Johnston et al., 2010; Salyer et al., 2012]. Each fecal sample included in this study represents a unique individual sampled only once.
We restricted our primate sampling to the three species that commonly inhabit both the park and the forest fragments: red colobus (Procolobus rufomitratus; n = 26 KNP, n = 98 fragment) and black-and-white colobus (Colobus guereza, n = 20 KNP, n = 80 fragment), both of which are folivorous colobines with ruminant-like chambered stomachs and fore-gut fermentation; and red-tailed guenons (Cercopithecus ascanius), an omnivorous cercopithecine with a simple stomach and hind-gut fermenting digestive system (n = 19, KNP, n = 92 fragment). For comparison, we also included 18 fecal samples from red colobus (P. rufomitratus) in Tana River National Primate Reserve (171 km2, 1°55′S, 40°5′E) in Kenya. Tana River is comprised of a series of 23 protected evergreen riparian forest fragments that occur along a 16-km2 stretch of the lower Tana River system [Owino & Oyugi, 2010] and is approximately 1,100 km from KNP.
Prior to the research, all procedures were reviewed and approved by the Uganda National Council for Science and Technology, the Uganda Wildlife Authority, the University of Wisconsin Institutional Review Board (IRB #H2009-0007), and the University of Wisconsin Research Animal Care and Use Committee (V01385-0-08-08). This research adhered to the American Society of Primatologists Principles for the Ethical Treatment of Non-Human Primates.
Analysis of Fecal Microbiomes
DNA from fecal samples was extracted using the Zymo Research (Irvine, CA) Fecal DNA MiniPrep Kit, with the following three modifications: (1) 4 μL of a Cy5-labelled internal amplification control from Primer Design Ltd (Southampton, UK) was added to the lysis buffer before addition of fecal samples, (2) samples were lysed for 3 min using a commercial “bead beater” homogenizer, and (3) the elution buffer was allowed to soak the filter for 2 min prior to final centrifugation. As an initial quality control step, internal amplification control real-time quantitative PCRs were performed on each sample according to Primer Design Ltd manufacturer instructions on a Bio-Rad (Hercules, CA) CFX96 instrument. Only samples that passed quality control (sigmoidal amplification curves and Ct values within normal ranges) were carried forward.
A microbial community fingerprint technique, automated ribosomal intergenic spacer analysis (ARISA), was then used to assess bacterial diversity based on operational taxonomic units (OTUs), which correspond roughly to species [Fisher & Triplett, 1999]. The highly variable bacterial intergenic spacer region between 16S and 23S rRNA genes was amplified using the primer set 1606f-FAM (16S FAM 5′-TGYACACACCGCCCGT-3′) and 23Sr (23S 5’-GGGTTBCCCCATTCRG-3’). PCR conditions were optimized using the FailSafe PCR system from Epicentre Biotechnologies (Madison, WI). Each 25 μL reaction contained 10 ng total DNA, 400 nM of each primer, 1× Epicentre FailSafe Pre-Mix B PCR Buffer, and 1 μL Epicentre FailSafe Enzyme Mix. A Bio-Rad C1000 thermocycler was used with the following cycling parameters: (1) initial denaturation for 2 min at 94°C, (2) 30× amplification cycles with 35 sec at 94°C, 45 sec at 55°C, and 2 min at 72°C, and (3) final extension for 2 min at 72°C. One microliter of PCR product diluted 1:1 in nuclease-free water was then mixed with 9.6 μL 10% formamide and 0.4 μL custom internal ROX-labeled size standard (100–2,000 bp; BioVentures, Murfreesboro, TN). The mixture was run for fragment analysis on ABI 3730xl DNA Analyzers (Life Technologies, Grand Island, NY) at the University of Wisconsin-Madison Biotechnology Center.
