Abstract
Growing evidence demonstrates that long non coding RNAs (lncRNAs) play an important role in cancer origination and progression. A novel lncRNA, TSLC1-AS1, is the antisense transcript of tumor suppressor TSLC1. The expression profile and function of TSLC1-AS1 in glioma were investigated using Real-Time Quantitative PCR and siRNA knockdown. The data showed that TSLC1-AS1 expression was down-regulated in tumor tissues compared with that in adjacent normal tissues, and negatively associated with the WHO criteria of the tumors. Overexpression of TSLC1-AS1 resulted in up-regulation of TSLC1 and significant inhibition of cell proliferation, migration and invasion in U87 cells, while knockdown of TSLC1-AS1 in SNB-19 cells showed the opposite effect. The expression of TSLC1-AS1 was also positively correlated with other tumor suppressors NF1, VHL, PIK3R1 and negatively correlated with the oncogene BRAF. The results suggested that TSLC1-AS1 was a tumor suppressor of glioma and a mediator of TSLC1 expression. LncRNA TSLC1-AS1 may serve as a potential biomarker and therapeutic target for glioma.
Keywords: Long non coding RNAs, tumor suppressor, glioma
Introduction
Glioma is the most common malignant tumor in central nervous system [1]. And a complex gene interaction and molecular modulation network is involved in the development of glioma. Increasing number of researches is focused on exploring the molecular modulation network and finding reliable diagnostic markers and effective therapeutic targets [2]. Recent studies revealed that numerous long non coding RNAs (lncRNAs) had extensive regulating activities in different levels of gene expression and crucial biological roles in cellular development and metabolism. And some lncRNA has been functional linked with cancer origination and progression [3-6].
ENST00000546273/RP11-713B9.1, termed TSLC1-AS1, is a novel one of lncRNAs, which is the antisense transcript of a protein coding gene TSLC1. It was found at the location of chromosome 11q23.2, that is frequently deleted in many malignant tumors [7]. Protein coding gene TSLC1 (also known as CADM1, IGSF4, SynCAM, SgIGSF and Necl-2) [8,9] has been identified as the tumor suppressor gene which is often down-regulated by hypermethylation in many tumors, including glioma, lung cancer, neuroblastoma, nasopharyngeal cancer, malignant melanoma, esophageal cancer, liver cancer, breast cancer, gastric cancer, pancreatic cancer, colorectal cancer, cervical cancer and prostate cancer [10-21].
This work aimed to determine the expression of TSLC1-AS1 in glioma, to explore its function and analyze the interaction of it and TSLC1.
Materials and methods
Patient samples
Patients with glioma (n = 46) who underwent initial surgery in Chinese PLA General Hospital from 2010 to 2012 were retrospectively selected for this study. No patients had received therapy before resection. Each patient participated after providing informed consent and the use of the tumor samples for research was approved by the ethical committee of Chinese PLA General Hospital. All tumors were classified on the basis of the WHO criteria for tumors of the central nervous system and quick frozen at the time of resection until analysis. The series consisted of 27 cases of low grade astrocytoma (WHO grade II), and 19 cases of high grade disease [10 cases of anaplastia astrocytoma (WHO grade III) and 9 cases of glioblastoma multiforme (WHO grade IV)].
Cancer cell lines
The human glioma cell lines U87, U251, SNB-19 were purchased from American Type Culture Collection (Manassas, VA, USA). The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) medium (Gibco, Carlsbad, CA) containing 10% fetal bovine serum (Invitrogen, Carlsbad, CA) at 37°C with 5% CO2.
Real-time quantitative PCR
Total RNA was isolated from glioma tumor tissue, adjacent normal tissue and glioma cell lines using the Trizol Total RNA Reagent (Invitrogen, Carlsbad CA). cDNA synthesis was performed with 2 μg total RNA using the RevertAidTM H Minus First Strand cDNA Synthesis Kit (Takara, Ohtsu, Japan). The primers were obtained from GenePharma (Shanghai, China) and the sequences were shown in Table 1. Quantitative PCR was performed using the SYBR PrimeScript RT-PCR kit (Takara, Ohtsu, Japan) in an Applied Biosystems 7500 Fluorescent Quantitative PCR System (Applied Biosystems, Foster City, CA). The reaction mixtures were incubated at 95°C for 30 s, followed by 40 amplification cycles of 95°C for 5 s and 60°C for 34 s. The quantification of gene expression was performed by using the ΔΔCT calculation with CT as the threshold cycle. The expression level of a target gene in a patient was calculated as the ratio: target in tumor tissue/target in nontumorous tissue [R (T/N)].
