Abstract
Background
Recently, more and more studies investigated the value of microRNA (miRNA) as a diagnostic or prognostic biomarker in various cancers. MiR-21 was found dysregulated in almost all types of cancers. While the prognostic role of miR-21 in many cancers has been studied, the results were not consistent.
Methods
We performed a meta-analysis to investigate the correlation between miR-21 and survival of general cancers by calculating pooled hazard ratios (HR) and 95% confidence intervals (CI).
Results
The pooled results of 63 published studies showed that elevated miR-21 was a predictor for poor survival of general carcinomas, with pooled HR of 1.91 (95%CI: 1.66–2.19) for OS, 1.42 (95% CI: 1.16–1.74) for DFS and 2.2 (95% CI: 1.64–2.96) for RFS/CSS. MiR-21 was also a prognostic biomarker in the patients who received adjuvant therapy, with pooled HR of 2.4 (95%CI: 1.18–4.9) for OS.
Conclusions
Our results showed that miR-21 could act as a significant biomarker in the prognosis of various cancers. Further studies are warranted before the application of the useful biomarker in the clinical.
Introduction
Due to the aging and growth of population as well as an increasing adoption of cancer-related lifestyle such as smoking and “westernized” diets, cancer has been a major public health problem all around the world [1]. Almost one in four deaths in the United States is related with cancer in 2012 [2]. Lack of efficiently diagnostic and prognostic biomarkers is responsible for the high mortality rates caused by cancer [3].
MicroRNAs (miRNAs), approximately 22 nucleotides in length, are a class of highly conserved RNAs that negatively regulate gene expression at post-transcriptional level by base pairing with the 3′-untranslated region of target mRNAs, resulting in either mRNA degradation or translational inhibition [4], [5]. Many studies have demonstrated that miRNAs play important roles in various biological processes, such as cellular development, differentiation, proliferation, cell death, angiogenesis and metabolism [6]–[9]. The success of utilizing miRNAs as diagnostic or prognostic markers from expression profiling has been reported in many studies.
MiR-21 was one of the most frequently studied cancer-related miRNAs and dysregulated in most cancers by acting as oncogene [10]–[14]. Up-regulated miR-21 could increase tumor growth, metastasis and invasion and reduce sensitivity to chemotherapy by its various targets [15]–[18]. Cancer patients with higher expression of miR-21 always had a worse prognostic outcome. But some studies represented inconsistent or even opposite results, such as the study of Valladares-Ayerbes et al. [19]. So we performed this meta-analysis to reveal the prognostic value of miR-21 in various cancers.
Material and Methods
Publication search and inclusion criteria
Medical subheading (Mesh) terms relating to miR-21 (e.g. “miR-21” or “microRNA-21”) in combination with words related to cancer (e.g. “cancer”, “tumor”, “carcinoma” or “neoplasm”) and terms to prognosis (e.g. “prognosis”, “survival”, “outcome” or “prognostic”) were searched on PubMed, EMBASE and WEB of science to retrieve eligible studies till December, 2013 .
We also carefully examined the references of articles and reviews to explore potentially additional studies. Studies were eligible if they met the following criteria: (a) studied patients with any type of cancers; (b) expression of miR-21 was measured; (c) the association between expression of miR-21 and clinical outcome was investigated; (d) full text articles in English. Studies were excluded based on the following criteria: (a) reviews, letters or laboratory studies; (b) studies had overlapping or duplicate data; (c) absence of key information for further analysis [20].
Data extraction
Data were evaluated and extracted independently from the eligible studies by two investigators (Zhou and Wang) under the guidelines of a critical review checklist of the Dutch Cochrane Centre proposed by Meta-analysis of Observational Studies in Epidemiology (MOOSE) [21]. The following items were recorded: first author's name, year of publication, country or area of origin, ethnicity, cancer type, sample type, TNM stage, method, total number of patients, cut-off value, follow ups and HRs of miR-21 for overall survival (OS), disease-free survival (DFS), recurrence-free survival (RFS) or cancer specific survival (CSS) with their 95% confidence intervals (CIs) and P value. If not available, data were extracted by the method of Tierney et al. [20]. When discrepancies existed between the two investigators, another investigator (Huang) was invited to discuss until a consensus was reached.
Statistical analysis
All the HRs with their 95% CIs were used to calculate pooled HRs. Cochran's Q test and Higgins I-squared statistic were used to check the heterogeneity of pooled results. A P<0.10 for Q-test suggested significant heterogeneity among studies, and the random-effects model (DerSimonian-Laird method) was applied to calculate the pooled HRs [22]. Otherwise, the fixed-effects model (Mantel-Haenszel method) was used [23]. Begg's funnel plot and the Egger's linear regression test were conducted to evaluate publication bias of literatures and a p<0.05 was considered significant [24]. Trim and fill method was applied to assess potential asymmetry in the funnel plot. Statistical analyses were performed in STATA software version 12.0 (STATA Corporation, College Station, TX, USA). All P values were two-sided.
Results
Study characteristics
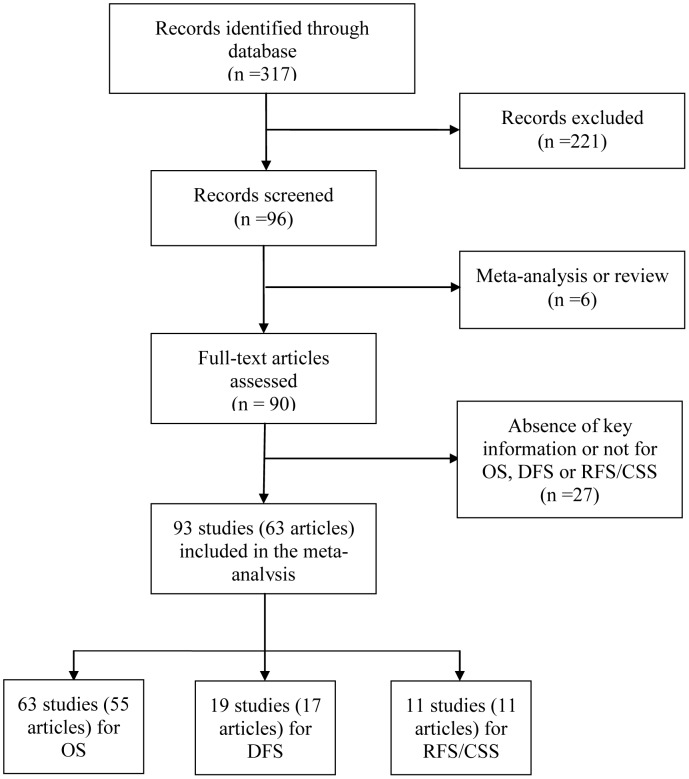
After careful read and selection, a total of 63 articles [19], were retrieved according to the inclusion and exclusion criteria. 55 of 63 articles investigated the prognostic role of miR-21 for OS, 17 for DFS, 8 for RFS and 3 for CSS. Schetter et al. [25], Hwang et al. [40] and Akagi et al. [79] presented separate HR by different ethnic background; Mathe et al. [33]_ENREF_34, Liu et al. [51], Toiyama et al. [80], Nielsen et al. [44] and Markou et al. [82] investigated the role of miR-21 in different type of samples; Voortman et al. [43] reported results from two centers. So the data from these studies were considered separately in our analysis. As there were only 3 studies for CSS, we combined the results for CSS and RFS together as RFS/CSS. Thus, a total of 63 studies including 6720 patients evaluating OS, 19 studies including 1965 cases for DFS and 11 studies including 1696 patients for RFS/CSS were considered in this analysis. The detailed screening process was shown in Figure 1.