As a final quality control step, peak profiles were examined visually; only samples with peak profiles indicative of successful ARISA were analyzed. For these samples, a matrix was created of the relative abundance of OTUs present. Peaks were called as previously described using an R-script and smoothing algorithm [Jones & McMahon, 2009]. A universal binning profile with a gradually scaled design was used to assign 172 OTUs according to amplicon size (300–399 bp: ±1 bp, 401–500 bp: ±1.5 bp, 501–600 bp: ±2 bp, 601–700 bp: ±3 bp, 701–800 bp: ±4 bp, 801–1,000 bp: ±10 bp).
Statistical Analyses
Operational taxonomic unit relative abundance matrices for each fecal sample were analyzed using non-metric multi-dimensional scaling to identify clustering of microbial communities [Ramette, 2007]. A Bray–Curtis similarity matrix was used to compare the OTU composition and abundance of all microbial communities. Statistical significance was assessed using analysis of similarity (ANOSIM) with the computer program PRIMER 6.1.9 (PRIMER-E Ltd, Plymouth, UK) to compare within- and between-group similarity. The ANOSIM R statistic varies from −1 to 1, with −1 indicating 100% between-group similarity, 1 indicating 100% within-group similarity, and 0 indicating random grouping. Although values are relative, R > 0.75 is generally considered strong evidence of separation, R > 0.5 suggests separation, and R < 0.25 suggests strong overlap [Clarke, 2001]. Unpaired Student’s t-tests were also used to compare average Bray–Curtis similarities among primate fecal microbiomes within KNP to average Bray–Curtis similarities among primate fecal microbiomes within the forest fragments, as a comparison of inter-individual diversity of microbiomes within each habitat type.
RESULTS
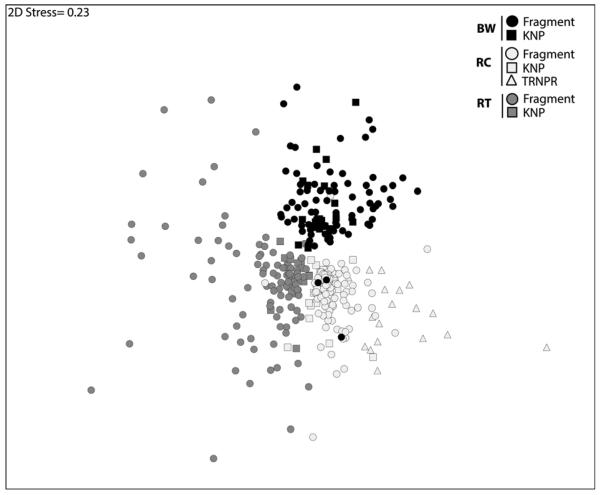
Fecal microbiomes of red colobus, black-and-white colobus, and red-tailed guenons within KNP sorted strongly according to host species (Table I; Fig. 1). These significant patterns persisted for the same species in the highly degraded forest fragments surrounding KNP (Table I; Fig. 1). When primates from KNP were compared directly to primates of the same species living in the fragments, no differences in fecal microbiome community composition were detectable between forest and fragment populations (Table I; Fig. 1). When microbiomes of red colobus from KNP were compared to those of geographically distant red colobus from Tana River National Primate Reserve in Kenya, strong separation was observed (Table I; Fig. 1); however, Tana River red colobus microbiomes still clustered most closely with Ugandan red colobus microbiomes. These results are based on 1,340 samples that passed quality control, with 118 samples excluded because of internal amplification control RT-PCR failure and 24 samples excluded because of failure to pass peak profile assessment. Within these samples, an average of 97 OTUs were present (range 28–129 OTUs).
TABLE I.
