Abstract
A microchip electrophoresis-mass spectrometric (MCE-MS) method was developed for fast chiral analysis. The proposed MCE-MS platform deployed a glass /PDMS hybrid microchip with an easy-to-fabricate monolithic nanoelectrospray emitter. Enantiomeric MCE separation was achieved by means of the partial filling technique. A novel chip design with an arm channel connecting to the middle of the MCE separation channel for delivering the chiral selector was tested and proven valid. Enantiomeric separation of 3.4-dihydroxyphenylalanine (DOPA), glutamic acid (Glu), and serine (Ser), the selected test compounds, were achieved within 130 s with resolution values (Rs) of 2.4, 1.1, and 1.0, respectively. The proposed chiral MCE-MS assay was sensitive and had detection limits of 43 nM for L-DOPA and 47 nM for D-DOPA. The analytical platform was well suited for studies of stereochemical preference in living cells because it integrated cell culture, sample injection, chiral separation, and MS detection into a single platform. Metabolism of DOPA in human SH-SY5Y neuronal cells was studied as a model system. On-chip incubation of SH-SY5Y cells with racemic DOPA was carried out, and the incubation solution was injected and in-line assayed at time intervals. It was found that L-DOPA concentration decreased gradually as incubation time increased while the concentration of coexisting D-DOPA remained constant. The results firmly indicated that SH-SY5Y cells metabolized L-DOPA effectively while left D-DOPA intact.
Keywords: Microchip electrophoresis, nano-electrospray ionization-mass spectrometry, chiral separation, partial filling technique, DOPA, serine
1. Introduction
Many important molecules in nature are chiral. These include proteins and their constituent amino acids which control most processes within biological systems. Chirality is, therefore, a property inherent to all biological systems. A large number of biochemical processes exhibit stereospecificity and involve chiral endogenous compounds and metabolites [1-4]. Recent studies have shown that invariant left-right (LR) patterning or chirality is critical for embryonic development. The loss or reversal of LR asymmetry is often associated with malformations and disease [5]. In many cases, drug metabolism with stereochemical or prochiral selectivity may also contribute to the toxicity or adverse effects of the drug therapy. One such example is 3.4-dihydroxyphenylalanine (DOPA). L-DOPA is a popular therapeutic drug to treat Parkinson's disease [6-8]. However, it's antipode, D-DOPA, is not only inactive, but also toxic [9-10]. Therefore, study of chirality in biological systems receives an increasing research attention, which has promoted method development for chiral analysis.
Analytical methods based on various instrumental techniques, including biosensors, high-performance liquid chromatography (HPLC), and capillary electrophoresis have been developed for chiral analysis [12-14]. HPLC enantiomeric separations with mass spectrometric detection (HPLC-MS) are the methods of choice for a wide range of biological applications [15-17]. They offer highly repeatable chiral separation and high sensitivity, detection versatility, and chemical specificity of MS detection. Chiral separations based on capillary electrophoresis (CE) are normally fast [18, 19]. Chiral CE coupled with mass spectrometry (CE-MS) has been reported for analysis of enantiomers [20-22]. Microchip electrophoresis (MCE), which can be regarded as a miniaturized version of classical CE performed on microchips, has been proven to be a speedy and highly efficient separation technique with many attractive microfluidic features such as precise control of flows, automation, and volumetric reduction of samples, reagents and waste. It has been applied in various biochemical and chemical applications since introduced by Manz et al [23-29]. Microchip electrophoresis with mass spectrometric detection (MCE-MS) has been intensively studied due to its great potentials in bioassays. Recent reviews are available [30, 31]. Although chiral MCE separations have been developed [32], few analytical methods based on chiral MCE with MS detection have not been reported so far [33].
The aim of this study was to develop a chiral MCE-MS method for fast separation and identification of enantiomers of biomedical interest. A new microchip design with an arm channel connecting to the middle of the MCE separation channel for delivering chiral selector was tested for performing partial filling chiral electrophoretic separations [34, 35]. To avoid any potential contamination from non-volatile chiral selector, negatively charged sulfated β-cyclodextrin (sulfated β-CD) was tested as the chiral selector because it migrated away from the MS detector [36, 37]. The proposed chiral MCE-MS analytical platform was evaluated by separating DOPA, glutamic acid (Glu), and serine (Ser) enantiomers as model analytes. It was then utilized to analyze on-chip incubation solutions of racemic DOPA with human SH-SY5Y neuronal cells to explore the stereochemical aspects of DOPA metabolism in the cells.
2. Experimental
2.1. Materials
Poly(dimethyl siloxane) (PDMS) prepolymer and the curing agent were purchased from Dow Corning (Sylgard 184 Kit, Midland, MI). Fused silica capillaries (254 μm ID, 360 μm OD) were obtained from Polymicro Technologies(Tucson, AZ). Glass slides were obtained from Silicon Valley Microelectronics (Santa Clara, CA). Hexamethyldisilazane (HMDS) was from Ultra Pure Solutions (Castroville, CA). Enantiomers of DOPA, Glu, and Ser were purchased from Sigma–Aldrich Chemical (St. Louis, MO). Milli-Q water was used throughout the work. All solutions were filtered through a nylon 0.22 μm syringe filter before use.
