Abstract
To exist in a wide range of environmental niches, bacteria must sense and respond to a myriad of external signals. A primary means by which this occurs is through two-component signal transduction pathways, typically comprised of a histidine kinase that receives the input stimuli and a response regulator that effects an appropriate change in cellular physiology. Histidine kinases and response regulators have an intrinsic modularity that separates signal input, phosphotransfer, and output response; this modularity has allowed bacteria to dramatically expand and diversify their signaling capabilities. Recent work has begun to reveal the molecular basis by which two-component proteins evolve. How and why do orthologous signaling proteins diverge? How do cells gain new pathways and recognize new signals? What changes are needed to insulate a new pathway from existing pathways? What constraints are there on gene duplication and lateral gene transfer? Here, we review progress made in answering these questions, highlighting how the integration of genome sequence data with experimental studies is providing major new insights.
Keywords: two-component signal transduction, evolution, duplication and divergence, cross-talk, histidine kinase, response regulator, lateral gene transfer
Overview
Two-component signal transduction systems are a predominant means by which bacteria sense and respond to their environments. These systems are generally comprised of a receptor histidine kinase that senses a specific signal and translates that input into a desired output through the phosphorylation of a cognate response regulator. The success of two-component signaling as a strategy for coupling changes in the environment to changes in cellular physiology is underscored by their prevalence throughout the bacterial kingdom. These signaling proteins have been found in the genomes of nearly all sequenced bacteria, with the majority of species encoding dozens, and sometimes hundreds, of two-component proteins. They have been uncovered in countless genetic screens and shown to respond to an enormous range of signals and stressors (for reviews, see (42; 77)).
While tremendous progress has been made in understanding the structure and function of some individual systems, two additional aspects of these pathways have garnered significant interest. First, how does a single cell coordinate so many highly-related signaling pathways? The kinases and regulators encoded by a given organism are often very similar, yet cells are able to match specific inputs with the desired output. How is unwanted cross-talk avoided? Do cells leverage the similarity of these proteins to integrate signals or diversify responses? The second, related, area of interest centers on understanding the evolution of these systems. How do two-component pathways evolve and how are new pathways introduced? How are new pathways insulated from one another? And finally, how do new pathways lead to new functions and new signaling capabilities?
Here we review recent progress in tackling these questions, focusing in particular on the evolution of two-component signaling proteins. Gene duplication and lateral gene transfer provide the raw materials for producing new pathways and, in either case, the introduction of new signaling proteins requires a flurry of changes if the new proteins are to be maintained over the course of evolution. The new pathway must gain a new function to provide a selective advantage and to warrant maintenance in the genome. Domain shuffling likely plays a critical role and recent work has begun to reveal how, at a mechanistic level, this process occurs. New pathways must also avoid cross-talk with other pathways, and vice versa, leading to changes in the specificity determinants of these pathways at multiple levels, including receptor dimerization and kinase-substrate partnering.
Although much of what is known about the evolution of two-component signaling is based on sequence and phylogenetic analyses, there are new efforts to integrate experimental approaches. Throughout the review we will highlight ways in which computational, phylogenetic, and experimental studies are being combined to provide new insights and we highlight areas ripe for future investigation. We begin with a brief review of the structure and function of two-component signaling; for more comprehensive reviews of the mechanistic aspects of two-component signaling, see (12; 24; 25; 77; 85).
The two-component signal transduction paradigm
The eponymous two-component signaling pathway contains a sensor histidine kinase and a cognate response regulator (Fig. 1a). Upon receipt of a stimulus, the histidine kinase typically catalyzes an autophosphorylation reaction on a conserved histidine residue. This phosphoryl group is then transferred to a conserved aspartate on a cognate response regulator. Phosphorylation of the regulator usually drives a conformational change that activates its output response, often leading to changes in gene expression. These systems thus represent versatile, powerful ways to couple changes in external or environmental conditions to corresponding changes in cellular physiology and gene expression. In most cases, histidine kinases are bifunctional such that, when not stimulated to autophosphorylate, they act as phosphatases for their cognate response regulators; thus it is ultimately the ratio of kinase to phosphatase activity that is responsible for modulating the output response (36; 93). In some cases, input signals may promote the phosphatase state rather than stimulating autophosphorylation (65).
Figure 1. Overview of two-component signal transduction.
(a) In the prototypical two-component pathway (left), the catalytic and ATPase (CA) domain of a histidine kinase binds ATP and autophosphorylates a conserved histidine in the dimerization and histidine phosphotransferase (DHp) domain. The phosphoryl group is then transferred to an aspartate in the receiver domain (RD) of the cognate response regulator, activating its output domain to effect cellular changes, frequently through changes in transcription. In a phosphorelay (right), a hybrid histidine kinase autophosphorylates and transfers its phosphoryl group intramolecularly to a receiver domain. A histidine phosphotransferase (HPT) then shuttles the phosphoryl group to a soluble response regulator that effects a pathway output. (b) Common domain organizations of histidine kinases and response regulators. For histidine kinases, the DHp and CA domains are shown with common intracellular domains, PAS, HAMP, and GAF. Note that some kinases have multiple copies of such domains. Two transmembrane domains (TM) are shown on the kinases, but kinases can harbor from 1-13 TM domains. A wide range of sensory domains (not shown) are often found in the periplasmic portions of membrane-bound histidine kinases. For response regulators, the conserved receiver domain is shown alone or with the common output domains, a DNA-binding domain (DBD), a AAA+ and DBD, a GGDEF domain involved in cyclic-di-GMP synthesis, or a CheB-like methyltransferase domain.
All histidine kinases contain two highly conserved domains, the dimerization and histidine phosphotransfer (DHp) domain harboring the conserved histidine that is the site of both the autophosphorylation and phosphotransfer reactions, and the catalytic and ATP-binding (CA) domain. Histidine kinases also usually contain at least one, and often several, additional domains N-terminal to the DHp domain (Fig. 1b). For the vast majority of kinases this includes 1–13 transmembrane domains (20) with signal recognition occurring primarily in the periplasmic or extracellular portion of the protein. Although some common domains have been noted, signal recognition domains tend to be more variable than the other domains. Most kinases also have at least one domain between the transmembrane and DHp domains, with PAS, HAMP, and GAF domains by far the most common (23). These domains can either relay signals from the periplasmic sensory domains to the DHp and CA domains or, in some cases, directly recognize cytoplasmic signals (52; 57).
Response regulators share a common, well-conserved receiver domain (RD) that catalyzes phosphotransfer from its cognate histidine kinase. Phosphorylation then promotes a conformational change in one face of the receiver domain, which in turn effects an output (24). In single domain response regulators, the conformational change in the receiver domain allows the protein to directly produce an output response. Most response regulators, however, contain a DNA binding output domain (21) (Fig. 1b). For these regulators, phosphorylation induces homodimerization of the receiver domain, stimulating DNA binding and leading to transcriptional changes. Other common output domains include diguanylate cyclases and methyltransferases.
A common variant of the two-component paradigm is the so-called phosphorelay (8) (Fig. 1a). These extended pathways typically initiate with a hybrid kinase, which is a histidine kinase with a receiver domain fused to its C-terminus. After autophosphorylation and an intramolecular phosphotransfer to the receiver domain, the phosphoryl group is shuttled to a histidine phosphotransferase, and from there to a terminal response regulator that effects an output. Nearly 25% of all histidine kinases are hybrids (15) suggesting that phosphorelays are common.
Evolution of genome content and gene number
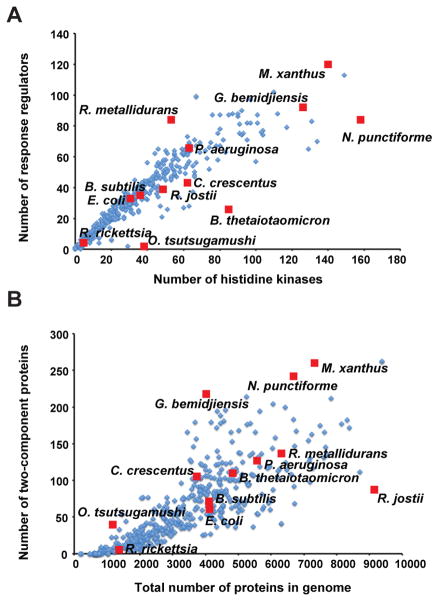
Two-component signaling proteins are among the most prevalent genes in bacteria and histidine kinases and response regulators constitute the two largest paralogous gene families in bacteria (20). Both kinases and regulators are easily identified by sequence homology, in contrast to many eukaryotic signaling systems in which protein kinases are easily identified but their substrates are not. Many histidine kinases are encoded in the same operon as their cognate regulators, allowing for cognate pairs to be identified through sequence analysis (Fig. 2a). Census-taking is thus straightforward and easily applied to fully sequenced bacterial genomes (Fig. 2a). Such analyses have revealed that the total number of two-component genes per genome typically grows as a square of the genome size (20) (Fig. 2b). In addition, the number of two-component genes appears to correlate strongly with ecological and environmental niche (1; 20; 23; 38). Bacteria that live primarily in constant environments typically encode relatively few two-component signaling genes, even taking into account their smaller genome sizes and characteristic reductive genome evolution. In the extreme, many obligate intracellular parasites and endosymbionts harbor only a few pathways or sometimes none at all, as with Mycoplasma and Amoebophilus. By contrast, bacteria that inhibit rapidly changing or diverse environments typically encode large numbers of these signaling proteins. Extreme cases include Myxococcus xanthas with 136 histidine kinases and 127 response regulators and Nostoc punctiforme with 160 kinases and 98 regulators (81) (Fig. 2). In some species, nearly 3% of the genome encodes for histidine kinases alone (20). These patterns of gene content strongly suggest that organisms expand their set of two-component signaling genes to help adapt to fluctuations in their environment.