Analysis of Similarity (ANOSIM) Comparing Microbiomes of Primates, People, and Livestock
| ANOSIM Global R | Significance (P) | |
|---|---|---|
| Among species | ||
| Within KNP: BWC vs. RC vs. RT | 0.63 | <0.001 |
| Within Fragment: BWC vs. RC vs. RT | 0.40 | <0.001 |
| Among locations | ||
| BWC: KNP vs. Fragment | −0.06 | 0.81 |
| RT: KNP vs. Fragment | −0.169 | 0.99 |
| RC: KNP vs. Fragment | 0.03 | 0.34 |
| RC: KNP vs. TRNPR | 0.82 | <0.001 |
| People among communities | 0.10 | <0.001 |
| Livestock among communities | 0.09 (BO) | 0.05 (BO) |
| 0.28 (SH) | <0.005 (SH) | |
| 0.22 (PIG) | 0.15 (PIG) | |
| 0.15 (GT) | <0.001 (GT) |
Note Primates living in Kibale National Park (KNP), in forest fragments (Fragment) and in Tana River National Primate Reserve, Kenya (TRNPR). “Communities” refers to the human settlements surrounding the forest fragments. RC, red colobus; BWC, black-and-white colobus; RT, red-tailed guenon; BO, cattle; SH, sheep; PIG, pigs; GT, goats.
Fig. 1.
Non-metric multi-dimensional scaling ordination based on Bray–Curtis similarity matrix for fecal microbiomes of three primate species from Kibale National Park (KNP) and surrounding forest fragments: red colobus (RC), black-and-white colobus (BW), and red-tailed guenon (RT). Red colobus from Tana River National Park, Kenya (TRNPR) are included for comparison. Each point represents the fecal microbiome of an individual primate. Two-dimensional stress value indicates moderate fidelity of original Bray–Curtis similarity matrix as represented in 2D space.
A Student’s t-test comparing average Bray–Curtis similarities of fecal microbiomes within KNP to average Bray–Curtis similarities of fecal microbiomes within nearby forest fragments indicated less intra-specific variation among microbiomes of primates in KNP than among primates in the forest fragments (Fig. 1). Red-tailed guenons (t = 5.90, df = 109, P < 0.0001) and black-and-white colobus (t = 3.56, df = 98, P < 0.001) exhibited significant differences in this regard; however, red colobus did not show such a difference (t = 0.84, df = 122, P < 0.40).
To assess whether observed differences in intraspecific microbiome variability resulted from physical location or membership in a particular social group, we analyzed microbiomes of primates within KNP according to social group. ANOSIM revealed a strong pattern of partitioning among black-and-white colobus social groups (ANOSIM: Global R = 0.48, P < 0.001), moderate partitioning among red colobus social groups (ANOSIM: Global R = 0.37, P < 0.001), and weaker partitioning among red-tailed guenon social groups (Global R = 0.27, P < 0.001).
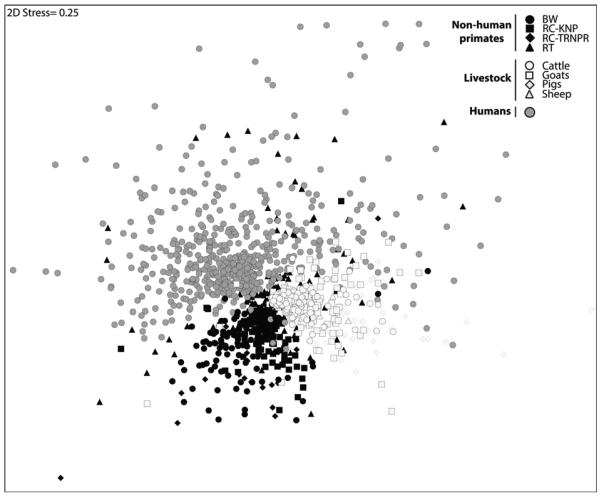
Species-specific patterns of microbial community composition generally persisted when livestock and human samples were included. Human microbiomes clustered together strongly and were separate from livestock and non-human primate microbiomes (Fig. 2). However, microbiomes of different species of livestock strongly overlapped (Table I; Fig. 2). When people and livestock were grouped according to location (i.e., the settlements surrounding forest fragments) no separation of microbiomes was observed (series of 4 nM DS plots not shown, Table I).
Fig. 2.
