Abstract
Site-2 Proteases (S2Ps) are a class of intramembrane metalloproteases named after the founding member of this protein family, human S2P, which cleaves Sterol Regulatory Element Binding Proteins which control cholesterol and fatty acid biosynthesis. S2Ps are widely distributed in bacteria and participate in diverse pathways that control such diverse functions as membrane integrity, sporulation, lipid biosynthesis, pheromone production, virulence, and others. The most common signaling mechanism mediated by S2Ps is the coupled degradation of transmembrane anti-Sigma factors to activate ECF Sigma factor regulons. However, additional signaling mechanisms continue to emerge as more prokaryotic S2Ps are characterized, including direct proteolysis of membrane embedded transcription factors and proteolysis of non-transcriptional membrane proteins or membrane protein remnants. In this review we seek to comprehensively review the functions of S2Ps in bacteria and bacterial pathogens and attempt to organize these proteases into conceptual groups that will spur further study.
Introduction
Site-2 proteases (S2Ps) are widely distributed in bacteria and participate in diverse pathways, all of which share the requirement for proteolysis of a transmembrane protein. For the purpose of this review, we define S2Ps as multipass transmembrane proteins with a conserved zinc metalloprotease active site (HExxH) within a transmembrane domain and an xDG motif within another transmembrane domain [1, 2]. Many bacterial S2Ps also have a centrally located PDZ domain [1]. Due to their prominence as model organisms, the S2Ps of E. coli (RseP) and B. subtilis (YluC and SpoIVFB) have been intensely studied and are the best understood in terms of upstream activating signals, signal transduction mechanism, and downstream regulons. Expanding investigation of S2Ps in bacterial pathogens has revealed roles for S2Ps in sensing host signals and regulating virulence gene expression during infection. In most cases, the signaling cascades in which S2Ps participate follow the same general paradigm. A site-1 protease (S1P) first cleaves the (usually) extracytoplasmic segment of the transmembrane substrate in response to specific inducing signal (e.g unfolded outer membrane proteins in the case of the S1P (DegS) in the E. coli SigE pathway). This site-1 cleavage is rapidly followed by S2P cleavage within the transmembrane segment of the substrate, thereby liberating the cytosolic fragment of the substrate. In many cases, the fragment released into the cytosol by the S2P cleavage is a transcriptional regulator. Although the tight coupling between S1P and S2P cleavage events is a hallmark of many of these signaling systems, the mechanisms that link S1P and S2P cleavage are still poorly understood.
Despite wide distribution of the S1P/S2P signaling paradigm, variations on this theme continue to emerge as more S2P signaling systems are studied. First, there is great diversity in both the signals that induce the S1P cleavage event and the physiologic consequences of pathway activation. In many cases, the S1P/S2P cleaved transmembrane protein is a transcriptional regulator, but the proteolytic destruction of the regulator has diverse effects depending on the system, in some cases activating gene expression and in some cases repressing. Finally, although the S1P/S2P paradigm of transmembrane signal transduction is widely distributed, exceptions to the rule continue to emerge, including examples in which S2P mediated cleavage occurs apparently independently of a S1P, examples in which S2P cleavage releases a extracellular signaling molecule rather than a cytoplasmic fragment, and functions for S2Ps in general cleavage of signal sequence remnants. In this review, we will systematically review S2P systems in bacteria and bacterial pathogens with the goal of highlighting canonical prokaryotic S2P systems and the deviations from the canon. This review benefits heavily from other recent outstanding reviews of this field, to which we refer the reader for additional details and perspectives [3–6].
Bacteria
Escherichia coli RseP
The E. coli sigma factor E (SigE) periplasmic stress response pathway is one the best studied S2P containing signaling systems. SigE is an alternative sigma factor of the extracytoplasmic function (ECF) sigma factor subclass, termed as such because these sigma factors typically respond to and regulate extracytoplasmic processes [7–9]. The SigE pathway of E. coli follows the common paradigm of a three component S1P/anti-sigma factor/S2P system (Figure 1A). The SigE ECF sigma factor is held inactive by the RseA transmembrane anti-sigma factor. The upstream activating signals for the pathway are the C-terminal hydrophobic amino acids of β-barrel outer membrane proteins (OMPs), which are ordinarily sequestered, but become exposed under conditions that unfold OMPs such as heat shock. The C-terminal peptides of OMPs bind to the DegS PDZ domain, thereby activating DegS to cleave the periplasmic domain of RseA [10–14]. RseB also controls DegS proteolysis of RseA by binding directly to the periplasmic domain of RseA [10]. DegS cleaved RseA is a substrate for RseP [15–17], although the mechanism of coupling of these two proteolytic events remains controversial [18, 19]. Following RseP cleavage, the RseA/SigE complex is released to the cytoplasm where further degradation of RseA [20] releases SigE to associate with RNA polymerase to activate target promoters. The SigE regulon, which is essential, includes genes involved in cell envelope remodeling, chaperones, and the heat shock response [21, 22]. The RseP pathway is one of the best understood signal transduction pathways involving a S2P and forms the paradigmatic example of a transmembrane signaling system in which an ECF sigma factor is activated by coupled proteolytic destruction of a transmembrane anti-sigma factor (Illustrated in Figure 1A).
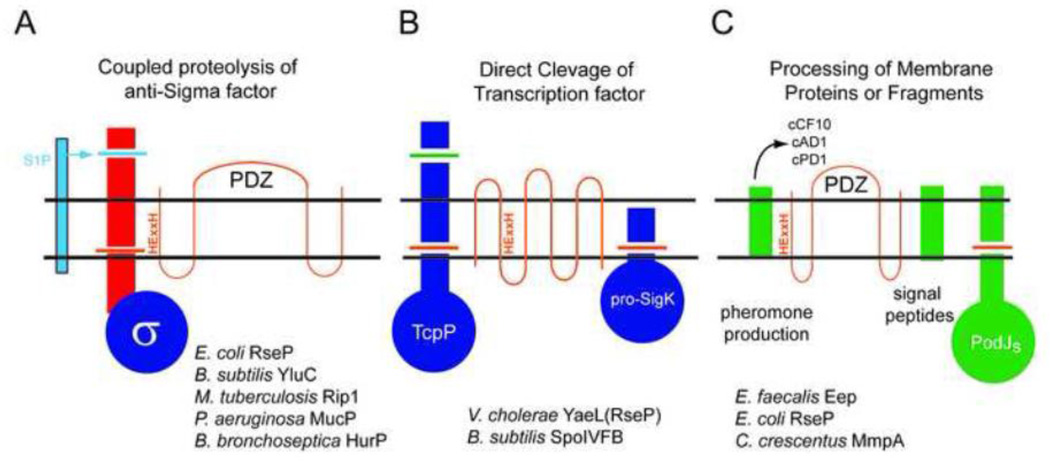
Figure 1. Conceptual Groupings of S2P mediated signaling Pathways.