Table 1.
Primers for real time PCR analysis
| Gene name | Forward | Reverse |
|---|---|---|
| β-actin | 5’-CCACTGGCATCGTGATGGA-3’ | 5’-CGCTCGGTGAGGATCTTCAT-3’ |
| TSLC1-AS1 | 5’-TGACAAAGGCAGGAGGTA-3’ | 5’-GCACTATGGCTGAGGAAA-3’ |
| TSLC1 | 5’-ATGGCGAGTGTAGTGCTGC-3’ | 5’-GATCACTGTCACGTCTTTCGT-3’ |
| NF1 | 5’-AGATGAAACGATGCTGGTCAAA-3’ | 5’-CCTGTAACCTGGTAGAAATGCGA-3’ |
| VHL | 5’-GCAGGCGTCGAAGAGTACG-3’ | 5’-CGGACTGCGATTGCAGAAGA-3’ |
| PIK3R1 | 5’-ACCACTACCGGAATGAATCTCT-3’ | 5’-GGGATGTGCGGGTATATTCTTC-3’ |
| BRAF | 5’-AATACACCAGCAAGCTAGATGC-3’ | 5’-AATCAGTTCCGTTCCCCAGAG-3’ |
Overexpression of TSLC1-AS1 in U87 cells
Plasmid cDNA-TSLC1-AS1 was constructed by introducing aBamHI-EcoRI fragment containing the TSLC1-AS1 cDNA into the same site in pcDNA3.1. The TSLC1-AS1 low expressed U87 cells were transfection with pcDNA-TSLC1-AS1 using Lipofectamine 2000 (Invitrogen, US) according to the manufacturer’s instructions. Cells were collected after transfection for RNA isolation, MTS cell proliferation assay, scratch wound healing assay and matrigel invasion assay.
Transfection of TSLC1-AS1 siRNA in SNB-19 cell line
For small interfering RNA (siRNA) analysis, siRNA for TSLC1-AS1 sequence and negative-control (NC) siRNA were obtained from GenePharma (Shanghai, China). The target sequence of TSLC1-AS1 siRNA was shown in Table 2. Approximately 5% SNB-19 cells were plated to each well of 12-well plates at least 24 h before transfection to achieve 30-50% confluency. SiRNA transfection was done with X-tremeGENE transfection reagent (Roche) according to the manufacturer’s instructions. The TSLC1-AS1 high expressed cell line SNB-19 were harvested or fixed 48 h after transfection for RNA isolation, MTS cell proliferation assay, scratch wound healing assay and matrigel invasion assay.
Table 2.
SiRNA sequences for TSLC1-AS1
| Name | The sequences | |
|---|---|---|
| T-AS1-si1 | Sense strand | 5’-rGrUrArCrCrUrCrCrUrGrCrCrUrUrUrGrUrCrArArGrCrCAA-3’ |
| Antisense strand | 5’-rUrUrGrGrCrUrUrGrArCrArArArGrGrCrArGrGrArGrGrUrArCrArA-3’ | |
| T-AS1-si2 | Sense strand | 5’-rGrArCrCrUrArUrCrGrArGrArArCrUrGrArGrArGrCrGrACA-3’ |
| Antisense strand | 5’-rUrGrUrCrGrCrUrCrUrCrArGrUrUrCrUrCrGrArUrArGrGrUrCrArG-3’ | |
Cell proliferation assay
After transfection, cell proliferation was assessed by MTS assay (Promega) according to the manufacturer’s protocol. SNB-19 cells (2,000 cells per well) in each group were plated in 96-well plates. 20 μl of the MTS reagent was added to each well containing 100 μl culture medium. The plate was incubated for 2 h at 37°C in a humidified, 5% CO2 atmosphere. The plate was then read at 490 nm using a plate reader.
Scratch wound healing assay
Uniform wounds were scraped in SNB-19 epithelial monolayers grown on plastic 6-well plates using a pipette tip before transfection. The initial gap length (0 h) and the residual gap length 24 h after wounding were calculated from photomicrographs.
Matrigel invasion assays
A cell invasion assay was carried out using modified Boyden Chambers consisting of transwell-precoated matrigel membrane filter inserts with 8 mm pores in 24-well tissue culture plates (BD Biosciences, Bedfold, MA, USA). DMEM containing 10% fetal bovine serum in the lower chamber served as the chemoattractant.