2.2. Microchip fabrication
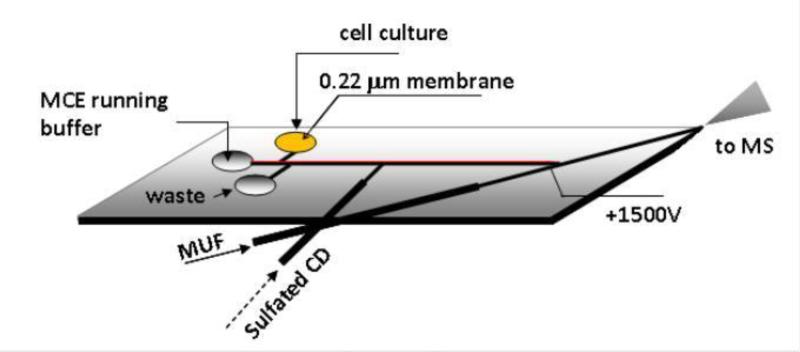
The chip design is shown in Figure 1. The microchip was composed of a glass substrate bearing the channel features and a PDMS cover. A corner of the microchip where the PDMS cover and the glass substrate were tapered into thin layers (< 200 μm in thickness combined) served as the nanoESI emitter. The procedure we described previously [38] was used with modifications to create the glass substrate. Briefly, the design on a photomask with microchannels was transferred onto the glass substrate by means of UV exposure. A corner of the glass substrate was then beveled by polishing it on a sanding paper before channels (60 μm wide and 20 μm deep) were etched into the substrate in a well-stirred bath containing an etching solution of HF:HNO3:H2O (10:20:70). The multilayer soft lithography technique [39-41] was used to fabricate the PDMS cover. A PDMS monopolymer solution prepared by mixing the PDMS prepolymer and curing agent at a 10:1 ratio was applied onto a silicon wafer that was coated with HMDS and spun at 2000rpm for 50s to obtain ~100μm thick PDMS film. A platinum electrode was embedded at its location as shown in Fig. 1A to make an electric contact. After 1 hour curing at 50 °C, a cofferdam was placed on top of the first layer of the PDMS cover, and filled with PDMS pre-polymer mixture, but leaving a ~5mm long section on the cover's edge uncovered. After 3 hour curing, the PDMS sheet was removed from the silicon wafer to yield a PDMS cover (~2mm in thickness) with a ~100μm thick section at the corner to serve as the upper layer of the nanoESI emitter. Access holes of 3-mm in diameter were drilled on the PDMS cover at channel terminals, forming the reservoirs. The microchip was made by bonding the glass substrate and the PDMS cover together through heating by means of an air plasma cleaner (10.5 W and 500mTorr, Harrick Plasma, Ithaca, NY). A piece of 0.22 μm membrane was placed at the bottom of the sample reservoir and fixed with PDMS prepolymer solution.
Fig 1.
Microchip design used in the proposed MCE-MS platform (channels were 60 μm wide × 20 μm deep).
2.3. MCE-nanoESI-MS system
The system consisted of an ion trap mass spectrometer (LCQ Deca, ThermoFinnigan, San Jose, CA), a microchip prepared above, a multichannel high voltage power supply, and two syringe pumps. One syringe pump was used for delivering the sulfated β-CD-containing buffer solution, and the other for MUF delivery. The microchip was fixed on a XYZ-translational stage and so positioned that the nanoESI emitter tip was about ~1.0 mm away from the MS orifice. Xcalibur software (ThermoFinnigan) was used to control the mass spectrometer and process MS data. House-written software was used for controlling the potentials applied to the microchip for MCE and nanoESI operations. MS detection conditions were optimized in positive mode and selected as follows: ion source voltage, 0V; relative collision energy of 25% was used for MS/MS experiments with an isolation width of 1.0 u and the activation time was set at 30 ms.
2.4. Chiral MCE-MS analysis
The two syringes were filled with the respective solutions and then connected to the respective capillaries assembled into the microchip (as shown in Fig. 1A). MCE running buffer was added to the buffer reservoir on the chip. The MCE channel was filled with the buffer by applying vacuum at the tip of the nano ESI emitter. MUF pump was tuned on to generate a MUF. The sulfated β-CD solution (about 35 nL) was infused to partially fill the MCE separation channel by turning on and off the syringe pump set at 250 nL/min. To inject a sample, potentials of 450 V, 500 V, 0 V, and 1000 V were applied at sample reservoir, running buffer reservoir, waste reservoir, and Pt wire (as labeled in Fig. 1) for 15 s, respectively. After sample injection, the potentials were changed to 3850V, float, float, and 1500V, respectively to start the MCE-MS assay. At the same time, MS data acquisition was started. When completed, the potentials were removed. MUF delivering syringe pump remained on for some time to clean the nanoESI emitter. MCE running buffer was a mixture of 15 mM ammonium acetate /acetic acid buffer (pH 5.5) and methanol (1:1). Make-up fluid was the MCE running buffer solution flowing at 100 nL /min. Sulfated β-CD solution was prepared by dissolving sulfated β-CD in the MCE running buffer at a concentration of 15 mM.