A. Coupled Proteolysis of transmembrane anti-Sigma factors. In this widely distributed mechanism of ECF Sigma activation, an activating signal triggers Site one proteolysis (S1P, light blue) of the periplasmic domain of the anti-Sigma, which is rapidly followed by S2P mediated cleavage in the transmembrane segment (orange cut). The mechanism of this tight coupling is still poorly understood. The S2Ps of this type contain PDZ domains and include the S2Ps listed below the figure.
B. Direct Cleavage of a transcriptional regulator. In this mechanism, the proteolytic target is a membrane bound transcriptional regulator, either TcpP or pro-SigmaK. In the case of TcpP, which is cleaved by YaeL(RseP), the cleavage event shutsoff gene expression. In the case of pro-SigK, proteolysis (by SpoIVFB) liberates the active transcription factor. Note that the transmembrane topology pictured is that of SpoIVFB, not RseP (which is represented in Panel A).
C. Cleavage of membrane proteins or protein remnants. In this grouping, the S2P processes a membrane protein that is not a transcription factor. Examples include the production of peptide pheromones in E. faecalis from lipoprotein signal sequences by Eep, the processing of signal peptides by RseP, and the destruction of PodJs by MmpA in C. crescentus. The constitutive cleavage of FtsL by YluC also may fall into this category. See text and Tables for details.
Bacillus subtilis
YluC/RasP
The B. subtilis SigW pathway controls the availability of ECF sigma factor SigW. SigW is held inactive by the transmembrane anti-Sigma factor RsiW. The SigW pathway of B. subtilis conforms to the common theme of sequential proteolytic destruction of a transmembrane antisigma factor. The transmembrane anti-sigma factor RsiW (anti-SigW) is first cleaved by the S1P PrsW (also called YpdC), which is a novel site-1 protease not homologous to DegS [23, 24]. Recent evidence also indicates that a second proteolytic event further shortens the PrsW cleaved RsiW [25]. The C-terminally shortened RsiW is then cleaved by the S2P YluC/RasP [26], which releases the cytoplasmic fragment of RsiW bound to SigW. The mechanism(s) by which the inducers of the SigW pathway are coupled to the activation of site-1 proteolysis of RsiW are not well understood. PrsW has also been implicated in the degradation of anti-sigma factors and virulence in C. difficile, although the S2P involved in this pathway has not been defined [27].
The SigW regulon is induced by a variety of signals, many of which perturb cell wall integrity. For example, induction of a SigW dependent promoter was observed with a variety of cell wall inhibiting antimicrobials, including Beta lactams and Vancomycin [28]. Similarly, antimicrobial peptides activate the SigW regulon [24], as does alkaline shock (pH 8.9) [29]. The SigW regulon consists of approximately 30 operons [30]. Consistent with its role in responding to perturbations in membrane integrity, one SigW-responsive promoter lies within the fabHafabF locus and is induced by alkaline shock, cell wall active antibiotics, and TritonX-100 [31]. Upregulation of this promoter increases membrane rigidity through upregulation of FabF and downregulation of FabHa, resulting in longer chain fatty acids, thereby increasing membrane rigidity [31].
A second substrate for YluC/RasP has been reported. FtsL, a transmembrane protein involved in cell division, is rapidly degraded in wild type cells but stabilized by inactivation of yluC [32]. The YluC-FtsL cleavage reaction was recapitulated in E. coli, supporting a direct proteolytic relationship between YluC and FtsL [32]. In addition to showing that the S2P YluC has two substrates, FtsL cleavage by YluC does not apparently require a site-1 cleavage event. The function of YluC in this pathway is constitutive degradation of FtsL, rather than proteolysis upon activation by an upstream protease. This latter feature may suggest that the mechanisms by which most S2Ps are held inactive until site-1 cleavage occurs may not be operative for the YluC/FtsL pair.
There is yet another predicted transmembrane anti-Sigma factor in B. subtilis, RsiV, which is paired with SigV. This pathway is induced by lysozyme, but the role of RsiV proteolysis in this cascade has not yet been examined [33, 34].
SpoIVFB-pro-SigK pathway
SpoIVFB is a founding member of the bacterial S2P metalloprotease family that plays a crucial role in the latter stages of the sporulation program [2, 35]. SpoIVFB cleaves pro-SigK, a membrane associated transcription factor, to a mature, active isoform, SigK, that governs gene expression in the mother cell after the forespore engulfment stage of Bacillus sporulation [2, 36]. While SigK ultimately controls the transcriptional program in the mother cell, the signal for pro-SigK activation originates in the forespore in the form of SigG-dependent expression [37] and subsequent secretion of serine protease, SpoIVB [38]. Like RseP and many other S2Ps, SpoIVFB is held inactive until the appropriate activation signal arrives, in this case SpoIVB from the forespore. Two proteins, BofA and SpoIVFA, form a stable complex with SpoIVFB in the outer forespore membrane (OFM) and inhibit cleavage of pro-SigK [36, 37, 39, 40]. SpoIVB is secreted from the forespore to the OFM where it degrades SpoIVFA, thereby relieving its inhibitory effect on SpoIVFB [41]. SpoIVB also activates a second forespore-derived protease, CtpB, which can also cleave SpoIVFA, providing a redundant mechanism to activate SpoIVFB processing of pro-SigK [41–43]. Once BofA and SpoIVFA inhibition have been relieved, SpoIVFB cleaves between residues 20 and 21 on pro-SigK, which associates with the OFM through an N-terminal transmembrane domain (residues 1–27), releasing soluble SigK [44]. This degradation is dependent upon the presence of ATP which may interact with SpoIVFB to alter its interaction with pro-SigK in a manner that facilitates pro-SigK cleavage [45]. SigK then directs gene expression in the mother cell in the latter stages of the developing sporangial cell, including genes for spore cortex, coat synthesis, and ultimately mother cell lysis and mature spore release [39]. Thus, SpoIVFB is the linchpin in the final sigma factor checkpoint between forespore and mother cell prior to the last stage of sporulation.
The B. subtilis SpoIVFB pathway differs from the aforementioned paradigmatic S1P/ Substrate/S2P systems in several important ways. In contrast to RseP, the SigK-SpoIVFB cascade provides an example a S2P cascade in which the intramembrane cleavage event occurs in the transcriptional regulator itself, rather than an anti-sigma factor (Illustrated in Figure 1B). Furthermore, there is no S1P that directly cleaves pro-SigK before SpoIVFB cleavage. However, a proteolytic cascade conceptually analogous to site-1 proteolysis does occur in the pro-SigK cascade. As has been suggested [41], one could think of the SpoIVB protease as a S1P that, instead of cleaving the S2P substrate to activate the cascade, cleaves an inhibitor in complex with the S2P. Finally, SpoIVFB also differs topologically from other S2Ps in its lack of a PDZ domain and the presence of six transmembrane domains (in contrast to the more common topology of 4 TMs) [46].