Figure 2. Diversity of two-component signaling gene content in bacterial genomes.
(a) Plot showing the number of histidine kinases and response regulators in a range of organisms. Generally, most genomes contain equal numbers of kinases and regulators, as pathways typically comprise a kinase and one cognate regulator. When the ratio is not 1:1, there are usually more kinases than regulators, suggesting that response regulators may sometimes integrate signals from multiple kinases. (b) Plot showing the number of two-component proteins as a function of genome size for the same organisms as in panel (a). Each plot is based on 504 bacterial genomes (22). A handful of well-studied and notable species are marked with red squares.
Although most abundant in the genomes of Gram-negative bacteria and cyanobacteria, two-component signaling genes are found in all three domains of life (38; 71). However, they are considerably less abundant in archaea and eukaryotes. The majority of systems found in eukaryotes involve hybrid kinases and phosphorelays; whether there is selective pressure against canonical systems is unknown. Many of the archaeal and eukaryotic systems likely originated through multiple, independent lateral gene transfers from bacteria (37; 38); plants likely gained two-component pathways through the integration of chloroplast genes into the nuclear genome (48). In plants, the two-component genes obtained through lateral transfer likely expanded through duplication and diversification and now play integral roles in diverse developmental pathways (69).
While two-component genes are found in yeasts, filamentous fungi, slime molds, and plants, they are conspicuously absent from higher eukaryotes and metazoans. The absence of two-component signaling proteins from humans, combined with their well-documented role in bacterial pathogenesis (28; 49), has made these proteins attractive new targets for antibiotic development (30). Evolutionarily, their absence begs the question as to why they were supplanted as the primary means of signaling by pathways employing serine, threonine, and tyrosine phosphorylation. Although a definitive answer is lacking, we speculate that the intrinsic lability of phosphoryl groups on aspartates may have contributed. In eukaryotes, a need for longer, more stable outputs may have been desirable, and perhaps necessary, for transmitting signals from the cell membrane to the nucleus without signal loss en route in the form of phosphoryl group hydrolysis. Consistent with this idea, many of the two-component pathways in eukaryotes do not regulate transcription and instead target other cytoplasmic proteins. For example, in S. cerevisiae, the Sln1-Ypd1-Ssk1 phosphorelay modulates the activity of a MAP kinase pathway that is also located in the cell membrane (61). Nevertheless, there are some cases of eukaryotic response regulators that directly affect transcription, particularly in plants. However, in these cases a histidine phosphotransferase typically shuttles phosphoryl groups from a cytoplasmic hybrid histidine kinase to a response regulator in the nucleus that is constitutively associated with the DNA (31; 34). Signal transmission may be successful in these cases because a histidyl-phosphate moiety is considerably more stable than an aspartyl-phosphate.
Where did two-component signaling pathways, in any organism, evolve from in the first place? Given their ancient origin, an unequivocal answer to this question may not be attainable. However, one clue is that histidine kinases share distant homology in their ATP-binding domains to Hsp90, the mismatch repair protein MutL, and type II topoisomerases (17; 18). These proteins, members of the so-called GHKL superfamily, are thought to bind ATP in similar ways and share significant structural similarities; in some cases this domain is used to drive ATP hydrolysis, while in the case of histidine kinases the gamma phosphoryl group is transferred to a histidine in the DHp domain. It is thus plausible that histidine kinases emerged from one of these ATPases. In contrast to histidine kinases, there are no such weak homologies for response regulators and their origin remains a mystery.
There are likely two sources of histidine phosphotransferases. Some, particularly those that are monomeric (80; 91), may have evolved de novo from a range of other proteins, as there are few structural and sequence requirements to function as a histidine phosphotransferase beyond a phosphorylatable histidine within an alpha-helical bundle. Others are dimeric and may have evolved through the degeneration of histidine kinases. For example, B. subtilis Spo0B has two domains with significant similarity to those in histidine kinases (83; 94). The domain that contains the crucial histidine is structurally similar to the DHp domain of histidine kinases, while the other is topologically and structurally similar to a CA domain but lacks key residues usually involved in ATP binding. A similar scenario of recruitment and degeneration of a histidine kinase may hold for the phosphotransferase ChpT in C. crescentus (7). In general, however, evolutionary analysis of histidine phosphotransferases has been limited by the difficulty of identifying these proteins from sequence alone, in contrast to histidine kinases and response regulators.
Mechanisms for evolving changes in two-component signaling gene content
Given the prevalence of two-component signaling pathways in bacterial genomes, it is natural to ask how new proteins and pathways arise. The possibilities fall into two broad categories: gene duplication and divergence, sometimes also referred to as lineage-specific expansion, and lateral (or horizontal) gene transfer (LGT). To assess the contributions made by these two mechanisms, one study systematically examined the origins of histidine kinases from 207 genomes, using BLAST to identify the closest homologs of each kinase (1). For those most closely related to a kinase within the same genome, gene duplication, or lineage-specific expansion, was inferred as the source. If the closest homolog was from a closely related species, and if a gene tree built from all homologs matched a species tree, the kinase was classified as ancient and vertically transmitted. If, however, the closest homolog for a given kinase was from a distantly related species, lateral gene transfer was invoked. This interpretation assumes that multiple gene losses is less parsimonious and hence less likely to have occurred. However, gene loss occurs at very high rates in bacteria. In addition, inferences of lateral transfer can be confounded by the inaccuracy of sequence-based distances and heterotachy, the notion that substitution rates in different lineages often vary significantly (41).
Nevertheless, lateral gene transfer of two-component pathways undoubtedly has occurred and these systematic studies provide a general sense of the frequency, both across all species and within individual genomes (1). Overall, lineage-specific expansion, or gene duplication, appears to explain the origin of the vast majority of kinases. However, the relative balance of duplication and lateral transfer varies substantially from species to species. For example, in Streptomyces coelicolor, essentially all of its 140 histidine kinases appear to be ancient or derived from lineage-specific expansions. By contrast, in Pseudomonas syringae and Ralstonia solanacearum, many of the recently derived kinases probably came from lateral transfer events.
The lateral transfer of genes in bacteria can occur in several ways, including through phage and plasmids, by direct conjugation, or by competence and the direct uptake of extracellular DNA. There are examples of two-component signaling genes encoded on plasmids, such as the VanR-VanS system found in Enterococci that senses and responds to vancomycin (2; 89). In R. solanacearum, many of the laterally-derived histidine kinases are encoded on a megaplasmid that may have moved laterally (70). There are also cases of two-component signaling proteins encoded on pathogeneicity islands, such as the SpiR-SsrB system in Salmonella, which frequently move through conjugation (16). However, for many chromosomally-encoded two-component genes derived by lateral transfer, the mechanism of transfer remains difficult to infer.
It should also be emphasized that both gene duplication and lateral transfer events have occurred more frequently than suggested by phylogenetic analysis. However, in most cases the newly introduced genes were likely eliminated from the genome, and thus are no longer present in extant species. Bacteria typically have high rates of gene loss through mutation and deletion. Indeed, histidine kinases and response regulators are among the most common pseudogenes present in bacterial genomes (45); these pseudogenes likely arose through relatively recent duplications or lateral transfers, and were then inactivated, but have not yet been removed from the genome. To be fixed in a population, duplicated or laterally transferred genes must provide a substantial selective advantage within a relatively short period of time, as gene loss and pseudogeneization occur rapidly in bacteria (33; 40).
The function of a particular two-component system can also influence its evolutionary history. For example, a recent analysis of six species of Xanthomonas compared the complement of signaling genes present in each genome and found extensive gene loss (63). Notably, those pathways involved in Xanthomonas pathogenesis were never lost or duplicated, while other, presumably less critical, pathways experienced more flux. Similarly, in C. crescentus, where two-component signaling proteins play important roles in cell cycle progression and development, those that are essential for viability are highly-conserved in other α-proteobacteria, whereas those that are non-essential in C. crescentus are less well-conserved (74). In most species there is probably a core set of two-component proteins that is maintained and relatively fixed, while an additional set can be lost, or modified, more easily.
This notion of fixed core signaling genes and malleable auxiliary factors has been well-characterized in the context of bacterial chemotaxis, which centers on a two-component pathway, CheA-CheY. In E. coli, where chemotaxis has been best studied, signal recognition requires a methyl-accepting chemoreceptor protein (MCP) and an adaptor protein CheW. Virtually all chemotactic bacteria encode orthologs of these core components: MCP, CheW, CheA, and CheY (90). In contrast, many of the auxiliary components, including the methyltransferase CheR and the methylesterase CheB that influence signal adaptation, are not universally conserved and are often missing or replaced by other types of regulators (90).