Figure 1. Methodological flow diagram of the review.
The main characteristics of eligible studies were listed in Table 1. Ethnicity background of patients were classified as Asian, Caucasian and mixed populations. Cancer types of cases were various, among which lung cancer, pancreatic cancer and gastrointestinal (GI) cancers were mostly investigated. Tissue samples including Frozen or formalin-fixed and paraffin-embedded (FFPE) tissues were used in 53 studies, while 11 studies used circulation samples (plasma, serum or blood) and one study by Ota et al. [47] applied bone marrow samples. Quantitative real-time PCR (qRT-PCR) was widely used in 57 studies and in situ hybridisation (ISH) assay was used in the other 6 studies. The most frequently used cut-off value was the median which was applied in 26 studies and the other values ranged from the mean to the highest quarter value.
Table 1. Main characteristics of eligible studies.
| Author | Year | Country | Ethnicity | Type | Sample | Stage | Number | Method | Endogenous control | cut-off | Results |
| Schetter | 2008 | USA/HK | Caucasian/Asian | Colon | Frozen tissue | I-IV | 197 | qRT-PCR | U6 | Highest tertile | OS |
| Dillhoff | 2008 | USA | Caucasian | Pancreatic | FFPE | NR | 80 | In Situ Hybridization | U6 | Highest score | OS |
| Markou | 2008 | Greece | Caucasian | NSCLC | Frozen tissue | I-IV | 48 | qRT-PCR | U6 | 2-fold | OS and DFS |
| Yan | 2008 | China | Asian | Breast | FFPE | I-III | 113 | qRT-PCR | U6 | Mean | OS |
| Qian | 2009 | Italy | Caucasian | Breast | Frozen tissue | I-IV | 301 | qRT-PCR | U6 | NR | OS and DFS |
| Busacca | 2010 | Italy | Caucasian | Malignant mesothelioma | FFPE | NR | 24 | qRT-PCR | U6 | Median | OS |
| Li | 2009 | China | Asian | Tongue | Frozen tissue | I-IV | 103 | qRT-PCR | U6 | Median | OS |
| Schetter | 2009 | HK/USA | Caucasian/Asian | Colon | Frozen tissue | I-IV | 196 | qRT-PCR | U6 | Highest tertile | CSS |
| Mathe | 2009 | USA,Canada/Japan | Caucasian/Asian | Esophageal | Tissue | I-IV | 170 | qRT-PCR | U66 | Median | OS |
| Avissar | 2009 | USA | Caucasian | HNSCC | Frozen tissue | I-IV | 169 | qRT-PCR | U48 | Highest quarter | OS |
| Zhi | 2010 | China | Asian | Astrocytoma | Frozen tissue | I-IV | 124 | qRT-PCR | miR-16 | Median | OS |
| Hu | 2011 | USA | Caucasian | Esophageal | FFPE | I-IV | 158 | In situ hybridization | NR | 1–3+/0–0.5 | OS and DFS |
| Gao | 2010 | China | Asian | NSCLC | Frozen tissue | I-III | 47 | qRT-PCR | U6 | Median | OS |
| Rossi | 2010 | USA | Caucasian | CLL | Blood | NR | 99 | qRT-PCR | U6 | Median | OS |
| Giovannetti | 2010 | Netherlands | Caucasian | Pancreatic | Tissue | I-IV | 59 | qRT-PCR | U43 | Median | OS and DFS |
| Hwang | 2010 | Korea/Italy | Asian/Caucasian | Pancreatic | Frozen tissue | II-IV | 82/45 | qRT-PCR | U66/U43 | Median | OS,DFS and RFS |
| Gao | 2011 | China | Asian | SCLC | Frozen tissue | I-III | 30 | qRT-PCR | U6 | Median | OS |
| Kulda | 2010 | Czech Republic | Caucasian | CRC | Frozen tissue | I-IV | 44 | qRT-PCR | U6 | NR | DFS |
| Voortman | 2010 | 14 countries | Mixed | NSCLC | FFPE | I-III | 631 | qRT-PCR/In situ hybridization | U66/U6 | Median | OS |
| Nielsen | 2011 | Denmark | Caucasian | Colon/rectum | FFPE | II | 129/67 | In Situ Hybridization | NR | 2-fold | DFS |
| Hamano | 2011 | Japan | Asian | Esophageal | FFPE | I-IV | 98 | qRT-PCR | U48 | Median | OS |
| Radojicic | 2011 | Greece | Caucasian | Breast | FFPE | NR | 49 | qRT-PCR | RNU5A/U6 | Median | OS and DFS |
| Ota | 2011 | Japan | Asian | Breast | Bone marrow | NR | 207 | qRT-PCR | U6 | 5.84 | OS and DFS |
| Walter | 2011 | USA | Caucasian | Breast | FFPE | NR | 25 | qRT-PCR | U6 | Median | OS |
| Saito | 2011 | USA/Norway/Japan | Caucasian/Asian | NSCLC | Frozen tissue | I-II | 126/191 | qRT-PCR | U66 | Median | CSS/RFS |
| Shibuya | 2010 | Japan | Asian | CRC | Frozen tissue | Dukes:A-D | 156 | qRT-PCR | U6 | Mean | OS and DFS |
| Liu | 2012 | China | Asian | NSCLC | Frozen tissue | I-IV | 70 | qRT-PCR | U6 | 2-fold | OS |
| Wang | 2011 | China | Asian | NSCLC | Serum | I-III | 88 | qRT-PCR | U6 | 5-fold | OS |
| Ayerbes | 2011 | Spain | Caucasian | Colon or rectum/gastric/pancreas | FFPE | I-IV | 32 | qRT-PCR | U6 | Mean | OS |
| Jiang | 2011 | China | Asian | Gastric | FFPE | III,IV | 55 | qRT-PCR | U44 | NR | OS |
