Abstract
Microsporidia in histologic sections are most often diagnosed by observing spores in host tissues. Spores are easy to identify if they occur in large aggregates or xenomas when sections are stained with hematoxylin and eosin (H&E). However, individual spores are not frequently detected in host tissues with conventional H&E staining, particularly if spores are scattered within the tissues, areas of inflammation or small spores in nuclei (i.e., Nucleospora salmonis). Hence, a variety of selective stains that enhance visualization of spores are recommended. We discovered that the Luna stain, used to highlight eosinophils, red blood cells and chitin in arthropods and other invertebrates, also stains spores of Pseudoloma neurophilia. We compared this stain to the Gram, Fite’s acid fast, Giemsa, and H&E stains on eight aquatic microsporidian organisms that were readily available in our two laboratories: Loma salmonae, Glugea anomala, Pseudoloma neurophilia, Pleistophora hyphessobryconis, Pleistophora vermiformis, Glugea sp., Steinhausia mytilovum and an unidentified microsporidian from E. sinensis, UK. Based on tinctorial properties and background staining, the Luna stain performed better for detection of 6 of the 8 microsporidia. Gram stain was superior for the two microsporidia from invertebrates, Steinhausia mytilovum and the unidentified microsporidian from E. sinensis.
Keywords: Luna stain, Selective stains - Microsporidia, Pseudoloma neurophilia, Gram stain, Chitin
INTRODUCTION
Microsporidian parasites are a reductionist group of eukaryotic obligate intracellular parasites related to the fungi that infect numerous different invertebrates and vertebrates, and infections are noted frequently in both arthropods and teleost fishes (Keeling and Fast 2002). These parasites affect not only wild or cultured fishes and aquatic invertebrates but laboratory fishes, such as zebrafish, as well (Matthews et al. 2001). Morphological diagnosis of these infections is usually based on visualization of the spore stage. Aside from this approach, in almost all diagnostic cases the detection of microsporidia relies upon the host organism’s infectious burden or obvious xenoma formation (Kotler et al. 1994). Spores are usually detected in wet tissue preparations, histological slides or tissue/fecal smears. One of the problems encountered in routine H&E sections is that spores can be obscured against a similarly staining background, retaining hematoxylin stain or may not stain at all. For this reason, a litany of selective stains have been employed to facilitate and enhance detection of spores in tissue sections or smears, which includes the Brown-Brenn and Brown-Hopps modified Gram, Gram chromotrope, Giemsa, Masson’s trichrome, Periodic-acid Schiff’s, Acridine orange, Gomori’s methenamine silver, Warthin-Starry, Ziehl-Nielsen and Fite’s acid-fast stains (Lamps et al. 1997, Moura et al. 1997, Weber et al. 1999, Garcia 2002, Joseph et al. 2006a, Joseph et al. 2006b). Many of the selective stains used to detect microsporidial organisms often give variable and inconsistent staining (Lamps et al. 1997, Garcia 2002). Fluorescent stains, such as the Calcofluor White and FungiFluor™, are also useful for detecting spores in smears or sections (Luna et al. 1995, Guzman et al. 2001, Garcia 2002, Kent and Bishop-Stewart JK 2003). Major difficulties encountered in the interpretation of fluorescent stains include distinguishing microsporidial spores from background, artifactual staining and determining the precise location of spores in tissue sections. PCR tests are also available for several microsporidia including those infecting fishes (Brown and Kent 2002, Whipps and Kent 2006). These tests have been adapted for in situ hybridization on tissue sections (Sanchez et al. 1999). Each of these selective stains has specific utility for detection of spores, including ease-of-use and interpretation, time and economy, degree of contrast with tissue elements and specificity, but also drawbacks for routine use, for example high background “noise”, variable staining, requirements for a fluorescent microscope or non-confirmatory, time-consuming and expensive reagents.