Non-metric multi-dimensional scaling ordination based on Bray–Curtis similarity matrix comparing the fecal microbiomes of people, livestock, and non-human primates (BW, black-and-white colobus; RC, red colobus; RT, red-tailed guenon) from the Kibale area. Red colobus from Tana River National Park, Kenya (TRNPR) are included for comparison. Each point represents the fecal microbiome of an individual. Two-dimensional stress value indicates moderate fidelity of original Bray–Curtis similarity matrix as represented in 2D space.
DISCUSSION
Our results suggest that species-specific processes strongly structure microbial communities and that geography plays a lesser role in this system. The dominant pattern of species-specific primate microbial community structure remained robust across habitat types. Although forest fragmentation in the Kibale area did not increase the similarity of microbiomes among different primate species, forest fragmentation had a modest, but discernible, effect on microbiome variation within species. These findings suggest that primate microbiomes are strongly constrained by host species, but that landscape-level factors, such as physical separation on the order of 1–10 km and/or habitat degradation, can lead to minor differentiation of microbiomes within species. Overall, our results suggest that primate microbial communities are more likely to be inherited than accumulated from the environment, conforming more closely to the “heirloom” paradigm than the “souvenir” paradigm.
Our results differ somewhat from those of other studies that have reported strong phylogenetic signal in primate microbiomes, albeit based on smaller sample sizes [Ley et al., 2008a; Ochman et al., 2010; Yeoman et al., 2011]. Despite evidence that species-Specific processes appear to dominate in the KNP system, the patterns of microbial community similarity that we document do not closely reflect host phylogeny. Specifically, although black-and-white colobus and red colobus are closely related [Perelman et al., 2011], microbiomes of these two colobine species were as different from each other as either was from the microbiomes of more distantly related red-tailed guenons. In fact, the degree of separation between red colobus and black-and-white colobus microbiomes was remarkably high considering the close phylogenetic relationship, similar digestive physiology, comparable ecology, and similar diets of the two species [Chapman & Pavelka, 2005; Chivers, 1994]. Because gastrointestinal microbial communities are known to partition into unique consortia along the gastrointestinal tract [Walter & Ley, 2011], we speculate that the greater-than-expected differentiation of microbiomes between red colobus and black-and-white colobus may reflect heretofore underappreciated differences in the digestive physiologies of the two species, perhaps reflecting “niche differentiation” of microbes within the gut. For example, we note that red colobus have an additional stomach compartment, the presaccus [Groves, 2007]; although the presaccus is an anatomic feature of the fore-gut, it could have “downstream” effects on fecal microbiomes.
Contrary to a similar study of black howler monkeys [Amato et al., 2013], species-specific differences among primate microbiomes in our study persisted even in the face of habitat degradation. Primates in the forest fragments near Kibale are rapidly going extinct [Chapman et al., 2012]. Indeed, many of our samples represent the last biological specimens collected prior to the extirpation of the population [Goldberg et al., 2008]. For primates in fragments, these conditions alter parasite dynamics and increase the potential for microbial transmission among species [Bonnell et al., 2010; Chapman et al., 2006; Rwego et al., 2008; Salyer et al., 2012; Salzer et al., 2007]. Furthermore, fragmentation decreases the ecological complexity of forest habitats, reducing the diversity of primate foods and preventing the degree of dietary niche partitioning that primates achieve in intact habitats [Chaves & Arroyo-Rodrigues, 2012]. The fact that microbiomes of primate species in fragments were no less differentiated than microbiomes of those same species in intact habitats re-enforces the idea that species-specific processes fundamentally shape and constrain primate microbiomes.
We note that forest fragmentation near KNP is a recent phenomenon, with some primate social groups becoming geographically isolated as recently as 10 years ago. Also, some fragments are as close as 1 km to the park edge, perhaps enabling primate movement across the park boundary. Finally, although ongoing habitat destruction in the fragments is occurring more rapidly than in KNP, the encroachment of human settlements along the increasingly abrupt national park boundary could influence primate social groups within KNP. Thus, our results are Specific to the spatial and temporal scale of our system, and considering fragments as isolated “islands” is probably an oversimplification [Weins, 1995].