| Nagao | 2012 | Japan | Asian | Pancreatic | FFPE | I-IV | 65 | qRT-PCR | U6 | Mean | OS |
| Jamieson | 2012 | UK | Caucasian | Pancreatic | Frozen tissue | II-III | 72 | qRT-PCR | U6 | Median | OS |
| Jiang | 2012 | China | Asian | Melanoma | Frozen tissue | I-IV | 86 | qRT-PCR | U6 | Median | OS and DFS |
| Liu | 2012 | China | Asian | Pancreatic | Serum | I-IV | 38 | qRT-PCR | NR | NR | OS |
| Karakatsanis | 2013 | Greece | Caucasian | Hepatocellular | FFPE | I-IV | 60 | qRT-PCR | U6 | Mean | OS |
| Gao | 2012 | China | Asian | NSCLC | Frozen tissue | I-III | 58 | qRT-PCR | U6 | Median | DFS |
| Lee | 2011 | Korea | Asian | Breast | FFPE | I-III | 109 | qRT-PCR | U6 | Mean | OS and DFS |
| Li | 2012 | China | Asian | Prostate | FFPE | II-III | 168 | in situ hybridization | NR | Score>1 | RFS |
| Faltejs kova | 2012 | Czech Republic | Caucasian | CRC | Frozen tissue | I-IV | 44 | qRT-PCR | U6 | Median | OS |
| Faragalla | 2012 | Canada | Caucasian | Renal | FFPE | I-III | 89 | qRT-PCR | U44 | NR | OS and DFS |
| Zaravinos | 2012 | Greece | Caucasian | Bladder | Tissue | NR | 77 | qRT-PCR | RNU1A1,5A and U6 | Median | OS and RFS |
| Jung | 2012 | USA | Caucasian | Oral | Frozen tissue | NR | 17 | qRT-PCR | U6 | Median | OS |
| Le | 2012 | China | Asian | Lung | Serum | I-IV | 82 | qRT-PCR | miR-16 | NR | OS |
| Xu | 2012 | China | Asian | Gastric | Frozen tissue | I-IV | 86 | qRT-PCR | Let-7a | ROC curve (AUC) | OS |
| Osawa | 2011 | Japan | Asian | Gastric | FFPE | I-IV | 37 | qRT-PCR | NR | T/N ratio >1.40 | OS |
| Papaconstantinou | 2013 | Greece | Caucasian | Pancreatic | FFPE | I-IV | 88 | qRT-PCR | U6 | Mean | OS |
| Frifeldt | 2012 | Denmark | Caucasian | Colon | FFPE | II | 520 | in situ hybridization | NR | Tertiles | OS and RFS |
| Hermansen | 2013 | Denmark | Caucasian | Gliomas | FFPE | NR | 189 | in situ hybridization | NR | NR | OS |
| Caponi | 2013 | UK/Italy | Caucasian | Pancreatic | FFPE | II-III | 81 | qRT-PCR | U6 | Median | OS and DFS |
| Wang | 2013 | China | Asian | Pancreatic | Serum | III-IV | 177 | qRT-PCR | U6 | Median | OS |
| Komatsu | 2013 | Japan | Asian | Gastric | Plasma | I-IV | 69 | qRT-PCR | NR | Median | CSS |
| Amankwah | 2013 | USA | Caucasian | Prostate | FFPE | I-IV | 65 | qRT-PCR | U6 | median | RFS |
| Chusorn | 2013 | Thailand | Asian | Cholangiocarcinoma | Frozen tissue | NR | 23 | qRT-PCR | U6 | Mean | OS |
| Huang | 2013 | China | Asian | Cholangiocarcinoma | FFPE | NR | 41 | qRT-PCR | U6 | NR | OS and RFS |
| Liu | 2013 | China | Asian | CRC | Serum | I-IV | 166 | qRT-PCR | MiR-16 | 0.0043 | OS |
| Akagi | 2013 | USA,Norway/Japan | Caucasian/Asian | Lung | Frozen tissue | I-II | 92/198 | qRT-PCR | NR | Median | OS and RFS |
| Toiyama | 2013 | Japan | Asian | CRC | FFPE/serum | I-IV | 166/188 | qRT-PCR | miR-16/Cel-miR-39 | Youden's index | OS |
| Bovell | 2013 | USA | Mixed | CRC | FFPE | IV | 55 | qRT-PCR | U6 | NR | OS |
| Markou | 2013 | Greece | Caucasian | NSCLC | FFPE/plasma | I-IV | 40/37 | qRT-PCR | miR-191/miR-16 | Median | OS and DFS |
| Chen | 2013 | Taiwan | Asian | CRC | Tissue | I-IV | 195 | qRT-PCR | U6 | Mean | OS |
| Ferrajoli | 2013 | USA | Caucasian | CLL | Blood | NR | 93 | qRT-PCR | miR-16 | 44th percentile | OS |
| Menendez | 2013 | Spain | Caucasian | CRC | Serum | I-IV | 102 | qRT-PCR | miR-16 | Relative expression>1 | OS and DFS |
| Kadera | 2013 | USA | Caucasian | Pancreatic | Tissue | I-IV | 147 | qRT-PCR | U6 | NR | OS |
NSCLC: non-small cell lung cancer; HNSCC: head and neck squamous cell carcinomas; CLL: chronic lymphocytic leukemia; SCLC: squamous cell lung carcinoma; CRC: colorectal carcinoma; ALL: acute lymphoblastic leukemia NR: not reported; FFPE: formalin-fixed and paraffin-embedded; OS: overall survival; DFS: disease-free survival; RFS: recurrence-free survival; CSS: cancer-specific survival.
Outcomes from eligible studies
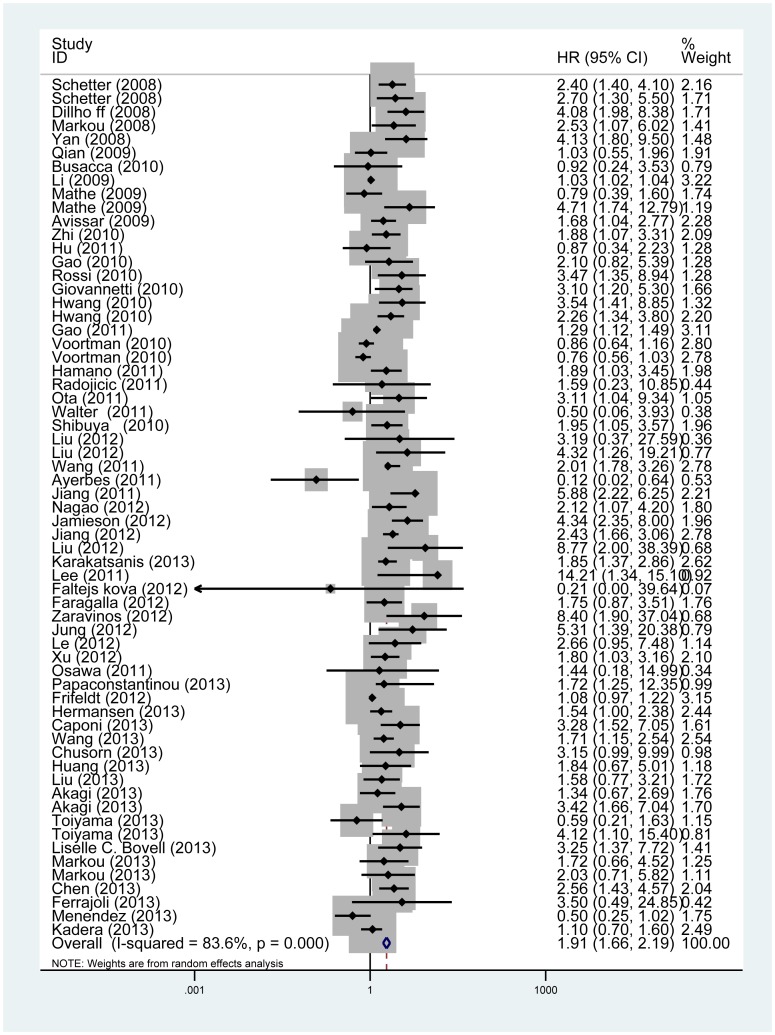
The main results of this meta-analysis are shown in Table 2. For 63 studies evaluating OS for miR-21, we found high expression of miR-21 predicting a worse outcome with the combined HR of 1.91 (95%CI: 1.66–2.19; Pheterogeneity<0.001; Figure 2). Similarly predictive roles of miR-21 for DFS and RFS/CSS were also investigated with pooled HR of 1.42 (95% CI: 1.16–1.74; Pheterogeneity = 0.001) and 2.2 (95% CI: 1.64–2.96; Pheterogeneity = 0.022), respectively.
