Abstract
Osteosarcoma is the most common primary malignancy of bone in adolescents and young adults. There is a shortage of tumorigenic and highly metastatic human osteosarcoma cell lines that can be used for metastasis study. Here we establish and characterize a highly metastatic human osteosarcoma cell line that is derived from Saos2 cell line based on bioluminescence. The occasional pulmonary metastatic cells developed from Saos2 were isolated, harvested, characterized and named Saos2-l. The parental Saos2 and Saos2-l cells were further characterized both in vitro and in vivo. Results showed that Saos2-l cells demonstrated increased cell adhesion, migration and invasion compared to the parental Saos2 cells. Conversely, Saos2-l cells grew at a slightly slower rate than that of the parental cells. When injected into nude mice, Saos2-l cells had a greater increase in developing pulmonary metastases compared to the parental Saos2 cells. Further transcriptional profiling analysis revealed that some gene expression were up-regulated or down-regulated in the highly metastatic Saos2-l cells, indicating possible influencing factors of metastasis. Thus, we have established and characterized a highly metastatic human osteosarcoma cell line that should serve as a valuable tool for future investigations on the pathogenesis, metastasis and potential treatments of human osteosarcoma.
Keywords: Animal model, osteosarcoma, metastasis, microarray
Introduction
Osteosarcoma (OS) is the most common primary malignancy of bone. Progressive pulmonary metastasis with respiratory failure is the most common cause of death. Although the standard neoadjuvant and adjuvant chemotherapy combined with surgery has improved the long-term survival of patients from approximately 20% (surgery alone) to over 60% (surgery with adjuvant chemotherapy) [1,2], approximately 30-40% of patients will subsequently develop recurrent disease in the form of distant metastases, and dose intensification of adjuvant systemic chemotherapy protocols achieves little [2]. Moreover, the prognosis for OS patients with the appearance of lung metastases is more disappointing with survival rates of less 20% within 5 years of diagnosis [3].
Though there are numerous cell lines and animal models of OS established [4], our understanding of molecular and cellular events leading to the development and metastasis of osteosarcoma remains limited until now. We tried to establish an osteosarcoma cell line that is highly metastatic. By employing the commonly used Saos2 human osteosarcoma cell line, we have established such a highly metastatic subline. The parental Saos2 cell line was first established from a primary osteosarcoma in an 11-year old Caucasian girl by Fogh et al. in 1973 [5]. We have established the xenograft model, isolated the occasional pulmonary metastatic cells in nude mice based on bioluminescence. Serially passages of these cells in vitro have been established and named a new subline Saos2-l. The two lines were characterized in vitro for cell proliferation, adhesion, migration and invasion. The two lines were also examined spontaneous pulmonary metastasis from orthotopic tumor in vivo. Furthermore, both cell lines were examined for their transcriptional profiling. Our results demonstrate that Saos2-l cells exhibit slightly decreased adhesion, increased cell adhesion, migration and invasion in vitro, and increased spontaneous pulmonary metastases in vivo. Furthermore, transcriptional profiling analysis revealed that some gene expression was up-regulated or down-regulated in the highly metastatic Saos2-l cells, which indicating possible influencing factors of metastasis. Thus, this newly established cell line should be a valuable tool for further basic and translational research on human osteosarcoma.
Materials and methods
Cell line
Human OS Saos2 cells were purchased from Chinese Academy of Sciences (Shanghai, China) and grown in α-MEM (Gibco, Grand Island, NY, USA) supplemented with 10% FBS (Hyclone, Tauranga, New Zealand) and antibiotics (100 U/ml penicillin, 100 μg/ml streptomycin) in 37°C humidified atmosphere with 5% CO2.
Labeling of Saos2 cells with luciferase
Saos2 cells were infected by lentivirus loading a firefly Luciferase gene and a neomycin selection cassette and then exposed to 500 μg/mL G418 (Invitrogen, Carlsbad, CA) for 4 weeks post-infection. Several luciferase expression clones were available; populations of these clones were expanded with lower G418 maintenance of 200 μg/ml to remove tumor cells that lose luciferase expression.
Animals and animal procedures
Four-week-old male nude mice (BALB/c, nu/nu; SIPPR-BK Laboratory Animal Co. Ltd, Shanghai, China) were housed under pathogen-free conditions at 26-28°C and 50-65% humidity. All animal operations were approved by the Animal Ethics Committee of Shanghai Jiaotong University School of Medicine.
For intra-tibia injections, Saos2 cells were harvested, counted and resuspended in PBS to a final concentration of 2 × 108 cells/ml. Trypan blue exclusion testing showed cells to be > 95% viable before injection. Animals were anesthetized with 10% chloral hydrate. The right knee of each nude mice were fixed beyond 90°; 1 × 107 cells resuspended in 50 μl of PBS were then injected into the proximal tibia using 25-gauge needle, as reported previously [6,7].
Bioluminescence assay
For in vivo imaging, mice were injected intraperitoneally with 200 μl of 15 mg/mL luciferin (Keyuandi, Shanghai, China) 7-8 mins before being anesthetized with isoflurane. Imaging was performed using an IVIS 200 imaging system and Living Image® software Version 3.0.4 (Xenogen, Hopkinton, MA, USA). After 5 secs of exposure, total flux of the ROI was recorded as photons/sec for each animal.
Isolation of the high metastatic Saos2-l cells
Mouse detected obvious pulmonary metastasis by bioluminescence was killed when moribund by CO2 inhalation. The lung was perfused with PBS, excised, finely minced and incubated for 1 hr in α-MEM with 150 IU/ml collagenase type IV (Sigma-Aldrich, St Louis, MO, USA). Single sell suspensions were prepared by repeatedly aspirating the mixture through a 10 ml syringe before filtering through a nylon mesh. Cells were then pelleted at 1000 r.p.m., washed in PBS, and plated on 6 cm tissue culture plates in complete media with 200 μg/ml of G418 to select for human cancer cells. Removed human cancer cells were allowed to outgrow and form colonies for a few weeks. Then the cells were passaged 40 times.