2.5. On-chip incubation of racemic DOPA with SH-SY5Y cells
The human neuronal cell line (SH-SY5Y) was purchased from The American Type Culture Collection (ATCC). Cells were cultured in Dulbecco's modified Eagle's medium supplemented with 100 units /mL penicillin, 10 mg /mL streptomycin, and 10% fetal bovine serum. Cells were routinely sub-cultured every 3 days. All experiments involving cell culture were conducted when cells were nearly confluent (normally 2-3 days after sub-culture). In the experiment day cells were harvested and suspended in PBS at a density of 2 × 10−6 cells /mL. A portion of the cell suspension (25 μL) was transferred to the sample reservoir and racemic DOPA was added at a final concentration of 100 μM. The sample reservoir was covered with a piece of paraffin membrane. The incubation solution was injected and in-line analyzed as described above at time intervals.
3. Results and discussion
3.1. Microchip design
To achieve effective MCE-MS coupling, various chip designs with distinct nanoESI emitters have been proposed and evaluated. These included a tapered capillary tip [42, 43], a liquid junction and a fused silica transfer capillary [44] a milled and pulled tiny cone enclosing centrically the exit of the MCE separation channe [45, 46], a microfabricated triangular tip in a SU-8 chip [47, 48], and a corner of a rectangular glass microchip made of thin glass substrates (300 μm thick combined) [49, 50]. All of these designs sought to keep the emitter's spraying area small in order to obtain a stable electrospray from a liquid flow at the nL /min level. In this work, a facile protocol to fabricate a nanoESI emitter in glass /PDMS hybrid microchips was proposed. The nanoESI emitter was constructed at a corner of the rectangular microchip. Rectangular glass substrates of 1 mm in thickness bearing channel features were used for their easy availability and good mechanical strength. One corner of the substrate was beveled by polishing it on a 400 grit sandpaper sheet. The PDMS cover was fabricated by using the multilayer soft lithography technique. The first layer was about 100 μm thick and the second casting made the cover ~2 mm thick except the corner for constructing the nanoESI emitter. When bonding the PDMS cover and the glass substrate together a monolithic nanoESI emitter was formed. Performance of this nanoESI emitter was evaluated by spraying the MUF flowing at 100nL /min. In a test of 2-hour continuous spraying, the TIC baseline was found stable with an RSD in MS background signal of 4.1%. The reproducibility of ionization efficiency was assessed by recording spray current values every 10 min, and the RSD was calculated to be 2.9% (n = 10). These results indicated that the nanoESI emitter prepared was effective and robust.
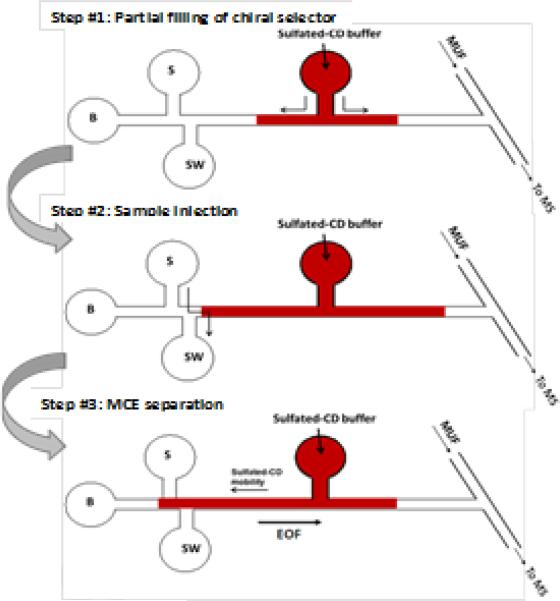
The partial filling technique in CE-MS analysis has been proposed previously to avoid the potential contamination of MS detector by non-volatile additives in the running buffer [34, 35]. Another advantage of this technique when used in chiral separation is that the chiral separation medium is freshened for each run, which improves the separation repeatability. The partial filling technique can be easily adopted in MCE because flows in a microfluidic chip can be conveniently and precisely controlled. In this work, a microchip design with an arm channel connecting to the middle of the MCE separation channel for delivering chiral selector (i.e. the sulfated β-CD solution) was tested. It was unknown how severely this opening in the middle of the separation channel would compromise the MCE separation efficiency. Two approaches were tested for the delivery: electrokinetically by applying a potential and hydrodynamically by using a syringe pump. The results obtained indicated that the approach with the use of a syringe pump was superior, which was reflected mainly in two ways: 1) repeatability of the migration times and 2) the width of the MCE-MS TIC peaks. In the hydrodynamical approach the volume of the liquid plugs delivered could be precisely controlled. In addition, there was no bias in the chemical composition of the plugs delivered each time. More importantly, the disturbance to MCE effluent by the opening to the arm channel in the middle of the separation channel was found insignificant likely due to the arm channel was connected to a gas tight syringe at the other end, which allowed sharp MCE peaks be obtained. All these factors combined contributed to achieving high efficient and reproducible chiral separations.