Statistical analysis
Differences between groups were analyzed using Student’s t test. Correlation between genes expression was studied by using Pearson’s correlation. Statistical analyses were performed using SPSS version 18.0 (SPSS, Chicago, IL). For all statistical analyses, P < 0.05 was considered statistically significant.
Results
TSLC1-AS1 and TSLC1 were both down-regulated in glioma tissue samples
The TSLC1-AS1 and TSLC1 expression levels were assessed in a panel paired specimen obtained from 46 patients with glioma. Both the expression levels of lncRNA TSLC1-AS1 and mRNA TSLC1 in glioma tumor tissues were significantly down-regulated compared with in matched nontumorous tissues (Figure 1A, 1B). The expression levels of TSLC1-AS1 and TSLC1 were significantly lower in tumors with higher WHO grades (III/IV) than that in tumors with lower WHO grades (II).
Figure 1.
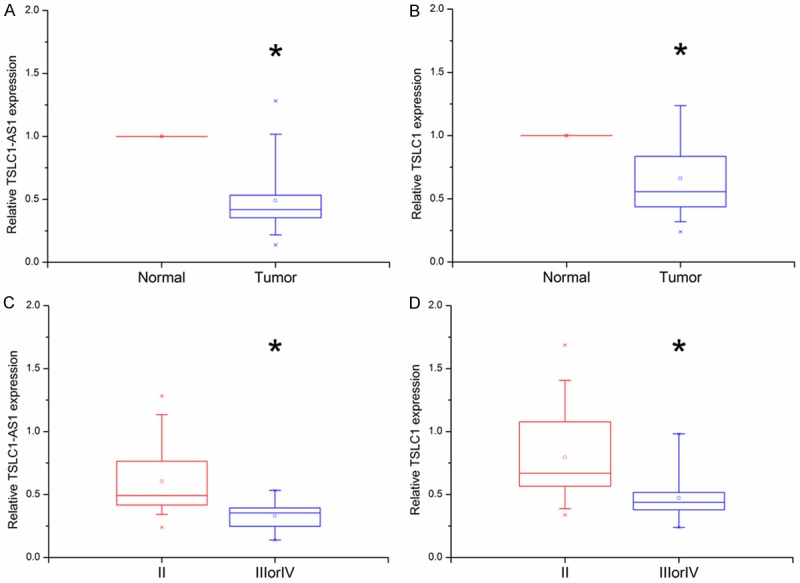
TSLC1-AS1 and TSLC1 expression levels were analyzed by real time PCR in 46 glioma tissue samples. A, B: Both of TSLC1-AS1 and TSLC1 expression levels were significantly lower in tumors tissues than that in normal tissues (*P < 0.05). C, D: TSLC1-AS1 and TSLC1 expression levels were significantly lower in tumors of higher WHO grades (III/IV) than those in tumors of lower WHO grades (II). (*P < 0.05).
Expression of TSLC1-AS1 correlated with that of TSLC1 and other tumor related genes
To identify the relationship between TSLC1-AS1 and TSLC1, we assessed the correlation of their expression levels using real time PCR. The result indicated that the TSLC1-AS1 expression was positively correlated with TSLC1 expression (R = 0.66, P < 0.01, Table 3).
Table 3.
Correlations of TSLC1-AS1 with TSLC1 and other tumor regulating genes in glioma
| Gene name | Correlation coefficients with TSLC1-AS1 |
|---|---|
| TSLC1 | R = 0.66, P < 0.01 |
| NF1 | R = 0.61, P < 0.01 |
| VHL | R = 0.52, P < 0.01 |
| PIK3R1 | R = 0.55, P < 0.01 |
| BRAF | R = -0.51, P < 0.01 |
The expression levels of TSLC1-AS1 and TSLC1 as well as three other cancer-related genes were analyzed by real time PCR in 46 glioma tissue samples. The expression of TSLC1-AS1 was positively correlated with TSLC1, NF1, VHL, PIK3R1 expression (P < 0.01), negatively correlated with BRAF expression (P < 0.01).