The Luna stain was originally developed to detect cytoplasmic granules within eosinophils, Negri bodies, erythrocytes and phagocytes (Luna 1968, Tomasi et al. 2008). It also has been used to highlight elastin in tissue sections (Kligman 1981). We discovered that spores of Pseudoloma neurophilia from zebrafish stain positive (brick red) with the Luna stain and have minimal background interference. We then evaluated several other microsporidia with this stain, comparing it with Fite’s acid fast, Giemsa and Gram stains which we have routinely used for the detection of microsporidia in our laboratories.
MATERIALS AND METHODS
Histologic samples representing the 8 different microsporidia were obtained from the case files of the Zebrafish International Resource Center (Eugene, OR), Oregon State University Department of Microbiology (Corvallis, OR), University of California-Davis School of Veterinary Medicine (Davis, CA) and the Centre for Environment, Fisheries and Aquaculture Science (Weymouth Laboratory, UK). The histologic samples were comprised of six different teleost fishes, Mitten crab (Eriocheir sinensis) and a mussel (Mytilus sp.) that had been previously diagnosed with microsporidian infections. 10% neutral buffered formalin and Dietrich’s fixative were used for preservation of fish tissues and sea water Davidson’s fixative for mussel and mitten crab. Five serial sections, cut at 4 µm and deparaffinized, were made from each of the selected tissue blocks. These tissue sections were then stained with H&E and a special stain panel of Luna, Gram, Fite’s acid-fast and Giemsa stains. Luna and Gram stains were performed using Luna’s method for erythrocytes and eosinophil granules (Luna 1968 pages 111–112) and the Accustain™ Gram stain for tissue kit (HT90T, Sigma-Aldrich). Fite’s acid-fast and Giemsa stains were employed using Fite’s method for acid fast organisms (Luna 1968 pages 217–218) and May-Grunwald Giemsa method (Luna 1968 pages 121–122). Slides were evaluated for routine histologic features, presence of microsporidian organisms, detection of microsporidian spores by special stains, fidelity of staining for each of the special stains, amount of background stain and artifacts.
RESULTS
Luna stain
The majority (approximately > 90%) of microsporidial spores in 6 of 8 cases, regardless of genus or species, stained brick red and were easily detected in tissue sections. The spores displayed exceptionally high contrast from the background stain with minimal to no interference from the few positively-staining tissue elements (Table 1, Figs 1 and 2). Spores were particularly evident in cases of diffuse multiorgan infections. With Nucleospora salmonis, a few small intranuclear spores were detected which stained deep brick red in contrast to blue-black stained nuclei (Fig. 1c). Spores of Steinhausia mytilovum from mussel oocytes did not stain, while the unidentified microsporidian from mitten crab enterocytes showed variable staining. With fish, other tissues that also stained red included erythrocytes, bone, lens of the eye, and eosinophilic granular cells of salmon.
Table 1.
Characteristics of microsporidian spores in histological sections with 274 special stains
| Microsporidia | Fixative | Host | Luna | Gram | Fite's Acid-Fast | Giemsa |
|---|---|---|---|---|---|---|
| Glugea anomala | Dietrich’s | Gasterosterus aculeatus | + | +/− | +/− | + |
| Loma salmonae | 10% neutral buffered formalin (10% NBF) | Oncorhynchus kisutch | + | +/− | +/− | + |
| Nucleospora salmonis | Davidson’s | Oncorhynchus tshawytscha | +/− | 0 | 0 | 0 |
| Pseudoloma neurophilia | Dietrich’s/ 10% NBF | Danio rerio | + | +/− | +/− | +/− |
| Pleistophora hyphessobryconis | Dietrich’s | Danio rerio | + | +/− | +/− | +/− |
| Pleistophora vermiformis | 10% NBF | Cottus gobio | + | + | 0 | + |
| Glugea stephani | 10% NBF | Pleuronectes platessa | + | + | 0 | +/− |
| Steinhausia mytilovum | Sea water Davidson’s | Mytilus edulis | 0 | +/− | 0 | 0 |
| Unidentified microsporidian | Sea water Davidson’s | Eriocheir sinensis | +/− | + | 0 | 0 |
+ = most or all spores were stained and distinct from background, +/− = variable, some spores stained, 0 = less than 10% of spores colored with special stain or spores not clearly distinguished from background.