Caulobacter crescentus
Caulobacter crescentus PodJ is a localization factor involved in polar morphogenesis which exists in two isoforms, PodJL and PodJs [47, 48]. The long form, PodJL, localizes to the swarmer cell pole in the predivisional cell. As the cell divides, PodJL is replaced by PodJs, a truncated form of PodJL that lacks the C terminus. The conversion of PodJL to PodJs occurs through proteolysis of the C terminal domain of PodJ by the periplasmic protease PerP at the time of cell division, and is regulated by cytokinesis signals via the DivJ-PleC-DivK system [49]. PodJs persists at the flagellated pole of the post divisional cell, where it is required for chemotaxis [48].
MmpA is a S2P in Caulobacter that contains the canonical zinc metalloprotease active site and a centrally located PDZ domain. In a ΔmmpA strain of Caulobacter, stability of PodJL is unaffected, but PodJs is stabilized [50], consistent with MmpA degrading PodJs in its transmembrane domain during the swarmer-to-stalked transition. In addition, MmpA can functionally complement a ΔyaeL strain of E. coli with respect to RseA degradation [50].
The Caulobacter PerP/PodJ/MmpA system in some ways follows the canonical S1P/TM substrate/S2P model, but differs in some important respects. In most of the S1P/S2P systems which cleave anti-sigma factors, the S1P/S2P cleavage events are tightly coupled and the S1P cleaved protein is a transient intermediate that only accumulates when the S2P is missing. In Caulobacter, PodJs, the product of the S1P (PerP) cleavage, is not only detectable, but has a distinct physiologic function in the cell [48]. These distinct functions for the two PodJ isoforms mandate that PerP and MmpA not be tightly coupled, as occurs in many other S1P/S2P systems. Another distinct feature of the Caulobacter system is the lack of apparent function of the cytoplasmic fragment of PodJ that is released into the cytoplasm by MmpA cleavage of PodJs, implying that the function of the S2P in this system is to eliminate the membrane bound PodJs, rather than supply an active soluble fragment of the protein.
Bacterial Pathogens
Mycobacterium tuberculosis Rip1
The M. tuberculosis (Mtb) genome encodes three putative S2Ps: Rv0359 (Rip2), Rv2625c (Rip3), and Rv2869c (Rip1), none are which are singularly essential for growth in vitro [51]. Initial examination of the Δrip1 strain of M. tuberculosis (but not Δrip2 or Δrip3) demonstrated defects in cording, a macroscopic colonial morphology of M. tuberculosis associated with virulence and cell envelope lipid composition. Indeed, the Δrip1 mutant is defective in the localization of the three principal mycolic acids species to the outer M. tuberculosis cell envelope and displays complex perturbation of multiple lipid biosynthetic and catabolic genes at the transcriptional level [51, 52]. In the mouse model of aerosol M. tuberculosis infection, deletion mutants in either rip2 or rip3 display similar growth kinetics to wild type M. tuberculosis (unpublished data) whereas M. tuberculosis Δrip1 is significantly attenuated in both the acute and chronic infection. Specifically, Δrip1 titers in mouse lung are 100 fold lower than wild type during acute infection and further decline 100 fold during chronic infection, indicating an important function for Rip1 in Mtb virulence [51].
Subsequent characterization of the Rip1 pathway has shown that, like many prokaryotic S2Ps, Rip1 cleaves the transmembrane anti-sigma factors for three ECF sigma factors: SigK, SigL, and SigM [52]. The S1Ps that cleave any of the anti-sigmas of the Rip1 pathway are unknown. A candidate gene approach that examined several transmembrane proteases as candidate S1Ps in M. tuberculosis failed to identify the S1Ps of the Rip1 pathway [52].
Dissection of the relationships between Rip1 and these three Sigma factors regulons has indicated Rip1 dependent/SigKLM dependent pathways and Rip1 dependent/SigKLM independent pathways. Analysis of the transcriptomes of Δrip1, ΔsigK, ΔsigL, and ΔsigM revealed that induction of the gene encoding the catalase-peroxidase KatG and its upstream iron dependent repressor FurA is dependent on Rip1, SigK and SigL [52]. By contrast, several other genes whose wild type expression pattern required Rip1 were not affected by loss of SigK, SigL, or SigM. The gene encoding the resuscitation-promoting factor C (rpfC), believed to play a role in dormancy, and the mycobacterial β-ketoacyl ACP synthase kasA, were both underexpressed in Δrip1 but unaffected by any sigma factor deletion [52]. These experiments strongly suggested that Rip1 controls pathways apart from the SigK, SigL, or SigM regulons, and by extension has additional substrates. This conclusion has been further substantiated by our recent finding that an M. tuberculosis ΔsigKΔsigLΔsigM triple mutant does not recapitulate the Δrip1 mutant phenotype in mice (M. Glickman, unpublished data).
One additional substrate of Rip1 has been identified in the literature, PBP3. PBP3 interacts with Wag31 (DivIVA), but when a mutant Wag31 protein is present that does not interact with PBP3, PBP3 is unstable and degraded by Rip1 under conditions of oxidative stress [53]. This may suggest that under certain circumstances in which the Wag31-PBP3 interaction is disrupted in wild type cells, Rip1 may degrade PBP3. This result also emphasizes the relative promiscuity of cleavage by Rip1, a feature shared with many S2Ps, which will cleave a variety of transmembrane segments without an apparent conserved cleavage site.
In summary, the Rip1 S2P controls an important virulence pathway in M. tuberculosis that regulates iron storage, lipid metabolism, oxidative stress defense (katG), dormancy (rpfC), and likely additional pathways that are yet to be defined. The major outstanding questions in the Rip1 pathway include the identity of the S1Ps, additional Rip1 substrates that contribute to the virulence phenotype of the Δrip1 strain, and the specific host signals that serve as the upstream activating signals of the Rip1 pathway in vivo.