3.2. Chiral MCE-MS of neuroactive test compounds
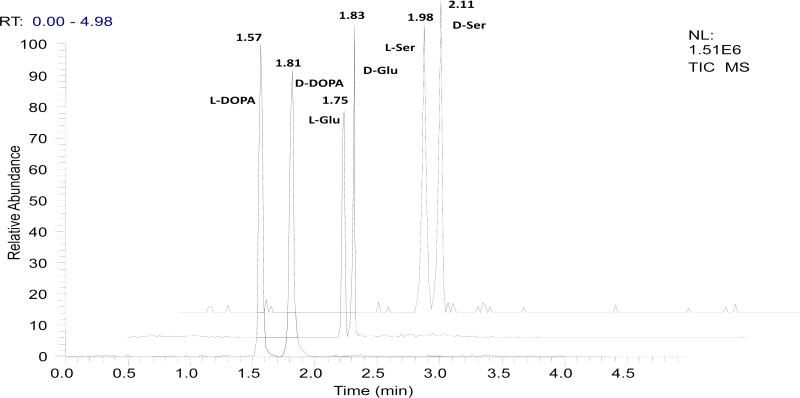
Three neuroactive compounds, i.e. DOPA, Glu, and Ser were included in the study. L-DOPA is a well known medicine used to treat Parkinson's symptoms [6-8]. L-Glu is a major excitatory amino acid in vaterbrate nervous system [51]. D-Ser has been recently identified as a gliotransmitter and is believed to be an endogenous co-agonist for synaptic N-methyl-D-aspartate receptors (NMDARs) [52]. To achieve chiral MCE-MS separation, a chiral selector must be introduced into the separation medium. In this work, we found that sulfated β-CD was an effective chiral selector for all the three pairs of enantiomers tested. For each run about 35 nL of a sulfated β-CD solution was infused into the separation channel that had a capacity of ~45 nL. It's worth noting that in the present MCE process the chiral solutes moved in a direction against that of the chiral selector, which enhanced the chiral resolution. This is illustrated in Fig. 2. Effects of sulfated β-CD concentration in the running buffer were investigated in the range from 2.0 to 50.0 mM. Resolution values for the three test pairs of enantiomers were calculated and listed in Table 1. Optimal separations in terms of enantiomeric resolution and migration time were obtained at 15.0 mM. A mixture of ammonium acetate /acetic acid buffer (pH 5.5) and methanol (1:1) was used as the MCE running buffer. Buffers containing ammonium acetate at concentrations ranging from 5 to 50 mM were tested. Best resolution of the enantiomers tested was achieved with a concentration greater than 10 mM. The electrophoretic current increased quickly with the increase in ammonium acetate concentration. Therefore, a running buffer containing 15 mM ammonium acetate was selected for further studies. Other electrophoretic parameters, such as pH of the running buffer (data are shown in Table 1), MCE separation voltage, injection method were also investigated. The selected conditions were as follows: running buffer, 15 mM NH4Ac /HAc buffer (pH 5.5) /methanol (1:1); chiral selector, the MCE running buffer containing 15 mM sulfated β-CD; separation voltage, 3850 V; nanoESI voltage, 1500 V. Under the selected conditions all the enantiomers tested were resolved within 130 seconds as shown in Fig. 3. The resolution values (calculated by Rs = 2 (tR2 – tR1) /(w2 +w1)) were 2.4 for L-/D-DOPA, 1.1 for L-/D-Glu, and 1.0 for L-/D-Ser. It should be pointed out that most chiral CE-based separations of amino acids previously reported involved pre-column derivatization to convert native amino acids into more separatable and detectable derivatives [18, 19, 53 - 56]. Enantiomeric separation of native amino acids, particularly those small ones such as Glu and Ser has been difficult to achieve. Actually, to our knowledge there has been no report on chiral electrophoretic separations of native (i.e. unlabeled) Ser or Glu. Taking the combined advantages of specific MS detection and highly efficient chiral MCE separations (with peak width of ~4 seconds as can be seen in Fig. 3) the present chiral MCE-MS method can be run in a successive manner to quantify enantiomers in samples such as cell cultures, offering a temporal resolution of <25 s, which will have a great potential in kinetic studies of biological systems.
Fig 2.
A schematic illustration of the proposed partial filling chiral MCE-MS analysis with sulfated β-CD as chiral selector. The approach can be easily executed in a microfluidic chip. Delivery of chiral selector is done preferably by using a gas-tight syringe (hydrodynamically) over by applying an electrical potential (electrokinetically).
Table 1.
