Abstract
We report a case of a 34-year-old Polish Caucasian male who was diagnosed with tinea manuum caused by Trichophyton rubrum var. raubitschekii. It would be the first described case of a dermatophytosis caused by this fungus in Poland and one of a few cases in Central Europe described so far. Admittedly, it would be the first case in Central Europe with no evidence pointing to African origin. The clinical condition improved after administering itraconazole (daily dose 100 mg orally) supplemented with a topical treatment, while the patient was totally cured after 2 months. The histopathological examination turned out to be highly useful in the diagnostic process. The genetic analysis of the urease gene pointed to a urease-positive T. rubrum rather than T. rubrum var. raubitschekii.
Keywords: Dermatophytes, Dermatophytosis, Epidemiology, Poland, Itraconazole, Diagnosis, Urease
Introduction
People suffering from fungal infections comprise a substantial group among dermatological patients. Mycoses of the glabrous skin are predominantly triggered by dermatophytes, while members of the Trichophyton genus, including Trichophyton rubrum, are among the most commonly diagnosed fungal pathogens in Poland. Superficial mycoses resulting from infections by T. rubrum var. raubitschekii are usually found in Africa, Southeast Asia, Australia and South America [1, 2]. In recent years, however, there have been a few cases in Germany and Italy; hence, the pathogen should be considered as undoubtedly present in Europe [3, 4]. To date, there has been no described case of an infection caused by this pathogen in Poland.
Case Report
A 34-year-old Polish Caucasian male was admitted to the Dermatology Clinic of the Poznan University of Medical Sciences due to the presence of an erythematous, exfoliating, clearly distinct lesion located on the index finger of the right hand, spreading onto surrounding areas (Figs. 1, 2).
Fig. 1.

Side of the index finger—hospital admission date
Fig. 2.
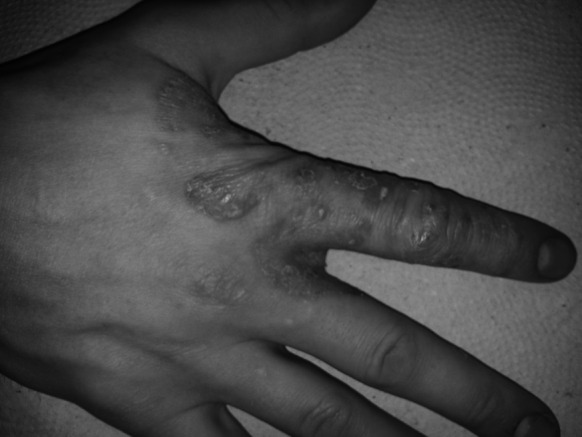
Top of the right hand—date of the second sample acquisition during the follow-up appointment at the Department of Medical Mycology, Poznan University of Medical Sciences
The first signs of the disease had appeared in August 2012, after a contact with raw fish—the patient is a cook. Due to a suspicion of erysipeloid, the patient underwent an antibiotic therapy (intravenous clindamycin, 2 × 600 mg daily, for the period of 5 days) coupled with steroids administered topically.
A series of diagnostic examinations were carried out during his hospitalization. A swab for bacteriological, skin biopsy for histopathological and a sample for mycological examinations were acquired. The bacterial culture was found to be positive for Enterococcus faecalis. The direct mycological examination was negative. Other examinations, such as the mycological culture and histopathology, were awaited for during the stay of the patient at the clinic. Due to the lack of significant improvement, the patient was discharged, while any further procedures were performed on an outpatient basis at the Dermatology Counseling Service of the clinic.