Moreover, to test the potential association between TSLC1-AS1 and other cancer regulating genes in glioma, we assessed the expression correlation of TSLC1-AS1 with other three glioma suppressors NF1, VHL, PIK3R1 and an oncogene BRAF. In Table 3, the results showed remarkable positive correlations of TSLC1-AS1 expression with three tumor suppressors, NF1 (R = 0.61, P < 0.01), VHL (R = 0.52, P < 0.01) and PIK3R1 (R = 0.55, P < 0.01), respectively, and negative correlation with oncogene BRAF (R = -0.51, P < 0.01).
TSLC1 expression was co-regulated together with the TSLC1-AS1 overexpression or knockdown in glioma cell lines
By real time PCR, we found that the expression of TSLC1-AS1 was the highest in SNB-19 and the lowest in U87 among the three human glioma cell lines (Figure 2A).
Figure 2.
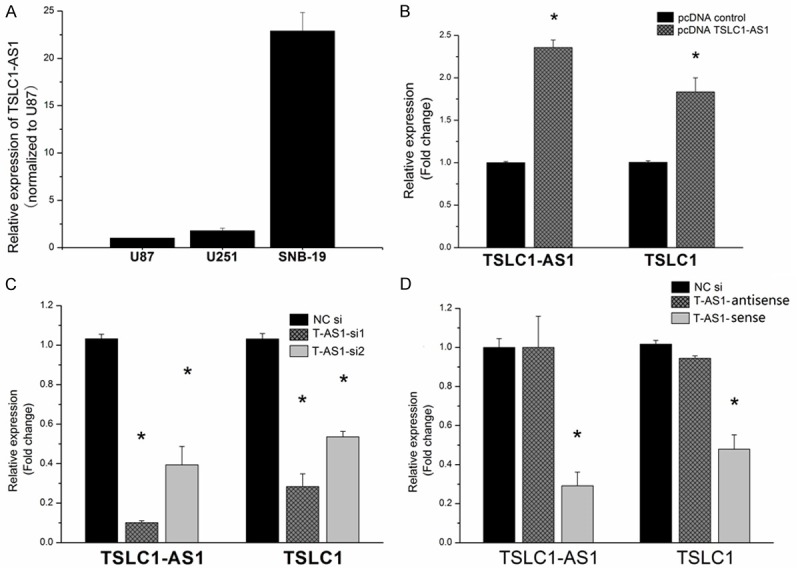
TSLC1 expression was co-regulated together with the TSLC1-AS1 overexpression or knockdown in glioma cell lines. A: The expression of TSLC1-AS1 was detected be qPCR in three glioma cell lines. B: The expression of TSLC1-AS1 and TSLC1 were measured in U87 cell line after transfection by plasmid pcDNA-TSLC1-AS1. TSLC1-AS1 lncRNA expression was markedly elevated by pcDNA-TSLC1-AS1, and TSLC1 mRNA expression level was also evidently up-regulated concomitantly (*P < 0.05). C: TSLC1 mRNA and TSLC1-AS1 lncRNA level knockdown were measured by real-time PCR in SNB-19 cells transfected with TSLC1-AS1 siRNA. Expression of TSLC1-AS1 and TSLC1 were significantly reduced in TSLC1-AS1 siRNA groups compared with negative control group (*P < 0.05). D: Only the sense strand of TSLC1-AS1 siRNA successfully knocked down the expression of both TSLC1-AS1 and TSLC1 (*P < 0.05), while the antisense strand had no effect on the TSLC1 expression (P > 0.05).
In the overexpression experiment, TSLC1-AS1 cDNA plasmid was constructed and stably transfected into U87 cells. TSLC1-AS1 expression was markedly elevated, and TSLC1 expression level was also evidently up-regulated by TSLC1-AS1 cDNA plasmid (Figure 2B).
In the knock-down experiment, TSLC1-AS1 siRNAs was transfected into SNB-19 cells. TSLC1-AS1 expression was markedly decreased. Quantification analysis showed that TSLC1-AS1 expression level was knocked down nearly 90% in TSLC1-AS1 siRNA group and TSLC1 expression level was also down-regulated by TSLC1-AS1 siRNA (Figure 2C). It suggested that TSLC1 expression may be modulated by TSLC1-AS1. Since our TSLC1-AS1 siRNAs were double strands RNAs, the sense or antisense single strand of TSLC1-AS1 siRNAs were transfected respectively into the SNB-19 cells to exclude target effect of TSLC1-AS1 siRNA antisense strand to TSLC1. It was found that only the sense strand of TSLC1-AS1 siRNA successfully knocked down the expression of both TSLC1-AS1 and TSLC1, while the antisense strand had no effect on the TSLC1 expression (Figure 2D).