Our results show that intra-specific variation in primate microbiomes was greater in forest fragments than in KNP. This finding may reflect the combined effects of each fragment’s unique history and ecology [Onderdonk & Chapman, 2000] or the nutritional and social stress that primates in fragments experience [Chapman et al., 2006], either or both of which could lead to changes in microbiome composition. Because, we were only able to analyze fecal microbiomes, which sample hind-gut bacteria [Eckburg et al., 2005], such effects could be even more pronounced for other compartments within the gastrointestinal tract.
Notably, when each primate social group within KNP was considered separately, significant microbiome partitioning was also observed. Previous studies of fecal microbiomes of wild chimpanzees revealed that gut microbial communities were partitioned according to social group and that such patterns persist even when animals migrate to new social groups [Degnan et al., 2012]. Our data support the conclusion that social group membership influences primate gastrointestinal microbial communities. Apparently, primate social structure can lead to sub-structuring of microbiomes even within geographically contiguous populations.
We observed strong evidence of microbiome partitioning between red colobus in the Kibale area and red colobus from Tana River in Kenya. However, Tana River red colobus microbiomes still clustered most closely with those of Kibale red colobus in our multi-species analysis, indicating that host species effects still predominate even across large geographic distances. This result was especially surprising given that diets and other ecological factors must vary considerably across such distances. We suggest that examining geographically disparate populations of related primates may offer valuable insights into the role of phylogeny versus geography in structuring primate microbiomes.
Previous work in Kibale has shown that forest fragmentation alters primate parasite dynamics and may increase the potential for disease transmission between people and wildlife [Bonnell et al., 2010; Chapman et al., 2006; Rwego et al., 2008; Salyer et al., 2012; Salzer et al., 2007]. However, the mechanisms underlying these dynamics are not well understood. People, domestic animals, and wildlife move freely across the permeable boundaries of forest fragments, creating ample opportunities for direct and environmentally mediated cross-species disease transmission [Goldberg et al., 2012]. The fact that our data showed microbial communities to sort strongly by species may indicate lack of resolution for detecting such effects when entire microbiomes are considered. We speculate that evidence for environmental effects and cross-species microbial transmission may emerge more clearly when individual microbial taxa are considered, especially those associated with disease.
Our study is based on ARISA, which provides only a general “fingerprint” of the microbiome but lacks the resolution of “deep sequencing.” DNA sequence data will allow identification of the microbial taxa underlying the trends we have documented and could reveal specific microbial taxa that are associated more strongly than others with environmental perturbation or cross-species transmission. Nevertheless, our findings support an emerging consensus in the literature that species-specific “heirloom” effects largely structure animal microbiomes [Arumugam et al., 2011; Muegge et al., 2011; Phillips et al., 2012; Smith et al., 2012; Walter & Ley, 2011]. “Souvenir” effects such as diet, environmental variation, and socio-demography can shift microbiomes slightly, but appear not to be major determinants of microbiome structure in this system. If primate microbiomes are generally highly constrained by species-specific processes, then this raises concerns about the ability of primates and their microbiomes to adapt to changing environments.
ACKNOWLEDGMENTS
This work was supported by the NIH grant TW009237 as part of the joint NIH-NSF Ecology of Infectious Disease program and the UK Economic and Social Research Council, by National Science Foundation grant no. 0935347, by the National Science Foundation IGERT Program (DGE-0549407), and by the National Science Foundation Graduate Research Fellowship Program. We thank Makerere University Biological Field Station, especially J. Lwanga and K. Innocent for logistical and field support. We also thank P. Omeja, as well as the Kibale EcoHealth Project and Kibale Fish and Monkey Project field staff for their assistance in the field. T. McMahon, G. Loudon Wolfe, and S. Jones provided invaluable training and support for laboratory and statistical methods. L. Naughton and J. Patz provided helpful comments on earlier drafts of this manuscript.
NIH
TW009237
National Science Foundation
0935347
National Science Foundation IGERT Program
DGE-0549407
National Science Foundation Graduate Research Fellowship Program
Footnotes
Conflicts of interest: The authors have no conflicts of interest to declare.