MTT assay
The MTT assay was done according to the method as reported previously [8] and the manufacturer’s instruction (Sigma-Aldrich, St Louis, MO, USA). Briefly, 1 × 103 Saos2 cells were seeded in a 96-well plate in 200 μl medium. At indicated times, MTT solution was added to each well and plates were incubated for 4 hr. Subsequently, DMSO (Sigma-Aldrich, St Louis, MO, USA) was added to each well for 15 mins. The plates were then read at 490 nm using an automated plate reader (Perkin-Elmer, Waltham, MA, USA).
Heterotypic adherence assay
A total of 5 × 104 cells were seeded in each of five 96-well plates precoated with 1:10 Matrigel (BD Biosciences, San Jose, CA). The plates were incubated at 37°C in 5% CO2 for 0.5, 1, 1.5, 12 or 24 hrs. After incubation, medium was carefully suctioned out of each well, and every well was washed 3 times with 200 μl PBS. Between each wash, plates were manually rocked back and forth three times. Subsequently, 0.1% crystal violet was added to the wells for 10 mins. Wells were then rinsed 6 times with PBS until no purple color was observed. Finally, the cells were extracted using 100 μl glacial acetic acid, and the absorbance was read at 490 nm to reflect the number of adherent cells.
Migration and invasion assays
1 × 105 Cells were seeded on a Matrigel-coated polycarbonate membrane insert (6.5 mm in diameter with 8.0 μm pores) in a Transwell apparatus (Costar, Cambridge, MA) and maintained in α-MEM containing 0.2% BSA. α-MEM containing 10% FBS was added to the lower chamber. After incubation for 8 h at 37°C in a CO2 incubator, the insert was washed with PBS, and cells on the top surface of the insert were removed by wiping with a cotton swab.
The invasion assay procedure was similar to the cell migration assay, except that the Transwell membrane was coated with 1:3 diluted Matrigel (BD Biosciences, San Jose, CA), and cells were incubated for 32 hr at 37°C. Cells that migrated to the bottom surface of the insert were fixed with 4% paraformaldehyde and stained by 0.1% crystal violet, and then subjected to microscopic inspection. Cells counts were based on five field digital images taken randomly at 200 × magnification.
In vivo tumor growth and metastasis analyses
After inoculation, OS volumes of each mouse were measured at one-week intervals using calipers until 4 weeks after orthotopic injection. Tumor volume was calculated by using a previously reported equation: volume = 0.2618 × L × W × (L + W) [9], in which W is an average of distances at the proximal tibia at the level of the knee joint in anterior–posterior and medial–lateral planes, and L is the distance between the most distal points of the distal and proximal tumor margins.
Total RNA isolation
Total RNA was extracted using trizol reagent according to manufacturer’s instructions (Invitrogen, Carlsbad, CA). Reverse transcription used 1 μg of each RNA sample in a final volume of 20 μl, and was carried out at 42°C for 1 hr and 70°C for 10 mins.
Microarray
For microarray studies, RNA quantity and quality were measured by NanoDrop ND-1000. RNA integrity was assessed by standard denaturing agarose gel electrophoresis. Double-strand cDNA (ds-cDNA) was synthesized from 5 μg of total RNA using an Invitrogen SuperScript ds-cDNA synthesis kit in the presence of 100 pmol oligo dT primers, ds-cDNA was cleaned and labeled in accordance with the NimbleGen Gene Expression Analysis protocol (NimbleGen Systems, Inc., USA). Microarrays were hybridized at 42°C during 16 to 20 h with 4 μg of Cy3 labeled ds-cDNA in NimbleGen hybridization buffer/hybridization component A in a hybridization chamber (Hybridization System-NimbleGen Systems, Inc., Madison, WI, USA). The slides were scanned using the Axon GenePix 4000B microarray scanner (Molecular Devices Corporation) piloted by GenePix Pro 6.0 software (Axon). Scanned images (TIFF format) were then imported into NimbleScan software (version 2.5) for grid alignment and expression data analysis. Expression data were normalized through quantile normalization and the Robust Multichip Average (RMA) algorithm included in the NimbleScan software. Differential expression was defined as a fourfold or greater difference in normalized fluorescence intensity between the Saos2 and Saos2-l cells. The differentially expressed genes identified from cDNA microarray comparisons were then assigned to six non-mutually exclusive metastasis-associated processes (proliferation/apoptosis, motility/cytoskeleton, invasion, adhesion, immune surveillance, and angiogenesis) using a PubMed search of the gene names as initially described by Khanna et al [10].
Real-time PCR
RT-PCR analysis was carried out as previously described. Real-time PCR used ABI 7500 Real Time PCR System (Applied Biosystems, Carlsbad, CA, USA) and SYBR Premix Ex Taq polymerase (Takara, Dalian, China) according to manufacturers’ instructions. The primer sequences are as follows: ATM, forward 5’-GACCGTGGAGAAGTAGAATCAATGG-3’ and reverse 5’-GGCTCTCTCCAGGTTCGTT-3’; CAV1, forward 5’- ACAGCCCAGGGAAACCTC-3’ and reverse 5’-GATGGGAACGGTGTAGAGATG-3’; Noggin, forward 5’-GCGCTGCGGCTGGAT-3’ and reverse 5’-AGCACTTGCACTCGGAAATGA-3’; JAG1, forward 5’-AGGCTGGATGGGCCCCGAAT-3’ and reverse 5’-TCCCGGGTGTGGGATGCACT-3’; MMP1, forward 5’-GCAAGAGGCTGGGAAGCCATCA-3’ and reverse 5’-GCAGCAGCAGCAGTGGAGGAAA-3’; GAPDH, forward 5’-CCTGCACCACCAACTGCTTA-3’ and reverse 5’-AGGCCATGCCAGTGAGCTT-3’. GAPDH was used as internal control. Each experiment was repeated three times independently, to ensure reproducibility of results.
Statistical analysis
Statistical significance was calculated using Student’s t-test for two-sample comparisons. The animal survival divergence was analyzed using Survival analysis according to Log-rank test. Statistical significance was analyzed for data from at least three independent experiments. P values < 0.05 were defined as significant. All data are presented as mean ± SD unless otherwise specified.