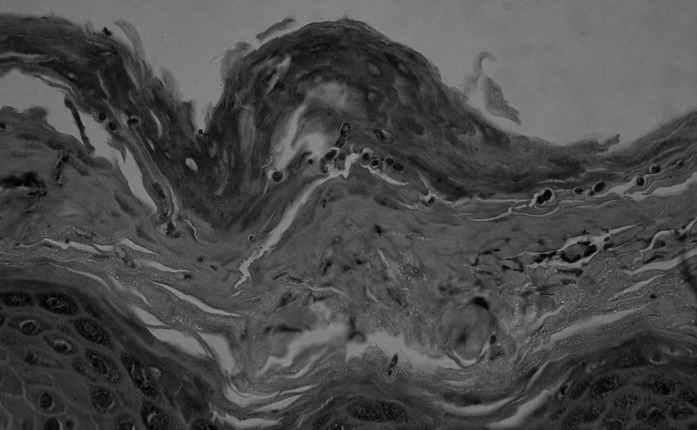
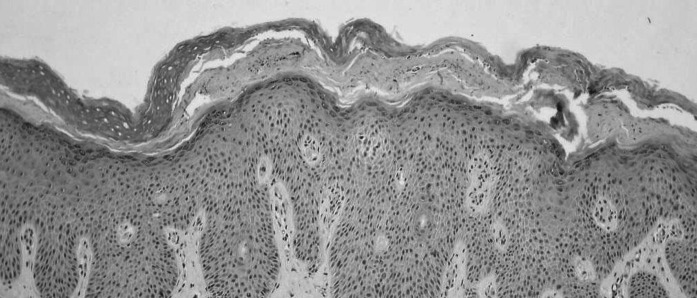
The results of the histopathological examination indicated a widened, swollen epidermis with a thickened stratum granulosum and a thick stratum corneum, in which there were numerous, round, fungal spores of 2.5 μm in diameter (Figs. 3, 4). On the other hand, the result of the mycological culture was negative. Due to discrepancies in the results, a second sample for mycological examination was acquired during a follow-up appointment. This time the results of the direct mycological examination were positive.
Fig. 3.
A thickened, swollen epidermis. A thick stratum corneum with numerous pathogenic fungi. Moderate, mixed inflammatory infiltrates present in the dermis. Hematoxylin and eosin stain, objective lens magnification: × 10
Fig. 4.
Numerous spores in the stratum corneum. Hematoxylin and eosin stain, objective lens magnification: × 60
Itraconazole (daily dose 100 mg) was administered orally. Additionally, an imidazole derivative was administered topically. The patient claimed an improvement in his dermatological condition after 2 weeks.
The mycological cultures were maintained for 20 days at 25 °C on two kinds of media: Sabo Sabouraud dextrose agar with chloramphenicol and Sabo Sabouraud dextrose agar with chloramphenicol and actidione. A fungal colony with a reddish brown color and a brownish reverse, strongly furrowed, suede-like surface and a raised center was observed (Fig. 5). A microscopic examination revealed numerous cigar-shaped and thin-walled septate macroconidia. Pyriform microconidia were located on short branches along hyphae (Fig. 6). A urease test, carried out on Christiansen’s medium, and dermatophyte test medium examination were both positive. Since the strain matched the original description, it was identified as T. rubrum var. raubitschekii [5].
Fig. 5.
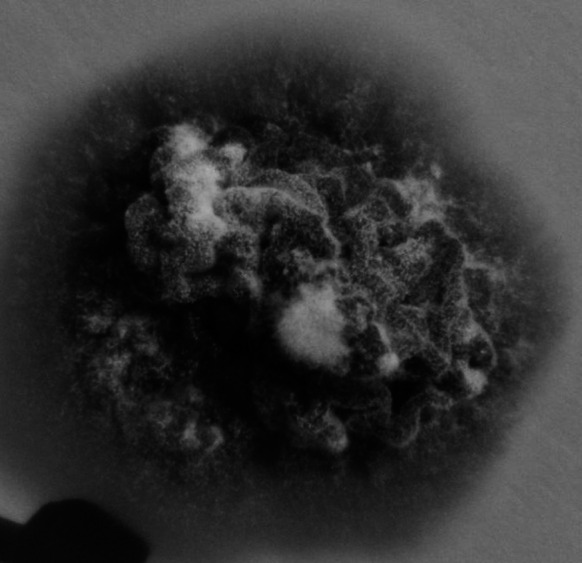
Trichophyton rubrum var. raubitscheki. A 20-day-old colony grown on Sabouraud agar medium
Fig. 6.