Gene fusions, rearrangements, and duplications
Many two-component genes are encoded in operons as cognate kinase-regulator pairs, allowing for the duplication or lateral transfer of an intact signaling pathway. It is rare to see operon shuffling and the mixing and matching of genes encoded in operons. Hence, for a given kinase-regulator pair, the orthologs are also usually found together in an operon and in the same relative order (38; 86). Fusions of kinases and regulators to create hybrid kinases also seem to be rare, but there are some examples. For instance, analysis of six species of Xanthomonas found that the individual domains of a hybrid histidine kinase in one species were most similar to, and likely derived from, an operonic kinase-regulator pair encoded as separate open reading frames in the closely related species. Such fusions probably occur through the mutation of stop codons in operons where the histidine kinase is upstream of the response regulator, although hybrid kinases may also form through the fusion of previously separated genes (63; 86; 96). As might be expected, fusion events that create hybrid kinases are rare for response regulators that contain DNA-binding output domains (15; 96). There are, however, examples of such hybrid kinases (75), but the mechanism by which these systems regulate transcription remains unclear.
Although E. coli encodes 55 of its 62 two-component genes in operons, many organisms encode a substantial fraction of their two-component genes as orphans. Frequently only one gene from an operon will be duplicated (or both are duplicated and one is lost) resulting in the production of orphan two-component signaling genes. An orphan kinase may, however, retain the ability to phosphorylate the regulator in the operon from which it was derived. Such duplication events, coupled with a change in kinase input domain, may be a primary mechanism for generating cross-regulated systems in which multiple, independent signals can trigger the same response. A classic example is in B. subtilis, where the five orphan kinases KinA/B/C/D/E, which likely evolved through duplication, can each phosphorylate Spo0F and initiate the sporulation phosphorelay (76). Similarly, duplication of just a response regulator can lead to a scenario in which a single sensor kinase can drive multiple outputs. For example, in cyanobacteria NblS-RpaB forms an essential two-component system. During divergence of the cyanobacteria in the clade including Synechococcus species, a duplication of RpaB produced a second response regulator SrrA. This regulator retained the ability to be phosphorylated by NblS, but appears to affect transcription in a different manner than RpaB (46).
Evolution of signaling protein structure and function
Gene duplication and lateral transfer ultimately provide the raw material for generating new two-component signaling pathways. But what happens immediately after new signaling genes are introduced? Due to large population sizes and selective pressure to minimize genome size (50), new signaling proteins presumably must quickly gain new functions to be retained. There are undoubtedly many mutations that must occur to produce a pathway that can respond to a new input or effect a new output. These mutations presumably include single amino acid substitutions, although rapid changes in function may rely heavily on larger scale rearrangements such as domain shuffling. Below we summarize our current understanding of how cells generate new signaling functions from duplicated genes, focusing on (i) changes in kinase sensory domains and pathway inputs, (ii) changes in response regulators and pathway outputs, and (iii) changes required to insulate new pathways from existing pathways.
Histidine kinase sensory domain evolution
After the duplication of a histidine kinase, whether alone or with a cognate response regulator, the duplicate histidine kinases must differentiate themselves and find new roles within the signaling network of a cell. One mechanism to accomplish this is through changes in the sensory domains of one or both kinases (14; 39). For most orthologous kinases, the sensory domains are less well-conserved than their catalytic domains. The ability to sense a new signal often arises via domain shuffling, which may occur coincident with, or shortly after, a duplication. Over 70% of recently duplicated histidine kinases show a different input domain structure than that of their closest paralog (1). Domain shuffling can occur between histidine kinases and other proteins. Sequence analyses indicate that the sensory domains of some histidine kinases are closely related to domains found on other types of proteins, including serine/threonine kinases (99), chemotaxis proteins, and diguanylate cyclases (97).
The domain shuffling observed in histidine kinases suggests that these proteins are intrinisically modular and, consequently, that the rational design of new kinases may be possible. Indeed, several groups have successfully fused the conserved phosphotransfer and catalytic domains from a histidine kinase to the sensory domain of another kinase, or even the sensory domain of completely unrelated proteins. The first such example, dubbed Taz, was a chimeric protein that fused the sensory domain of the aspartate chemoreceptor Tar with the DHp and CA domains of the model histidine kinase EnvZ, producing an aspartate-responsive kinase (82). In addition to demonstrating the fundamental modularity of histidine kinases, the Taz protein has been used to dissect the functions and activities of EnvZ in vivo (19; 36; 98). Other functional chemoreceptor-EnvZ constructs have also been made (4; 68).
How does domain shuffling, either during evolution or during rational construction of chimeric proteins, produce successful, signal-responsive proteins? Is there a particular way in which sensory domains must be fused to the catalytic domains to function? This question was recently examined in the context of a chimeric protein that fused a light-sensing PAS domain, taken from the B. subtilis protein YtvA (which is not a histidine kinase), with the DHp and CA domains of the histidine kinase FixL from Bradyrhizobium japonicum. Successful fusions of the PAS domain to FixL led to light-responsive changes in FixL signaling and FixL-FixJ-dependent gene expression (51). Intriguingly, successful fusions had linkers, which form coiled coils, separating the PAS and DHp domains that differed in length by exactly seven amino acids. Inspection of other histidine kinases containing PAS domains further revealed that the linkers are of variable lengths, but often differ by multiples of seven. Together, these results suggest that maintaining the heptad periodicity of the coiled-coil linker may be critical to the construction of functional chimeras, either during evolution or for rational engineering purposes. Further work demonstrated that, by following similar rules, multiple PAS domains could be engineered into the same kinase, allowing it to integrate multiple signals (53). Naturally occurring histidine kinases also often have multiple input domains, suggesting that partial gene duplications, where only a single input domain is duplicated, may be a common mechanism for generating input diversity. In sum, these efforts to engineer novel proteins are not only producing valuable tools, but are providing important new insights into how domain shuffling occurs and how it contributes to the origin of new two-component signaling pathways in nature.
An additional mechanism for acquiring new input signals is through accumulated substitutions in a sensory domain rather than its complete replacement. A prime example comes from the NarX and NarQ sensor kinases in E. coli. A gene duplication event led to the emergence of these two related kinases, although which is more ancestral is unclear. Nevertheless, studies of signal recognition have demonstrated that NarQ responds to both nitrate and nitrite whereas NarX responds preferentially to nitrate (64). Although the periplasmic domains of NarQ and NarX are significantly diverged, they do share substantial similarity, particularly in a region critical to ligand binding (13). Notably, a single point mutation in this region of NarX that substitutes a lysine with an isoleucine, as found at the equivalent position in NarQ, reduced the ability of NarX to discriminate between nitrate and nitrite, rendering a more NarQ-like response pattern (87). This study highlights how the accumulation of single point mutations is a plausible means of rapidly generating new and different inputs to two-component signaling pathways.
Divergence and evolution of pathway outputs
Within a two-component signaling pathway, the response regulator is the ultimate arbiter of physiological change. How does the output of a response regulator evolve, and how are new output responses generated by response regulators after they emerge through duplication or following lateral transfer? As the majority of response regulators direct changes in gene expression, the evolution of pathway outputs can be easily studied by following changes in target genes.
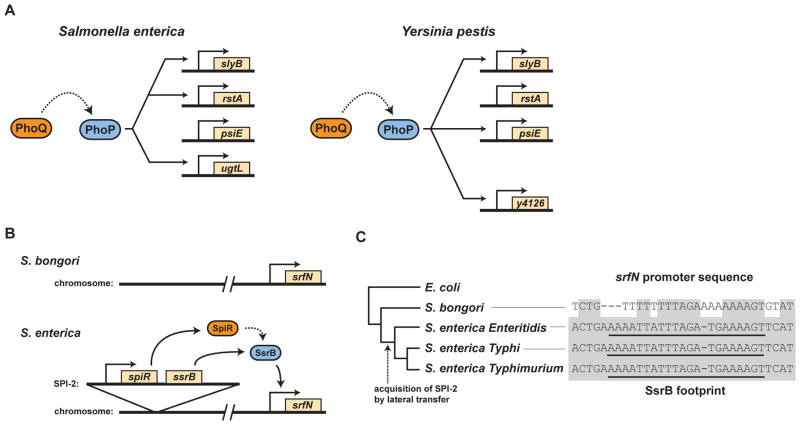
One of the best-studied examples is the PhoQ-PhoP system found in the Enterobacteriaceae. In response to low extracellular concentrations of Mg++, the histidine kinase PhoQ drives phosphorylation of PhoP, which then regulates gene expression. The direct regulon of PhoP has been mapped in both Salmonella enterica serovar Typhimurium and Yersinia pestis (60), which probably shared a common ancestor ~200 million years ago. Strikingly, only three genes were directly regulated by PhoP in both species: the autoregulated phoQ and phoP genes and slyB, which encodes a lipoprotein thought to be a critical regulator of PhoQ activity (Fig. 3a). There were also some genes, such as pbgP and ugd, that were directly regulated in one species, but indirectly regulated in the other; the overall regulatory logic for these genes was thus conserved, but the precise mechanism has changed. Despite these examples, the vast majority of genes directly regulated by PhoP in each organism were not conserved. Instead, transcriptional rewiring appears to have been prevalent since the divergence of Salmonella and Yersinia, leading to the gain and loss of PhoP-regulated genes in each species (Fig. 3a). It is tempting to speculate that these changes have tailored the response of each species to magnesium limitation.
Figure 3. Evolution of transcriptional circuits controlled by two-component pathways.