Pseudomonas aeruginosa
P. aeruginosa is a gram negative opportunistic pathogen that causes a variety of clinical syndromes, including chronic lung infections in cystic fibrosis patients. P. aeruginosa can undergo mucoid conversion, a morphologic colony phenotype that is caused by overproduction of the polysaccharide alginate. Alginate production is controlled by an anti-sigma/sigma factor system in which the transmembrane anti-sigma (MucA) holds the sigma factor (AlgU) inactive. AlgU activates transcription of the genes encoding alginate biosynthetic enzymes [54–57]. MucA is subject to proteolytic destruction through sequential proteolysis by a S1P/S2P that closely resembles the E. coli RseA system. For a comprehensive review of this system, please see [4]. In P. aeruginosa, the S1P is AlgW [58] and the S2P is MucP. Accumulation of envelope proteins (MucE) activates AlgW (S1P) to cleave MucA in a manner similar to DegS cleaving RseA [58]. MucP then cleaves MucA within its transmembrane domain, releasing the MucA/AlgU cytoplasmic complex [58–60]. The upstream activating signals of the alginate pathway include unfolded membrane proteins (similar to the E. coli SigE pathway) [58] and cellwall active antibiotics [61].
Although the AlgW/MucA/MucP cascade follows the standard model of other S1P/Anti-Sigma/S2P systems, there is evidence that MucA proteolysis by MucP can occur independently of AlgW under certain conditions. Pseudomonas strains isolated from cystic fibrosis patients often display a constitutive mucoid phenotype. Some of these strains have a nonsense mutation in MucA which truncates the protein. This truncated MucA (MucA22) does not require AlgW proteolysis, but may require MucP degradation [4]. Similarly, in the absence of MucD, MucA activation is AlgW independent but MucP dependent [59]. These findings are thus similar to the S1P-independent proteolysis of anti-sigma factors that occurs when C-terminally truncated forms are expressed in M. tuberculosis and E. coli [16, 52]. In addition, some evidence suggests that MucP can directly degrade MucA in the absence of AlgW in wild type cells [4, 59, 62].
There is also evidence that MucP participates in other signaling systems apart from Alginate biosynthesis. One report [63] suggests that MucP in Pseudomonas cleaves FpvR, an anti-sigma factor for the sigma factors PvdS and FpvI. PvdS and FpvI regulate in iron uptake through siderophore biosynthesis [63]. These results indicate that the Pseudomonas MucP S2P has multiple substrates that presumably use independent S1Ps for activation.
Bordetella bronchiseptica
B. bronchiseptica is a respiratory pathogen that infects agricultural animals and occasionally humans and is related to the major human pathogen B. pertussis, the cause of Whooping Cough. B. bronchiseptica hurP encodes an S2P necessary for heme utilization [64]. hurP can complement the V. cholerae yaeL null mutant, which cannot degrade TcpP [64]. Though hurP is not essential in Bordetella, it is required for heme utilization as the sole source of nutrient iron through induction of the outer membrane Heme receptor BhuR. The substrate of HurP that controls this pathway has not been directly identified, but is likely HurR, a predicted transmembrane protein that may act as the anti-Sigma factor for HurI, and ECF sigma factor [64]. The S1P in this system has also not been identified, although a candidate gene approach excluded 4 candidate S1Ps [64].
Salmonella enterica
RseP, the S2P in Salmonella enterica s. typhimurium, plays a role in the sigE stress response system that is similar to the E. coli DegS/RseA/RseP system. In contrast to E. coli, Salmonella sigE is not essential in vitro, but plays an essential role in pathogenesis [65, 66]. The SigE pathway also has a distinct role in response to acid stress. Acid conditions induce transcription of SigE target genes, a response that is DegS independent but RseP dependent [67]. Similar to the E. coli system, heat stress induction of sigE requires DegS. Also in contrast to OMP induction of the SigE pathway, RseP lacking its PDZ domain (RsePΔPDZ) constitutively cleaves RseA, but acid stress induction of SigE targets is abolished [67]. This study implies that acid stress directly activates cleavage of RseA at the S2P cleavage step and is independent of a S1P.
Vibrio cholerae
In contrast to many of the systems discussed thus far, in which a transcriptional regulator is held inactive by a transmembrane anti-Sigma factor, two transmembrane regulators of V. cholera toxT transcription, TcpP and ToxR, are active as transcriptional regulators when membrane bound. TcpP is required for the induction of the toxT gene, which in turn directly activates cholera toxin (ctxAB) and the toxin-coregulated pilus (tcp operon, including TcpA). The TcpP protein is unstable in the absence of another protein TcpH. Vibrio cholera YaeL (S2P), the homolog of E. coli YaeL/RseP, was identified in a screen for mutants that stabilized TcpP in a tcpH null strain [68]. This study also showed that this negative regulation of virulence gene expression by YaeL proteolysis is operative in wild type cells when Vibrio cholerae is placed in conditions that turns virulence genes off (pH 8.5, 37°C) [68]. Although there appears to be a site one cleavage event of TcpP that precedes YaeL degradation, DegS was not required for this degradation and the S1P has not yet been identified [68, 69]. In contrast, DegS and YaeL appear to both be required for induction of the V. cholera rpoE response as judged by the similar sensitivity of the rpoE, yaeL and degS mutants to 3%Ethanol [68], suggesting that YaeL (RseP) has at least two substrates (TcpP and RseA) in V. cholerae and both positively and negatively influences gene expression based on proteolysis of either a negative (RseA) or positive(TcpP) regulator.
Enterococcus faecalis
E. faecalis is a gram positive, naturally competent bacterium that causes urinary tract infections, endocarditis, and infections of indwelling catheters. E. faecalis has a pheromone-inducible plasmid transfer system in which plasmid free cells secrete a pheromone called cCF10 (or other similar pheromones such as cAD1 or cPD1), which induces transfer of the pCF10 plasmid from plasmid bearing cells. cCF10 is an octapeptide pheromone that is encoded within the signal sequence of a lipoprotein. After cleavage of the lipoprotein signal peptide by signal peptidase, the membrane embedded signal peptide is further processed by the S2P Eep (for enhanced expression of pheromone) [70–72]. This function of Eep is remarkable because it implicates a S2P in production of an extracellular diffusible signal, and raises interesting questions about whether and how Eep recognizes signal sequences that encode pheromones and differentiates these proteins from other signal sequence remnants [73]. In this regard, the function of Eep is reminiscent of the recently proposed function of E. coli RseP and B. subtilis RasP in cleaving remnant signal peptides [74]. In addition to its role in pheromone production, Eep was also recently identified in an in vivo screen for E. faecalis promoters induced in an abscess model [75]. The eep promoter was upregulated in this model and a Δeep strain was highly attenuated in an endocarditis model [75]. Importantly, this virulence function of Eep is apparently independent of its function in enhancing plasmid conjugation as this screen was performed in a strain lacking a conjugative plasmid [75]. The Eep substrates mediating the virulence phenotype have not yet been identified.