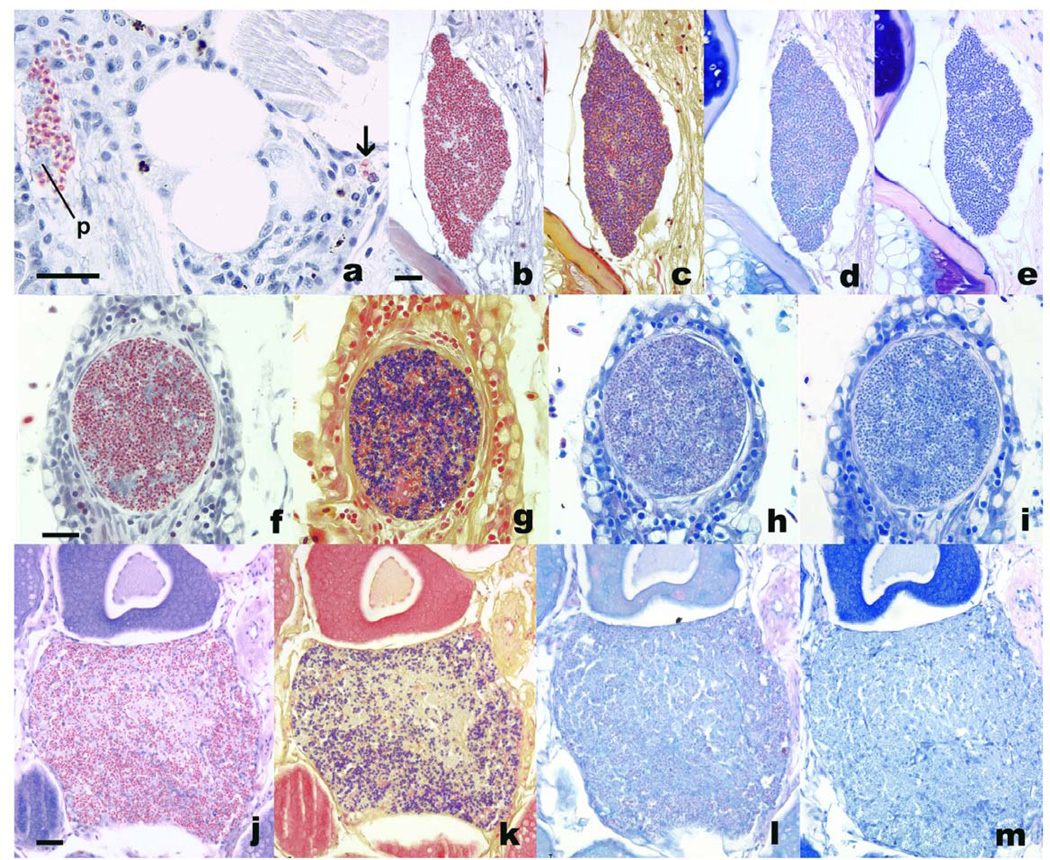
Figure 1.
Microsporidia in histologic stains from various fishes. Bars = 25 µm. a. Pseudoloma neurophilia individual spores (arrow) and aggregate with presporogonic stages (p) associated with chronic inflammation in the nerve and skeletal muscle of Danio rerio. Luna. b – e. P. neurophilia xenoma in spinal cord of zebrafish. b, Luna; c, Gram; d, Fite’s acid fast; e, Giemsa. f-i, Loma salmonae xenoma in Oncorhynchus kistuch gill. f, Luna; g, Gram; h, Fite’s acid fast; i, Giemsa. j-m. Glugea stephani spores, within background of chronic inflammation, in ovarian follicle of Pleuronectes platessa. j, Luna;; k, Gram;; l, Fite’s acid fast;; m, Giemsa.