(a) Examples of genes directly regulated by the two-component pathway PhoQ-PhoP in S. enterica and Y. pestis. The gene slyB is conserved, and directly regulated by PhoP, in both species. The genes rstA and psiE are conserved, but directly regulated by PhoP in only one of the two species. The directly regulated genes ugtL and y4126 are unique to S. enterica and Y. pestis, respectively. (b) Schematic of S. bongori and S. enterica chromosomes, each harboring a srfN ortholog. The horizontally-acquired SpiR-SsrB system, encoded on Salmonella pathogenicity island-2 (SPI-2) in S. enterica but not S. bongori, evolved to transcriptionally activate srfN. (b) De novo evolution of a response regulator binding site. SPI-2 encodes the two-component pathway SpiR-SsrB, which was acquired after the divergence of S. enterica from S. bongori. The gene srfN, ancestral to the Salmonella lineage, accumulated promoter mutations that enabled activation by SsrB, a transcriptional link that contributes to Salmonella virulence. The relevant portion of the srfN promoter is shown with conserved positions shaded grey and the region bound by SsrB in S. enterica underlined.
Notably, the change in PhoP regulons between Salmonella and Yersinia may not always result from a simple gain or loss of PhoP binding sites. In some cases, regulon differences may reflect changes in (i) the orientation of, and distance between, a PhoP binding site and the transcriptional start site and (ii) concomitant changes in how PhoP recruits RNA polymerase. For instance, the promoter of mgtC in Yersinia is located in a position and orientation that enables gene activation by Yersinia PhoP, but not by Salmonella PhoP, despite the fact that Salmonella PhoP can bind the mgtC promoter (59). The ability to change the targets of a response regulator without necessarily changing DNA binding sites is also seen in Desulfovibrio, where two recently duplicated response regulators share DNA binding motifs but regulate non-overlapping target genes (66). Point mutations in OmpR have also been identified which allow it to activate the kdpABC operon, usually activated by KdpE, not by changing DNA binding but by changing the ability to interact with RNA polymerase while bound to the promoter (55). Thus with a single point mutation, and without any changes needed in the promoters of target genes, a duplicated response regulator can regulate a new set of target genes. Collectively, these studies demonstrate that two-component pathway outputs can evolve through changes in the DNA-binding sites of response regulators or through changes in how response regulators interact with RNA polymerase. They also highlight the critical need to couple computational analyses of binding sites with experimental studies to reveal the functional and evolutionary consequences of binding site conservation or loss.
Changes in response regulator outputs may also frequently occur after duplication or lateral transfer events. For gene duplication, changes in the output response of one or both regulators is likely a critical step in the establishment of new functions and, consequently, the maintenance of the duplicated proteins. For instance, in E. coli, a duplication event likely gave rise to the paralogous systems NarX-NarL and NarQ-NarP which respond to nitrate and nitrite in anaerobic conditions (64). While the regulators NarP and NarL share significant similarity and even recognize highly similar consensus binding sites, divergent evolution has enabled each response regulator to recognize different promoter architectures and to activate different genes (62). The duplication of the Nar two-component system has thus led to an increase in complexity of the transcriptional control of genes necessary for growth in anaerobic conditions.
The evolution of response regulator outputs in response to lateral gene transfer has also been explored recently. A particularly illuminating example comes from studies of Salmonella pathogenicity island-2, SPI-2, which encodes a two-component signaling system called SpiR-SsrB (Fig. 3b–c). In addition to regulating the expression of other SPI-2-encoded genes, the response regulator SsrB directly regulates the expression of genes outside SPI-2 (88), indicating that SsrB-binding sites probably evolved de novo within the promoters of these genes. This hypothesis was recently tested by examining the evolution of a Salmonella gene called srfN (56). This gene is ancestral to the Salmonella lineage and present in both S. enterica and S. bongori. By contrast, SPI-2 and SsrB are found in S. enterica but not S. bongori (Fig. 3b). A comparison of the cis-regulatory regions of srfN indicated that the binding site for SsrB was not present in S. bongori meaning it likely arose in the lineage leading to S. enterica (Fig. 3c). Importantly, this recruitment of an ancestral gene into the regulon of a horizontally-acquired response regulator provided S. enterica with an adaptive advantage as a pathogen. When the promoter of S. enterica srfN was replaced with that found in S. bongori, cells were rendered significantly less virulent compared to the wild-type.
Conversely, the genes encoded on SPI-2 have evolved to be regulated by ancestral two-component pathways. A case in point is the expression of ssrB and spiR, which are themselves regulated by OmpR and PhoP, two response regulators found throughout the γ-proteobacteria (6; 44). By controlling spiR and ssrB, these ancestral regulators likely help to ensure that virulence genes are maximally expressed when Salmonella enters host cells. For instance, the PhoQ-PhoP system is activated by the low magnesium conditions that Salmonella experiences inside host macrophages; the consequent activation of ssrB and spiR would then drive the expression of virulence genes.
Although we have only discussed a few cases, it is clear that response regulator outputs can, and do, change rapidly. The changes to transcriptional circuitry observed suggest that bacteria are resilient to, and capable of, transcriptional rewiring (58). This notion was recently tested systematically by artificially rewiring transcriptional connections; promoters for 26 different sigma and transcription factors (including some response regulators) were combined with the open reading frames of 23 of these transcriptional regulators and introduced into E. coli cells on a high-copy plasmid (35). Strikingly, over 95% of these constructs, many of which led to substantial transcriptional rewiring, were tolerated, with little to no growth defect under standard laboratory conditions. One implication of this study is that after a new DNA-binding response regulator is introduced by gene duplication or lateral transfer, there is time to “scan” different regulatory possibilities. A new combination that yields even a slight benefit could then be selected and rapidly fixed in a population. Finally, the evolvability of response regulators and their outputs may also benefit from the fact that most prokaryotic transcription factors regulate only a few genes, either directly or indirectly (47), decreasing the number of binding sites that would need to co-evolve with the DNA-binding domain of a response regulator, thereby increasing the likelihood that they can change (67).
Evolution of phosphotransfer specificity and the insulation of pathways
The flow of information through two-component signaling pathways depends critically on the transfer of phosphoryl groups from a histidine kinase to its cognate response regulator. Despite early suggestions of rampant cross-talk, there is little evidence for such promiscuity in vivo with most kinases having one response regulator substrate, or occasionally two or three (43; 74; 92). This in vivo preference is mirrored in vitro, with histidine kinases harboring a strong kinetic preference for phosphotransfer to their in vivo partner. For example, a systematic, global study of phosphotransfer from E. coli EnvZ to each of the 32 response regulators in E. coli demonstrated that OmpR was the preferred substrate. EnvZ only transferred to other substrates after extended incubation times (74). These in vitro studies demonstrate that the specificity of two-component signaling pathways is based primarily on molecular recognition rather than relying on scaffolds or other cellular strategies. This observation further suggests that the information necessary for promoting the “correct”, or desired, interaction and preventing “incorrect” interactions is encoded at the sequence level (10; 73).
A consequence of relying on molecular recognition for specificity is that, during the course of evolution, any mutation in a residue contributing to a kinase-regulator interaction may disrupt signaling and place cells at a strong fitness disadvantage. Survival would then depend on reversion of the mutation or a compensatory mutation in the partner protein. Consistently, computational analyses of large sets of cognate kinase-regulator pairs have revealed extensive amino acid coevolution (9; 10; 73; 84). Conspicuously, the most significantly coevolving pairs of residues map to the molecular interface formed during phosphotransfer (11) suggesting they mediate the specificity of this protein-protein interaction (Fig. 4a-b). Using E. coli EnvZ as a model kinase, a subset of these residues was shown to be sufficient, when mutated, to reprogram substrate specificity both in vitro and in vivo (73). For example, mutating as few as three residues in EnvZ to match those found at equivalent positions in RstB led EnvZ to preferentially phosphorylate RstA instead of OmpR (Fig. 4a-b, also see Fig. 5b). Similarly, response regulators, including OmpR from E. coli and CheY from R. sphaeroides, have been rationally rewired to interact with non-cognate kinases by mutating the coevolving, specificity-determining residues (5; 10). Directed evolution has also been used to rewire two-component specificity. For example, mutants of the E. coli kinase CpxA were selected that phosphorylate and dephosphorylate OmpR; many of the mutated residues were also identified in the studies of kinase-regulator coevolution (72).
Figure 4. Amino-acid coevolution in two-component signaling proteins.
(a) Residues that coevolve in cognate pairs of histidine kinases and response regulators are shown with spacefilling on the crystal structure of the T. maritima kinase TM0853, shown in blue, bound to its cognate regulator TM0468, shown in green. The histidine and the aspartate that are involved in phosphotransfer are shown in purple. Coevolving residues on the histidine kinase and response regulator are shown with spacefilling and colored orange and red, respectively. Residues in histidine kinases that coevolve strongly with other kinase residues are shown with spacefilling and colored cyan. (b–c) Coevolving residues from panel (a) are shown on a sequence alignment of TM0853 with three E. coli kinases, EnvZ, RstB, and CpxA (b), and an alignment of TM0468 with three E. coli regulators, OmpR, RstA, and CpxR (c). Highly conserved residues are shaded grey. Secondary structure elements are indicated beneath the primary sequence.