Results
Establishment of the highly metastatic Saos2-l cells
We labeled the Saos2 cell line with firefly luciferase via lentivirus transfection. The stable clones were sorted by antibiotic selection. As shown in Figure 1, the pooled populations of stable clones were injected into the proximal tibia of nude mice. After several weeks, as the orthotopic tumor grew, the occasional pulmonary metastases are formed according to bioluminescence. The lung with occasional metastases was separated and digested under sterile condition, and the cells were harvested. The cells were then selected by their drug resistance, expanded and characterized. The highly metastatic Saos2-l cell line was established after 40 serial passages in vitro.
Figure 1.
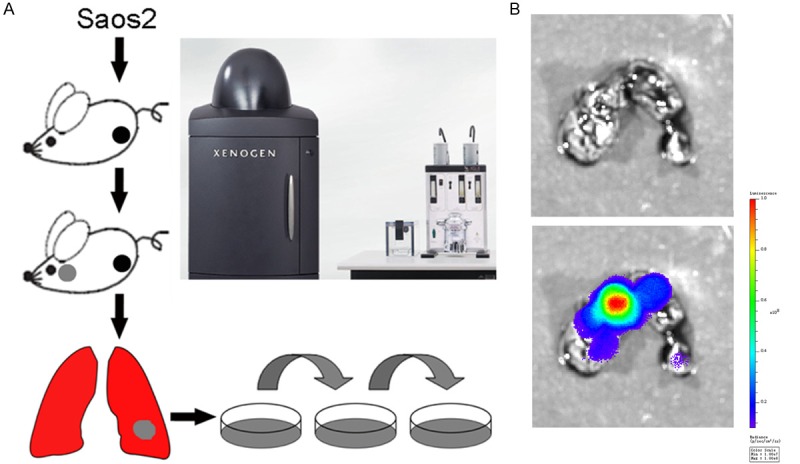
Establishment of the new highly metastatic osteosarcoma cell line. A: Diagram shown the selection process for the establishment of the highly metastatic Saos2-l cells. The parental Saos2 cells labeled with luciferase were injected into the proximal tibia of nude mice. The lungs were harvested when pulmonary metastases were observed by bioluminescence, digested with collagenase, expanded and passaged. B: Lung metastases observed by bioluminescence imaging.
Highly metastatic Saos2-l cells exhibited decreased cell proliferation and increased cell adhesion, migration and invasion ability
We next sought to characterize the Saos2-l cells for in vitro phenotypes important for tumorigenesis and metastasis [11]. We first examined Saos2 and Saos2-l cells for their in vitro proliferation. Qualitatively, we noticed that the Saos2-l cells tended to grow at a slightly slower rate than the parental ones. Cells were plated in triplicate and on subsequent days, the cells were fixed, stained and analyzed. As shown in Figure 2A, the highly metastatic Saos2-l cells exhibited a slightly lower in vitro proliferation rate than that of the parental ones (P value < 0.05). Then Saos2-l cells and Saos2 cells were plated in 96-well plate with matrigel coating (to mimic extracellular matrix) and investigated their adhesion ability. The result indicated that Saos2-l cells had higher adhesion ability in relative to the parental ones after 24-hour plating (Figure 2B). Transwell in combination with matrigel was employed to investigate the migration and invasion of Saos2-l cells. As Figure 2C and 2D shown, Saos2-l cells displayed higher migration and invasion ability compared with the parental ones.
Figure 2.
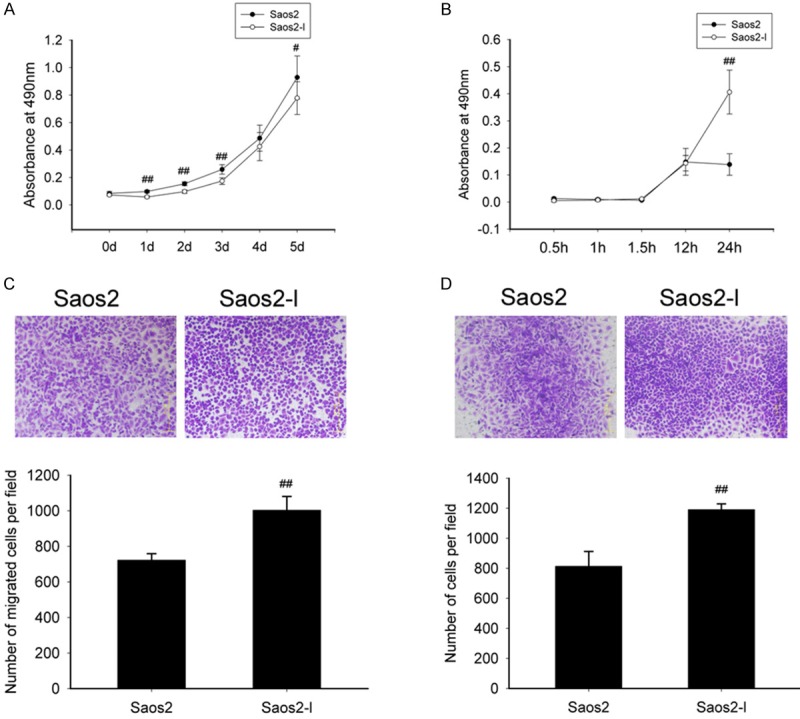
In vitro proliferation, cell adhesion, cell migration and cell invasion. A: MTT assay to evaluate proliferation of Saos2-l cells (Saos2-l vs Saos2 cells). B: Adhesion assay using matrigel coating to mimic extracellular matrix. #: P < 0.05 (Saos2-l vs Saos2 cells). C: Transwell assay to evaluate migration ability of Saos2-l cells. ##: P < 0.01 (Saos2-l vs Saos2 cells). D: Transwell assay using matrigel to evaluate invasion ability of Saos2-l cells. ##: P < 0.01 (Saos2-l vs Saos2 cells). All numerical data are presented as mean ± SD. Data are from at least three independent experiments.