Summary of the results from investigating chiral MCE conditions
| Resolution values, Rs* |
|||
|---|---|---|---|
| DOPA | Glu | Ser | |
| Sulfated CD Conc. (mM) | |||
| 2.0 | 0.82 | 0.11 | 0 |
| 5.0 | 1.5 | 0.58 | 0.25 |
| 10 | 2.3 | 1.1 | 0.88 |
| 15 | 2.4 | 1.1 | 1.0 |
| 25 | 2.4 | 1.1 | 1.0 |
| 50 | 2.3 | 1.3 | 1.0 |
| Running Buffer pH | |||
| 3.5 | 1.9 | 0.72 | 0.16 |
| 4.5 | 2.0 | 0.94 | 0.84 |
| 5.5 | 2.4 | 1.1 | 1.0 |
| 6.5 | 2.2 | 0.77 | 0.94 |
Rs = 2 (tR2 – tR1) /(w2 +w1)), means of three runs.
Fig 3.
TIC electropherograms from the proposed chiral MCE-MS separation of neuroactive compounds, i.e. DOPA, glutamic acid (Glu), and serine (Ser). [test compound] = 100 μM (each enantiomer). Chiral MCE conditions: MCE separation channel, 4 cm long × 60 μm wide × 20 μm deep; electrokinetic sample injection, 15 s at 600V; 35nL chiral selector solution infused by a syringe pump; separation voltage, 3850V; electrospray voltage,1500V; MCE running buffer, 15mM ammonium acetate/ acetic acid buffer (pH 5.5) /methanol (1:1); chiral selector solution, the MCE running buffer containing 15 mM sulfated β-CD; MUF, the MCE running buffer at a flow rate of 100 nL /min.
Analytical figures of merit were studied using DOPA as the model analyte for the proposed chiral MCE-MS method. Quantitation was carried out by using m/z 198 → 152 SRM MS/MS detection mode. Standard curves were prepared by analyzing a series of standard enantiomeric DOPA solutions. The calibration curves based on peak height versus analyte concentration showed a good linearity with correlation coefficient > 0.992 for both enantiomers. The linear range was 0.12 - 25 μM. Detection limits (S/N =3) were estimated to be 0.043 μM and 0.047 μM for L-DOPA and D-DOPA, respectively. Compared with the CE-MS analytical results [57], the present MCE-nanoESI-MS method was 10-times more sensitive (LODs: 0.04 μM versus 0.50 μM). The increase in assay sensitivity can be attributed to at least two factors: 1) less dilution of the MCE effluent by MUF liquid, and 2) use of nanoESI in the present system. Assay reproducibility was determined by repeatedly analyzing a solution of L-/D-DOPA (1.0 μM each enantiomer) for six times. Relative standard deviations (RSD) were 3.51% for L-DOPA and 4.13% for D-DOPA.
3.3. Enantioselective metabolism of DOPA in SH-SY5Y cells
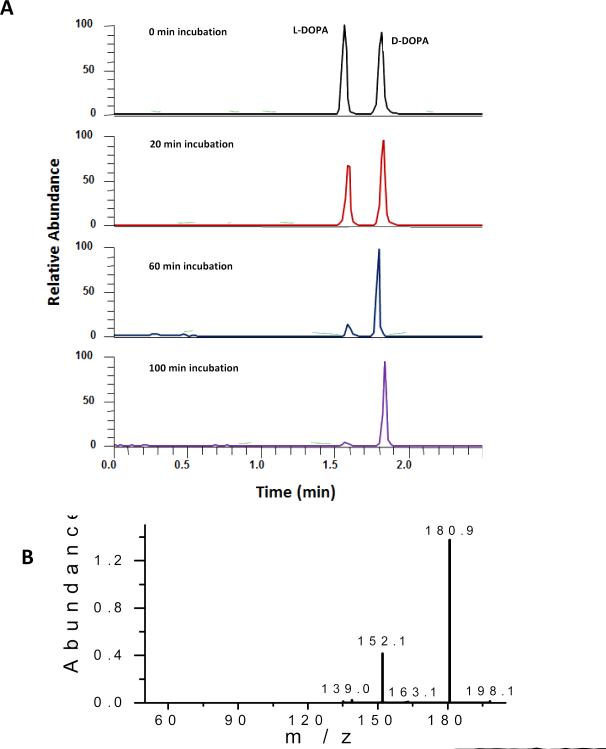
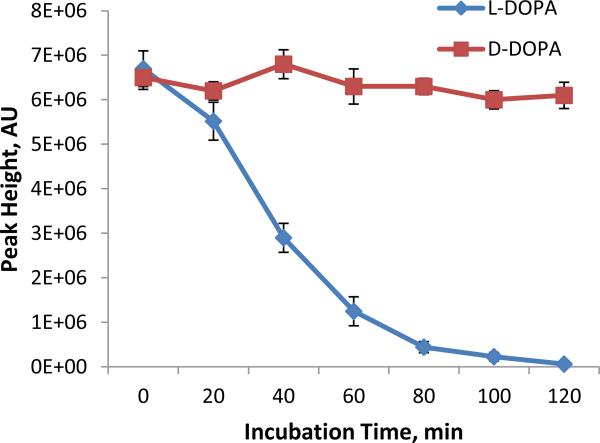
It has been known for long that L-DOPA is effective for treating Parkinson's symptoms because it's metabolized by decarboxylase, producing dopamine in the central nervous system. However, little in vitro study has been done on DOPA metabolism using neuronal cells. In this work, the proposed chiral MCE-MS platform was used to study DOPA metabolism in SH-SY5Y cells, a widely used neuronal model. Racemic DOPA (50 μM) was on-chip incubated with SH-SY5Y cells in the sample reservoir. Cell density was estimated to be 2 × 106 cells /mL. The incubation solution was electrokinetically injected into the MCE channel for enantiomeric quantification of L- and D-DOPA every 20 min after incubation started. Figure 4A shows the electropherograms from the analyses. L-DOPA and D-DOPA enantiomers were well separated. Peak identities were confirmed by the MS2 spectra showing ion transitions m/z 198 → 181, 152, and 139 (Fig. 4B). As can be seen the peak height for L-DOPA decreased gradually with the increase in incubation time while that for D-DOPA remained unchanged. The analytical data are summarized in Fig. 5. These results firmly indicated that SH-SY5Y cells metabolized L-DOPA effectively, but left D-DOPA intact. Previous studies [58, 59] have shown that L- DOPA, unlike D-DOPA, is the precursor of dopamine because only L-DOPA can be converted into dopamine by DOPA decarboxylase. Data obtained in this work were consistent with the previous publications, validating the proposed chiral MCE-MS analytical method.