REFERENCES
- Adams DA, Riggs MM, Donskey CJ. Effect of fluoroquinolone treatment on growth of and toxin production by epidemic and nonepidemic Clostridium difficile strains in the cecal contents of mice. Antimicrob Agents Chemother. 2007;51:2674–2678. doi: 10.1128/AAC.01582-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato KR, Yeoman CJ, Kent A, et al. Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. ISME J. 2013;7:1344–1353. doi: 10.1038/ismej.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks JC, Cary SC, Hogg ID. The phylogeography of Adelie penguin faecal flora. Environ Microbiol. 2009;11:577–588. doi: 10.1111/j.1462-2920.2008.01816.x. [DOI] [PubMed] [Google Scholar]
- Bonnell TR, Sengupta RR, Chapman CA, Goldberg TL. An agent-based model of red colobus resources and disease dynamics implicates key resource sites as hot spots of disease transmission. Ecol Model. 2010;221:2491–2500. [Google Scholar]
- Caporaso JG, Lauber CL, Costello EK, et al. Moving pictures of the human microbiome. Genome Biol. 2011;12:R50. doi: 10.1186/gb-2011-12-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman CA, Pavelka MS. Group size in folivorous primates: ecological constraints and the possible influence of social factors. Primates. 2005;46:1–9. doi: 10.1007/s10329-004-0093-9. [DOI] [PubMed] [Google Scholar]
- Chapman CA, Wasserman MD, Gillespie TR, et al. Do food availability, parasitism, and stress have synergistic effects on red colobus populations living in forest fragments? Am J Phys Anthropol. 2006;131:525–534. doi: 10.1002/ajpa.20477. [DOI] [PubMed] [Google Scholar]
- Chapman CA, Ghai R, Jacob A, et al. Going, going, gone: a 15-year history of the decline of primates in forest fragments near Kibale National Park, Uganda. In: Marsh LK, editor. Primates in fragments. Springer; New York: 2012. [Google Scholar]
- Chaves KE, Arroyo-Rodrigues VA. Differences in diet between spider monkey groups living in forest fragments and continuous forest in Mexico. Biotropica. 2012;44:105–113. [Google Scholar]
- Chivers DJ. Functional anatomy of the gastrointestinal tract. In: Davies AG, Oates JF, editors. Colobine monkeys: Their ecology, behaviour and evolution. Cambridge University Press; Cambridge: 1994. pp. p 205–228. [Google Scholar]
- Clarke RN. PRIMER v5: user manual/tutorial, PRIMER-E. Plymouth; UK: 2001. p. p 91. [Google Scholar]
- Costello EK, Lauber CL, Hamady M, et al. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croswell A, Amir E, Teggatz P, Barman M, Salzman NH. Prolonged impact of antibiotics on intestinal microbial ecology and susceptibility to enteric Salmonella infection. Infect Immun. 2009;77:2741–2753. doi: 10.1128/IAI.00006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan PH, Pusey AE, Lonsdorf EV, et al. Factors associated with the diversification of the gut microbial communities within chimpanzees from Gombe National Park. Proc Natl Acad Sci USA. 2012;109:13034–13039. doi: 10.1073/pnas.1110994109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon RJ, Vennard CT, Buckling A, Charnley AK. Diversity of locust gut bacteria protects against pathogen invasion. Ecol Lett. 2005;8:1291–1298. [Google Scholar]
- Dominguez-Bello MG, Blaser MJ, Ley RE, Knight R. Development of the human gastrointestinal microbiota and insights from high-throughput sequencing. Gastroenterology. 2011;140:1713–1719. doi: 10.1053/j.gastro.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, urdom E, Dethlefesen L, Sargent M, Gill SR, Nelon KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezenwa VO, Gerardo NM, Inouye DW, Medina M, Xavier JB. Animal behavior and the microbiome. Science. 2012;338:198–199. doi: 10.1126/science.1227412. [DOI] [PubMed] [Google Scholar]