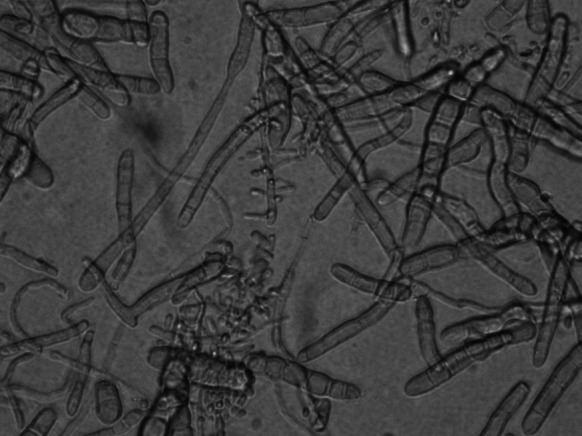
Trichophyton rubrum var. raubitschekii. Microscopic image (magnification— × 400, lactophenol cotton blue). Numerous macro- and microconidia
The following 3 weeks brought a further improvement in the dermatological status (Fig. 7). It was decided to continue the itraconazole treatment while the topical drug was switched to a pyridinone derivative. The patient received itraconazole for a period of 2 months, yet due to low tolerance to the topical drug, the pyridinone derivative was no longer administered. Afterward, the patient showed no signs of the infection.
Fig. 7.
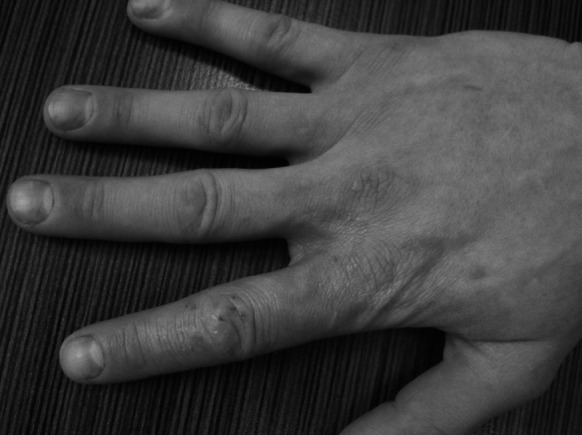
Top of the right hand—after 3 weeks of itraconazole treatment
In order to reassess the diagnosis, we analyzed a ribosomal RNA genomic fragment [6] and the urease gene of this isolate as described by Hiruma et al. [7]. Because the isolate ceased to grow on any new media, we isolated its DNA from a plate deposited for long-term storage at our clinic. One hundred and twenty milligrams of the colony was acquired from the deposited agar plate and ground in liquid nitrogen. DNA was isolated with the use of the column-based GeneMATRIX Plant & Fungi DNA Purification Kit (EURx, Poland) that includes both RNase A and proteinase K.
Firstly, we analyzed a fragment containing partial 18S rRNA gene, ITS1, 5.8S rRNA gene, ITS2 and partial 28S rRNA gene with the use of common ITS1 and ITS4 primers for PCR and sequencing. PCR was performed with the following ingredients: 25 pmol of ITS1 and ITS4 primers each [6], 0.5 µl of FastStart Taq DNA Polymerase (2.5 U, Roche), 10 µl of 5 × GC-rich solution, 5 µl of 10 × PCR buffer with 20 mM MgCl2, 1 µl of dNTPs (10 mM each), 5 ng of DNA sample, all in a volume of 50 µl. PCR was performed with the following program: pre-denaturation—4 min, 95 °C; amplification—35 × [denaturation: 1 min, 95 °C; annealing: 1 min, 55 °C; elongation: 1 min, 72 °C]; cooling: 1 min, 40 °C. Gel electrophoresis and paired-end sequencing revealed a 692-bp-long fragment, presenting a 100 % coverage with multiple GenBank T. rubrum entries and with our own T. rubrum reference strain. This result was expected as T. rubrum and T. rubrum var. raubitschekii were found to be identical in this genomic region [8].
Secondly, urease gene fragment was amplified by PCR with the following ingredients: 25 pmol of TrURE1S and TrURE1R primers each [7], 0.5 µl of FastStart Taq DNA Polymerase (2.5 U, Roche), 10 µl of 5 × GC-rich solution, 5 µl of 10 × PCR buffer with 20 mM MgCl2, 1 µl of dNTPs (10 mM each), 0.5 µl of DMSO, 5 ng of DNA sample, all in a volume of 50 µl. Touchdown PCR was performed with the following program: pre-denaturation—4 min, 95 °C; amplification—35 × [denaturation: 1 min, 95 °C; annealing 1 min, 69–63 °C, step size 0.5 °C, step delay 0; elongation: 1 min, 72 °C]; cooling—1 min, 40 °C. Gel electrophoresis revealed that the product was approximately 330 bp long, suggesting a urease-positive T. rubrum rather than T. rubrum var. raubitschekii (Fig. 8). According to Hiruma et al. [7], the genomic sequence of a raubitschekii urease amplicon should also include a 68-bp intron and a 9-bp-long coding insertion (DDBJ AB719058, CBS 100084). The PCR product was sequenced from both ends. This analysis revealed a sequence of 329 bp (including primers) presenting a 100 % coverage with XM_003233873 (T. rubrum CBS 118892 urease (TERG_05790) mRNA) and with the corresponding genomic sequence (NW_003456423.1, Gene ID: 10376003). Both the 68-bp intron and the 9-bp insertion, specific to the CBS 100084 raubitschekii strain, were missing. Based on these results, it is more likely that the isolate was, in fact, a urease-positive T. rubrum.