Fig 4.
Electropherograms obtained from studying DOPA metabolism in SH-SY5Y neuronal cells: (A) TICs of m/z 198 from incubation solution injected at different times, and (B) MS/MS spectrum of the peaks, confirming DOPA identity. MCE-MS conditions were as in Fig. 3. Peak height of L-DOPA decreases gradually as incubation time increases while that of co-existing D-DOPA remains unchanged.
Fig 5.
Metabolic trend curves of L-DOPA and D-DOPA (at 50 μM each) in incubation with SH-SY5Y neuronal cells as measured by the proposed chiral MCE-MS method (mean ± SD, n=3).
4. Conclusions
The first chiral MCE-MS analytical method was developed in this work. An innovative microchip design with an arm channel connecting to the middle of the MCE separation channel for delivering chiral selector was tested for performing partial filling chiral electrophoretic separation and proven valid. Enantiomers of three neuroactive compounds tested, i.e. DOPA, Glu, and Ser were separated within 130 s with resolution values of 2.4, 1.1 and 1.0, respectively. To our knowledge, no chiral electrophoretic separations of native (i.e. un-labeled) Glu or Ser have been reported previously. The proposed MCE-MS analytical platform was shown to be useful for studying stereochemical preference in living cells. Operations of on-chip cell culture, injection of the culture solution, chiral MCE separation, and specific MS detection were integrated into the single platform and programmed under control by a computer. Study on DOPA metabolism in SH-SY5Y cells by using the platform showed that the cells metabolized only L-DOPA and left con-existing D-DOPA intact.
Highlights.
A chiral microchip electrophoresis-mass spectrometric method employing the partial filling technique
Fast separation and identification of enantiomers by MCE-MS
Chiral electrophoretic separation of underivatized amino acids
Stereochemical selectivity in drug metabolism by neuronal cells
Acknowledgement
Financial support from US NIH (GM089557) and partially from a NSF grant (CHE 0840450) to YML is gratefully acknowledged. Assistance from Dr. Sharonda Harris for cell culture is also appreciated.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Taniguchi K, Maeda R, Ando T, Okumura T, Nakazawa N, Hatori R, Nakamura M, Hozumi S, Fujiwara H, Matsuno K. Science. 2011;333:339. doi: 10.1126/science.1200940. [DOI] [PubMed] [Google Scholar]
- 2.Wan LQ, Ronaldson K, Park M, Taylor G, Zhang Y, Gimble JM, Vunjak-Novakovic G. Proc Natl Acad Sci U S A. 2011;108:12295. doi: 10.1073/pnas.1103834108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zask A, Murphy J, Ellestad GA. Chirality. 2013;25:265. doi: 10.1002/chir.22145. [DOI] [PubMed] [Google Scholar]
- 4.Caglioti L, Micskei K, Palyi G. Chirality. 2011;23:65. doi: 10.1002/chir.20796. [DOI] [PubMed] [Google Scholar]
- 5.Wan LQ, Ronaldson K, Guirguis M, Vunjak-Novakovic G. Stem Cell Res Ther. 2013;4:24. doi: 10.1186/scrt172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huot P, Johnston TH, Koprich JB, Fox SH, Brotchie JM. Neuropharmacology. 2012;63:829. doi: 10.1016/j.neuropharm.2012.06.012. [DOI] [PubMed] [Google Scholar]
- 7.Alachkar A, Brotchie JM, Jones OT. Neurosci Res. 2010;67:245. doi: 10.1016/j.neures.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 8.Alam MR, Yoshizawa F, Sugahara K. Neurosci Lett. 2011;495:126. doi: 10.1016/j.neulet.2011.03.053. [DOI] [PubMed] [Google Scholar]