Highly metastatic Saos2-l cells developed orthotopic tumors and lung metastases with higher efficiency in animal models
To further characterize the tumorigenic and metastatic phenotypes, we then determined the ability of the Saos2 and Saos2-l cells to form orthotopic tumors and produce spontaneous pulmonary metastases. Here, the Saos2 and Saos2-l cells were injected into the proximal tibia of nude mice, animals were serially observed, and pulmonary metastases were recorded at each time point. As shown in Figure 3A-C, both two cells developed comparatively equal tumors compared to each other not only in tumor volume but also in intensity of tumor bioluminescence. We next compared the metastatic potential of the parental Saos2 and the Saos2-l cells. According to bioluminescence, we were able to tracing pulmonary metastases by imaging twice a week. As shown in Figure 3D, 40% mice were detected pulmonary metastases in the Saos2-l group at 3 weeks post-injection. While mice injected Saos2 cells were beginning to detected pulmonary metastases only at five and a half weeks post-injection. Further survival analysis revealed that the survival time of Saos2-l bearing mice was much shorter than that of Saos2 bearing mice (P value < 0.0001) (Figure 3F). In Saos2 group, the 25th percentile, median and 75th percentile of survival time is 51 days, 52 days and 52 days, respectively. In Saos2-l group, the 25th percentile, median and 75th percentile of survival time is 29 days, 34 days and 40 days, respectively.
Figure 3.
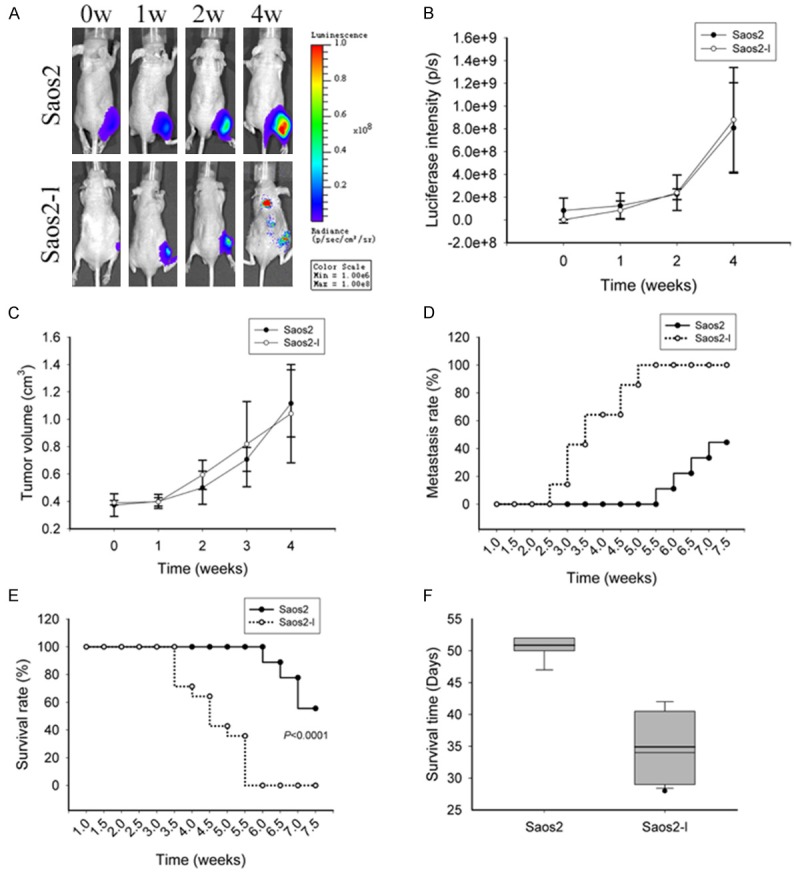
Osteosarcoma tumorigenesis and lung metastasis of Saos2-l cells and Saos2 cells in nude mice. A: Saos2-l cells and Saos2 cells were injected into the tibial marrow cavities of nude mice; at indicated time points, tumor growth and metastasis was monitored in vivo through IVIS system. B: Comparison of orthotopic tumor luminescence intensity between Saos2-l group and Saos2 group. C: Comparison of orthotopic tumor volumes between Saos2-l group and Saos2 group at each indicated time point. D: Comparison of lung metastasis rate between Saos2-l group and Saos2 group at each indicated time point. E: Comparison of mice survival rate between Saos2-l group and Saos2 group at each indicated time point. F: Survival analysis of Saos2-l bearing group and Saos2 bearing group.
Gene expression profile differences between Saos2 and Saos2-l cells
To have an exhaustive investigation on gene expression differences between Saos2-l and the parental Saos2 cells, we then did microarray. From the results, 132 of those genes identified were upregulated greater than fourfold. Conversely, 62 genes were downregulated greater than fourfold in Saos2-l compared with Saos2 cells. These genes were further classified according to their functions such as angiogenesis, cell motility, apoptosis, immune surveillance, cell adhesion and signal transduction (Tables 1 and 2). Most of these genes were used to create the non-supervised hierarchical cluster shown in Figure 4, color intensity is proportional to the log2 of the sample to reference intensity ratio and varies from -3 to +3. Some interested gene expression differences were verified by Real-time PCR (Figure 5). MMP1 and JAG1 were found down-regulation in the highly metastatic Saos2-l cells, with CAV1 and Noggin up-regulation. These genes are all important intermediate molecules in signaling transduction.
Table 1.
List of downregulated genes in Saos2-l categorized in metastasis-associated functional groups
| Biologic function | Gene title | Symbol | Fold change |
|---|---|---|---|
| Cell adhesion | Endothelial cell adhesion molecule | ESAM | 16.86 |
| Melanoma cell adhesion molecule | MCAM | 6.77 | |
| Cell motility | Palladin, cytoskeletal associated protein | PALLD | 9.30 |
| Matrix metallopeptidase 1 | MMP1 | 13.20 | |
| Apoptosis | SMAD family member 7 | SMAD7 | 4.87 |
| Immune surveillance | Desmoplakin | DSP | 13.91 |
| Signal transduction | Growth factor receptor-bound protein 10 | GRB10 | 14.60 |
| Jagged 1 | JAG1 | 9.65 | |
| Mitogen-activated protein kinase kinase kinase 15 | MAP3K15 | 6.23 | |
| Fibroblast growth factor 5 | FGF5 | 4.53 |
Table 2.