U87 cell proliferation, migration and invasion were inhibited by TSLC1-AS1 overexpression
To further identify the role of TSLC1-AS1 in U87 cells, functional assay was performed by transfecting plasmid cDNA TSLC1-AS1 and negative control plasmid. The results showed that the U87 cells in pcDNA-TSLC1-AS1 group grew significantly slower compared with the cells in the pcDNA control group (Figure 3A). The wound-healing assay showed remarkable cell migration retardation in pcDNA-TSLC1-AS1 group compared with in the pcDNA control group (Figure 3B). The matrigel invasion assay also showed significant cell invasion inhibition in the pcDNA-TSLC1-AS1 group compared with the pcDNA control group (Figure 3C).
Figure 3.
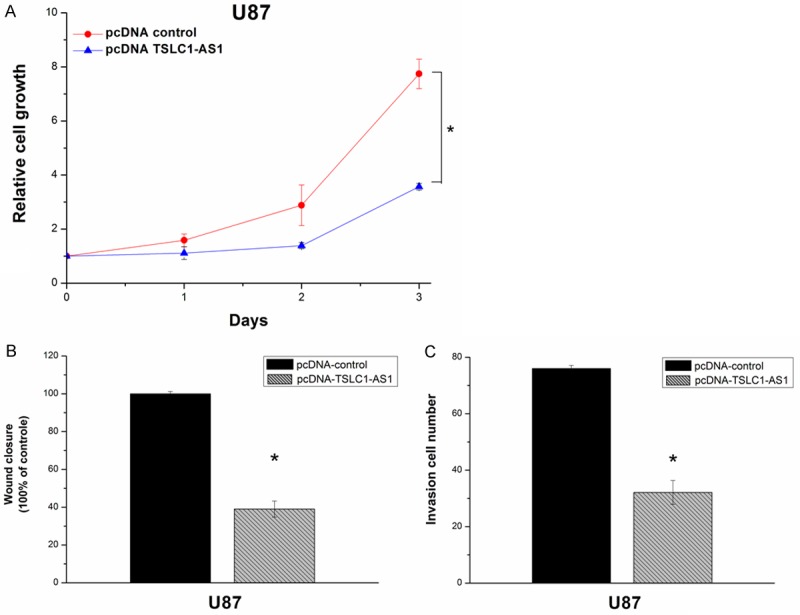
Cell growth, migration and invasion were inhibited by pcDNA-TSLC1-AS1 in U87 cells. A: Cell proliferation fold increase was tested using MTS with 1 day intervals. PcDNA-TSLC1-AS1 group showed decreased growth rates compared with pcDNA control group in U87 cells (*P < 0.05). B, C: Overexpression of TSLC1-AS1 inhibited the cell migration and invasion. Scratch wound healing assay and matrigel invasion assay showed that the cell migration and invasion capacities in pcDNA-TSLC1-AS1 group were remarkably inhibited (*P < 0.05).
SNB-19 cell proliferation, migration and invasion were enhanced by TSLC1-AS1 knockdown
The results showed that the SNB-19 cells in TSLC1-AS1 siRNA transfected groups grew significantly faster compared with the cells in the negative controls group (Figure 4A). The scratch wound healing assay and matrigel invasion assay were performed also using SNB-19 cells. The wound-healing assay showed remarkable cell migration elevation in TSLC1-AS1 siRNA group compared with in the NC group (Figure 4B). The matrigel invasion assay also showed significant cell invasion elevation in the TSLC1-AS1 siRNA groups compared with the NC group (Figure 4C).
Figure 4.