Table 2. Meta-analysis results.
| Outcome | Variables | Number of studies | Model | HR (95% CI) | Pheterogeneity |
| OS | ALL | 63 | Random | 1.91(1.66,2.19) | <0.001 |
| Cancer type | |||||
| GI | 15 | Random | 1.68(1.12,2.52) | <0.001 | |
| Pancreas | 11 | Random | 2.53(1.82,3.51) | 0.003 | |
| Lung | 13 | Random | 1.59(1.2,2.1) | <0.001 | |
| Breast | 6 | Random | 2.55(1.04,6.29) | 0.002 | |
| Oral | 2 | Random | 2.02(0.41,9.88) | 0.016 | |
| Esophagus | 4 | Random | 1.53(0.74,3.15) | 0.018 | |
| Liver | 3 | Fixed | 1.93(1.39,2.69) | 0.688 | |
| Ethnicity | |||||
| Asian | 29 | Random | 2.19(1.76,2.73) | <0.001 | |
| Caucasian | 29 | Random | 1.86(1.46,2.37) | <0.001 | |
| Sample | |||||
| Tissue | 51 | Random | 1.87(1.61,2.16) | <0.001 | |
| FFPE | 25 | Random | 1.68(1.29,2.18) | <0.001 | |
| Frozen tissue | 23 | Random | 1.99(1.59,2.49) | <0.001 | |
| Circulation | 11 | Random | 2.06(1.42,2.99) | 0.008 | |
| Serum | 8 | Random | 1.94(1.25,3.03) | 0.003 | |
| Therapy | |||||
| Adjuvant therapy | 7 | Random | 2.4(1.18,4.9) | <0.001 | |
| Mixed | 56 | Random | 1.85(1.61,2.13) | <0.001 | |
| DFS | ALL | 19 | Random | 1.42(1.16,1.74) | 0.001 |
| Cancer type | |||||
| GI | 5 | Random | 1.12(0.81,1.55) | 0.01 | |
| Pancreas | 3 | Fixed | 2.87(1.89,4.35) | 0.524 | |
| Lung | 4 | Fixed | 2.05(1.32,3.18) | 0.839 | |
| Breast | 4 | Fixed | 1.1(0.82,1.49) | 0.919 | |
| Ethnicity | |||||
| Asian | 6 | Random | 1.62(1.06,2.47) | 0.008 | |
| Caucasian | 14 | Random | 1.37(1.07,1.76) | 0.006 | |
| RFS/CSS | ALL | 11 | Random | 2.2(1.64,2.96) | 0.022 |
| Cancer type | |||||
| GI | 3 | Random | 2.5(1.1,5.71) | 0.005 | |
| Lung | 3 | Fixed | 2.25(1.57,3.23) | 0.605 | |
| Prostate | 2 | Fixed | 2.04(1.17,3.54) | 0.957 | |
| Ethnicity | |||||
| Asian | 5 | Fixed | 2.17(1.52,3.09) | 0.322 | |
| Caucasian | 5 | Random | 2.1(1.34,3.27) | 0.065 | |
OS: overall survival; DFS: disease-free survival; RFS: recurrence-free survival; CSS: cancer-specific survival; GI: gastrointestinal; FFPE: formalin-fixed and paraffin-embedded.
Figure 2. Forrest plots of studies evaluating hazard ratios (HRs) of miR-21 for overall survival.
Subgroup analyses by cancer type showed that elevated miR-21 yielded a worse OS in GI cancers (HR = 1.68, 95%CI: 1.12–2.52; Pheterogeneity<0.001), lung cancer (HR = 1.59, 95%CI: 1.2–2.1; Pheterogeneity<0.001), breast cancer (HR = 2.55, 95%CI: 1.04–6.29; Pheterogeneity = 0.002), pancreatic cancer (HR = 2.53, 95%CI: 1.82–3.51; Pheterogeneity = 0.003) and liver cancer (HR = 1.93, 95%CI: 1.39–2.69; Pheterogeneity = 0.688); a worse DFS in lung cancer (HR = 2.05, 95%CI: 1.32–3.18; Pheterogeneity = 0.839) and pancreatic cancer (HR = 2.87, 95%CI: 1.89–4.35; Pheterogeneity = 0.524); a poorer RFS/CSS in GI cancers (HR = 2.5, 95%CI: 1.1–5.71; Pheterogeneity = 0.005), lung cancer (HR = 2.25, 95%CI: 1.57–3.23; Pheterogeneity = 0.605) and prostate cancer (HR = 2.04, 95%CI: 1.17–3.54; Pheterogeneity = 0.957).
In the subgroup analyses by ethnicity, we found that no matter the cases were Asian or Caucasian, the high expression of miR-21 was still a significantly poor predictor for OS (Asian: HR = 2.19, 95%CI: 1.76–2.73; Pheterogeneity<0.001; Caucasian: HR = 1.86, 95%CI: 1.46–2.37; Pheterogeneity<0.001), DFS (Asian: HR = 1.62, 95%CI: 1.06–2.47; Pheterogeneity = 0.008; Caucasian: HR = 1.37, 95%CI: 1.07–1.76; Pheterogeneity = 0.006) and RFS/CSS (Asian: HR = 2.17, 95%CI: 1.52–3.09; Pheterogeneity = 0.322; Caucasian: HR = 2.1, 95%CI: 1.34–3.27; Pheterogeneity = 0.065).
Further analyses of studies evaluating OS by sample type also revealed that high expression of miR-21 remained to be a worse prognostic marker regardless of sample source (tissue sample: HR = 1.87, 95%CI: 1.61–2.16; Pheterogeneity<0.001; circulation sample: HR = 2.06, 95%CI: 1.42–2.99; Pheterogeneity = 0.008). In addition, high miR-21 in FFPE (HR = 1.68, 95%CI: 1.29–2.18; Pheterogeneity<0.001) and frozen tissue (HR = 1.99, 95%CI: 1.59–2.49; Pheterogeneity<0.001) showed consistent results. Pooled results of 8 studies that explored serum miR-21 also revealed negative prognostic role of increased miR-21 (HR = 1.94, 95%CI: 1.25–3.03; Pheterogeneity = 0.003)
A total of seven studies [27], [39], [40], [43], [53], [82] investigated the prognostic role of miR-21 in the patients who received adjuvant therapy which yielded a significantly pooled HR of 2.4 (95%CI: 1.18–4.9; Pheterogeneity<0.001).
Publication bias
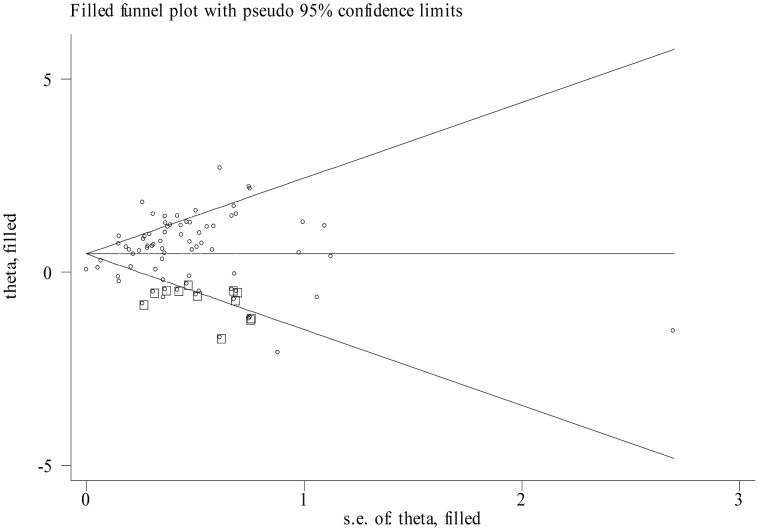
Begg's funnel plot and the Egger's linear regression test were used to assess publication bias. However, the funnel plots were asymmetric and the P values of Egger's test for OS, DFS and RFS/CSS were <0.001, 0.011 and 0.003, respectively. Thus, a trim and fill method was conducted and pooled HRs were recalculated with hypothetically non-published studies to evaluate the asymmetry in the funnel plots. The recalculated HRs did not change significantly for OS (HR = 1.61, 95%CI: 1.41–1.83; Pheterogeneity<0.001;Figure 3) and RFS/CSS (HR = 2.01, 95%CI: 1.54–2.77; Pheterogeneity = 0.018). But the prognostic role of high expression of miR-21 for DFS was weaken with a recalculated HR of 1.11 (95%CI: 0.9–1.38; Pheterogeneity<0.001).