- Fisher MM, Triplett EW. Automated approach for ribosomal intergenic spacer analysis of microbial diversity and its application to freshwater bacterial communities. Appl Environ Microb. 1999;65:4630–4636. doi: 10.1128/aem.65.10.4630-4636.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey JC, Rothman JM, Pell AN, et al. Fecal bacterial diversity in a wild gorilla. Appl Environ Microb. 2006;72:3788–3792. doi: 10.1128/AEM.72.5.3788-3792.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg TL, Gillespie TR, Rwego IB, Estoff EL, Chapman CA. Forest fragmentation and bacterial transmission among nonhuman primates, humans, and livestock, Uganda. Emerg Infect Dis. 2008;14:1375. doi: 10.3201/eid1409.071196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg TL, Paige SB, Chapman CA. The Kibale EcoHealth Project: exploring connections among human health, animal health, and landscape dynamics in western Uganda. In: Aguirre AA, Daszak P, Ostfeld RS, editors. New directions in conservation medicine: applied cases of ecological health. Oxford University Press; New York: 2012. pp. p 452–465. [Google Scholar]
- Groves CP. The taxonomic diversity of the Colobinae of Africa. J Anthropol Sci. 2007;85:7–34. [Google Scholar]
- Johnston AR, Gillespie TR, Rwego IB, et al. Molecular epidemiology of cross-species Giardia duodenalis transmission in Western Uganda. PLoS Negl Trop Dis. 2010;4:e683. doi: 10.1371/journal.pntd.0000683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SE, McMahon KD. Species-sorting may explain an apparent minimal effect of immigration on freshwater bacterial community dynamics. Environ Microbiol. 2009;11:905–913. doi: 10.1111/j.1462-2920.2008.01814.x. [DOI] [PubMed] [Google Scholar]
- Kliks MM. Helminths as heirlooms and souvenirs—a review of New-World paleoparasitology. Parasitol Today. 1990;6:93–100. doi: 10.1016/0169-4758(90)90223-q. [DOI] [PubMed] [Google Scholar]
- Koch H, Schmid-Hempel P. Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proc Natl Acad Sci USA. 2011;108:19288–19292. doi: 10.1073/pnas.1110474108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig JE, Spor A, Scalfone N, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci US A. 2011;108:4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Hamady M, Lozupone C, et al. Evolution of mammals and their gut microbes. Science. 2008a;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008b;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai MJ. Unseen forces: the influence of bacteria on animal development. Dev Biol. 2002;242:1–14. doi: 10.1006/dbio.2001.0522. [DOI] [PubMed] [Google Scholar]
- McKenna P, Hoffmann C, Minkah N, et al. The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis. PLoS Pathog. 2008;4:e20. doi: 10.1371/journal.ppat.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie VJ, Bowers RM, Fierer N, Knight R, Lauber CL. Co-habiting amphibian species harbor unique skin bacterial communities in wild populations. ISME J. 2012;6:588–596. doi: 10.1038/ismej.2011.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- Muegge BD, Kuczynski J, Knights D, et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332:970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Worobey M, Kuo CH, et al. Evolutionary relationships of wild hominids recapitulated by gut microbial communities. PLoS Biol. 2010;8:e1000546. doi: 10.1371/journal.pbio.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onderdonk DA, Chapman CA. Coping with forest fragmentation: the primates of Kibale National Park, Uganda. Int J Primatol. 2000;21:587–611. [Google Scholar]
- Owino AO, Oyugi JO. Assessment of forest patches’ extents and land cover changes in the Tana River Primate National Reserve, 1994–2004. Afr J Ecol. 2010;48:546–550. [Google Scholar]