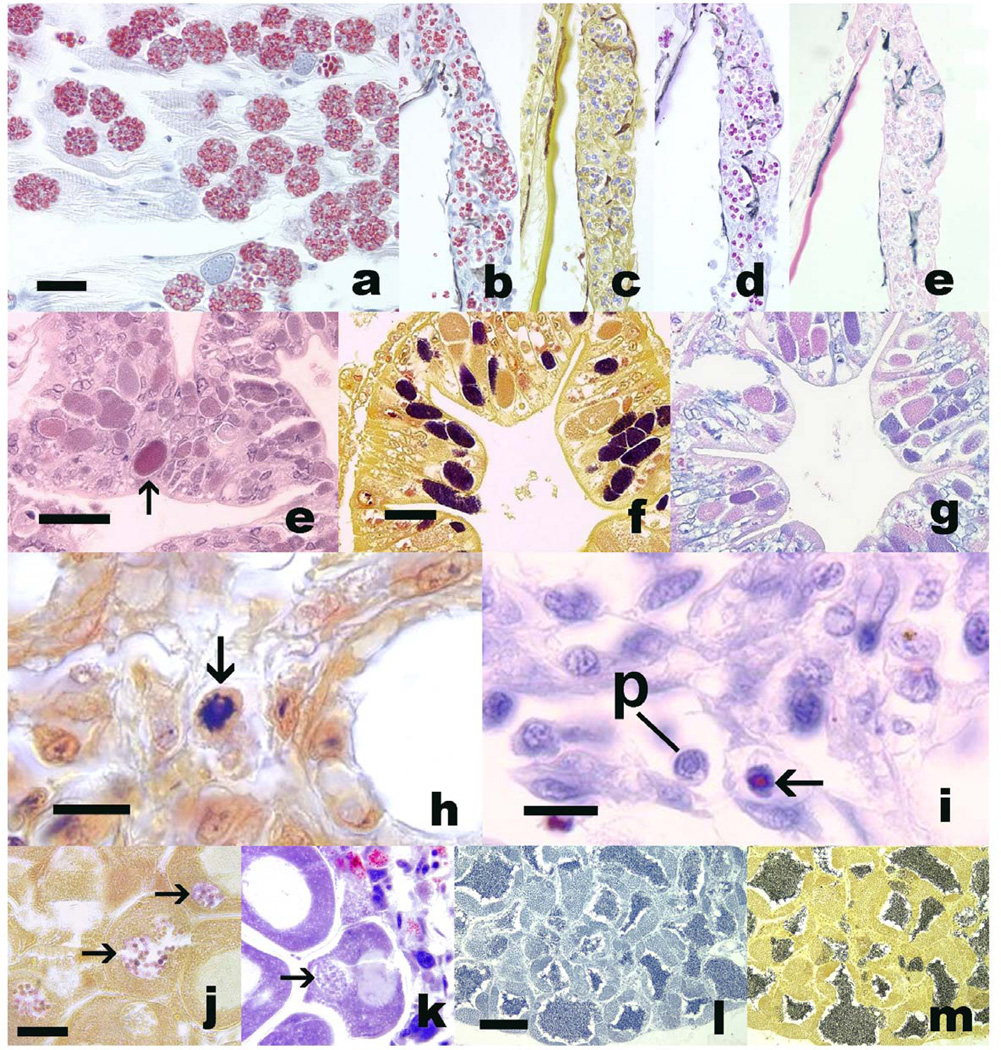
Figure 2.