List of upregulated genes in Saos2-l categorized in metastasis-associated functional groups
| Biologic function | Gene title | Symbol | Fold change |
|---|---|---|---|
| Angiogenesis | Thrombospondin 1 | THBS1 | 8.06 |
| TIMP metallopeptidase inhibitor 2 | TIMP2 | 5.96 | |
| Neuropilin 1 | NRP1 | 4.30 | |
| Hypoxia inducible factor 1, alpha subunit | HIF1A | 4.01 | |
| Cell motility | Asp (abnormal spindle) homolog, microcephaly associated (Drosophila) | ASPM | 5.57 |
| Myosin 1B | MYO1B | 6.11 | |
| Kinesin family member C3 | KIFC3 | 4.43 | |
| Apoptosis | Stearoyl-CoA desaturase (delta-9-desaturase) | SCD | 5.57 |
| Protein kinase C, epsilon | PRKCE | 4.53 | |
| Immune surveillance | Peroxidasin homolog (Drosophila) | PXDN | 14.77 |
| Neuropilin 1 | NRP1 | 4.30 | |
| Signal transduction | Interleukin 7 receptor | IL7R | 9.19 |
| Noggin | NOG | 9.09 | |
| Dickkopf WNT signaling pathway inhibitor 1 | DKK1 | 6.87 | |
| Insulin-like growth factor binding protein 4 | IGFBP4 | 5.91 | |
| Transforming growth factor, alpha | TGFA | 4.89 | |
| Neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase | NEDD4L | 4.95 |
Figure 4.
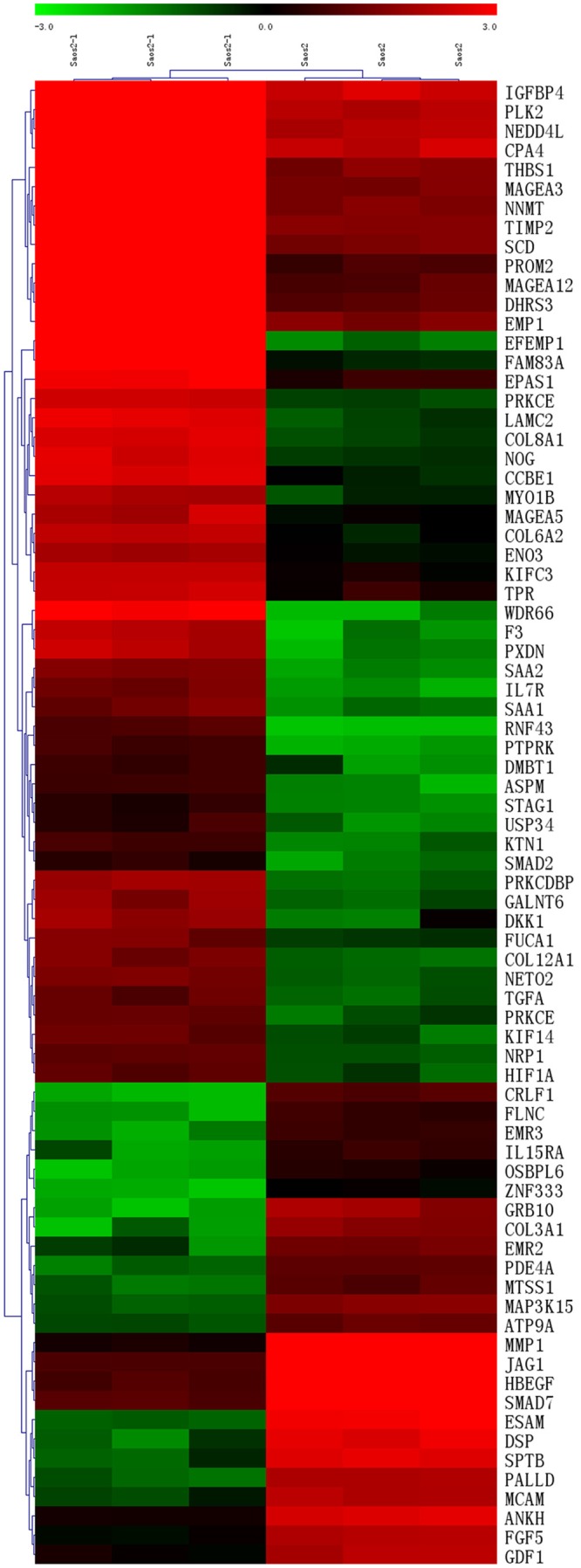
Non-supervised hierarchical clustering on differentially expressed genes. Color intensity correlates with the sample to reference signal intensity ratio. In green are those genes in which expression was down-regulated, and in red are those in which the expression was up-regulated. Sample names are on the top. On the right are the gene symbols.
Figure 5.
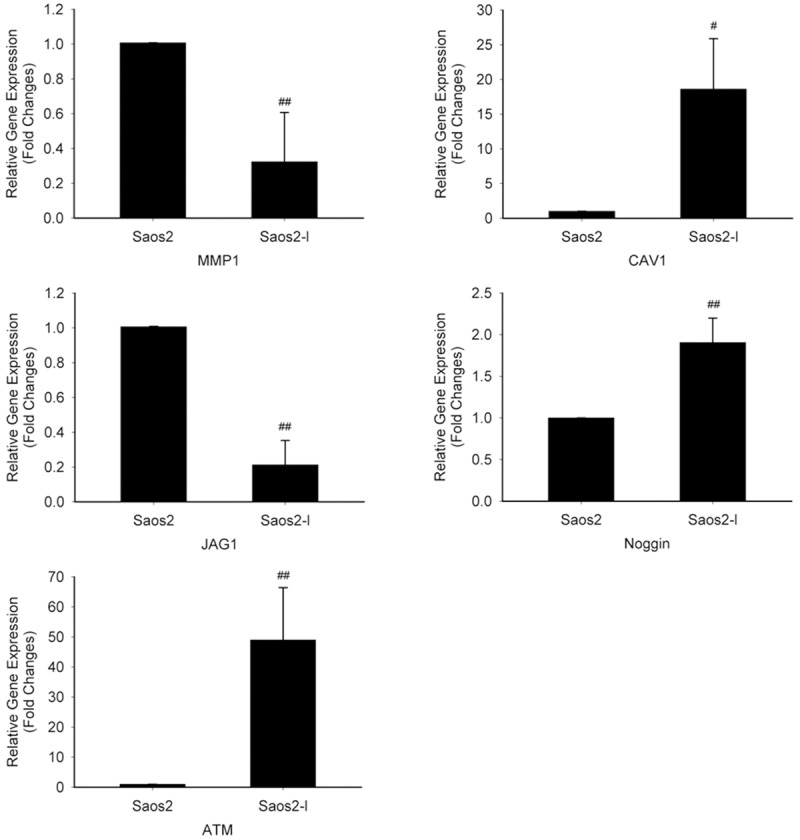
Interested gene expression verified by Real-time PCR. Results are expressed as fold changes of Saos2-l cells relative to Saos2 cells. GAPDH was used as loading control. MMP1 and JAG1 were found down-regulation in the highly metastatic Saos2-l cells, with CAV1 and Noggin up-regulation. #: P < 0.05 (Saos2-l vs Saos2 cells). ##: P < 0.01 (Saos2-l vs Saos2 cells).