Cyanobacteria
Several S2Ps have been identified recently in Cyanobacteria, but their full functions are still being elucidated. The Synechocystis S2P Slr0643 is required for acid resistance and may mediate this effect through control of the SigH pathway in that organism [76]. Anabaena variabilis encodes five putative S2Ps, all of which can cleave B. subtilis pro-SigmaK when coexpressed in E. coli [77], but their substrates and physiologic function in Anabaena await further study.
Summary and common themes in bacterial S2P pathways
Based on the functions of prokaryotic S2Ps presented above, we have derived three major S2P functional groups, which we illustrate in Figure 1. The first group is the most abundant and illustrated in Figure 1A. These are the systems similar to the RseP pathway of E. coli, which use coupled proteolytic destruction of a transmembrane anti-sigma factor to active a soluble sigma factor. The second group, which is illustrated in Figure 1B, are systems in which the proteolytic target of the S2P is itself a transmembrane transcription factor. In the case of B. subtilis SpoIVFB, the substrate is a transmembrane precursor of SigK, which is inactive when membrane bound. In V. cholerae the substrate is TcpP, which is active in the membrane and therefore its proteolysis inhibits transcription. The third group is illustrated in Figure 1C and consists of systems in which the S2P acts to degrade a membrane protein without a downstream transcriptional function. Examples of these systems include the destruction of PodJs by MmpA and maturation of Enterococcal pheromones by Eep from lipoprotein signal peptides. We anticipate that further variations on these themes will continue to emerge as more S2P systems are studied in additional bacterial systems.
Table 1.
Signaling systems in bacteria mediated by S2Ps. Listed is the bacterial species, the S1P (if known or required), the substrate of the S2P, and the S2P itself. If the S2P signaling system controls a transcriptional regulator, it is listed along with the physiologic function of the pathway and relevant references.
| Bacterium | S1P | Substrate | S2P | Transcriptional regulator |
Activating Signal/Pathway |
Ref |
|---|---|---|---|---|---|---|
| E. coli | DegS | RseA | RseP | SigE | C termini of unfolded OMPs/Cell envelope stress response | [10, 12–14, 16, 17, 21] |
| B. subtilis | PrsW+ TSP | RsiW | YluC/RasP | SigW | Alkaline Shock, antimicrobial peptides, cell wall active antimicrobials/Membrane rigidity | [23–26, 28, 30, 31] |
| B. subtilis | SpoIVB, (conceptually, see text) | pro-SigK | SpoIVFB | SigK | Sigma G activation in forespore / Latter stages of sporulation developmental program in mother cell | [2, 35, 41] |
| Caulobacter crescentus | PerP | PodJL, -PodJs | MmpA | none | Cytokinesis/Polar cell division | [49, 50] |
Table 2.
Signaling systems in bacterial pathogens mediated by S2Ps. Listed is the bacterial pathogen, the S1P (if known or required), the substrate of the S2P, and the S2P itself. If the S2P signaling system controls a transcriptional regulator, it is listed along with the activating signal/physiologic function of the pathway.
| Pathogen | S1P | Substrate | S2P | Transcriptional regulator |
Activating Signal/Pathway |
Ref |
|---|---|---|---|---|---|---|
| Vibrio cholerae | ? | TcpP | YaeL | TcpP | Shuts off toxin production in conditions that limit virulence gene expression (Ph 8.5) | [68] |
| Vibrio cholerae | DegS | RseA | YaeL | SigE | Unfolded OMPs/Cell envelope stress response | [68] |
| Salmonella enterica | DegS | RseA | RseP | SigE | Unfolded OMPs/Cell envelope stress response | [67] |
| Salmonella enterica | none | RseA | RseP | SigE | Acid/Acid Resistance | [67] |
| M. tuberculosis | ? | RskA, RslA, RsmA, ? | Rip1 | SigK, SigL, SigM | Host Signals/Cell envelope remodeling, catalase-peroxidase, resuscitation promoting factor, growth and persistence in the mouse. | [51, 52] |
| M. tuberculosis | ? | PBP3 | Rip1 | none | Oxidative Stress | [53] |
| Enterococcus faecalis | SPase II | cCF10, cAD1, cPD1 | Eep | none | Peptide Pheromone production | [70–72, 78, 79] |
| Enterococcus faecalis | ? | ? | Eep | ? | ?/Virulence in Endocarditis Model independent of conjugation function | [75] |
| Pseudomonas aeruginosa | AlgW | MucA | MucP | AlgU | Mucoid transformation via alginate production | [57, 59, 61, 62] |
| Pseudomonas aeruginosa | ? | FvpR, FoxR, FiuR | MucP | PvdS, FpvI | Siderophore biosynthesis and uptake | [63] |
| Bordetella bronchiseptica | ? | ? HurR | HurP | ?HurI | Iron Uptake | [64] |
Highlights.
Comprehensive Review of the present state of knowledge in the bacterial S2P field
Both Model organisms and bacterial pathogens are reviewed
Common themes of S2P systems are emphasized.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Kinch LN, Ginalski K, Grishin NV. Site-2 protease regulated intramembrane proteolysis: sequence homologs suggest an ancient signaling cascade. Protein science : a publication of the Protein Society. 2006;15:84–93. doi: 10.1110/ps.051766506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rudner DZ, Fawcett P, Losick R. A family of membrane-embedded metalloproteases involved in regulated proteolysis of membrane-associated transcription factors. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:14765–14770. doi: 10.1073/pnas.96.26.14765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heinrich J, Wiegert T. Regulated intramembrane proteolysis in the control of extracytoplasmic function sigma factors. Research in microbiology. 2009;160:696–703. doi: 10.1016/j.resmic.2009.08.019. [DOI] [PubMed] [Google Scholar]
- 4.Damron FH, Goldberg JB. Proteolytic regulation of alginate overproduction in Pseudomonas aeruginosa. Molecular microbiology. 2012;84:595–607. doi: 10.1111/j.1365-2958.2012.08049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Urban S. Making the cut: central roles of intramembrane proteolysis in pathogenic microorganisms. Nature reviews. Microbiology. 2009;7:411–423. doi: 10.1038/nrmicro2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ho TD, Ellermeier CD. Extra cytoplasmic function sigma factor activation. Current opinion in microbiology. 2012;15:182–188. doi: 10.1016/j.mib.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Helmann JD. The extracytoplasmic function (ECF) sigma factors. Advances in microbial physiology. 2002;46:47–110. doi: 10.1016/s0065-2911(02)46002-x. [DOI] [PubMed] [Google Scholar]