a – e. Pleistophora hyphessobryconis in skeletal muscle (a) and skin (b – e) of Danio rerio. Bar = 25 µm. a, b. Luna; c, Gram; d, Fite’s acid fast; e, Giemsa. e –g, unidentified microsporidian within enterocytes of Eriocheir sinensis. Bar = 25 µm. e, Luna, Arrow = red staining aggregate of intra-enterocytic spores. f, Gram, with numerous Gram-positive spore aggregates within enterocytes; g, Fite’s acid fast. h, i, Nucleospora salmonis in nuclei of Chinook salmon leukocytes. h, Gram, with spore (arrow) in deep blue staining nucleus. i, Luna. Note red staining spore in nucleus (arrow). p = presporogonic stage within nucleus. j - k, Steinhausia mytilovum spores (arrows) in oocyte nuclei from Mytilus edulis. Note blue staining of spores with Gram (j), whereas spores do not stain red with Luna (k). Bar = 20 µm. l, m. Testis stained with Luna (l) or Gram (m). Note with both stains spermatids stain dark blue to black. Bar = 100 µm.
Gram stain
All microsporidial spores tested with the Gram stain showed approximately 50 to 75 % positive staining (defined as staining Gram positive), with many individual spores having a high degree of variable staining and in some cases, almost no positive staining. The background for this stain is yellow and contrasts well with the deep blue to purple color of spores. For most microsporidia, spores were easily distinguished from the surrounding tissue elements. However, the Gram stain was much less sensitive for Nucleospora salmonis as the nuclear chromatin also stained deep blue to purple (Fig. 2h). Steinhausia mytilovum spores were variable with this stain, but some aggregates were distinctly Gram positive. Spores of the unidentified microsporidian from E. sinensis were intensely positive (deep blue). Gram stains of testes from zebrafish revealed variably strong dark blue to purple staining of intracystic spermatids.
Fite’s acid-fast stain
There was extensive variability among microsporidial spore staining, from no spores staining positive to approximately 25–50% of spores in any given tissue section staining positive. In many of the tissue sections the spores were incompletely stained with minimal background contrast or did not stain positive at all.
Giemsa stain
Spores consistently stained blue to deep blue with this stain. However, there was essentially no contrast with surrounding tissues as they stained the same color.
DISCUSSION
Microsporidian infections in histologic sections are most often diagnosed by observation of spores in host tissues. Spores are easy to identify when they occur as large aggregates or xenomas in tissue sections stained with hematoxylin and eosin. However, spores do not stain consistently with either dye or may stain with hematoxylin, and thus individual spores or small numbers of spores in tissue and areas of inflammation as well as small intranuclear spores (i.e., N. salmonis) are difficult to detect. Hence, a variety of selective histologic stains that highlight spores are recommended.
Our study comparing the performance of four special stains on 8 different microsporidia (6 from fish and 2 from aquatic invertebrates) demonstrated that the Luna stain was optimal or at least comparable to the Gram stain for most specimens. Positive background staining with the Luna stain was confined to erythrocytes, bone, the lens of the eye and the chitin capsule of brine shrimp nauplii (within the gastrointestinal tract of zebrafish), all of which tended to be variably stained. None of these specific host tissues were infected by microsporidia. A novel finding with respect to teleost tissues was the intense positive staining of eosinophilic granular cells, noted in the tissue sections of salmonid gills. One key question is which microsporidian cellular component or components have an affinity for the stain. The endospore wall of microsporidia contains chitin (Vavra and Larsson 1999, Bigliardi and Sacchi 2001, Keeling and Fast 2002) and the active dye (1% Biebrich scarlet) of the Luna stain specifically complexes with chitosaccharides (Holler et al. 1975). Historically, Biebrich scarlet dye has been used to identify chitinous structures of invertebrates (Joffe and Hepburn 1973) and this was confirmed in our study by the positive staining of brine shrimp exoskeleton seen within the intestinal lumen of zebrafish. Chitin is a long chain polymer of N-acetylglucosamine. A reasonable hypothesis is that the charged sulphur trioxide of the dye binds with the nitrogen of the amine in chitin, as this is accessible. Each dye molecule contains two charged sulphur trioxides so there is the possibility that these two sulphur trioxides bind to the two adjacent amines on the polymer chain, making the binding tighter. Therefore, we conclude that the Luna stain binds to and stains chitin in microsporidian spores. The Gram stain was also very effective for staining spores in most of the samples, with the dramatic differentiation between blue staining spores and a brilliant yellow background. Indeed, we used this stain recently to identify spores of Pleistophora hyphessobryconis (Sanders et al. 2010), and variations of the Gram stain have been used by many pathologists to highlight a wide variety of microsporidian spores in tissue sections (Rupstra et al. 1988, Moura et al. 1996). In fact, this stain was superior to the Luna stain for the two invertebrate species. The Gram stain forms an insoluble moiety with peptidoglycan rather than chitin. Spores that are Gram positive but Luna negative may contain little to no chitin or are less fully developed. However, the majority of infected enterocytes contained mature spores of the unidentified microsporidian from E. sinensis.