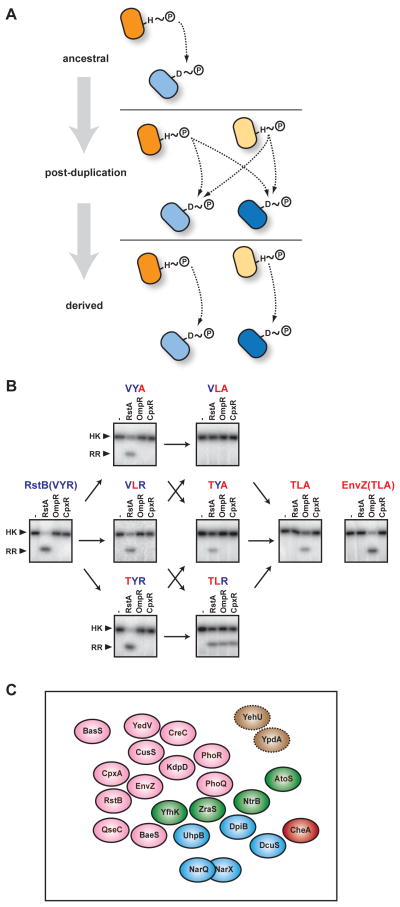
Figure 5. Insulation of two-component pathways following gene duplication.
(a) Schematic of major steps in the insulation of two pathways following a duplication event. The duplication of an ancestral pathway initially produces two identical pathways that cross-talk at the level of phosphotransfer. Through the accumulation of mutations in specificity-determining residues, the two pathways can become insulated. A similar process must occur, but is not shown, at the levels of kinase and regulator homodimerization. (b) Phosphotransfer specificity of EnvZ, RstB, and various RstB mutants. Each kinase was autophosphorylated and tested for transfer to each of three response regulators, RstA, OmpR, and CpxR. Data are from (10). The wild-type RstB is shown at far left. The phosphotransfer specificity can be converted to that of EnvZ, shown at far right, by mutating three of its six specificity- determining residues to match those found in EnvZ (the other three sites are already identical between EnvZ and RstB; see Fig. 4b). This triple mutant of RstB as well as each single and double mutant intermediate are labeled based on the identity of the three specificity residues with blue text indicating identity with the wild-type RstB and red indicating identity with the wild-type EnvZ. Notably, some intermediates do not phosphorylate any of the regulators whereas some phosphorylate all three. (c) Schematic summarizing the distribution of histidine kinases in the sequence space defined by their specificity-determining residues. Each sphere represents the set of response regulators that a given kinase phosphorylates. With the exception of NarQ and NarX, these spheres are presented as non-overlapping to reflect the minimal cross-talk between pathways. The relative positions of spheres is based on the ability of individual kinases to phosphorylate the cognate response regulators of other kinases after extended times in vitro (74; 92). Positions are approximate and the diagram is only intended to convey a general sense of how kinases are distributed in sequence space. Spheres are colored according to the subfamily of each kinase’s response regulator: red, CheY/receiver domain only; pink, OmpR/winged helix-turn-helix; green, NtrC/AAA+ and FIS domains; blue, NarL/GerE helix-turn-helix; brown, LytR. Spheres with dashed outlines indicate kinases for which no data existed to infer relative positions. Note, hybrid histidine kinases are excluded.
Although specificity-determining residues do coevolve, these correlated changes appear to be rare events as the specificity residues of many kinase-regulator systems are nearly invariant over relatively long time scales. So when and why do specificity residues change, and coevolve? One strong possibility is that substitutions occur following gene duplication, helping to insulate the duplicate kinase-regulator pairs from each other (Fig. 5a). That is, a series of mutations presumably must occur to prevent cross-talk between two duplicated pathways, while maintaining the interaction within each pair. Such an accumulation of changes in specificity residues is, however, inherently risky business for a bacterium. Due to large population sizes, even slightly deleterious mutations are likely to be quickly removed from the population. Hence, for a new kinase-regulator pair to be maintained in the genome, the mutational intermediates between its initial state and its final, insulated state must be neutral, or nearly neutral. In other words, cognate kinase-regulator pairs must retain their ability to interact as the specificity residues coevolve and find a region of sequence space in which they are insulated from other two-component proteins within the cell. Similarly, after a lateral gene transfer event involving two-component signaling genes, the newly introduced kinase-regulator pair may need to accumulate substitutions in phosphotransfer specificity residues to avoid cross-talk with existing systems, thereby maintaining the fidelity of information flow within the cell.
What mutational trajectories do two-component proteins follow during evolution? In particular, how do duplicated proteins move through the sequence space defined by the specificity-determining residues of histidine kinases and response regulators? Answering these questions through sequence analysis alone is problematic as transient intermediates may not be captured in extant sequences and the behavior of ancestral or intermediate states is difficult to infer from sequence alone. To circumvent these issues, one recent study experimentally examined all possible specificity intermediates between two E. coli histidine kinases, EnvZ and RstB (10). These kinases likely arose through duplication and divergence, and, as noted above, the specificity of each can be converted to that of the other by just three mutations (10; 73) (Fig. 5b). Similarly, their cognate response regulators, OmpR and RstA, can be rewired, with respect to partner specificity, through a small number of mutations. It was thus feasible to build all possible mutational intermediates and characterize their phosphotransfer specificity in vitro.
The results of this systematic study demonstrated that a cognate kinase-regulator pair can, in fact, move in sequence space from the region occupied by EnvZ-OmpR to that of RstB-RstA while (i) introducing only one mutation at a time, (ii) maintaining the interaction between the kinase and the regulator, and (iii) avoiding the introduction of cross-talk to other closely related pathways such as CpxA-CpxR (10) (Fig. 5b). Notably though, only a small fraction of all possible mutational trajectories satisfy these criteria, indicating that the evolution of signaling proteins post-duplication may be fundamentally constrained. These studies further suggest that kinase-regulator pairs occupying relatively isolated regions of specificity sequence space may be easier to duplicate and retain. In addition, for cases where a severely limited number of trajectories are accessible, there may be cases of convergent evolution in which independent duplication events have led to similar mutational trajectories.
The notion of insulation, or orthogonality, in sequence space can be extended from individual, recently duplicated pairs of signaling proteins to the entire complement of two-component signaling proteins in a given organism. For example, all 29 histidine kinases in E. coli ultimately arose through some combination of gene duplication and lateral transfer. The net result is a system of signaling pathways that are, with a few exceptions, insulated from one another in sequence space and with respect to phosphotransfer, as observed by global phosphotransfer profiling (Fig. 5c). This system-wide insulation suggests that negative selection and the avoidance of cross-talk are powerful forces influencing the evolution of two-component signaling proteins. Negative selection has been suggested to influence other paralogous signaling protein families, such as SH3-domain-containing proteins found in eukaryotes (95). One notable exception to the orthogonality of phosphotransfer specificity in E. coli is the kinases NarQ and NarX, which share significant similarity in terms of phosphotransfer specificity residues and, consistently, both phosphorylate the response regulators NarL and NarP, although with different kinetic preferences (54).
Much remains to be understood about how kinase-regulator interactions evolve. Has the distribution of signaling proteins in specificity sequence space been optimized? How dense is sequence space and how does this impact mutational trajectories, both in the absence of duplication and post-duplication? Is there a fundamental limit to the number of pathways an organism can have while avoiding cross-talk? Does gene loss lead to relaxed selection on the specificity residues of the remaining signaling proteins? Answering many of these questions will benefit from a systematic mutational analysis of kinase and regulator specificity residues. Most of what is known about kinase-regulator interactions comes from limited alanine-scanning or targeted mutagenesis (10; 79); the application of new, systematic mutagenesis techniques (32) promises to shed significant new light on the specificity and evolution of phosphotransfer in two-component signaling.
Dimerization specificity
The generation of new pathways by duplication and divergence also requires changes to the residues that mediate homodimerization of histidine kinases and response regulators. To establish new and insulated pathways, substitutions are needed that eliminate heterodimerization of the diverging paralogous proteins while maintaining homodimerization.
Most, if not all, histidine kinases form homodimers in order to autophosphorylate. There is almost no evidence of physiologically-relevant heterodimerization, with one exception in Pseudomonas aeruginosa (29), indicating that histidine kinases harbor a set of amino acids that enforce homodimerization. Many of these residues are likely to reside in the DHp domain, although upstream domains, such as PAS and HAMP domains, could also contribute to dimerization specificity and stability. To better pinpoint the residues mediating specificity, one recent study looked for coevolving residues in a set of more than 15,000 histidine kinase sequences (3). As with the kinase-regulator interaction, this approach revealed a small set of strongly coevolving residues that mapped primarily to the DHp domain and mostly within the lower half of the four-helix bundle. These dimerization specificity residues map to the same general region as the phosphotransfer specificity residues, but with the dimerization specificity residues buried in the four-helix bundle and kinase-regulator specificity residues in solvent-exposed positions (Fig. 4). As with kinase-regulator interactions, homodimerization specificity could be changed through directed mutagenesis of these homodimerization specificity residues (3).
Nearly 50% of response regulators form homodimers upon phosphorylation, including all members of the OmpR family (26). Homodimerization is often crucial for producing an output response as many response regulators have DNA-binding domains and recognize tandem or inverted repeat elements within target promoters. A systematic study of the 17 OmpR-family response regulators from E. coli demonstrated that essentially all of them specifically homodimerize (27). Although intermolecular interactions on the dimer interface involves highly conserved residues within the receiver domain, some interfacial residues do vary, perhaps providing a mechanism for ensuring homodimerization and excluding heterodimerization (78). As with kinase dimerization and kinase-regulator interaction, amino acid coevolution studies have identified a subset of interfacial residues that may help enforce homodimerization and prevent heterodimerization (84). These residues are likely to change following gene duplication as a means of insulating paralogous response regulators from one another, thereby enabling distinct outputs to result from the phosphorylation of each regulator.