Figure 3. Funnel plot adjusted with trim and fill method for overall survival.
Circles: included studies. Diamonds: presumed missing studies.
Discussion
MiR-21, a well-known onco-miR, is up-regulated in most malignancies. Acting on various target genes such as PTEN [87] and PDCD4 [18], miR-21 plays an important role in the process of cell proliferation, migration, invasion, drug resistance [88] and so on. It has been reported that miR-21 could regulate Ras/MEK/ERK pathway so to influence the tumor formation. Moreover the incidence of lung tumors is higher in miR-21 overexpression mice, while lower in miR-21 knockout mice [89]. Additionally, miR-21 has been proposed as a marker of cancers for diagnosis in circulation [90], [91], stool [92] and sputum [93], prediction in therapy response [59] and prognosis of patients.
Nair et al. [94] systematically reviewed and synthesized that miRNAs showed promising associations with outcomes of various cancers. As the first meta-analysis [95] of miR-21 related to outcomes of various cancers, Fu et al. retrieved 17 studies and found higher level of miR-21 might be associated with poorer clinical outcome, especially in subgroup of head and neck squamous cell carcinoma and digestive carcinoma. Recently, Wang et al. [96] analyzed the value of circulating miR-21 and yielded a conclusion that circulating miR-21 might act as a significantly prognostic biomarker but not be suitable for a sensitive diagnostic biomarker. However, the number of studies included in these analyses was relatively small and the obtained results might not be powerful. In terms of this, we performed this updated meta-analysis including 63 articles and demonstrated that high expression of miR-21 was a significant marker for predicting worse outcomes of various cancers (HR was 1.91, 2.2 and 1.42 for OS, RFS/CSS and DFS, respectively). Subgroup analyses revealed that high expression of miR-21 could predict a worse OS in GI tumors, pancreatic cancer, lung cancer, breast cancer and liver cancer, a worse DFS in pancreatic cancer and lung cancer and poor RFS/CSS in GI tumors, lung cancer and prostate cancer. Regardless of the ethnicity background or sample source, high expression level of miR-21 was a significantly negative prognostic marker for various malignancies. As publication bias was observed, a trim and fill method was adopted to calculate the adjusted HRs. The results for OS and RFS/CSS did not change, but the results for DFS were altered.
Recently, many studies demonstrated that miRNAs including miR-21 had great potential as biomarkers for various cancers. However, several problems should be well solved before utilizing them as diagnostic or prognostic biomarkers in the clinical. As is known, non-invasive circulation sample (plasma/serum) or body fluid sample could be obtained more conveniently than tissue sample. However, studies using different types of samples may yield different results [51]. Tsujiura et al. [91] found that some individuals might even have opposite tendency of the expression levels of miRNAs in tumor tissue and plasma. Now, many studies have investigated the clinical impact of miRNAs from exosomes which were small membrane vesicles containing proteins and nucleotides [97]. In our study, it is pleasing that high expression of miR-21 in the tissue (FFPE/frozen tissue) or circulation both predicted poor outcomes. Thus, we might assume that patients with high expression of miR-21 from any type of sample might suffer worse clinical outcomes. Yet, normalization among different studies was not consistent. The internal controls used for tissue samples are relatively consistent ranging from U6 to U44, while there is no consensus on suitable small RNA reference genes for circulation or body fluid sample. MiR-16 was used as a reference gene in some studies [66], [78]. But the optimal way for miRNA normalization in circulation or body fluid sample is probably the spiked-in normalization method [98]. Therefore, future studies focusing on the consistent normalization are warranted. In addition, as biomarkers, a panel of miRNAs might be more sensitive and specific than a single miRNA [99], [100]. The combination of miR-21 and some specific miRNAs might elevate its predictive power. Finally, methods for detecting miRNAs were diverse, among which RT-PCR was one of the most widely used approaches. Nevertheless many new methodologies emerged, such as the next-generation sequencing approach [101] and the electrochemical approach [102]. In short, a proper method for clinical application should be less expensive, reproducible, stable and with high sensitivity and specificity. Accordingly, great efforts should be made in the future to apply miRNAs including miR-21 as reliable biomarkers in the clinical.
Several limitations of this study should be considered. First, the studies retrieved in our study were limited in English, which might partially contribute to the observed publication bias. By conducting the trim and fill method, we found that the pooled results did not change significantly except for DFS. Thus, attention should be paid to the prognostic role of miR-21 for DFS. Second, different countries, cancer types, methods and other variables might contribute to the relatively large heterogeneity in this study. Third, the number of studies investigating some special types of cancer was small. For instance, there was only one study focusing on mesothelioma [30]. More studies on these cancers are needed in the future.
In conclusion, the evidence from the meta-analysis revealed that high expression level of miR-21 was a negative predictor for survival in various cancers, especially for OS and RFS/CSS. However, our results should be considered with caution due to the limitations listed above. To better understand and use miRNAs as biomarkers in the clinical, more large-scale and standard investigations are worth conducting.
Supporting Information
PRISMA Checklist.