Fig. 8.

Gel electrophoresis of the urease gene PCR amplicon. Left lane approximately 329-bp fragment, right lane pUC19 DNA/MspI marker (Fermentas)
Discussion
Trichophyton rubrum var. raubitschekii is an anthropophilic, pathogenic dermatophyte, rarely found in Europe. Initially, it had been described as a separate species—Trichophyton raubitschekii [5, 9], yet molecular studies revealed it to be a variant of T. rubrum [4, 8]. It was described to differ from other T. rubrum variants by a limited growth, reddish brown color, the presence of numerous macro- and microconidia and the ability to produce urease [5, 8, 10]. The dermatophyte isolated at the clinic demonstrated all of these characteristics. Noteworthy, the Polish taxonomic key in operation states that the urease activity is the criterion for differentiation between T. rubrum var. raubitschekii and T. rubrum [11]. The characterization of urease-positive T. rubrum that are distinct from T. rubrum var. raubitschekii has clearly complicated the diagnostic process [2, 7].
The vast majority of T. rubrum var. raubitschekii cases originate in Asia, Africa and South America [1, 2, 12]. The aforementioned Polish patient had not traveled to these regions of the world. Furthermore, he did not claim contacts with anyone traveling to these regions, nor with people showing lesions that might be mycotic.
There has been a T. rubrum var. raubitschekii case of tinea faciei in Germany in a female patient born in Angola [12]. Furthermore, Brasch et al. [13] described a different German case, involving a 32-year-old male Cameroonian student with well distinct, erythematous, exfoliating lesions located on the abdomen as well as on both hands. The clinical picture suggested tinea manuum. A culture grown on Sabouraud medium at 26 °C and a urease test led to the diagnosis of T. rubrum var. raubitschekii. Both the Cameroonian and the described Polish patients responded well to their oral itraconazole treatments. To date, there have been eight described cases in Central Europe. All of them were Africans residing in Germany [3, 12–14]. The described Polish case is exceptional, as it appears not to be linked with Africa in any way.
Arabatzis et al. [15] described a study involving a group of patients from southern Europe. Cultures and urease tests were performed on samples originating from different parts of the skin of 95 Greek and 10 Bulgarian patients. T. rubrum var. raubitschekii was identified in five Greeks and one Bulgarian. The pathogen originated from lesions of tinea manuum (one case), tinea corporis (one case), tinea cruris (one case) and tinea unguium (three cases). The group originated from and resided in Greece and Bulgaria at the time of the study. They all had not traveled to the aforementioned endemic regions nor abroad for the past 6 months.
The molecular characterization of the urease gene of T. rubrum var. raubitschekii will probably become the method of choice for distinguishing between the quite alike urease-positive T. rubrum and T. rubrum var. raubitschekii. The procedure is straightforward and can easily be modified for probe or melting curve assays. For the time being, it has to be approached with caution as only one T. rubrum var. raubitschekii isolate (CBS 100084) has been analyzed in this way so far [7]. At the moment, public whole genome sequencing projects include multiple T. rubrum and two T. rubrum var. raubitschekii isolates: CBS 100084 and CBS 202.88. The results of these projects may answer the question whether the urease allele of the CBS 100084 strain is the specific marker of all T. rubrum var. raubitschekii isolates, as suggested by Hiruma et al. In the meantime, we encourage other researchers to reassess their T. rubrum var. raubitschekii isolates with this molecular approach.