- 8.Lonetto MA, Brown KL, Rudd KE, Buttner MJ. Analysis of the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of eubacterial RNA polymerase sigma factors involved in the regulation of extracytoplasmic functions. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:7573–7577. doi: 10.1073/pnas.91.16.7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Missiakas D, Raina S. The extracytoplasmic function sigma factors: role and regulation. Molecular microbiology. 1998;28:1059–1066. doi: 10.1046/j.1365-2958.1998.00865.x. [DOI] [PubMed] [Google Scholar]
- 10.Chaba R, Alba BM, Guo MS, Sohn J, Ahuja N, Sauer RT, Gross CA. Signal integration by DegS and RseB governs the σ E-mediated envelope stress response in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:2106–2111. doi: 10.1073/pnas.1019277108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sohn J, Sauer RT. OMP peptides modulate the activity of DegS protease by differential binding to active and inactive conformations. Molecular cell. 2009;33:64–74. doi: 10.1016/j.molcel.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sohn J, Grant RA, Sauer RT. Allosteric activation of DegS, a stress sensor PDZ protease. Cell. 2007;131:572–583. doi: 10.1016/j.cell.2007.08.044. [DOI] [PubMed] [Google Scholar]
- 13.Walsh NP, Alba BM, Bose B, Gross CA, Sauer RT. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell. 2003;113:61–71. doi: 10.1016/s0092-8674(03)00203-4. [DOI] [PubMed] [Google Scholar]
- 14.Ades SE, Connolly LE, Alba BM, Gross CA. The Escherichia coli sigma(E)-dependent extracytoplasmic stress response is controlled by the regulated proteolysis of an anti-sigma factor. Genes & development. 1999;13:2449–2461. doi: 10.1101/gad.13.18.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koide K, Ito K, Akiyama Y. Substrate recognition and binding by RseP, an Escherichia coli intramembrane protease. The Journal of biological chemistry. 2008;283:9562–9570. doi: 10.1074/jbc.M709984200. [DOI] [PubMed] [Google Scholar]
- 16.Akiyama Y, Kanehara K, Ito K. RseP (YaeL), an Escherichia coli RIP protease, cleaves transmembrane sequences. The EMBO journal. 2004;23:4434–4442. doi: 10.1038/sj.emboj.7600449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanehara K, Ito K, Akiyama Y. YaeL (EcfE) activates the sigma(E) pathway of stress response through a site-2 cleavage of anti-sigma(E), RseA. Genes & development. 2002;16:2147–2155. doi: 10.1101/gad.1002302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hizukuri Y, Akiyama Y. PDZ domains of RseP are not essential for sequential cleavage of RseA or stress-induced sigma(E) activation in vivo. Molecular microbiology. 2012;86:1232–1245. doi: 10.1111/mmi.12053. [DOI] [PubMed] [Google Scholar]
- 19.Li X, Wang B, Feng L, Kang H, Qi Y, Wang J, Shi Y. Cleavage of RseA by RseP requires a carboxyl-terminal hydrophobic amino acid following DegS cleavage. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:14837–14842. doi: 10.1073/pnas.0903289106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flynn JM, Levchenko I, Sauer RT, Baker TA. Modulating substrate choice: the SspB adaptor delivers a regulator of the extracytoplasmic-stress response to the AAA+ protease ClpXP for degradation. Genes & development. 2004;18:2292–2301. doi: 10.1101/gad.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhodius VA, Suh WC, Nonaka G, West J, Gross CA. Conserved and variable functions of the sigmaE stress response in related genomes. PLoS biology. 2006;4:e2. doi: 10.1371/journal.pbio.0040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raivio TL, Silhavy TJ. Periplasmic stress and ECF sigma factors. Annual review of microbiology. 2001;55:591–624. doi: 10.1146/annurev.micro.55.1.591. [DOI] [PubMed] [Google Scholar]
- 23.Heinrich J, Wiegert T. YpdC determines site-1 degradation in regulated intramembrane proteolysis of the RsiW anti-sigma factor of Bacillus subtilis. Molecular microbiology. 2006;62:566–579. doi: 10.1111/j.1365-2958.2006.05391.x. [DOI] [PubMed] [Google Scholar]
- 24.Ellermeier CD, Losick R. Evidence for a novel protease governing regulated intramembrane proteolysis and resistance to antimicrobial peptides in Bacillus subtilis. Genes & development. 2006;20:1911–1922. doi: 10.1101/gad.1440606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heinrich J, Hein K, Wiegert T. Two proteolytic modules are involved in regulated intramembrane proteolysis of Bacillus subtilis RsiW. Molecular microbiology. 2009;74:1412–1426. doi: 10.1111/j.1365-2958.2009.06940.x. [DOI] [PubMed] [Google Scholar]
- 26.Schobel S, Zellmeier S, Schumann W, Wiegert T. The Bacillus subtilis sigmaW antisigma factor RsiW is degraded by intramembrane proteolysis through YluC. Molecular microbiology. 2004;52:1091–1105. doi: 10.1111/j.1365-2958.2004.04031.x. [DOI] [PubMed] [Google Scholar]
- 27.Ho TD, Ellermeier CD. PrsW is required for colonization, resistance to antimicrobial peptides, and expression of extracytoplasmic function sigma factors in Clostridium difficile. Infection and immunity. 2011;79:3229–3238. doi: 10.1128/IAI.00019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao M, Wang T, Ye R, Helmann JD. Antibiotics that inhibit cell wall biosynthesis induce expression of the Bacillus subtilis sigma(W) and sigma(M) regulons. Molecular microbiology. 2002;45:1267–1276. doi: 10.1046/j.1365-2958.2002.03050.x. [DOI] [PubMed] [Google Scholar]
- 29.Wiegert T, Homuth G, Versteeg S, Schumann W. Alkaline shock induces the Bacillus subtilis sigma(W) regulon. Molecular microbiology. 2001;41:59–71. doi: 10.1046/j.1365-2958.2001.02489.x. [DOI] [PubMed] [Google Scholar]
- 30.Cao M, Kobel PA, Morshedi MM, Wu MF, Paddon C, Helmann JD. Defining the Bacillus subtilis sigma(W) regulon: a comparative analysis of promoter consensus search, runoff transcription/macroarray analysis (ROMA), and transcriptional profiling approaches. Journal of molecular biology. 2002;316:443–457. doi: 10.1006/jmbi.2001.5372. [DOI] [PubMed] [Google Scholar]
- 31.Kingston AW, Subramanian C, Rock CO, Helmann JD. A sigmaW-dependent stress response in Bacillus subtilis that reduces membrane fluidity. Molecular microbiology. 2011;81:69–79. doi: 10.1111/j.1365-2958.2011.07679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bramkamp M, Weston L, Daniel RA, Errington J. Regulated intramembrane proteolysis of FtsL protein and the control of cell division in Bacillus subtilis. Molecular microbiology. 2006;62:580–591. doi: 10.1111/j.1365-2958.2006.05402.x. [DOI] [PubMed] [Google Scholar]