Discussion
Osteosarcoma is the most common malignant bone tumor in childhood and adolescence. It represents 15% of all primary bone tumors and 0.2% of all malignant tumors in children. There are slightly more boys affected than girls (1.5:1). The peak incidence is in the second decade of life [12,13]. About 80% of osteosarcomas occur in the extremities, with the most common sites being the distal femur, the proximal tibia and the proximal humerus. About 80% of cases have localized tumor at presentation whereas the remainder present most commonly with pulmonary metastasis. The incidence of osteosarcoma has been increasing by about 1.4% per year [14].
A large number of osteosarcoma cell lines are currently available in the American Tissue Culture Center. Metastatic osteosarcoma has proven more valuable for osteosarcoma study. Though there exists several murine osteosarcoma cell lines with a higher in vivo metastatic potential currently, including UMR 106-01, K7M2, K12, Dunn and LM8 [15,16], human osteosarcoma cell lines are more clinically relevant to clinical study of osteosarcoma. Establishing new highly metastatic human osteosarcoma cells is quite useful for revealing metastatic mechanism of osteosarcoma and helpful for exploring new clinical treatment. Saos2 is one of the best-characterized human osteosarcoma cell lines in vitro [17-19].
In this report, we established a new highly metastatic human osteosarcoma cell line from a parental cell line Saos2. Animals injected with the Saos2-l cells readily form spontaneous pulmonary metastases.
As expected, the highly metastatic Saos2-l cells had slightly increased cell adhesion, high migration and invasion while cell proliferation was slightly decreased relative to the parental Saos2 cells. Cell migration, invasion, and adhesion are all important phenotypes for early events in the metastatic cascade [20,21]. Alterations in these phenotypes facilitate osteosarcoma cells to overcome local adhesive forces, migrate towards the microvasculature, and invade the vessels, which result in that Saos2-l bearing mice make pulmonary metastasis earlier and survive much shorter than that of the parental Saos2 cells.
Potentially, the highly metastatic Saos2-l cell line established here can be used as an investigative tool to examine tumor formation and metastasis in mice. It is conceivable that gene expression profiling between the highly metastatic Saos2-l and parental Saos2 cells may be used to identify novel genes that modulate metastasis. The newly identified genes can then be examined for their effects on metastasis in the animal model.
There have been some reports about establishment and characterization of new highly metastatic osteosarcoma cells. Jia et al. reported a series of cell lines separated from pulmonary metastases of Saos2 xenograft models. After the approach of 6 repeated cycling of tumor cells in mice, researchers isolated a new cell line named Saos-LM6 [22]. The new cell line was then confirmed displaying higher metastasis ability. Su et al. established another new highly metastatic osteosarcoma subline named MG63.2 from the parental MG63 cells [23]. While both the highly metastatic subline were unknown about their gene expression profiling differences contrast to the parental cells. Here, we also established a new highly metastatic subline from Saos2 cells, which may lose important information in etiology study especially metastasis mechanism. We further investigated gene expression differences between new highly metastatic subline and the parental cells. We found that there really exist some gene expression differences between Saos2-l and the parent Saos2 cells. These genes were classified according to their functions such as angiogenesis, cell motility, apoptosis, immune surveillance, cell adhesion and signal transduction.
ESAM, which is a member of the immunoglobulin superfamily, is selectively expressed in cultured human vascular endothelial cells and revealed high level expression in lung and heart and low level expression in kidney and skin. Hirate et al. reported that Chinese hamster ovary (CHO) cells expressing mouse ESAM formed large cell clusters in suspension culture [24]. Cangara et al. reported that ESAM regulated tumor metastasis through endothelial cell migration and tube formation in metastatic nodules, inhibition of ESAM may inhibit tumor metastasis by inhibiting the angiogenic processes [25]. Despite our findings of decreased ESAM expression, the Saos2-l cells demonstrated greater metastasis ability in vivo compared to the parental Saos2 cells. PALLD is a gene that encodes a component of the cytoskeleton that controls cell shape and motility [26]. It is likely that the phenotype of Saos2-l is independent of ESAM and PALLD expression. Desmosomes are intercellular junctions that confer strong cell-cell adhesion, thus conferring resistance against mechanical stress on epithelial tissues. A body of evidence indicates that decreased expression of desmosomal proteins is associated with poor prognosis in various cancers. As is a key component of desmosomal plaque proteins, desmoplakin has been reported acting as a tumor suppressor by inhibition of the Wnt/β-catenin signaling pathway in human lung cancer [27]. Neuropilin-1 (NRP1) and Neuropilin-2 (NRP2) are transmembrane glycoproteins which interact with VEGF to prevent tumor cell apoptosis and regulate angiogenesis. Preclinical data suggest that blockade of NRP1 suppresses tumor growth by inhibiting angiogenesis, in addition to directly inhibiting tumor cell proliferation in certain models [28]. NRP2 has been reported as a prognostic factor in osteosarcoma [29]. High HIF1α transcription level in osteosarcoma cell lines has been reported [30], which coincide with our microarray result. ASPM has been reported to correlate with tumor grade and survival in epithelial ovarian cancer [31].