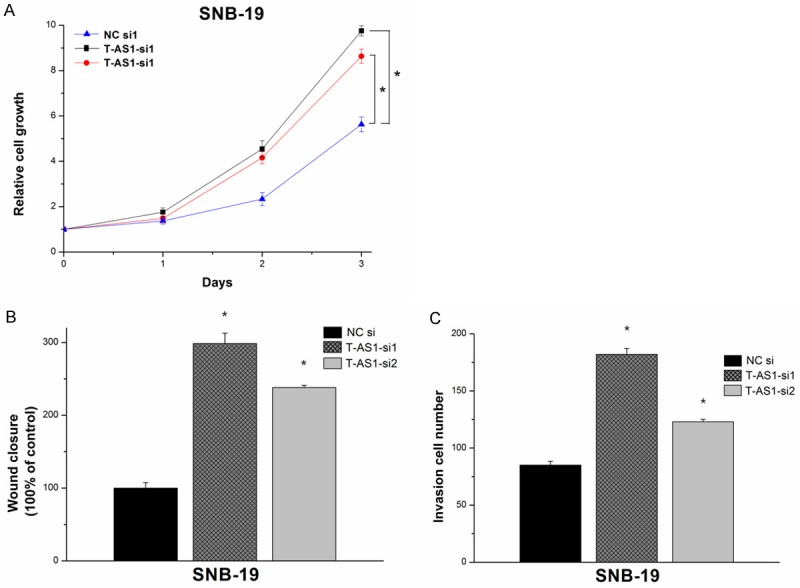
Cell growth, migration and invasion were elevated by TSLC1-AS1 siRNA in SNB-19 cells. A: Cell proliferation fold increase was tested using MTS with 1 day intervals. TSLC1-AS1 siRNA groups showed elevated growth rates compared with negative control groups in SNB-19 cells (*P < 0.05). B, C: Knockdown of TSLC1-AS1 enhanced the cell migration and invasion. Scratch wound healing assay and matrigel invasion assay showed that the cell migration and invasion capacities in TSLC1-AS1 siRNA group were remarkably elevated (*P < 0.05).
Discussion
This study provides the first evidence linking the expression of a novel lncRNA TSLC1-AS1 with the diagnosis, clinic-pathological characteristics and in vitro cellular growth behaviors of glioma. More specifically, we found that TSLC1-AS1 expression was evidently decreased in glioma compared with normal tissues, and reversely linked with the WHO grade of the tumor. Through down-regulating the TSLC1-AS1 expression in a high TSLC1-AS1 expressing cell line, the proliferation and invasion abilities of the glioma cells could be remarkably increased, while the overexpression of TSLC1-AS1 showed the opposite effect.
TSLC1 has been identified as a critical tumor suppressor in a variety of different cancers, including glioma, which is mainly controlled by promoter hypermethylation [10-21]. TSLC1-AS1, as a new found lncRNA, is the antisense partner of TSLC1. Nevertheless, to our knowledge there are no studies on the literature regarding the expression profile and functional role of TSLC1-AS1.
Besides TSLC1, thousands of protein-coding genes in the human genome have their antisense transcript partners, most of which are noncoding. This kind of sense-antisense RNA duplex formation can result in multiple outcomes. And the antisense lncRNA can modulate the sense mRNA either in a discordant manner or a concordant manner [22-24]. For instance, HIF1α together with its antisense partner aHIF has been recognized as a concordant sense-antisense RNA couple [25].
In the present study, we showed that the TSLC1-AS1 was positively correlated with the TSLC1 mRNA expression. And TSLC1-AS1 overexpression or knockdown resulted in concomitant TSLC1 sense-transcript increase or reduction. Since it has been proved that the regulation of TSLC1 mRNA expression can also be conducted without promoter hypermethylation [26], we speculated TSLC1-AS1 may be a new modulator of TSLC1 mRNA expression. Our observations of elevated proliferation, migration and invasion abilities of the SNB-19 cells after TSLC1-AS1 siRNA transfection are consistent with reduced TSLC1 mRNA expression, allowing increased tumor progression. While overexpression of TSLC1-AS1 in U87 cells showed the opposite effect. These findings demonstrated that the TSLC1-AS1 functions as a tumor suppressor in glioma.
Recent studies also reported some new genetic mutations that activate oncogenes, such as BRAF [27], and result in loss of function in tumor suppressors, such as NF1, VHL, PIK3R1 in glioma [28-30]. Interestingly, we found that the expression of TSLC1-AS1 was also positively correlated with tumor suppressors NF1, VHL and PIK3R1, and negatively correlated with the oncogene BRAF.
A plausible explanation is that the TSLC1-AS1 may be a pivotal modulating molecular in glioma, communicating with many other cancer-related genes and regulating the expression of its sense partner TSLC1.
Future studies should clarify the exact modulating mechanism between TSLC1-AS1 and TSLC1, and seek additional pathways that may also contribute to this relationship.
Disclosure of conflict of interest
None.