One major drawback of the Gram stain was for detection of intranuclear spores of Nucleospora salmonis as nuclear material also stained dark blue to deep purple. The Warthin-Starry silver stain has been used to detect microsporidian spores (Guzman et al. 2001) and Kent et al. (1995) previously used this stain for detecting spores of N. salmonis. Spores were positive, but there was considerable nuclear background staining that frequently confounded detection and interpretation of the intranuclear spores. Furthermore, we are investigating vertical transmission of Pseudoloma neurophilia. The Gram stain is effective for demonstrating these spores in zebrafish ovaries and eggs. Similar to the Warthin-Starry stain, intense deep basophilic staining of nuclear material and developing sperm makes detection and interpretation of spores difficult within testes.
Acid fast stains have been used for highlighting microsporidian spores and we previously used the Ziehl-Nielsen stain to demonstrate spores of P. neurophilia in tissue sections (Ramsay et al. 2009). In our experience with both the Ziehl-Nielsen and Fite’s stains, we found that results were quite variable between laboratories and between runs, largely based on the amount of decolorization as spores do not retain carbol fuschin stain as well as mycobacteria. As positive staining of spores may be reversed by over-decolorizing, Ramsay et al. (2009) recommended reducing the time of this step. However, this results in more background staining. In other words, the correct balance between excessive background staining and over-decolorization of spores is required to obtain optimal contrast between spores and surrounding tissues.
The Giemsa stain is commonly used to highlight protozoan and myxozoan parasites and has been used for detection of microsporidia (Rupstra et al. 1988, Awadalla et al. 1998). We found that spores from all microsporidia species in our study did stain positive but could not be readily distinguished from surrounding tissues, as similarly reported by Awadalla et al. (1998). Therefore, the Giemsa stain would still be better than H&E, which usually does not highlight spores well.
In conclusion, the Luna stain is superior to the Gram, acid fast, and Giemsa stains for staining microsporidian spores with the least amount of background interference and also allows individual spores to be readily detected in tissues. Moreover, it is a relatively simple stain that involves only two principle dyes and is readily available in most histology laboratories.
Acknowledgements
The authors wish to thank Virginia Watral, Jayde Ferguson, John Bignell, Justin Sanders, Dr. Grant Stentiford, Dr. Katy Murray and Dr. Ronald Hedrick for contributing case material and technical assistance for this paper. We would also like to thank Kristin Berkenkamp and the Oregon State University Veterinary Diagnostic Laboratory for excellent histotechnical assistance. We thank Dr. Stephen Irving for discussion on chemistry of the Luna stain. This work was supported by NIH NCRR grants 5R24RR017386-02 and 1 T32 RR023917 and UK Department for Environment, Food and Rural Affairs contract FB001 (SWF).