(DOC)
Funding Statement
This work was supported by National Natural Science Foundation of China (Grant number: 81171908). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, et al. (2011) Global cancer statistics. CA Cancer J Clin 61: 69–90. [DOI] [PubMed] [Google Scholar]
- 2. Siegel R, Naishadham D, Jemal A (2012) Cancer statistics, 2012. CA Cancer J Clin 62: 10–29. [DOI] [PubMed] [Google Scholar]
- 3. Iorio MV, Croce CM (2012) MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med 4: 143–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854. [DOI] [PubMed] [Google Scholar]
- 5. Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- 6. Carrington JC, Ambros V (2003) Role of microRNAs in plant and animal development. Science 301: 336–338. [DOI] [PubMed] [Google Scholar]
- 7. Suarez Y, Sessa WC (2009) MicroRNAs as novel regulators of angiogenesis. Circ Res 104: 442–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xu P, Guo M, Hay BA (2004) MicroRNAs and the regulation of cell death. Trends Genet 20: 617–624. [DOI] [PubMed] [Google Scholar]
- 9. Bartel DP, Chen CZ (2004) Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet 5: 396–400. [DOI] [PubMed] [Google Scholar]
- 10. Schee K, Lorenz S, Worren MM, Gunther CC, Holden M, et al. (2013) Deep Sequencing the MicroRNA Transcriptome in Colorectal Cancer. PLoS One 8: e66165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Capodanno A, Boldrini L, Proietti A, Ali G, Pelliccioni S, et al. (2013) Let-7g and miR-21 expression in non-small cell lung cancer: Correlation with clinicopathological and molecular features. Int J Oncol 43: 765–774. [DOI] [PubMed] [Google Scholar]
- 12. Gombos K, Horvath R, Szele E, Juhasz K, Gocze K, et al. (2013) miRNA expression profiles of oral squamous cell carcinomas. Anticancer Res 33: 1511–1517. [PubMed] [Google Scholar]
- 13. Li T, Li D, Sha J, Sun P, Huang Y (2009) MicroRNA-21 directly targets MARCKS and promotes apoptosis resistance and invasion in prostate cancer cells. Biochem Biophys Res Commun 383: 280–285. [DOI] [PubMed] [Google Scholar]
- 14. Kumarswamy R, Volkmann I, Thum T (2011) Regulation and function of miRNA-21 in health and disease. RNA Biol 8: 706–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Soria-Valles C, Gutierrez-Fernandez A, Guiu M, Mari B, Fueyo A, et al. (2013) The anti-metastatic activity of collagenase-2 in breast cancer cells is mediated by a signaling pathway involving decorin and miR-21. Oncogene. [DOI] [PubMed] [Google Scholar]
- 16. Seca H, Lima RT, Lopes-Rodrigues V, Guimaraes JE, Almeida GM, et al. (2013) Targeting miR-21 Induces Autophagy and Chemosensitivity of Leukemia Cells. Curr Drug Targets. [DOI] [PubMed] [Google Scholar]
- 17. Liu S, Fang Y, Shen H, Xu W, Li H (2013) Berberine sensitizes ovarian cancer cells to cisplatin through miR-21/PDCD4 axis. Acta Biochim Biophys Sin (Shanghai). [DOI] [PubMed] [Google Scholar]
- 18. Zhou L, Yang ZX, Song WJ, Li QJ, Yang F, et al. (2013) MicroRNA-21 regulates the migration and invasion of a stem-like population in hepatocellular carcinoma. Int J Oncol 43: 661–669. [DOI] [PubMed] [Google Scholar]
- 19. Valladares-Ayerbes M, Blanco M, Haz M, Medina V, Iglesias-Diaz P, et al. (2011) Prognostic impact of disseminated tumor cells and microRNA-17-92 cluster deregulation in gastrointestinal cancer. Int J Oncol 39: 1253–1264. [DOI] [PubMed] [Google Scholar]
- 20. Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR (2007) Practical methods for incorporating summary time-to-event data into meta-analysis. Trials 8: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stroup DF, Berlin JA, Morton SC, Olkin I, Williamson GD, et al. (2000) Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA 283: 2008–2012. [DOI] [PubMed] [Google Scholar]
- 22. DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7: 177–188. [DOI] [PubMed] [Google Scholar]
- 23. Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22: 719–748. [PubMed] [Google Scholar]
- 24. Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, et al. (2008) MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA 299: 425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dillhoff M, Liu J, Frankel W, Croce C, Bloomston M (2008) MicroRNA-21 is overexpressed in pancreatic cancer and a potential predictor of survival. J Gastrointest Surg 12: 2171–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Markou A, Tsaroucha EG, Kaklamanis L, Fotinou M, Georgoulias V, et al. (2008) Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non-small cell lung cancer by quantitative real-time RT-PCR. Clin Chem 54: 1696–1704. [DOI] [PubMed] [Google Scholar]
- 28. Yan LX, Huang XF, Shao Q, Huang MY, Deng L, et al. (2008) MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA 14: 2348–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Qian B, Katsaros D, Lu L, Preti M, Durando A, et al. (2009) High miR-21 expression in breast cancer associated with poor disease-free survival in early stage disease and high TGF-beta1. Breast Cancer Res Treat 117: 131–140. [DOI] [PubMed] [Google Scholar]
- 30. Busacca S, Germano S, De Cecco L, Rinaldi M, Comoglio F, et al. (2010) MicroRNA signature of malignant mesothelioma with potential diagnostic and prognostic implications. Am J Respir Cell Mol Biol 42: 312–319. [DOI] [PubMed] [Google Scholar]
- 31. Li J, Huang H, Sun L, Yang M, Pan C, et al. (2009) MiR-21 indicates poor prognosis in tongue squamous cell carcinomas as an apoptosis inhibitor. Clin Cancer Res 15: 3998–4008. [DOI] [PubMed] [Google Scholar]
- 32. Schetter AJ, Nguyen GH, Bowman ED, Mathe EA, Yuen ST, et al. (2009) Association of inflammation-related and microRNA gene expression with cancer-specific mortality of colon adenocarcinoma. Clin Cancer Res 15: 5878–5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mathe EA, Nguyen GH, Bowman ED, Zhao Y, Budhu A, et al. (2009) MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: associations with survival. Clin Cancer Res 15: 6192–6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Avissar M, McClean MD, Kelsey KT, Marsit CJ (2009) MicroRNA expression in head and neck cancer associates with alcohol consumption and survival. Carcinogenesis 30: 2059–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhi F, Chen X, Wang S, Xia X, Shi Y, et al. (2010) The use of hsa-miR-21, hsa-miR-181b and hsa-miR-106a as prognostic indicators of astrocytoma. Eur J Cancer 46: 1640–1649. [DOI] [PubMed] [Google Scholar]
- 36. Hu Y, Correa AM, Hoque A, Guan B, Ye F, et al. (2011) Prognostic significance of differentially expressed miRNAs in esophageal cancer. Int J Cancer 128: 132–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gao W, Yu Y, Cao H, Shen H, Li X, et al. (2010) Deregulated expression of miR-21, miR-143 and miR-181a in non small cell lung cancer is related to clinicopathologic characteristics or patient prognosis. Biomed Pharmacother 64: 399–408. [DOI] [PubMed] [Google Scholar]
- 38. Rossi S, Shimizu M, Barbarotto E, Nicoloso MS, Dimitri F, et al. (2010) microRNA fingerprinting of CLL patients with chromosome 17p deletion identify a miR-21 score that stratifies early survival. Blood 116: 945–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Giovannetti E, Funel N, Peters GJ, Del Chiaro M, Erozenci LA, et al. (2010) MicroRNA-21 in pancreatic cancer: correlation with clinical outcome and pharmacologic aspects underlying its role in the modulation of gemcitabine activity. Cancer Res 70: 4528–4538. [DOI] [PubMed] [Google Scholar]