References
- 1.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classifcation of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maher EA, Furnari FB, Bachoo RM, Rowitch DH, Louis DN. Malignant glioma: genetics and biology of a grave matter. Genes Dev. 2001;15:1311–1333. doi: 10.1101/gad.891601. [DOI] [PubMed] [Google Scholar]
- 3.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Hum Mol Genet. 2010;19:R152–161. doi: 10.1093/hmg/ddq353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31:4577–4587. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71:3–7. doi: 10.1158/0008-5472.CAN-10-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nowacki S, Skowron M, Oberthuer A, Fagin A, Voth H, Brors B, Westermann F, Eggert A, Hero B, Berthold F, Fischer M. Expression of the tumour suppressor gene CADM1 is associated with favourable outcome and inhibits cell survival in neuroblastoma. Oncogene. 2008;27:3329–3338. doi: 10.1038/sj.onc.1210996. [DOI] [PubMed] [Google Scholar]
- 8.Fischer M, Oberthuer A, Brors B, Kahlert Y, Skowron M, Voth H, Warnat P, Ernestus K, Hero B, Berthold F. Differential expression of neuronal genes defines subtypes of disseminated neuroblastoma with favorable and unfavorable outcome. Clin Cancer Res. 2006;12:5118–5128. doi: 10.1158/1078-0432.CCR-06-0985. [DOI] [PubMed] [Google Scholar]
- 9.Watabe K, Ito A, Koma YI, Kitamura Y. IGSF4: a new intercellular adhesion molecule that is called by three names, TSLC1, SgIGSF and SynCAM, by virtue of its diverse function. Histol Histopathol. 2003;18:1321–1329. doi: 10.14670/HH-18.1321. [DOI] [PubMed] [Google Scholar]
- 10.Houshmandi SS, Surace EI, Zhang HB, Fuller GN, Gutmann DH. Tumor suppressor in lung cancer-1 (TSLC1) functions as a glioma tumor suppressor. Neurology. 2006;67:1863–1866. doi: 10.1212/01.wnl.0000244472.56198.84. [DOI] [PubMed] [Google Scholar]
- 11.Murakami Y. Involvement of a cell adhesion molecule, TSLC1/IGSF4, in human oncogenesis. Cancer Sci. 2005;96:543–552. doi: 10.1111/j.1349-7006.2005.00089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuramochi M, Fukuhara H, Nobukuni T, Kanbe T, Maruyama T, Ghosh HP, Pletcher M, Isomura M, Onizuka M, Kitamura T, Sekiya T, Reeves RH, Murakami Y. TSLC1 is a tumor suppressor gene in human non-small cell lung cancer. Nat Genet. 2001;27:427–430. doi: 10.1038/86934. [DOI] [PubMed] [Google Scholar]
- 13.Lung HL, Cheung AK, Xie D, Cheng Y, Kwong FM, Murakami Y, Guan XY, Sham JS, Chua D, Protopopov AI, Zabarovsky ER, Tsao SW, Stanbridge EJ, Lung ML. TSLC1 is a tumor suppressor gene associated with metastasis in nasopharyngeal carcinoma. Cancer Res. 2006;66:9385–9392. doi: 10.1158/0008-5472.CAN-06-0590. [DOI] [PubMed] [Google Scholar]
- 14.Heller G, Geradts J, Ziegler B, Newsham I, Filipits M, Markis-Ritzinger EM, Kandioler D, Berger W, Stiglbauer W, Depisch D, Pirker R, Zielinski CC, Zöchbauer-Müller S. Downregulation of TSLC1 and DAL-1 expression occurs frequently in breast cancer. Breast Cancer Res Treat. 2007;103:283–291. doi: 10.1007/s10549-006-9377-7. [DOI] [PubMed] [Google Scholar]
- 15.Ito T, Shimada Y, Hashimoto Y, Kaganoi J, Kan T, Watanabe G, Murakami Y, Imamura M. Involvement of TSLC1 in progression of esophageal squamous cell carcinoma. Cancer Res. 2003;63:6320–6326. [PubMed] [Google Scholar]
- 16.Honda T, Tamura G, Waki T, Jin Z, Sato K, Motoyama T, Kawata S, Kimura W, Nishizuka S, Murakami Y. Hypermethylation of the TSLC1 gene promoter in primary gastric cancers and gastric cancer cell lines. Jpn J Cancer Res. 2002;93:857–860. doi: 10.1111/j.1349-7006.2002.tb01329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams YN, Masuda M, Sakurai-Yageta M, Maruyama T, Shibuya M, Murakami Y. Cell adhesion and prostate tumorsuppressor activity of TSLL2/IGSF4C, an immunoglobulin superfamily molecule homologous to TSLC1/IGSF4. Oncogene. 2006;25:1446–1453. doi: 10.1038/sj.onc.1209192. [DOI] [PubMed] [Google Scholar]