LITERATURE CITED
- Awadalla HN, el-Naga IF, el-Temsahi MM, Negm AY. Detection of Microsporidia by different staining techniques. J Egyptian Soc Parasitol. 1998;28(3):729–738. [PubMed] [Google Scholar]
- Bigliardi E, Sacchi L. Cell biology and invasion of the microsporidia. Microbes and Infection. 2001;3:373–379. doi: 10.1016/s1286-4579(01)01393-4. [DOI] [PubMed] [Google Scholar]
- Brown AMV, Kent ML. Molecular diagnostics for Loma salmonae and Nucleospora salmonis (Microsporidia) In: Cunningham C, editor. Molecular diagnosis of salmonid diseases. England: Kluwer Acad. Publ., London; 2002. pp. 267–283. [Google Scholar]
- Garcia LS. Laboratory identification of the microsporidia. J Clin Microbiol. 2002;40(6):1892–1901. doi: 10.1128/JCM.40.6.1892-1901.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman O, Enriquez R, Schottelius J. Detection of spores of Nucleospora salmonis in frozen tissue material from Atlantic salmon. Bull European Assoc Fish Pathol. 2001;21:81–83. [Google Scholar]
- Holler E, Rupley JA, Hess GP. Productive and unproductive lysozyme-chitosaccharide complexes. Equilibrium measurements. Biochemistry. 1975 Mar 11;14(5):1088–1094. doi: 10.1021/bi00676a032. [DOI] [PubMed] [Google Scholar]
- Joffe I, Hepburn HR. Observations on regenerated chitin films. J Materials Sci. 1973;8:1751–1754. [Google Scholar]
- Joseph J, Murthy S, Garg P, Sharma S. Use of different stains for microscopic evaluation of corneal scrapings for diagnosis of microsporidial keratitis. J Clin Microbiol. 2006a;44(2):583–585. doi: 10.1128/JCM.44.2.583-585.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph J, Vemuganti GK, Garg P, Sharma S. Histopathological evaluation of ocular microsporidiosis by different stains. BMC Clin Pathol. 2006b;6(6):1–8. doi: 10.1186/1472-6890-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ, Fast NM. Microsporidia: Biology and evolution of highly reduced intracellular parasites. Annual Rev Microbiol. 2002;56:93–116. doi: 10.1146/annurev.micro.56.012302.160854. [DOI] [PubMed] [Google Scholar]
- Kent ML, Rantis V, Bagshaw JW, Dawe SC. Enhanced detection of Enterocytozoon salmonis (Microspora), an intranuclear microsporean of salmonid fishes, with the Warthin-Starry stain combined with hematoxylin and eosin. Dis Aquat Org. 1995;23:235–237. [Google Scholar]
- Kent ML, Bishop-Stewart JK. Transmission and tissue distribution of Pseudoloma neurophilia (Microsporidia) of zebrafish, Danio rerio (Hamilton) J Fish Dis. 2003;26:423–426. doi: 10.1046/j.1365-2761.2003.00467.x. [DOI] [PubMed] [Google Scholar]
- Kligman LH. Luna’s technique: A beautiful stain for elastin. Am J Dermatopathol. 1981;3(2):199–200. doi: 10.1097/00000372-198100320-00014. [DOI] [PubMed] [Google Scholar]
- Kotler DP, Giang TT, Garro ML, et al. Light microscopic diagnosis of microsporidiosis in patients with AIDS. Am J Gastroenterol. 1994;89:540–544. [PubMed] [Google Scholar]
- Lamps LW, Bronner MP, Venecak-Jones CL, Tham KT, Mertz HR, Scott MA. Optimal screening and diagnosis of microsporidia in tissue sections – a comparison of polarization, special stains and molecular techniques. Microbiol Infect Dis. 1997;109(4):404–410. doi: 10.1093/ajcp/109.4.404. [DOI] [PubMed] [Google Scholar]