Final perspective
Bacteria can survive and thrive in a bewildering array of environments and in the face of enormous competition. They have, consequently, evolved elegant mechanisms for sensing and responding to their environments, often involving two-component signal transduction pathways. The prevalence and diversity of these signaling proteins in the bacterial kingdom underscores the roles they have played in the adaptation of bacteria to a range of ecological niches. Studies of how two-component signaling pathway evolve are only in their infancy, but promise to be an active area of exploration in the coming years. The diversity and depth of genome sequences, coupled with the ability to experimentally manipulate bacteria, should help to further reveal how, at a detailed molecular level, these pathways are created, lost, rearranged, and integrated into complex regulatory circuits.
Summary Points.
Two-component signal transduction proteins are prevalent throughout the bacterial kingdom and found in some archaea, plants, and lower eukaryotes, but were lost in metazoans.
Most bacteria encode dozens, and sometimes hundereds, of these signaling proteins. The number of proteins encoded in individual genomes typically scales with genome size and diversity of the environment in which organisms live.
Bacteria expand their repertoires of two-component signaling proteins through a combination of gene duplication and lateral gene transfer.
Domain shuffling is rampant among two-component signaling proteins, particularly after gene duplication events, enabling the rapid gain of new sensory and regulatory functions.
Studies of how domain shuffling occurs at the molecular level are enabling the rational design of new sensor kinases.
The transcriptional outputs of two-component signaling pathways show enormous plasticity with gain and loss of cis-regulatory elements often driving rapid diversification of a response regulator’s transcriptional program.
Cognate histidine kinases and response regulators coevolve to maintain their interaction and to avoid cross-talk with other pathways. Studies of amino-acid coevolution in large sets of cognate kinase-regulator pairs have revealed the key specificity-determining residues.
Specificity residues may change to accomodate the emergence of new pathways that arise through duplication or lateral transfer; the reconstruction of ancestral and intermediate states of specificity residues in model kinase-regulator pairs is shedding new light on the mutational trajectories that occurred and the constraints that influenced them.
Acknowledgments
Support was provided by the National Institutes of Health and the National Science Foundation. M.T.L is an Early Career Scientist at the Howard Hughes Medical Institute.
Mini-glossary
- autophosphorylation
A reaction in which a protein kinase covalently attaches the gamma phosphoryl group from ATP to itself; for histidine kinases, the phosphoryl group is transferred to a conserved histidine residue
- phosphotransfer
The transfer of a phosphoryl group from an autophosphorylated histidine kinase to a conserved aspartate on a response regulator
- phosphatase
Some histidine kinases stimulate the dephosphorylation of a phosphorylated response regulator and are therefore deemed bifunctional as they have both autokinase and phosphatase activity
- phosphorelay
A variant of the canonical two-component signaling pathway in which a phosphoryl group is transferred successively from a histidine kinase to a response regulator to a histidine phosphotransferase, and finally to a second, terminal, response regulator containing an output domain
- cross-talk
Detrimental communication between two different signaling pathways
- cross-regulation
Communication between distinct signaling pathways that provides a physiological benefit to the organism
- gene duplication
A process which produces two copies of a gene or set of genes
- vertical inheritence
A process in which genetic material is transmitted from an organism to its offspring or progeny
- lineage-specific expansion
The growth of a paralogous gene family through duplication and subsequent vertical inheritence of the duplicated genes
- lateral (horizontal) gene transfer
A process in which genetic material is transferred from one organism to another, but not through vertical inheritence
Ackronyms
- HK
histidine kinase
- RR
response regulator
- HPT
histidine phosphotransferase
- DHp
dimerization and histidine phosphotransfer
- CA
catalytic and ATPase
- PAS
Per Arnt Sim
- GAF
cGMP-specific phosphodiesterases, adenylyl cyclases and FhlA
- HAMP
histidine kinases, adenyl cyclases, methyl-accepting proteins and phosphatases
- RD
receiver domain
Literature Cited
- 1.Alm E, Huang K, Arkin A. The evolution of two-component systems in bacteria reveals different strategies for niche adaptation. PLoS Comput Biol. 2006;2:e143. doi: 10.1371/journal.pcbi.0020143. Computational inference of lateral transfer and duplication rates of histidine kinase genes across 207 genomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur M, Molinas C, Courvalin P. The VanS-VanR two-component regulatory system controls synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1992;174:2582–91. doi: 10.1128/jb.174.8.2582-2591.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashenberg O, Rozen-Gagnon K, Laub MT, Keating AE. Determinants of homodimerization specificity in histidine kinases. J Mol Biol. 2011;413:222–35. doi: 10.1016/j.jmb.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumgartner JW, Kim C, Brissette RE, Inouye M, Park C, Hazelbauer GL. Transmembrane signalling by a hybrid protein: communication from the domain of chemoreceptor Trg that recognizes sugar-binding proteins to the kinase/phosphatase domain of osmosensor EnvZ. J Bacteriol. 1994;176:1157–63. doi: 10.1128/jb.176.4.1157-1163.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell CH, Porter SL, Strawson A, Stuart DI, Armitage JP. Using structural information to change the phosphotransfer specificity of a two-component chemotaxis signalling complex. PLoS Biol. 2010;8:e1000306. doi: 10.1371/journal.pbio.1000306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bijlsma JJ, Groisman EA. The PhoP/PhoQ system controls the intramacrophage type three secretion system of Salmonella enterica. Mol Microbiol. 2005;57:85–96. doi: 10.1111/j.1365-2958.2005.04668.x. [DOI] [PubMed] [Google Scholar]
- 7.Biondi EG, Reisinger SJ, Skerker JM, Arif M, Perchuk BS, et al. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature. 2006;444:899–904. doi: 10.1038/nature05321. [DOI] [PubMed] [Google Scholar]
- 8.Burbulys D, Trach KA, Hoch JA. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell. 1991;64:545–52. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- 9.Burger L, van Nimwegen E. Accurate prediction of protein-protein interactions from sequence alignments using a Bayesian method. Mol Syst Biol. 2008;4:165. doi: 10.1038/msb4100203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Capra EJ, Perchuk BS, Lubin EA, Ashenberg O, Skerker JM, Laub MT. Systematic dissection and trajectory-scanning mutagenesis of the molecular interface that ensures specificity of two-component signaling pathways. PLoS Genet. 2010;6:e1001220. doi: 10.1371/journal.pgen.1001220. Experimental analysis of mutational trajectories separating extant two-component signaling pathways. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casino P, Rubio V, Marina A. Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell. 2009;139:325–36. doi: 10.1016/j.cell.2009.08.032. The first crystal structure of a histidine kinase in complex with its cognate response regulator. [DOI] [PubMed] [Google Scholar]
- 12.Casino P, Rubio V, Marina A. The mechanism of signal transduction by two-component systems. Curr Opin Struct Biol. 2010;20:763–71. doi: 10.1016/j.sbi.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 13.Cheung J, Hendrickson WA. Structural analysis of ligand stimulation of the histidine kinase NarX. Structure. 2009;17:190–201. doi: 10.1016/j.str.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheung J, Hendrickson WA. Sensor domains of two-component regulatory systems. Current Opinion in Microbiology. 2010;13:116–23. doi: 10.1016/j.mib.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cock PJ, Whitworth DE. Evolution of prokaryotic two-component system signaling pathways: gene fusions and fissions. Mol Biol Evol. 2007;24:2355–7. doi: 10.1093/molbev/msm170. [DOI] [PubMed] [Google Scholar]
- 16.Deiwick J, Nikolaus T, Erdogan S, Hensel M. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol Microbiol. 1999;31:1759–73. doi: 10.1046/j.1365-2958.1999.01312.x. [DOI] [PubMed] [Google Scholar]
- 17.Dutta R, Inouye M. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem Sci. 2000;25:24–8. doi: 10.1016/s0968-0004(99)01503-0. [DOI] [PubMed] [Google Scholar]
- 18.Dutta R, Qin L, Inouye M. Histidine kinases: diversity of domain organization. Mol Microbiol. 1999;34:633–40. doi: 10.1046/j.1365-2958.1999.01646.x. [DOI] [PubMed] [Google Scholar]
- 19.Dutta R, Yoshida T, Inouye M. The critical role of the conserved Thr247 residue in the functioning of the osmosensor EnvZ, a histidine Kinase/Phosphatase, in Escherichia coli. J Biol Chem. 2000;275:38645–53. doi: 10.1074/jbc.M005872200. [DOI] [PubMed] [Google Scholar]