While there are still some discrepancies between our profiling results and existed reports. Engin et al. reported that the invasive potential of osteosarcoma is associated with increased Notch signaling, and elevated expression of JAG1 is found in human osteosarcoma [32]. And overexpression of JAG1 was observed in osteosarcoma biopsy specimens contrast to normal bone [33]. In our study, highly metastatic Saos2-l cells displayed more invasive behavior with downregulation of JAG1. One explanation is both the conclusions are based on taking the normal bone as a control. But within the overall osteosarcoma cell line, there may be some correlations between JAG1 expression and malignancy among different tumor sublines. Cantiani et al. reported that the majority of primary osteosarcoma showed significantly lower levels of CAV1 than normal osteoblasts [34]. Their further studies demonstrated that osteosarcoma cell lines forced to overexpress CAV1 showed reduced malignancy in vitro, and CAV1 overexpression abrogates the metastatic ability of osteosarcoma cells in vivo. These conclusions may indicate another more important molecular mechanism about the malignancy of osteosarcoma.
In conclusion, we have established a highly metastatic human cell line from Saos2 cell line. We believe that this line can be used as a valuable tool in investigating the basic mechanism in the progression of osteosarcoma as well as novel therapies.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (81172549, 81302341) the Shanghai Science and Technology Development Fund (12140901300, 11XD1403300), and the fund for the Doctoral Program of Higher Education of China (20100073110078).
Disclosure of conflict of interest
None.
References
- 1.Bacci G, Picci P, Ferrari S, Avella M, Prever BA, Ruggieri P, Casadei R, Lari S, Monti C, Cazzola A, et al. Neoadjuvant chemotherapy for nonmetastatic osteosarcoma of the extremities: the recent experience at the Rizzoli Institute. Cancer Treat Res. 1993;62:299–308. doi: 10.1007/978-1-4615-3518-8_36. [DOI] [PubMed] [Google Scholar]
- 2.Ferrari S, Palmerini E. Adjuvant and neoadjuvant combination chemotherapy for osteogenic sarcoma. Curr Opin Oncol. 2007;19:341–346. doi: 10.1097/CCO.0b013e328122d73f. [DOI] [PubMed] [Google Scholar]
- 3.Marina N, Gebhardt M, Teot L, Gorlick R. Biology and therapeutic advances for pediatric osteosarcoma. Oncologist. 2004;9:422–441. doi: 10.1634/theoncologist.9-4-422. [DOI] [PubMed] [Google Scholar]
- 4.Fan TM. Animal models of osteosarcoma. Expert Rev Anticancer Ther. 2010;10:1327–1338. doi: 10.1586/era.10.107. [DOI] [PubMed] [Google Scholar]
- 5.Fogh J, Fogh JM, Orfeo T. One hundred and twenty-seven cultured human tumor cell lines producing tumors in nude mice. J Natl Cancer Inst. 1977;59:221–226. doi: 10.1093/jnci/59.1.221. [DOI] [PubMed] [Google Scholar]
- 6.Du L, Xu WT, Fan QM, Tu B, Shen Y, Yan W, Tang TT, Wang Y. Tumorigenesis and spontaneous metastasis by luciferase-labeled human xenograft osteosarcoma cells in nude mice. Chin Med J (Engl) 2012;125:4022–4030. [PubMed] [Google Scholar]
- 7.Tu B, Du L, Fan QM, Tang Z, Tang TT. STAT3 activation by IL-6 from mesenchymal stem cells promotes the proliferation and metastasis of osteosarcoma. Cancer Lett. 2012;325:80–88. doi: 10.1016/j.canlet.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 8.Bian ZY, Fan QM, Li G, Xu WT, Tang TT. Human mesenchymal stem cells promote growth of osteosarcoma: involvement of interleukin-6 in the interaction between human mesenchymal stem cells and Saos-2. Cancer Sci. 2010;101:2554–2560. doi: 10.1111/j.1349-7006.2010.01731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu WT, Bian ZY, Fan QM, Li G, Tang TT. Human mesenchymal stem cells (hMSCs) target osteosarcoma and promote its growth and pulmonary metastasis. Cancer Lett. 2009;281:32–41. doi: 10.1016/j.canlet.2009.02.022. [DOI] [PubMed] [Google Scholar]
- 10.Khanna C, Khan J, Nguyen P, Prehn J, Caylor J, Yeung C, Trepel J, Meltzer P, Helman L. Metastasis-associated differences in gene expression in a murine model of osteosarcoma. Cancer Res. 2001;61:3750–3759. [PubMed] [Google Scholar]
- 11.Yoshida BA, Sokoloff MM, Welch DR, Rinker-Schaeffer CW. Metastasis-suppressor genes: a review and perspective on an emerging field. J Natl Cancer Inst. 2000;92:1717–1730. doi: 10.1093/jnci/92.21.1717. [DOI] [PubMed] [Google Scholar]
- 12.Picci P. Osteosarcoma (osteogenic sarcoma) Orphanet J Rare Dis. 2007;2:6. doi: 10.1186/1750-1172-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartford CM, Wodowski KS, Rao BN, Khoury JD, Neel MD, Daw NC. Osteosarcoma among children aged 5 years or younger: the St. Jude Children’s Research Hospital experience. J Pediatr Hematol Oncol. 2006;28:43–47. [PubMed] [Google Scholar]
- 14.Caudill JS, Arndt CA. Diagnosis and management of bone malignancy in adolescence. Adolesc Med State Art Rev. 2007;18:62–78. ix. [PubMed] [Google Scholar]
- 15.Fisher JL, Mackie PS, Howard ML, Zhou H, Choong PF. The expression of the urokinase plasminogen activator system in metastatic murine osteosarcoma: an in vivo mouse model. Clin Cancer Res. 2001;7:1654–1660. [PubMed] [Google Scholar]