- Luna LG. Manual of histologic staining methods of the armed forces institute of pathology. 3rd Ed. New York: McGraw-Hill; 1968. pp. 111–112. [Google Scholar]
- Luna VA, Stewart BK, Bergeron Dl, Clausen CR, Plorde JJ, Fritsche TR. Use of fluorochrome calcofluor white in the screening of stool specimens for spores of microsporidia. Am J Clin Pathol. 1995;103:656–659. doi: 10.1093/ajcp/103.5.656. [DOI] [PubMed] [Google Scholar]
- Matthews JL, Brown AM, Larison K, Bishop-Stewart JK, Rogers P, Kent ML. Pseudoloma neurophilia n.g., n. sp., a new microsporidium from the central nervous system of the zebrafish (Danio rerio) J Eukaryot Microbiol. 2001;48(2):227–233. doi: 10.1111/j.1550-7408.2001.tb00307.x. [DOI] [PubMed] [Google Scholar]
- Moura H, Nunes Da Silva JL, Sodra FC, Brasil P, Wallmo K, Wahlquist S, Wallace S, Croppo GP, Visvesvara GS. Gram-chromotrope: a new technique that enhances detection of microsporidial spores in clinical samples. J Eukaryot Microbiol Sept-Oct. 1996;43(5):94S–95S. doi: 10.1111/j.1550-7408.1996.tb05019.x. [DOI] [PubMed] [Google Scholar]
- Moura H, Schwartz DA, Bornay-Llinares F, Sodré FC, Wallace S, Visvesvara GS. A new and improved "quick-hot Gram-chromotrope" technique that differentially stains microsporidian spores in clinical samples, including paraffin-embedded tissue sections. Arch Pathol Lab Med. 1997;121(8):888–893. [PubMed] [Google Scholar]
- Ramsay JM, Watral V, Schreck CB, Kent ML. Pseudoloma neurophilia infections in zebrafish Danio rerio : effects of stress on survival, growth and reproduction. Dis Aquat Org. 2009;88:69–84. doi: 10.3354/dao02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupstra AC, Canning EU, Van Ketel RJ, Eeftinck Schattenkerk JKM, Laarman JJ. Use of light microscopy to diagnose small-intestinal microsporidiosis in patients with AIDS. J Infect Dis Apr. 1988;157(4):827–831. doi: 10.1093/infdis/157.4.827. [DOI] [PubMed] [Google Scholar]
- Sanchez JG, Speare DJ, Markham RJF. Nonisotopic detection of Loma salmonae (Microspora) in rainbow trout (Oncorhynchus mykiss) gills by in situ hybridization. Vet Pathol. 1999;36:610–612. doi: 10.1354/vp.36-6-610. [DOI] [PubMed] [Google Scholar]
- Sanders JL, Lawrence C, Nichols DK, Brubaker JF, Peterson TS, Murray KN, Kent ML. Pleistophora hyphessobryconis (Microsporidia) infecting zebrafish Danio rerio in research facilities. Dis Aquat Org. 2010;91:47–56. doi: 10.3354/dao02245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi VH, Perez MA, Itoiz ME. Modification of Luna’s technique for staining eosinophils in the hamster cheek pouch. Biotechnic and Histochem. 2008;83(3–4):147–151. doi: 10.1080/10520290802352424. [DOI] [PubMed] [Google Scholar]
- Vavra J, Larsson JIR. Structure of the microsporidia. In: Wittner M, Weis LM, editors. The microsporidians and microsporidiosis. Washington, DC: ASM Press; 1999. pp. 7–84. [Google Scholar]
- Weber R, Schwartz DA, Deplazes P. Laboratory diagnosis of microsporidiosis. In: Wittner M, Weiss LM, editors. The microsporidia and microsporidiosis. Washington, D.C.: ASM Press; 1999. pp. 315–362. [Google Scholar]
- Whipps CM, Kent ML. Polymerase chain reaction detection of Pseudoloma neurophilia, a common microsporidian of zebrafish (Danio rerio) reared in research laboratories. J Am Assoc Lab Anim Sci. 2006 Jan;45(1):36–39. [PMC free article] [PubMed] [Google Scholar]