- 20.Galperin MY. A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol. 2005;5:35. doi: 10.1186/1471-2180-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galperin MY. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J Bacteriol. 2006;188:4169–82. doi: 10.1128/JB.01887-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galperin MY, Higdon R, Kolker E. Interplay of heritage and habitat in the distribution of bacterial signal transduction systems. Mol Biosyst. 2010;6:721–8. doi: 10.1039/b908047c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galperin MY, Nikolskaya AN, Koonin EV. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol Lett. 2001;203:11–21. doi: 10.1111/j.1574-6968.2001.tb10814.x. [DOI] [PubMed] [Google Scholar]
- 24.Gao R, Mack TR, Stock AM. Bacterial response regulators: versatile regulatory strategies from common domains. Trends Biochem Sci. 2007;32:225–34. doi: 10.1016/j.tibs.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao R, Stock AM. Biological insights from structures of two-component proteins. Annu Rev Microbiol. 2009;63:133–54. doi: 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao R, Stock AM. Molecular strategies for phosphorylation-mediated regulation of response regulator activity. Curr Opin Microbiol. 2010;13:160–7. doi: 10.1016/j.mib.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao R, Tao Y, Stock AM. System-level mapping of Escherichia coli response regulator dimerization with FRET hybrids. Mol Microbiol. 2008;69:1358–72. doi: 10.1111/j.1365-2958.2008.06355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gooderham WJ, Hancock RE. Regulation of virulence and antibiotic resistance by two-component regulatory systems in Pseudomonas aeruginosa. FEMS Microbiol Rev. 2009;33:279–94. doi: 10.1111/j.1574-6976.2008.00135.x. [DOI] [PubMed] [Google Scholar]
- 29.Goodman AL, Merighi M, Hyodo M, Ventre I, Filloux A, Lory S. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev. 2009;23:249–59. doi: 10.1101/gad.1739009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gotoh Y, Eguchi Y, Watanabe T, Okamoto S, Doi A, Utsumi R. Two-component signal transduction as potential drug targets in pathogenic bacteria. Curr Opin Microbiol. 2010;13:232–9. doi: 10.1016/j.mib.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 31.Grefen C, Harter K. Plant two-component systems: principles, functions, complexity and cross talk. Planta. 2004;219:733–42. doi: 10.1007/s00425-004-1316-4. [DOI] [PubMed] [Google Scholar]
- 32.Hietpas RT, Jensen JD, Bolon DN. Experimental illumination of a fitness landscape. Proc Natl Acad Sci U S A. 2011;108:7896–901. doi: 10.1073/pnas.1016024108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hooper SD, Berg OG. On the nature of gene innovation: duplication patterns in microbial genomes. Mol Biol Evol. 2003;20:945–54. doi: 10.1093/molbev/msg101. [DOI] [PubMed] [Google Scholar]
- 34.Imamura A, Yoshino Y, Mizuno T. Cellular localization of the signaling components of Arabidopsis His-to-Asp phosphorelay. Biosci Biotechnol Biochem. 2001;65:2113–7. doi: 10.1271/bbb.65.2113. [DOI] [PubMed] [Google Scholar]
- 35.Isalan M, Lemerle C, Michalodimitrakis K, Horn C, Beltrao P, et al. Evolvability and hierarchy in rewired bacterial gene networks. Nature. 2008;452:840–5. doi: 10.1038/nature06847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin T, Inouye M. Ligand binding to the receptor domain regulates the ratio of kinase to phosphatase activities of the signaling domain of the hybrid Escherichia coli transmembrane receptor, Taz1. J Mol Biol. 1993;232:484–92. doi: 10.1006/jmbi.1993.1404. [DOI] [PubMed] [Google Scholar]
- 37.Kim D, Forst S. Genomic analysis of the histidine kinase family in bacteria and archaea. Microbiology. 2001;147:1197–212. doi: 10.1099/00221287-147-5-1197. [DOI] [PubMed] [Google Scholar]
- 38.Koretke KK, Lupas AN, Warren PV, Rosenberg M, Brown JR. Evolution of two-component signal transduction. Mol Biol Evol. 2000;17:1956–70. doi: 10.1093/oxfordjournals.molbev.a026297. [DOI] [PubMed] [Google Scholar]
- 39.Krell T, Lacal J, Busch A, Silva-Jimenez H, Guazzaroni ME, Ramos JL. Bacterial sensor kinases: diversity in the recognition of environmental signals. Annual Review of Microbiology. 2010;64:539–59. doi: 10.1146/annurev.micro.112408.134054. [DOI] [PubMed] [Google Scholar]
- 40.Kuo CH, Ochman H. The extinction dynamics of bacterial pseudogenes. PLoS Genet. 2010:6. doi: 10.1371/journal.pgen.1001050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kurland CG, Canback B, Berg OG. Horizontal gene transfer: a critical view. Proc Natl Acad Sci U S A. 2003;100:9658–62. doi: 10.1073/pnas.1632870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laub MT. The role of two-component signal transduction systems in bacterial stress responses. In: Storz G, Hengge R, editors. Bacterial Stress Responses. Washington, D.C: ASM Press; 2011. [Google Scholar]
- 43.Laub MT, Goulian M. Specificity in two-component signal transduction pathways. Annu Rev Genet. 2007;41:121–45. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- 44.Lee AK, Detweiler CS, Falkow S. OmpR regulates the two-component system SsrA-ssrB in Salmonella pathogenicity island 2. J Bacteriol. 2000;182:771–81. doi: 10.1128/jb.182.3.771-781.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y, Harrison PM, Kunin V, Gerstein M. Comprehensive analysis of pseudogenes in prokaryotes: widespread gene decay and failure of putative horizontally transferred genes. Genome Biol. 2004;5:R64. doi: 10.1186/gb-2004-5-9-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lopez-Redondo ML, Moronta F, Salinas P, Espinosa J, Cantos R, et al. Environmental control of phosphorylation pathways in a branched two-component system. Mol Microbiol. 2010;78:475–89. doi: 10.1111/j.1365-2958.2010.07348.x. [DOI] [PubMed] [Google Scholar]
- 47.Madan Babu M, Teichmann SA. Evolution of transcription factors and the gene regulatory network in Escherichia coli. Nucleic Acids Res. 2003;31:1234–44. doi: 10.1093/nar/gkg210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin W, Rujan T, Richly E, Hansen A, Cornelsen S, et al. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc Natl Acad Sci U S A. 2002;99:12246–51. doi: 10.1073/pnas.182432999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miller SI, Kukral AM, Mekalanos JJ. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc Natl Acad Sci U S A. 1989;86:5054–8. doi: 10.1073/pnas.86.13.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mira A, Ochman H, Moran NA. Deletional bias and the evolution of bacterial genomes. Trends Genet. 2001;17:589–96. doi: 10.1016/s0168-9525(01)02447-7. [DOI] [PubMed] [Google Scholar]
- 51.Moglich A, Ayers RA, Moffat K. Design and signaling mechanism of light-regulated histidine kinases. J Mol Biol. 2009;385:1433–44. doi: 10.1016/j.jmb.2008.12.017. Construction of a light-sensing histidine kinase through rational domain shuffling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moglich A, Ayers RA, Moffat K. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure. 2009;17:1282–94. doi: 10.1016/j.str.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moglich A, Ayers RA, Moffat K. Addition at the molecular level: signal integration in designed Per-ARNT-Sim receptor proteins. J Mol Biol. 2010;400:477–86. doi: 10.1016/j.jmb.2010.05.019. [DOI] [PubMed] [Google Scholar]
- 54.Noriega CE, Lin HY, Chen LL, Williams SB, Stewart V. Asymmetric cross-regulation between the nitrate-responsive NarX-NarL and NarQ-NarP two-component regulatory systems from Escherichia coli K-12. Mol Microbiol. 2010;75:394–412. doi: 10.1111/j.1365-2958.2009.06987.x. Demonstration of cross-regulation between two signaling pathways at the level of phosphotransfer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ohashi K, Yamashino T, Mizuno T. Molecular basis for promoter selectivity of the transcriptional activator OmpR of Escherichia coli: isolation of mutants that can activate the non-cognate kdpABC promoter. J Biochem. 2005;137:51–9. doi: 10.1093/jb/mvi006. [DOI] [PubMed] [Google Scholar]
- 56.Osborne SE, Walthers D, Tomljenovic AM, Mulder DT, Silphaduang U, et al. Pathogenic adaptation of intracellular bacteria by rewiring a cis-regulatory input function. Proc Natl Acad Sci U S A. 2009;106:3982–7. doi: 10.1073/pnas.0811669106. Evidence that promoter mutations led to regulation of an ancestral gene by a horizontally-acquired two-component system. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parkinson JS. Signaling mechanisms of HAMP domains in chemoreceptors and sensor kinases. Annu Rev Microbiol. 2010;64:101–22. doi: 10.1146/annurev.micro.112408.134215. [DOI] [PubMed] [Google Scholar]
- 58.Perez JC, Groisman EA. Evolution of transcriptional regulatory circuits in bacteria. Cell. 2009;138:233–44. doi: 10.1016/j.cell.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perez JC, Groisman EA. Transcription factor function and promoter architecture govern the evolution of bacterial regulons. Proc Natl Acad Sci U S A. 2009;106:4319–24. doi: 10.1073/pnas.0810343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perez JC, Shin D, Zwir I, Latifi T, Hadley TJ, Groisman EA. Evolution of a bacterial regulon controlling virulence and Mg(2+) homeostasis. PLoS Genet. 2009;5:e1000428. doi: 10.1371/journal.pgen.1000428. Mapped the evolution of a response regulator’s regulon from Salmonella to Yersinia. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Posas F, Wurgler-Murphy SM, Maeda T, Witten EA, Thai TC, Saito H. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell. 1996;86:865–75. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- 62.Price MN, Dehal PS, Arkin AP. Horizontal gene transfer and the evolution of transcriptional regulation in Escherichia coli. Genome Biol. 2008;9:R4. doi: 10.1186/gb-2008-9-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qian W, Han ZJ, He C. Two-component signal transduction systems of Xanthomonas spp.: a lesson from genomics. Mol Plant Microbe Interact. 2008;21:151–61. doi: 10.1094/MPMI-21-2-0151. [DOI] [PubMed] [Google Scholar]