- 40. Hwang JH, Voortman J, Giovannetti E, Steinberg SM, Leon LG, et al. (2010) Identification of microRNA-21 as a biomarker for chemoresistance and clinical outcome following adjuvant therapy in resectable pancreatic cancer. PLoS One 5: e10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gao W, Shen H, Liu L, Xu J, Shu Y (2011) MiR-21 overexpression in human primary squamous cell lung carcinoma is associated with poor patient prognosis. J Cancer Res Clin Oncol 137: 557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kulda V, Pesta M, Topolcan O, Liska V, Treska V, et al. (2010) Relevance of miR-21 and miR-143 expression in tissue samples of colorectal carcinoma and its liver metastases. Cancer Genet Cytogenet 200: 154–160. [DOI] [PubMed] [Google Scholar]
- 43. Voortman J, Goto A, Mendiboure J, Sohn JJ, Schetter AJ, et al. (2010) MicroRNA expression and clinical outcomes in patients treated with adjuvant chemotherapy after complete resection of non-small cell lung carcinoma. Cancer Res 70: 8288–8298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nielsen BS, Jorgensen S, Fog JU, Sokilde R, Christensen IJ, et al. (2011) High levels of microRNA-21 in the stroma of colorectal cancers predict short disease-free survival in stage II colon cancer patients. Clin Exp Metastasis 28: 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hamano R, Miyata H, Yamasaki M, Kurokawa Y, Hara J, et al. (2011) Overexpression of miR-200c induces chemoresistance in esophageal cancers mediated through activation of the Akt signaling pathway. Clin Cancer Res 17: 3029–3038. [DOI] [PubMed] [Google Scholar]
- 46. Radojicic J, Zaravinos A, Vrekoussis T, Kafousi M, Spandidos DA, et al. (2011) MicroRNA expression analysis in triple-negative (ER, PR and Her2/neu) breast cancer. Cell Cycle 10: 507–517. [DOI] [PubMed] [Google Scholar]
- 47. Ota D, Mimori K, Yokobori T, Iwatsuki M, Kataoka A, et al. (2011) Identification of recurrence-related microRNAs in the bone marrow of breast cancer patients. Int J Oncol 38: 955–962. [DOI] [PubMed] [Google Scholar]
- 48. Walter BA, Gomez-Macias G, Valera VA, Sobel M, Merino MJ (2011) miR-21 Expression in Pregnancy-Associated Breast Cancer: A Possible Marker of Poor Prognosis. J Cancer 2: 67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Saito M, Schetter AJ, Mollerup S, Kohno T, Skaug V, et al. (2011) The association of microRNA expression with prognosis and progression in early-stage, non-small cell lung adenocarcinoma: a retrospective analysis of three cohorts. Clin Cancer Res 17: 1875–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shibuya H, Iinuma H, Shimada R, Horiuchi A, Watanabe T (2010) Clinicopathological and prognostic value of microRNA-21 and microRNA-155 in colorectal cancer. Oncology 79: 313–320. [DOI] [PubMed] [Google Scholar]
- 51. Liu XG, Zhu WY, Huang YY, Ma LN, Zhou SQ, et al. (2012) High expression of serum miR-21 and tumor miR-200c associated with poor prognosis in patients with lung cancer. Med Oncol 29: 618–626. [DOI] [PubMed] [Google Scholar]
- 52. Wang ZX, Bian HB, Wang JR, Cheng ZX, Wang KM, et al. (2011) Prognostic significance of serum miRNA-21 expression in human non-small cell lung cancer. J Surg Oncol 104: 847–851. [DOI] [PubMed] [Google Scholar]
- 53. Jiang J, Zheng X, Xu X, Zhou Q, Yan H, et al. (2011) Prognostic significance of miR-181b and miR-21 in gastric cancer patients treated with S-1/Oxaliplatin or Doxifluridine/Oxaliplatin. PLoS One 6: e23271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nagao Y, Hisaoka M, Matsuyama A, Kanemitsu S, Hamada T, et al. (2012) Association of microRNA-21 expression with its targets, PDCD4 and TIMP3, in pancreatic ductal adenocarcinoma. Mod Pathol 25: 112–121. [DOI] [PubMed] [Google Scholar]
- 55. Jamieson NB, Morran DC, Morton JP, Ali A, Dickson EJ, et al. (2012) MicroRNA molecular profiles associated with diagnosis, clinicopathologic criteria, and overall survival in patients with resectable pancreatic ductal adenocarcinoma. Clin Cancer Res 18: 534–545. [DOI] [PubMed] [Google Scholar]
- 56. Jiang L, Lv X, Li J, Li X, Li W, et al. (2012) The status of microRNA-21 expression and its clinical significance in human cutaneous malignant melanoma. Acta Histochem 114: 582–588. [DOI] [PubMed] [Google Scholar]
- 57. Liu R, Chen X, Du Y, Yao W, Shen L, et al. (2012) Serum microRNA expression profile as a biomarker in the diagnosis and prognosis of pancreatic cancer. Clin Chem 58: 610–618. [DOI] [PubMed] [Google Scholar]
- 58. Karakatsanis A, Papaconstantinou I, Gazouli M, Lyberopoulou A, Polymeneas G, et al. (2013) Expression of microRNAs, miR-21, miR-31, miR-122, miR-145, miR-146a, miR-200c, miR-221, miR-222, and miR-223 in patients with hepatocellular carcinoma or intrahepatic cholangiocarcinoma and its prognostic significance. Mol Carcinog 52: 297–303. [DOI] [PubMed] [Google Scholar]
- 59. Gao W, Lu X, Liu L, Xu J, Feng D, et al. (2012) MiRNA-21: a biomarker predictive for platinum-based adjuvant chemotherapy response in patients with non-small cell lung cancer. Cancer Biol Ther 13: 330–340. [DOI] [PubMed] [Google Scholar]
- 60. Lee JA, Lee HY, Lee ES, Kim I, Bae JW (2011) Prognostic Implications of MicroRNA-21 Overexpression in Invasive Ductal Carcinomas of the Breast. J Breast Cancer 14: 269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Li T, Li RS, Li YH, Zhong S, Chen YY, et al. (2012) miR-21 as an independent biochemical recurrence predictor and potential therapeutic target for prostate cancer. J Urol 187: 1466–1472. [DOI] [PubMed] [Google Scholar]
- 62. Faltejskova P, Besse A, Sevcikova S, Kubiczkova L, Svoboda M, et al. (2012) Clinical correlations of miR-21 expression in colorectal cancer patients and effects of its inhibition on DLD1 colon cancer cells. Int J Colorectal Dis 27: 1401–1408. [DOI] [PubMed] [Google Scholar]
- 63. Faragalla H, Youssef YM, Scorilas A, Khalil B, White NM, et al. (2012) The clinical utility of miR-21 as a diagnostic and prognostic marker for renal cell carcinoma. J Mol Diagn 14: 385–392. [DOI] [PubMed] [Google Scholar]
- 64. Zaravinos A, Radojicic J, Lambrou GI, Volanis D, Delakas D, et al. (2012) Expression of miRNAs involved in angiogenesis, tumor cell proliferation, tumor suppressor inhibition, epithelial-mesenchymal transition and activation of metastasis in bladder cancer. J Urol 188: 615–623. [DOI] [PubMed] [Google Scholar]
- 65. Jung HM, Phillips BL, Patel RS, Cohen DM, Jakymiw A, et al. (2012) Keratinization-associated miR-7 and miR-21 regulate tumor suppressor reversion-inducing cysteine-rich protein with kazal motifs (RECK) in oral cancer. J Biol Chem 287: 29261–29272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Le HB, Zhu WY, Chen DD, He JY, Huang YY, et al. (2012) Evaluation of dynamic change of serum miR-21 and miR-24 in pre- and post-operative lung carcinoma patients. Med Oncol 29: 3190–3197. [DOI] [PubMed] [Google Scholar]
- 67. Xu Y, Sun J, Xu J, Li Q, Guo Y, et al. (2012) miR-21 Is a Promising Novel Biomarker for Lymph Node Metastasis in Patients with Gastric Cancer. Gastroenterol Res Pract 2012: 640168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Osawa S, Shimada Y, Sekine S, Okumura T, Nagata T, et al. (2011) MicroRNA profiling of gastric cancer patients from formalin-fixed paraffin-embedded samples. Oncol Lett 2: 613–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Papaconstantinou IG, Manta A, Gazouli M, Lyberopoulou A, Lykoudis PM, et al. (2013) Expression of microRNAs in patients with pancreatic cancer and its prognostic significance. Pancreas 42: 67–71. [DOI] [PubMed] [Google Scholar]