- 33.Guariglia-Oropeza V, Helmann JD. Bacillus subtilis sigma(V) confers lysozyme resistance by activation of two cell wall modification pathways, peptidoglycan O-acetylation and D-alanylation of teichoic acids. Journal of bacteriology. 2011;193:6223–6232. doi: 10.1128/JB.06023-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ho TD, Hastie JL, Intile PJ, Ellermeier CD. The Bacillus subtilis extracytoplasmic function sigma factor sigma(V) is induced by lysozyme and provides resistance to lysozyme. Journal of bacteriology. 2011;193:6215–6222. doi: 10.1128/JB.05467-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu YT, Kroos L. Evidence that SpoIVFB is a novel type of membrane metalloprotease governing intercompartmental communication during Bacillus subtilis sporulation. Journal of bacteriology. 2000;182:3305–3309. doi: 10.1128/jb.182.11.3305-3309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Resnekov O, Alper S, Losick R. Subcellular localization of proteins governing the proteolytic activation of a developmental transcription factor in Bacillus subtilis. Genes to cells : devoted to molecular & cellular mechanisms. 1996;1:529–542. doi: 10.1046/j.1365-2443.1996.d01-262.x. [DOI] [PubMed] [Google Scholar]
- 37.Cutting S, Driks A, Schmidt R, Kunkel B, Losick R. Forespore-specific transcription of a gene in the signal transduction pathway that governs Pro-sigma K processing in Bacillus subtilis. Genes & development. 1991;5:456–466. doi: 10.1101/gad.5.3.456. [DOI] [PubMed] [Google Scholar]
- 38.Gomez M, Cutting S, Stragier P. Transcription of spoIVB is the only role of sigma G that is essential for pro-sigma K processing during spore formation in Bacillus subtilis. Journal of bacteriology. 1995;177:4825–4827. doi: 10.1128/jb.177.16.4825-4827.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cutting S, Oke V, Driks A, Losick R, Lu S, Kroos L. A forespore checkpoint for mother cell gene expression during development in B. subtilis. Cell. 1990;62:239–250. doi: 10.1016/0092-8674(90)90362-i. [DOI] [PubMed] [Google Scholar]
- 40.Rudner DZ, Losick R. A sporulation membrane protein tethers the pro-sigmaK processing enzyme to its inhibitor and dictates its subcellular localization. Genes & development. 2002;16:1007–1018. doi: 10.1101/gad.977702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Campo N, Rudner DZ. A branched pathway governing the activation of a developmental transcription factor by regulated intramembrane proteolysis. Molecular cell. 2006;23:25–35. doi: 10.1016/j.molcel.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 42.Campo N, Rudner DZ. SpoIVB and CtpB are both forespore signals in the activation of the sporulation transcription factor sigmaK in Bacillus subtilis. Journal of bacteriology. 2007;189:6021–6027. doi: 10.1128/JB.00399-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou R, Kroos L. Serine proteases from two cell types target different components of a complex that governs regulated intramembrane proteolysis of pro-sigmaK during Bacillus subtilis development. Molecular microbiology. 2005;58:835–846. doi: 10.1111/j.1365-2958.2005.04870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prince H, Zhou R, Kroos L. Substrate requirements for regulated intramembrane proteolysis of Bacillus subtilis pro-sigmaK. Journal of bacteriology. 2005;187:961–971. doi: 10.1128/JB.187.3.961-971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou R, Cusumano C, Sui D, Garavito RM, Kroos L. Intramembrane proteolytic cleavage of a membrane-tethered transcription factor by a metalloprotease depends on ATP. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:16174–16179. doi: 10.1073/pnas.0901455106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Green DH, Cutting SM. Membrane topology of the Bacillus subtilis pro-sigma(K) processing complex. Journal of bacteriology. 2000;182:278–285. doi: 10.1128/jb.182.2.278-285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hinz AJ, Larson DE, Smith CS, Brun YV. The Caulobacter crescentus polar organelle development protein PodJ is differentially localized and is required for polar targeting of the PleC development regulator. Molecular microbiology. 2003;47:929–941. doi: 10.1046/j.1365-2958.2003.03349.x. [DOI] [PubMed] [Google Scholar]
- 48.Viollier PH, Sternheim N, Shapiro L. Identification of a localization factor for the polar positioning of bacterial structural and regulatory proteins. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:13831–13836. doi: 10.1073/pnas.182411999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen JC, Hottes AK, McAdams HH, McGrath PT, Viollier PH, Shapiro L. Cytokinesis signals truncation of the PodJ polarity factor by a cell cycle-regulated protease. The EMBO journal. 2006;25:377–386. doi: 10.1038/sj.emboj.7600935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen JC, Viollier PH, Shapiro L. A membrane metalloprotease participates in the sequential degradation of a Caulobacter polarity determinant. Molecular microbiology. 2005;55:1085–1103. doi: 10.1111/j.1365-2958.2004.04443.x. [DOI] [PubMed] [Google Scholar]
- 51.Makinoshima H, Glickman MS. Regulation of Mycobacterium tuberculosis cell envelope composition and virulence by intramembrane proteolysis. Nature. 2005;436:406–409. doi: 10.1038/nature03713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sklar JG, Makinoshima H, Schneider JS, Glickman MS. M. tuberculosis intramembrane protease Rip1 controls transcription through three anti-sigma factor substrates. Molecular microbiology. 2010;77:605–617. doi: 10.1111/j.1365-2958.2010.07232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mukherjee P, Sureka K, Datta P, Hossain T, Barik S, Das KP, Kundu M, Basu J. Novel role of Wag31 in protection of mycobacteria under oxidative stress. Molecular microbiology. 2009;73:103–119. doi: 10.1111/j.1365-2958.2009.06750.x. [DOI] [PubMed] [Google Scholar]
- 54.Deretic V, Schurr MJ, Boucher JC, Martin DW. Conversion of Pseudomonas aeruginosa to mucoidy in cystic fibrosis: environmental stress and regulation of bacterial virulence by alternative sigma factors. Journal of bacteriology. 1994;176:2773–2780. doi: 10.1128/jb.176.10.2773-2780.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chitnis CE, Ohman DE. Genetic analysis of the alginate biosynthetic gene cluster of Pseudomonas aeruginosa shows evidence of an operonic structure. Molecular microbiology. 1993;8:583–593. doi: 10.1111/j.1365-2958.1993.tb01602.x. [DOI] [PubMed] [Google Scholar]