- Perelman P, Johnson WE, Roos C, Seuanez HN, Horvath JE, Moreira MA, Kessing B, Pontius J, Roelke M, Rumpler Y, Schneider MP, Silva A, O’Brien SJ, Pecon-Slattery J. A molecular phylogeny of living primates. PLoS Genet. 2011;7:e1001342. doi: 10.1371/journal.pgen.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips CD, Phelan G, Dowd SE, et al. Microbiome analysis among bats describes influences of host phylogeny, life history, physiology and geography. Mol Ecol. 2012;21:2617–2627. doi: 10.1111/j.1365-294X.2012.05568.x. [DOI] [PubMed] [Google Scholar]
- Ramette A. Multivariate analyses in microbial ecology. FEMS Microbiol Ecol. 2007;62:142–160. doi: 10.1111/j.1574-6941.2007.00375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson CJ, Bohannan BJM, Young VB. From structure to function: the ecology of host-associated microbial communities. Microbiol Mol Biol Rev. 2010;74:453. doi: 10.1128/MMBR.00014-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rwego IB, Gillespie TR, Isabirye-Basuta G, Goldberg TL. High rates of Escherichia coli transmission between livestock and humans in rural Uganda. J Clin Microbiol. 2008;46:3187–3191. doi: 10.1128/JCM.00285-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salyer SJ, Gillespie TR, Rwego IB, Chapman CA, Goldberg TL. Epidemiology and molecular relationships of Cryptosporidium spp. in people, primates, and livestock from Western Uganda. PLoS Negl Trop Dis. 2012;6:e1597. doi: 10.1371/journal.pntd.0001597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzer JS, Rwego IB, Goldberg TL, Kuhlenschmidt MS, Gillespie TR. Giardia sp and Cryptosporidium sp infections in primates in fragmented and undisturbed forest in Western Uganda. J Parasitol. 2007;93:439–440. doi: 10.1645/GE-970R1.1. [DOI] [PubMed] [Google Scholar]
- Shendure J, Ji HL. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- Smith P, Siddharth J, Pearson R, et al. Host genetics and environmental factors regulate ecological succession of the mouse colon tissue-associated microbiota. PLoS ONE. 2012;7:e30273. doi: 10.1371/journal.pone.0030273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol. 2011;9:279–290. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- Szekely BA, Singh J, Marsh TL, et al. Fecal bacterial diversity of human-habituated wild chimpanzees (Pantroglodytes schweinfurthii) at Mahale Mountains National Park, Western Tanzania. Am J Primatol. 2010;72:566–574. doi: 10.1002/ajp.20809. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Hamady M, et al. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uenishi G, Fujita S, Ohashi G, et al. Molecular analyses of the intestinal microbiota of chimpanzees in the wild and in captivity. Am J Primatol. 2007;69:367–376. doi: 10.1002/ajp.20351. [DOI] [PubMed] [Google Scholar]
- Ursell LK, Metcalf JL, Parfrey LW, Knight R. Defining the human microbiome. Nutr Rev. 2012;70:S38–S44. doi: 10.1111/j.1753-4887.2012.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Ley R. The human gut microbiome: ecology and recent evolutionary changes. In: Gottesman S, Harwood CS, editors. Annual review of microbiology. Annual Reviews; Palo Alto: 2011. 65. pp. p 411–429. [DOI] [PubMed] [Google Scholar]
- Weins JA. Habitat fragmentation: island v landscape perspectives on bird conservation. IBIS. 1995;137:S97–S104. [Google Scholar]
- Xu B, Huang ZX, Wang XY, et al. Phylogenetic analysis of the fecal flora of the wild pygmy loris. Am J Primatol. 2010;72:699–706. doi: 10.1002/ajp.20826. [DOI] [PubMed] [Google Scholar]
- Yeoman CJ, Chia N, Yildirim S, et al. Towards an evolutionary model of animal-associated microbiomes. Entropy. 2011;13:570–594. [Google Scholar]
- Yildirim S, Yeoman CJ, Sipos M, et al. Characterization of the fecal microbiome from non-human wild primates reveals species specific microbial communities. PLoS ONE. 2010;5:e13963. doi: 10.1371/journal.pone.0013963. [DOI] [PMC free article] [PubMed] [Google Scholar]