- 70. Kjaer-Frifeldt S, Hansen TF, Nielsen BS, Joergensen S, Lindebjerg J, et al. (2012) The prognostic importance of miR-21 in stage II colon cancer: a population-based study. Br J Cancer 107: 1169–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hermansen SK, Dahlrot RH, Nielsen BS, Hansen S, Kristensen BW (2013) MiR-21 expression in the tumor cell compartment holds unfavorable prognostic value in gliomas. J Neurooncol 111: 71–81. [DOI] [PubMed] [Google Scholar]
- 72. Caponi S, Funel N, Frampton AE, Mosca F, Santarpia L, et al. (2013) The good, the bad and the ugly: a tale of miR-101, miR-21 and miR-155 in pancreatic intraductal papillary mucinous neoplasms. Ann Oncol 24: 734–741. [DOI] [PubMed] [Google Scholar]
- 73. Wang P, Zhuang L, Zhang J, Fan J, Luo J, et al. (2013) The serum miR-21 level serves as a predictor for the chemosensitivity of advanced pancreatic cancer, and miR-21 expression confers chemoresistance by targeting FasL. Mol Oncol 7: 334–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Komatsu S, Ichikawa D, Tsujiura M, Konishi H, Takeshita H, et al. (2013) Prognostic impact of circulating miR-21 in the plasma of patients with gastric carcinoma. Anticancer Res 33: 271–276. [PubMed] [Google Scholar]
- 75. Amankwah EK, Anegbe E, Park H, Pow-Sang J, Hakam A, et al. (2013) miR-21, miR-221 and miR-222 expression and prostate cancer recurrence among obese and non-obese cases. Asian J Androl 15: 226–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Chusorn P, Namwat N, Loilome W, Techasen A, Pairojkul C, et al. (2013) Overexpression of microRNA-21 regulating PDCD4 during tumorigenesis of liver fluke-associated cholangiocarcinoma contributes to tumor growth and metastasis. Tumour Biol 34: 1579–1588. [DOI] [PubMed] [Google Scholar]
- 77. Huang Q, Liu L, Liu CH, You H, Shao F, et al. (2013) MicroRNA-21 Regulates the Invasion and Metastasis in Cholangiocarcinoma and May Be a Potential Biomarker for Cancer Prognosis. Asian Pac J Cancer Prev 14: 829–834. [DOI] [PubMed] [Google Scholar]
- 78. Liu GH, Zhou ZG, Chen R, Wang MJ, Zhou B, et al. (2013) Serum miR-21 and miR-92a as biomarkers in the diagnosis and prognosis of colorectal cancer. Tumour Biol 34: 2175–2181. [DOI] [PubMed] [Google Scholar]
- 79. Akagi I, Okayama H, Schetter AJ, Robles AI, Kohno T, et al. (2013) Combination of Protein Coding and Noncoding Gene Expression as a Robust Prognostic Classifier in Stage I Lung Adenocarcinoma. Cancer Res 73: 3821–3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Toiyama Y, Takahashi M, Hur K, Nagasaka T, Tanaka K, et al. (2013) Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst 105: 849–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Bovell LC, Shanmugam C, Putcha BD, Katkoori VR, Zhang B, et al. (2013) The Prognostic Value of MicroRNAs Varies with Patient Race/Ethnicity and Stage of Colorectal Cancer. Clin Cancer Res 19: 3955–3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Markou A, Sourvinou I, Vorkas PA, Yousef GM, Lianidou E (2013) Clinical evaluation of microRNA expression profiling in non small cell lung cancer. Lung Cancer 81: 388–396. [DOI] [PubMed] [Google Scholar]
- 83. Chen TH, Chang SW, Huang CC, Wang KL, Yeh KT, et al. (2013) The prognostic significance of APC gene mutation and miR-21 expression in advanced stage colorectal cancer. Colorectal Dis. [DOI] [PubMed] [Google Scholar]
- 84. Ferrajoli A, Shanafelt TD, Ivan C, Shimizu M, Rabe KG, et al. (2013) Prognostic value of miR-155 in individuals with monoclonal B-cell lymphocytosis and patients with B chronic lymphocytic leukemia. Blood 122: 1891–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Menendez P, Padilla D, Villarejo P, Palomino T, Nieto P, et al. (2013) Prognostic implications of serum microRNA-21 in colorectal cancer. J Surg Oncol. [DOI] [PubMed] [Google Scholar]
- 86. Kadera BE, Li L, Toste PA, Wu N, Adams C, et al. (2013) MicroRNA-21 in Pancreatic Ductal Adenocarcinoma Tumor-Associated Fibroblasts Promotes Metastasis. PLoS One 8: e71978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Bao L, Yan Y, Xu C, Ji W, Shen S, et al. (2013) MicroRNA-21 suppresses PTEN and hSulf-1 expression and promotes hepatocellular carcinoma progression through AKT/ERK pathways. Cancer Lett 337: 226–236. [DOI] [PubMed] [Google Scholar]
- 88. Roy S, Yu Y, Padhye SB, Sarkar FH, Majumdar AP (2013) Difluorinated-Curcumin (CDF) Restores PTEN Expression in Colon Cancer Cells by Down-Regulating miR-21. PLoS One 8: e68543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, et al. (2010) Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell 18: 282–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Asaga S, Kuo C, Nguyen T, Terpenning M, Giuliano AE, et al. (2011) Direct serum assay for microRNA-21 concentrations in early and advanced breast cancer. Clin Chem 57: 84–91. [DOI] [PubMed] [Google Scholar]
- 91. Tsujiura M, Ichikawa D, Komatsu S, Shiozaki A, Takeshita H, et al. (2010) Circulating microRNAs in plasma of patients with gastric cancers. Br J Cancer 102: 1174–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Wu CW, Ng SS, Dong YJ, Ng SC, Leung WW, et al. (2012) Detection of miR-92a and miR-21 in stool samples as potential screening biomarkers for colorectal cancer and polyps. Gut 61: 739–745. [DOI] [PubMed] [Google Scholar]
- 93. Yu L, Todd NW, Xing L, Xie Y, Zhang H, et al. (2010) Early detection of lung adenocarcinoma in sputum by a panel of microRNA markers. Int J Cancer 127: 2870–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Nair VS, Maeda LS, Ioannidis JP (2012) Clinical outcome prediction by microRNAs in human cancer: a systematic review. J Natl Cancer Inst 104: 528–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Fu X, Han Y, Wu Y, Zhu X, Lu X, et al. (2011) Prognostic role of microRNA-21 in various carcinomas: a systematic review and meta-analysis. Eur J Clin Invest 41: 1245–1253. [DOI] [PubMed] [Google Scholar]
- 96. Wang Y, Gao X, Wei F, Zhang X, Yu J, et al. (2014) Diagnostic and prognostic value of circulating miR-21 for cancer: a systematic review and meta-analysis. Gene 533: 389–397. [DOI] [PubMed] [Google Scholar]
- 97. Tanaka Y, Kamohara H, Kinoshita K, Kurashige J, Ishimoto T, et al. (2013) Clinical impact of serum exosomal microRNA-21 as a clinical biomarker in human esophageal squamous cell carcinoma. Cancer 119: 1159–1167. [DOI] [PubMed] [Google Scholar]
- 98. Kroh EM, Parkin RK, Mitchell PS, Tewari M (2010) Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods 50: 298–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Mar-Aguilar F, Mendoza-Ramirez JA, Malagon-Santiago I, Espino-Silva PK, Santuario-Facio SK, et al. (2013) Serum circulating microRNA profiling for identification of potential breast cancer biomarkers. Dis Markers 34: 163–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Li X, Zhang Y, Ding J, Wu K, Fan D (2010) Survival prediction of gastric cancer by a seven-microRNA signature. Gut 59: 579–585. [DOI] [PubMed] [Google Scholar]
- 101. Yang Q, Lu J, Wang S, Li H, Ge Q, et al. (2011) Application of next-generation sequencing technology to profile the circulating microRNAs in the serum of preeclampsia versus normal pregnant women. Clin Chim Acta 412: 2167–2173. [DOI] [PubMed] [Google Scholar]
- 102. Lusi EA, Passamano M, Guarascio P, Scarpa A, Schiavo L (2009) Innovative electrochemical approach for an early detection of microRNAs. Anal Chem 81: 2819–2822. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PRISMA Checklist.
(DOC)