- 16.Khanna C, Prehn J, Yeung C, Caylor J, Tsokos M, Helman L. An orthotopic model of murine osteosarcoma with clonally related variants differing in pulmonary metastatic potential. Clin Exp Metastasis. 2000;18:261–271. doi: 10.1023/a:1006767007547. [DOI] [PubMed] [Google Scholar]
- 17.Murrills RJ, Matteo JJ, Samuel RL, Andrews JL, Bhat BM, Coleburn VE, Kharode YP, Bex FJ. In vitro and in vivo activities of C-terminally truncated PTH peptides reveal a disconnect between cAMP signaling and functional activity. Bone. 2004;35:1263–1272. doi: 10.1016/j.bone.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 18.Orimo H, Shimada T. Regulation of the human tissue-nonspecific alkaline phosphatase gene expression by all-trans-retinoic acid in SaOS-2 osteosarcoma cell line. Bone. 2005;36:866–876. doi: 10.1016/j.bone.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 19.Yu Y, Harris RI, Yang JL, Anderson HC, Walsh WR. Differential expression of osteogenic factors associated with osteoinductivity of human osteosarcoma cell lines. J Biomed Mater Res A. 2004;70:122–128. doi: 10.1002/jbm.a.30072. [DOI] [PubMed] [Google Scholar]
- 20.Geiger TR, Peeper DS. Metastasis mechanisms. Biochim Biophys Acta. 2009;1796:293–308. doi: 10.1016/j.bbcan.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 21.Skubitz AP. Adhesion molecules. Cancer Treat Res. 2002;107:305–329. doi: 10.1007/978-1-4757-3587-1_15. [DOI] [PubMed] [Google Scholar]
- 22.Jia SF, Worth LL, Kleinerman ES. A nude mouse model of human osteosarcoma lung metastases for evaluating new therapeutic strategies. Clin Exp Metastasis. 1999;17:501–506. doi: 10.1023/a:1006623001465. [DOI] [PubMed] [Google Scholar]
- 23.Su Y, Luo X, He BC, Wang Y, Chen L, Zuo GW, Liu B, Bi Y, Huang J, Zhu GH, He Y, Kang Q, Luo J, Shen J, Chen J, Jin X, Haydon RC, He TC, Luu HH. Establishment and characterization of a new highly metastatic human osteosarcoma cell line. Clin Exp Metastasis. 2009;26:599–610. doi: 10.1007/s10585-009-9259-6. [DOI] [PubMed] [Google Scholar]
- 24.Hirata K, Ishida T, Penta K, Rezaee M, Yang E, Wohlgemuth J, Quertermous T. Cloning of an immunoglobulin family adhesion molecule selectively expressed by endothelial cells. J Biol Chem. 2001;276:16223–16231. doi: 10.1074/jbc.M100630200. [DOI] [PubMed] [Google Scholar]
- 25.Cangara HM, Ishida T, Hara T, Sun L, Toh R, Rikitake Y, Kundu RK, Quertermous T, Hirata K, Hayashi Y. Role of endothelial cell-selective adhesion molecule in hematogeneous metastasis. Microvasc Res. 2010;80:133–141. doi: 10.1016/j.mvr.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pogue-Geile KL, Chen R, Bronner MP, Crnogorac-Jurcevic T, Moyes KW, Dowen S, Otey CA, Crispin DA, George RD, Whitcomb DC, Brentnall TA. Palladin mutation causes familial pancreatic cancer and suggests a new cancer mechanism. PLoS Med. 2006;3:e516. doi: 10.1371/journal.pmed.0030516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang L, Chen Y, Cui T, Knosel T, Zhang Q, Albring KF, Huber O, Petersen I. Desmoplakin acts as a tumor suppressor by inhibition of the Wnt/beta-catenin signaling pathway in human lung cancer. Carcinogenesis. 2012;33:1863–1870. doi: 10.1093/carcin/bgs226. [DOI] [PubMed] [Google Scholar]
- 28.Jubb AM, Strickland LA, Liu SD, Mak J, Schmidt M, Koeppen H. Neuropilin-1 expression in cancer and development. J Pathol. 2012;226:50–60. doi: 10.1002/path.2989. [DOI] [PubMed] [Google Scholar]
- 29.Handa A, Tokunaga T, Tsuchida T, Lee YH, Kijima H, Yamazaki H, Ueyama Y, Fukuda H, Nakamura M. Neuropilin-2 expression affects the increased vascularization and is a prognostic factor in osteosarcoma. Int J Oncol. 2000;17:291–295. doi: 10.3892/ijo.17.2.291. [DOI] [PubMed] [Google Scholar]
- 30.El Naggar A, Clarkson P, Zhang F, Mathers J, Tognon C, Sorensen PH. Expression and stability of hypoxia inducible factor 1alpha in osteosarcoma. Pediatr Blood Cancer. 2012;59:1215–1222. doi: 10.1002/pbc.24191. [DOI] [PubMed] [Google Scholar]
- 31.Bruning-Richardson A, Bond J, Alsiary R, Richardson J, Cairns DA, McCormack L, Hutson R, Burns P, Wilkinson N, Hall GD, Morrison EE, Bell SM. ASPM and microcephalin expression in epithelial ovarian cancer correlates with tumour grade and survival. Br J Cancer. 2011;104:1602–1610. doi: 10.1038/bjc.2011.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Engin F, Bertin T, Ma O, Jiang MM, Wang L, Sutton RE, Donehower LA, Lee B. Notch signaling contributes to the pathogenesis of human osteosarcomas. Hum Mol Genet. 2009;18:1464–1470. doi: 10.1093/hmg/ddp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanaka M, Setoguchi T, Hirotsu M, Gao H, Sasaki H, Matsunoshita Y, Komiya S. Inhibition of Notch pathway prevents osteosarcoma growth by cell cycle regulation. Br J Cancer. 2009;100:1957–1965. doi: 10.1038/sj.bjc.6605060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cantiani L, Manara MC, Zucchini C, De Sanctis P, Zuntini M, Valvassori L, Serra M, Olivero M, Di Renzo MF, Colombo MP, Picci P, Scotlandi K. Caveolin-1 reduces osteosarcoma metastases by inhibiting c-Src activity and met signaling. Cancer Res. 2007;67:7675–7685. doi: 10.1158/0008-5472.CAN-06-4697. [DOI] [PubMed] [Google Scholar]