- 64.Rabin RS, Stewart V. Dual response regulators (NarL and NarP) interact with dual sensors (NarX and NarQ) to control nitrate- and nitrite-regulated gene expression in Escherichia coli K-12. J Bacteriol. 1993;175:3259–68. doi: 10.1128/jb.175.11.3259-3268.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Raivio TL, Silhavy TJ. Transduction of envelope stress in Escherichia coli by the Cpx two-component system. J Bacteriol. 1997;179:7724–33. doi: 10.1128/jb.179.24.7724-7733.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rajeev L, Luning EG, Dehal PS, Price MN, Arkin AP, Mukhopadhyay A. Systematic mapping of two component response regulators to gene targets in a model sulfate reducing bacterium. Genome Biol. 2011;12:R99. doi: 10.1186/gb-2011-12-10-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rajewsky N, Socci ND, Zapotocky M, Siggia ED. The evolution of DNA regulatory regions for proteo-gamma bacteria by interspecies comparisons. Genome Res. 2002;12:298–308. doi: 10.1101/gr.207502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rampersaud A, Utsumi R, Delgado J, Forst SA, Inouye M. Ca2(+)- enhanced phosphorylation of a chimeric protein kinase involved with bacterial signal transduction. J Biol Chem. 1991;266:7633–7. [PubMed] [Google Scholar]
- 69.Ren B, Liang Y, Deng Y, Chen Q, Zhang J, et al. Genome-wide comparative analysis of type-A Arabidopsis response regulator genes by overexpression studies reveals their diverse roles and regulatory mechanisms in cytokinin signaling. Cell Res. 2009;19:1178–90. doi: 10.1038/cr.2009.88. [DOI] [PubMed] [Google Scholar]
- 70.Salanoubat M, Genin S, Artiguenave F, Gouzy J, Mangenot S, et al. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature. 2002;415:497–502. doi: 10.1038/415497a. [DOI] [PubMed] [Google Scholar]
- 71.Schaller GE, Shiu SH, Armitage JP. Two-component systems and their co-option for eukaryotic signal transduction. Curr Biol. 2011;21:R320–30. doi: 10.1016/j.cub.2011.02.045. [DOI] [PubMed] [Google Scholar]
- 72.Siryaporn A, Perchuk BS, Laub MT, Goulian M. Evolving a robust signal transduction pathway from weak cross-talk. Mol Syst Biol. 2010;6:452. doi: 10.1038/msb.2010.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Skerker JM, Perchuk BS, Siryaporn A, Lubin EA, Ashenberg O, et al. Rewiring the specificity of two-component signal transduction systems. Cell. 2008;133:1043–54. doi: 10.1016/j.cell.2008.04.040. Identified specificity-determining residues in two-component proteins by analyzing amino-acid coevolution and rationally rewiring histidine kinases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Skerker JM, Prasol M, Perchuk BS, Biondi EG, Laub MT. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol. 2005;3:e334. doi: 10.1371/journal.pbio.0030334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sonnenburg ED, Sonnenburg JL, Manchester JK, Hansen EE, Chiang HC, Gordon JI. A hybrid two-component system protein of a prominent human gut symbiont couples glycan sensing in vivo to carbohydrate metabolism. Proc Natl Acad Sci U S A. 2006;103:8834–9. doi: 10.1073/pnas.0603249103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stephenson K, Hoch JA. Evolution of signalling in the sporulation phosphorelay. Mol Microbiol. 2002;46:297–304. doi: 10.1046/j.1365-2958.2002.03186.x. [DOI] [PubMed] [Google Scholar]
- 77.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 78.Toro-Roman A, Wu T, Stock AM. A common dimerization interface in bacterial response regulators KdpE and TorR. Protein Sci. 2005;14:3077–88. doi: 10.1110/ps.051722805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tzeng YL, Hoch JA. Molecular recognition in signal transduction: the interaction surfaces of the Spo0F response regulator with its cognate phosphorelay proteins revealed by alanine scanning mutagenesis. J Mol Biol. 1997;272:200–12. doi: 10.1006/jmbi.1997.1226. [DOI] [PubMed] [Google Scholar]
- 80.Ulrich DL, Kojetin D, Bassler BL, Cavanagh J, Loria JP. Solution structure and dynamics of LuxU from Vibrio harveyi, a phosphotransferase protein involved in bacterial quorum sensing. J Mol Biol. 2005;347:297–307. doi: 10.1016/j.jmb.2005.01.039. [DOI] [PubMed] [Google Scholar]
- 81.Ulrich LE, Zhulin IB. The MiST2 database: a comprehensive genomics resource on microbial signal transduction. Nucleic Acids Res. 2010;38:D401–7. doi: 10.1093/nar/gkp940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Utsumi R, Brissette RE, Rampersaud A, Forst SA, Oosawa K, Inouye M. Activation of bacterial porin gene expression by a chimeric signal transducer in response to aspartate. Science. 1989;245:1246–9. doi: 10.1126/science.2476847. [DOI] [PubMed] [Google Scholar]
- 83.Varughese KI, Madhusudan, Zhou XZ, Whiteley JM, Hoch JA. Formation of a novel four-helix bundle and molecular recognition sites by dimerization of a response regulator phosphotransferase. Mol Cell. 1998;2:485–93. doi: 10.1016/s1097-2765(00)80148-3. [DOI] [PubMed] [Google Scholar]
- 84.Weigt M, White RA, Szurmant H, Hoch JA, Hwa T. Identification of direct residue contacts in protein-protein interaction by message passing. Proc Natl Acad Sci U S A. 2009;106:67–72. doi: 10.1073/pnas.0805923106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.West AH, Stock AM. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem Sci. 2001;26:369–76. doi: 10.1016/s0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- 86.Whitworth DE, Cock PJ. Evolution of prokaryotic two-component systems: insights from comparative genomics. Amino Acids. 2009;37:459–66. doi: 10.1007/s00726-009-0259-2. [DOI] [PubMed] [Google Scholar]
- 87.Williams SB, Stewart V. Discrimination between structurally related ligands nitrate and nitrite controls autokinase activity of the NarX transmembrane signal transducer of Escherichia coli K-12. Mol Microbiol. 1997;26:911–25. doi: 10.1046/j.1365-2958.1997.6262002.x. [DOI] [PubMed] [Google Scholar]
- 88.Worley MJ, Ching KH, Heffron F. Salmonella SsrB activates a global regulon of horizontally acquired genes. Mol Microbiol. 2000;36:749–61. doi: 10.1046/j.1365-2958.2000.01902.x. [DOI] [PubMed] [Google Scholar]
- 89.Wright GD, Holman TR, Walsh CT. Purification and characterization of VanR and the cytosolic domain of VanS: a two-component regulatory system required for vancomycin resistance in Enterococcus faecium BM4147. Biochemistry. 1993;32:5057–63. doi: 10.1021/bi00070a013. [DOI] [PubMed] [Google Scholar]
- 90.Wuichet K, Zhulin IB. Origins and diversification of a complex signal transduction system in prokaryotes. Sci Signal. 2010;3:ra50. doi: 10.1126/scisignal.2000724. Comprehensive survey of the evolution of chemotaxis systems across bacteria. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xu Q, Carlton D, Miller MD, Elsliger MA, Krishna SS, et al. Crystal structure of histidine phosphotransfer protein ShpA, an essential regulator of stalk biogenesis in Caulobacter crescentus. J Mol Biol. 2009;390:686–98. doi: 10.1016/j.jmb.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yamamoto K, Hirao K, Oshima T, Aiba H, Utsumi R, Ishihama A. Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli. J Biol Chem. 2005;280:1448–56. doi: 10.1074/jbc.M410104200. [DOI] [PubMed] [Google Scholar]
- 93.Yang Y, Inouye M. Requirement of both kinase and phosphatase activities of an Escherichia coli receptor (Taz1) for ligand-dependent signal transduction. J Mol Biol. 1993;231:335–42. doi: 10.1006/jmbi.1993.1286. [DOI] [PubMed] [Google Scholar]
- 94.Zapf J, Sen U, Madhusudan, Hoch JA, Varughese KI. A transient interaction between two phosphorelay proteins trapped in a crystal lattice reveals the mechanism of molecular recognition and phosphotransfer in signal transduction. Structure. 2000;8:851–62. doi: 10.1016/s0969-2126(00)00174-x. [DOI] [PubMed] [Google Scholar]
- 95.Zarrinpar A, Park SH, Lim WA. Optimization of specificity in a cellular protein interaction network by negative selection. Nature. 2003;426:676–80. doi: 10.1038/nature02178. [DOI] [PubMed] [Google Scholar]
- 96.Zhang W, Shi L. Distribution and evolution of multiple-step phosphorelay in prokaryotes: lateral domain recruitment involved in the formation of hybrid-type histidine kinases. Microbiology. 2005;151:2159–73. doi: 10.1099/mic.0.27987-0. [DOI] [PubMed] [Google Scholar]
- 97.Zhang Z, Hendrickson WA. Structural characterization of the predominant family of histidine kinase sensor domains. J Mol Biol. 2010;400:335–53. doi: 10.1016/j.jmb.2010.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhu Y, Inouye M. Analysis of the role of the EnvZ linker region in signal transduction using a chimeric Tar/EnvZ receptor protein, Tez1. J Biol Chem. 2003;278:22812–9. doi: 10.1074/jbc.M300916200. [DOI] [PubMed] [Google Scholar]
- 99.Zhulin IB, Nikolskaya AN, Galperin MY. Common extracellular sensory domains in transmembrane receptors for diverse signal transduction pathways in bacteria and archaea. J Bacteriol. 2003;185:285–94. doi: 10.1128/JB.185.1.285-294.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]