- 56.DeVries CA, Ohman DE. Mucoid-to-nonmucoid conversion in alginate-producing Pseudomonas aeruginosa often results from spontaneous mutations in algT, encoding a putative alternate sigma factor, and shows evidence for autoregulation. Journal of bacteriology. 1994;176:6677–6687. doi: 10.1128/jb.176.21.6677-6687.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martin DW, Holloway BW, Deretic V. Characterization of a locus determining the mucoid status of Pseudomonas aeruginosa: AlgU shows sequence similarities with a Bacillus sigma factor. Journal of bacteriology. 1993;175:1153–1164. doi: 10.1128/jb.175.4.1153-1164.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qiu D, Eisinger VM, Rowen DW, Yu HD. Regulated proteolysis controls mucoid conversion in Pseudomonas aeruginosa. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:8107–8112. doi: 10.1073/pnas.0702660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Damron FH, Yu HD. Pseudomonas aeruginosa MucD regulates the alginate pathway through activation of MucA degradation via MucP proteolytic activity. Journal of bacteriology. 2011;193:286–291. doi: 10.1128/JB.01132-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wood LF, Ohman DE. Use of cell wall stress to characterize sigma 22 (AlgT/U) activation by regulated proteolysis and its regulon in Pseudomonas aeruginosa. Molecular microbiology. 2009;72:183–201. doi: 10.1111/j.1365-2958.2009.06635.x. [DOI] [PubMed] [Google Scholar]
- 61.Wood LF, Leech AJ, Ohman DE. Cell wall-inhibitory antibiotics activate the alginate biosynthesis operon in Pseudomonas aeruginosa: Roles of sigma (AlgT) and the AlgW and Prc proteases. Molecular microbiology. 2006;62:412–426. doi: 10.1111/j.1365-2958.2006.05390.x. [DOI] [PubMed] [Google Scholar]
- 62.Boucher JC, Martinez-Salazar J, Schurr MJ, Mudd MH, Yu H, Deretic V. Two distinct loci affecting conversion to mucoidy in Pseudomonas aeruginosa in cystic fibrosis encode homologs of the serine protease HtrA. Journal of bacteriology. 1996;178:511–523. doi: 10.1128/jb.178.2.511-523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Draper RC, Martin LW, Beare PA, Lamont IL. Differential proteolysis of sigma regulators controls cell-surface signalling in Pseudomonas aeruginosa. Molecular microbiology. 2011;82:1444–1453. doi: 10.1111/j.1365-2958.2011.07901.x. [DOI] [PubMed] [Google Scholar]
- 64.King-Lyons ND, Smith KF, Connell TD. Expression of hurP, a gene encoding a prospective site 2 protease, is essential for heme-dependent induction of bhuR in Bordetella bronchiseptica. Journal of bacteriology. 2007;189:6266–6275. doi: 10.1128/JB.00629-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Testerman TL, Vazquez-Torres A, Xu Y, Jones-Carson J, Libby SJ, Fang FC. The alternative sigma factor sigmaE controls antioxidant defences required for Salmonella virulence and stationary-phase survival. Molecular microbiology. 2002;43:771–782. doi: 10.1046/j.1365-2958.2002.02787.x. [DOI] [PubMed] [Google Scholar]
- 66.Humphreys S, Stevenson A, Bacon A, Weinhardt AB, Roberts M. The alternative sigma factor, sigmaE, is critically important for the virulence of Salmonella typhimurium. Infection and immunity. 1999;67:1560–1568. doi: 10.1128/iai.67.4.1560-1568.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Muller C, Bang IS, Velayudhan J, Karlinsey J, Papenfort K, Vogel J, Fang FC. Acid stress activation of the sigma(E) stress response in Salmonella enterica serovar Typhimurium. Molecular microbiology. 2009;71:1228–1238. doi: 10.1111/j.1365-2958.2009.06597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Matson JS, DiRita VJ. Degradation of the membrane-localized virulence activator TcpP by the YaeL protease in Vibrio cholerae. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:16403–16408. doi: 10.1073/pnas.0505818102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Matson JS, Withey JH, DiRita VJ. Regulatory networks controlling Vibrio cholerae virulence gene expression. Infection and immunity. 2007;75:5542–5549. doi: 10.1128/IAI.01094-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.An FY, Clewell DB. Identification of the cAD1 sex pheromone precursor in Enterococcus faecalis. Journal of bacteriology. 2002;184:1880–1887. doi: 10.1128/JB.184.7.1880-1887.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.An FY, Sulavik MC, Clewell DB. Identification and characterization of a determinant (eep) on the Enterococcus faecalis chromosome that is involved in production of the peptide sex pheromone cAD1. Journal of bacteriology. 1999;181:5915–5921. doi: 10.1128/jb.181.19.5915-5921.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chandler JR, Dunny GM. Characterization of the sequence specificity determinants required for processing and control of sex pheromone by the intramembrane protease Eep and the plasmid-encoded protein PrgY. Journal of bacteriology. 2008;190:1172–1183. doi: 10.1128/JB.01327-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Denham EL, Ward PN, Leigh JA. Lipoprotein signal peptides are processed by Lsp and Eep of Streptococcus uberis. Journal of bacteriology. 2008;190:4641–4647. doi: 10.1128/JB.00287-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Saito A, Hizukuri Y, Matsuo E, Chiba S, Mori H, Nishimura O, Ito K, Akiyama Y. Post-liberation cleavage of signal peptides is catalyzed by the site-2 protease (S2P) in bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:13740–13745. doi: 10.1073/pnas.1108376108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Frank KL, Barnes AM, Grindle SM, Manias DA, Schlievert PM, Dunny GM. Use of recombinase-based in vivo expression technology to characterize Enterococcus faecalis gene expression during infection identifies in vivo-expressed antisense RNAs and implicates the protease Eep in pathogenesis. Infection and immunity. 2012;80:539–549. doi: 10.1128/IAI.05964-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang X, Chen G, Qin C, Wang Y, Wei D. Slr0643, an S2P homologue, is essential for acid acclimation in the cyanobacterium Synechocystis sp. PCC 6803. Microbiology. 2012;158:2765–2780. doi: 10.1099/mic.0.060632-0. [DOI] [PubMed] [Google Scholar]
- 77.Chen K, Gu L, Xiang X, Lynch M, Zhou R. Identification and characterization of five intramembrane metalloproteases in Anabaena variabilis. Journal of bacteriology. 2012;194:6105–6115. doi: 10.1128/JB.01366-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dunny GM, Johnson CM. Regulatory circuits controlling enterococcal conjugation: lessons for functional genomics. Current opinion in microbiology. 2011;14:174–180. doi: 10.1016/j.mib.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dunny GM, Antiporta MH, Hirt H. Peptide pheromone-induced transfer of plasmid pCF10 in Enterococcus faecalis: probing the genetic and molecular basis for specificity of the pheromone response. Peptides. 2001;22:1529–1539. doi: 10.1016/s0196-9781(01)00489-2. [DOI] [PubMed] [Google Scholar]