- 18.Jansen M, Fukushima N, Rosty C, Walter K, Altink R, Heek TV, Hruban R, Offerhaus JG, Goggins M. Aberrant methylation of the 5-CpG island of TSLC1 is common in pancreatic ductal adenocarcinoma and is first manifest in high-grade PanlNs. Cancer Biol Ther. 2002;1:293–296. doi: 10.4161/cbt.84. [DOI] [PubMed] [Google Scholar]
- 19.Chen K, Wang G, Peng L, Liu S, Fu X, Zhou Y, Yu H, Li A, Li J, Zhang S, Bai Y, Zhang Y. CADM1/TSLC1 inactivation by promoter hypermethylation is a frequent event in colorectal carcinogenesis and correlates with late stages of the disease. Int J Cancer. 2011;128:266–273. doi: 10.1002/ijc.25356. [DOI] [PubMed] [Google Scholar]
- 20.Yang YX, Yang AH, Yang ZJ, Wang ZR, Xia XH. Involvement of tumor suppressor in lung cancer 1 gene expression in cervical carcinogenesis. Int J Gynecol Cancer. 2006;16:1868–1872. doi: 10.1111/j.1525-1438.2006.00656.x. [DOI] [PubMed] [Google Scholar]
- 21.You Y, Ma L, You M, Li X, Wang S, Li H, Wu D, Yang H, Li ZY. TSLC1 gene silencing in cutaneous melanoma. Melanoma Res. 2010;20:179–183. doi: 10.1097/CMR.0b013e32833413c0. [DOI] [PubMed] [Google Scholar]
- 22.Wahlestedt C. Natural antisense and noncoding RNA transcripts as potential drug targets. Drug Discov Today. 2006;11:503–508. doi: 10.1016/j.drudis.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 23.Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;1:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yelin R, Dahary D, Sorek R, Levanon EY, Goldstein O, Shoshan A, Diber A, Biton S, Tamir Y, Khosravi R, Nemzer S, Pinner E, Walach S, Bernstein J, Savitsky K, Rotman G. Widespread occurrence of antisense transcription in the human genome. Nat Biotechnol. 2003;21:379–386. doi: 10.1038/nbt808. [DOI] [PubMed] [Google Scholar]
- 25.Thrash-Bingham CA, Tartof KD. aHIF: a natural antisense transcript overexpressed in human renal cancer and during hypoxia. J Natl Cancer Inst. 1999;91:143–151. doi: 10.1093/jnci/91.2.143. [DOI] [PubMed] [Google Scholar]
- 26.Ando K, Ohira M, Ozaki T, Nakagawa A, Akazawa K, Suenaga Y, Nakamura Y, Koda T, Kamijo T, Murakami Y, Nakagawara A. Expression of TSLC1, a candidate tumor suppressor gene mapped to chromosome 11q23, is downregulated in unfavorable neuroblastoma without promoter hypermethylation. Int J Cancer. 2008;123:2087–2094. doi: 10.1002/ijc.23776. [DOI] [PubMed] [Google Scholar]
- 27.Deimling AV, Korshunov A, Hartmann C. The next generation of glioma biomarkers: MGMT methylation, BRAF fusions and IDH1 mutations. Brain Pathol. 2011;21:74–87. doi: 10.1111/j.1750-3639.2010.00454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weber GL, Parat MO, Binder ZA, Gallia GL, Riggins GJ. Abrogation of PIK3CA or PIK3R1 reduces proliferation, migration, and invasion in glioblastoma multiforme cells. Oncotarget. 2011;2:833–849. doi: 10.18632/oncotarget.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dasgupta B, Wen L, Perry A, David H. Gutmann1. glioma formation in neurofibromatosis 1 reflects preferential activation of K-RAS in astrocytes. Cancer Res. 2005;65:236–245. [PubMed] [Google Scholar]
- 30.Chen LC, Han L, Zhang KL, Shi ZD, Zhang JX, Zhang AL. VHL regulates the effects of miR-23b on glioma survival and invasion via suppression of HIF-1α/VEGF and β-catenin/Tcf-4 signaling. Neuro Oncol. 2012;14:1026–1036. doi: 10.1093/neuonc/nos122. [DOI] [PMC free article] [PubMed] [Google Scholar]