We have described a possible case of an infection with T. rubrum var. raubitschekii in Poland. Irrespective of whether it was truly a T. rubrum var. raubitschekii or a urease-positive T. rubrum, there seems to be a necessity of including both into the Polish and other Central European diagnostic schemes, as their identification obviously poses a challenge at the moment. Moreover, it is noteworthy that the histopathological examination turned out to be very useful, if not necessary, in the diagnostic process. It enabled the adequate treatment to be implemented even before the results of the mycological cultures and molecular studies were available.
Conflict of interest
None declared.
References
- 1.Ellis D, Davis S, Alexiou H, Handke R, Bartley R. Descriptions of medical fungi. 2nd ed. Adelaide: Nexus Print Solutions; 2007. p. 158–161.
- 2.Hiruma M, Kano R, Sugita T, Mochizuki T, Hasegawa A, Hiruma M. Epidemiological aspects of Trichophyton rubrum var. raubitschekii in Japan. J Dermatol. 2012;39:1000–1001. doi: 10.1111/j.1346-8138.2012.01635.x. [DOI] [PubMed] [Google Scholar]
- 3.Brasch J. Var. raubitschekii of Trichophyton rubrum as a cause of tinea in Germany. Mycoses. 2007;50(Suppl 2):2–5. doi: 10.1111/j.1439-0507.2007.01423.x. [DOI] [PubMed] [Google Scholar]
- 4.Papini M, Greco C, Pileri F. Onychomycosis caused by an isolate conforming to the description of Trichophyton raubitschekii. Med Mycol. 2004;42:273–276. doi: 10.1080/13693780310001602794. [DOI] [PubMed] [Google Scholar]
- 5.Kane J, Salkin IF, Weitzman I, Smitka CM. Trichophyton raubitschekii sp. nov. Mycotaxon. 1981;13:259–266. [Google Scholar]
- 6.Jackson CJ, Barton RC, Evans EG. Species identification and strain differentiation of dermatophyte fungi by analysis of ribosomal-DNA intergenic spacer regions. J Clin Microbiol. 1999;37:931–936. doi: 10.1128/jcm.37.4.931-936.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hiruma M, Kano R, Sugita T, Mochizuki T, Hasegawa A, Hiruma M. Urease gene of Trichophyton rubrum var. raubitschekii. J Dermatol. 2013;40:111–113. doi: 10.1111/1346-8138.12017. [DOI] [PubMed] [Google Scholar]
- 8.Graser Y, Kuijpers AF, Presber W, de Hoog GS. Molecular taxonomy of the Trichophyton rubrum complex. J Clin Microbiol. 2000;38:3329–3336. doi: 10.1128/jcm.38.9.3329-3336.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kane J, Krajden S, Summerbell RC, Sibbald RG. Infectious caused by Trichophyton raubitschekii: clinical and epidemiological features. Mycoses. 1990;33:499–506. doi: 10.1111/myc.1990.33.9-10.499. [DOI] [PubMed] [Google Scholar]
- 10.Gill D, Marks R. A review of the epidemiology of tinea unguium in the community. Australas J Dermatol. 1999;40:6–13. doi: 10.1046/j.1440-0960.1999.00308.x. [DOI] [PubMed] [Google Scholar]
- 11.Baran E, editor. Zarys mikologii lekarskiej, Volumed. Wroclaw, Poland; 1998.
- 12.Brasch J, Jensen JM. Tinea faciei caused by Trichophyton rubrum var. raubitschekii in Germany. Mikol Lek. 2008;15:109–111. [Google Scholar]
- 13.Brasch J, Gräser Y. Die Variante Raubitschekii von Trichophyton rubrum hat Deutschland erreicht. Der Hautarz. 2005;56:473–477. doi: 10.1007/s00105-004-0821-5. [DOI] [PubMed] [Google Scholar]
- 14.Tietz HJ, Hopp M, Gräser Y. First isolation of Trichophyton raubitschekii (syn. T. rubrum) in Europe. Mycoses. 2002;45:10–14. doi: 10.1046/j.0933-7407.2001.00717.x. [DOI] [PubMed] [Google Scholar]
- 15.Arabatzis M, Velegraki A, Kantardjiev T, Stavrakieva V, Rigopoulos D, Katsambas D. First report on autochthonous urease-positive Trichophyton rubrum (T. raubitschekii) from South-east Europe. Br J Dermatol. 2005;153:178–182. doi: 10.1111/j.1365-2133.2005.06615.x. [DOI] [PubMed] [Google Scholar]