- 9.Andreou D, Saetre P, Werge T, Andreassen OA, Agartz I, Sedvall GC, Hall H, Terenius L, Jonsson EG. Eur Arch Psychiatry Clin Neurosci. 2012;262:549. doi: 10.1007/s00406-012-0313-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beaulieu JM, Gainetdinov RR. Pharmacol Rev. 2011;63:182. doi: 10.1124/pr.110.002642. [DOI] [PubMed] [Google Scholar]
- 11.Leung D, Kang SO, Anslyn EV. Chem Soc Rev. 2012;41:448. doi: 10.1039/c1cs15135e. [DOI] [PubMed] [Google Scholar]
- 12.Lämmerhofer M. J. Chromatogr. A. 2010;1217:814. doi: 10.1016/j.chroma.2009.10.022. [DOI] [PubMed] [Google Scholar]
- 13.Xiao Y, Ng SC, Thatt T, Tan Y, Wang Y. J. Chromatogr. A. 2012;1269:52. doi: 10.1016/j.chroma.2012.08.049. [DOI] [PubMed] [Google Scholar]
- 14.Wolf C, Bentley KW. Chem Soc Rev. 2013;42:5408. doi: 10.1039/c3cs35498a. [DOI] [PubMed] [Google Scholar]
- 15.Pérez-Fernández V, García MÁ, Marina ML. J. Chromatogr. A. 2011;1218:6561. doi: 10.1016/j.chroma.2011.07.084. [DOI] [PubMed] [Google Scholar]
- 16.Barclay VK, Tyrefors NL, Johansson IM, Pettersson CE. J Chromatogr A. 2012;1269:208. doi: 10.1016/j.chroma.2012.09.090. [DOI] [PubMed] [Google Scholar]
- 17.Moon K, Lim C, Kim S, Oh DC. J Chromatogr A. 2013;1272:141. doi: 10.1016/j.chroma.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 18.Gübitz G, Schmid MG. J. Chromatogr. A. 2008;1204:140. doi: 10.1016/j.chroma.2008.07.071. [DOI] [PubMed] [Google Scholar]
- 19.Amin NC, Blanchin MD, Aké M, Fabre H. J. Chromatogr. A. 2012;1264:1. doi: 10.1016/j.chroma.2012.09.057. [DOI] [PubMed] [Google Scholar]
- 20.Simó C, García-Cañas V, Cifuentes A. Electrophoresis. 2010;31:1442. doi: 10.1002/elps.200900673. [DOI] [PubMed] [Google Scholar]
- 21.Bonvin G, Schappler J, Rudaz S. J. Chromatogr. A. 2012;1267:17. doi: 10.1016/j.chroma.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 22.Dominguez-Vega E, Crego AL, Marina ML. Methods Mol Biol. 2013;970:429. doi: 10.1007/978-1-62703-263-6_27. [DOI] [PubMed] [Google Scholar]
- 23.Manz A, Harrison DJ, Verpoorte EM, Fettinger JC, Paulus A, Ludi H, Widmer HM. J. Chromatogr. 1992;593:253. [Google Scholar]
- 24.El-Ali J, Sorger PK, Jensen KF. Nature. 2006;442:403. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- 25.Sims CE, Allbritton NL. Lab Chip. 2007;7:423. doi: 10.1039/b615235j. [DOI] [PubMed] [Google Scholar]
- 26.Hulvey MK, Frankenfeld CN, Lunte SM. Anal Chem. 2010;82:1608. doi: 10.1021/ac902821v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagl S, Schulze P, Ludwig M, Belder D. Electrophoresis. 2009;30:2765. doi: 10.1002/elps.200900153. [DOI] [PubMed] [Google Scholar]
- 28.Chambers AG, Mellors JS, Henley WH, Ramsey JM. Anal Chem. 2011;83:842. doi: 10.1021/ac102437z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Breadmore MC. J Chromatogr A. 2012;1221:42. doi: 10.1016/j.chroma.2011.09.062. [DOI] [PubMed] [Google Scholar]
- 30.Kitagawa F, Otsuka K. J Pharm Biomed Anal. 2011;55:668. doi: 10.1016/j.jpba.2010.11.013. [DOI] [PubMed] [Google Scholar]
- 31.Ohla S, Belder D. Curr Opin Chem Biol. 2012;16:453. doi: 10.1016/j.cbpa.2012.05.180. [DOI] [PubMed] [Google Scholar]
- 32.Nagl S, Schulze P, Ohla S, Beyreiss R, Gitlin L, Belder D. Anal Chem. 2011;83:3232. doi: 10.1021/ac200150w. [DOI] [PubMed] [Google Scholar]
- 33.Fritzsche S, Ohla S, Glaser P, Giera DS, Sickert M, Schneider C, Belder D. Angew Chem Int Ed Engl. 2011;50:9467. doi: 10.1002/anie.201102331. [DOI] [PubMed] [Google Scholar]
- 34.Nelson WM, Lee CS. Anal Chem. 1996;68:3265. doi: 10.1021/ac951137o. [DOI] [PubMed] [Google Scholar]
- 35.Tanaka Y, Kishimoto Y, Terabe S. J. Chromatogr. A. 1998;802:83. [Google Scholar]
- 36.Liang RP RP, Liu CM, Meng XY, Wang JW, Qiu JD. J. Chromatogr A. 2012;1266:95. doi: 10.1016/j.chroma.2012.09.101. [DOI] [PubMed] [Google Scholar]
- 37.Jac P, Scriba GK. J Sep Sci. 2013;36:52. doi: 10.1002/jssc.201200836. [DOI] [PubMed] [Google Scholar]
- 38.Zhao S, Li X, Liu YM. Anal Chem. 2009;81:3873. doi: 10.1021/ac900391u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cellar NA, Kennedy RT. Lab Chip. 2006;6:1205. doi: 10.1039/b603561b. [DOI] [PubMed] [Google Scholar]
- 40.Johann RM, Baiotto C, Renaud P. Biomed Microdevices. 2007;9:475. doi: 10.1007/s10544-007-9054-6. [DOI] [PubMed] [Google Scholar]
- 41.Wang W, Zhao S, Pan T. Lab Chip. 2009;9:1133. doi: 10.1039/b816287e. [DOI] [PubMed] [Google Scholar]
- 42.Tachibana Y, Otsuka K, Terabe S, Arai A, Suzuki K, Nakamura S. J Chromatogr A. 2003;1011:181. doi: 10.1016/s0021-9673(03)01181-6. [DOI] [PubMed] [Google Scholar]
- 43.Zhang B, Liu H, Karger BL, Foret F. Anal Chem. 1999;71:3258. doi: 10.1021/ac990090u. [DOI] [PubMed] [Google Scholar]
- 44.Tachibana Y, Otsuka K, Terabe S, Arai A, Suzuki K, Nakamura S. J Chromatogr A. 2004;1025:287. doi: 10.1016/j.chroma.2003.10.103. [DOI] [PubMed] [Google Scholar]
- 45.Hoffmann P, Hausig U, Schulze P, Belder D. Angew Chem Int Ed Engl. 2007;46:4913. doi: 10.1002/anie.200605152. [DOI] [PubMed] [Google Scholar]
- 46.Hoffmann P, Eschner M, Fritzsche S, Belder D. Anal Chem. 2009;81:7256. doi: 10.1021/ac9015038. [DOI] [PubMed] [Google Scholar]
- 47.Sikanen T, Tuomikoski S, Ketola RA, Kostiainen R, Franssila S, Kotiaho T. Anal Chem. 2007;79:9135. doi: 10.1021/ac071531+. [DOI] [PubMed] [Google Scholar]
- 48.Nordman N, Sikanen T, Moilanen ME, Aura S, Kotiaho T, Franssila S, Kostiainen R. J Chromatogr A. 2011;1218:739. doi: 10.1016/j.chroma.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 49.Mellors JS, Gorbounov V, Ramsey RS, Ramsey JM. Anal Chem. 2008;80:6881. doi: 10.1021/ac800428w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mellors JS, Jorabchi K, Smith LM, Ramsey JM. Anal Chem. 2010;82:967. doi: 10.1021/ac902218y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meldrum BS. J Nutr. 2000;130:1007S. doi: 10.1093/jn/130.4.1007S. [DOI] [PubMed] [Google Scholar]
- 52.Mothet JP, Parent AT, Wolosker H, Brady RO, Jr., Linden DJ, Ferris CD, Rogawski MA, Snyder SH. Proc Natl Acad Sci U S A. 2000;97:4926. doi: 10.1073/pnas.97.9.4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kirschner DL, Jaramillo M, Green TK. Anal Chem. 2007;79:736. doi: 10.1021/ac061725+. [DOI] [PubMed] [Google Scholar]
- 54.Kostal V, Katzenmeyer J, Arriaga EA. Anal Chem. 2008;80:4533. doi: 10.1021/ac8007384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun X, Li D, Lee ML. Anal Chem. 2009;81:6278. doi: 10.1021/ac9001832. [DOI] [PubMed] [Google Scholar]
- 56.Ilisz I, Aranyi A, Péter A. J. Chromatogr. A. 2013;1296:119. doi: 10.1016/j.chroma.2013.03.034. [DOI] [PubMed] [Google Scholar]
- 57.Yuan B, Wu H, Sanders T, McCullum C, Zheng Y, Tchounwou PB, Liu YM. Anal Biochem. 2011;416:191. doi: 10.1016/j.ab.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Biedler JL, Roffler-Tarlov S, Schachner M, Freedman LS. Cancer Res. 1978;38:3751. [PubMed] [Google Scholar]
- 59.Ou XM, Partoens PM, Wang JM, Walker JH, Danks K, Vaughan PF, De Potter WP. Int J Mol Med. 1998;1:105. doi: 10.3892/ijmm.1.1.105. [DOI] [PubMed] [Google Scholar]