Abstract
Bovine respiratory syncytial virus (BRSV) and human respiratory syncytial virus (HRSV) are major causes of respiratory disease in calves and children, respectively, and are priorities for vaccine development. We previously demonstrated that an experimental vaccine, BRSV-immunostimulating complex (ISCOM), is effective in calves with maternal antibodies. The present study focuses on the antigenic characterization of this vaccine for the design of new-generation subunit vaccines. The results of our study confirmed the presence of membrane glycoprotein (G), fusion glycoprotein (F), and nucleoprotein (N) proteins in the ISCOMs, and this knowledge was extended by the identification of matrix (M), M2-1, phosphoprotein (P), small hydrophobic protein (SH) and of cellular membrane proteins, such as the integrins αVβ1, αVβ3, and α3β1. The quantity of the major protein F was 4- to 5-fold greater than that of N (∼77 μg versus ∼17 μg/calf dose), whereas G, M, M2-1, P, and SH were likely present in smaller amounts. The polymerase (L), M2-2, nonstructural 1 (NS1), and NS2 proteins were not detected, suggesting that they are not essential for protection. Sera from the BRSV-ISCOM-immunized calves contained high titers of IgG antibody specific for F, G, N, and SH. Antibody responses against M and P were not detected; however, this does not exclude their role in protective T-cell responses. The absence of immunopathological effects of the cellular proteins, such as integrins, needs to be further confirmed, and their possible contribution to adjuvant functions requires elucidation. This work suggests that a combination of several surface and internal proteins should be included in subunit RSV vaccines and identifies absent proteins as potential candidates for differentiating infected from vaccinated animals.
INTRODUCTION
The effective control of bovine respiratory syncytial virus (BRSV) is a priority for European farmers and animal health organizations. This pneumovirus in the Paramyxoviridae family is a key player in the bovine respiratory disease complex, which is one of the most economically important and outstanding welfare issues in industrialized beef cattle production (1–3).
Young calves need to be immunized before their first encounter with BRSV. However, the development of vaccine-induced immune responses is hampered by BRSV-specific maternally derived antibodies (MDA). Vaccinated calves are, in general, no longer protected when the MDA decline to subprotective levels (4–7). Therefore, vaccines with a rapid, strong, and durable effect in calves with MDA are needed for vaccination before transport to calf-rearing herds where the virus is often circulating.
Moreover, none of the currently available commercial BRSV vaccines (n = 81; Vetvac database [http://vetvac.org/]) enables the differentiation of infected from vaccinated animals (DIVA), which is another desired characteristic. Vaccination that enables DIVA (DIVA vaccination) would allow for the protection and surveillance of BRSV-negative herds, which would provide animals to the market that are BRSV-free but immune. It would also enable vaccine safety and efficacy monitoring in the field and seroepidemiological studies in vaccinated areas. To design such vaccines, it is essential to identify which virus proteins are dispensable and indispensable for protection and which indispensable proteins are immunogenic. A new-generation DIVA vaccine might contain selected viral proteins produced by genetic engineering that are adjusted for large-scale production (8). A rational composition of several proteins rather than one would likely induce a more multifaceted and longer immunity, and it would probably have a better chance of circumventing MDA.
The RNA genome of BRSV encodes 11 proteins: the membrane proteins fusion glycoprotein (F), membrane glycoprotein (G), and small hydrophobic protein (SH), the nucleocapsid proteins polymerase (L), nucleoprotein (N), and phosphoprotein (P), the matrix protein (M), the polymerase cofactors M2-1 and M2-2, as well as the nonstructural proteins (NS1 and NS2) (9). Produced using different protein expression systems (vector viruses, DNA vaccines, or Escherichia coli), F, G, and N, but not M2-1, have each separately induced significant protection against BRSV challenge of gnotobiotic and/or seronegative calves. This protection is believed to be mediated through neutralizing antibodies (for F, and at low levels for G), priming of T cells (for F, G, and N), and rapid IgA responses after challenge (for F) (10–17). Besides F protein, the N, M2-1, and P proteins are the main proteins recognized by memory CD8+ T lymphocytes from BRSV-infected calves, and since CD8 T cells are important in the clearance of BRSV from calves, specific immunity to these proteins may contribute to protection (10), (G. Taylor, unpublished data).
We previously produced and evaluated an experimental vaccine, BRSV-immunostimulating complexes (BRSV-ISCOMs), which induced rapid protection against virulent challenge in calves with MDA, likely partly mediated through mucosal IgA, and that seemed safe with regard to any toxic and disease-enhancing effects (18, 19). ISCOMs are spherical particles that contain proteins from purified solubilized virus as well as lipids and Quillaja saponin (Quil A) (20). The presence of F, G, and N proteins in bovine and human RSV-ISCOM formulations produced by different methods has been reported (21–25), but comprehensive data on protein content and the relative quantities of the proteins are missing. The main aim of the present work was therefore to characterize BRSV-ISCOMs, especially with regard to protein content, for the future design of new-generation vaccines.
MATERIALS AND METHODS
Production of BRSV-ISCOMs and controls.
BRSV-ISCOMs were produced based on purified solubilized BRSV (strain 9402022, Denmark [26], most likely belonging to BRSV genetic subgroup II and antigenic subgroup AB [27] and propagated in Vero cells). Briefly, BRSV was purified by (i) centrifugation of frozen and thawed infected cell cultures at 200 × g and 5°C for 5 min; (ii) the resulting supernatants were centrifuged at ∼53,900 × g and 5°C for 5 h; (iii) the resulting pellets were dissolved in phosphate-buffered saline (PBS) and centrifuged at 200 × g and 5°C for 5 min, and the new pellets were treated in a similar manner five times; (iv) those supernatants were centrifuged over 20% sucrose at ∼50,000 × g and 5°C for 20 h; and (v) the resulting pellets were dissolved as described in iii. The proteins in the supernatants collected at this stage were solubilized and separated over a sucrose gradient by centrifugation, and one sample fraction was selected for further inclusion in the ISCOMs, as described previously (19). The ISCOMs were prepared through the addition of cholesterol, phosphatidylcholine, and Quil A (QPUF300; Desert King, Chile), dialysis, and purification, as described previously (19). Control ISCOMs (Vero-ISCOMs) were similarly prepared from uninfected Vero cells that underwent the virus purification and solubilization protocols. BRSV protein controls (i.e., similar protein extracts as in BRSV-ISCOMs but that were not formulated as ISCOMs) were likewise produced from infected cells, excluding the addition of lipids and adjuvant before dialysis. The commercial adjuvant used as a control in calves, AbISCO-300, was made of the same batches of cholesterol, phosphatidylcholine, and Quillaja saponin as used in the BRSV-ISCOMs.
Vaccine characterization.
The experiments for vaccine characterization were carried out on concentrated vaccine preparations. The protein contents of BRSV-ISCOMs, BRSV proteins, and Vero-ISCOMs used for this purpose were 1.46 mg/ml, 1.43 mg/ml, and 0.73 mg/ml, respectively.
Determination of morphology and size and exclusion of contaminant live BRSV.
The ISCOM formulations were analyzed using negative-stain transmission electron microscopy (FEI Tecnai G2 microscope), and the images were recorded on a Morada CCD camera (2,000 by 2,000 pixels) using the iTEM control software (Olympus Soft Imaging Solutions GmbH, Vironova AB, Sweden). The presence of live BRSV in the BRSV-ISCOMs was analyzed by attempting virus isolation in cell culture. In total, 50 μl BRSV-ISCOM preparation was diluted 1:20 to dilute the toxic effects of Quil A on cell culture and inoculated onto 25-cm2 sections of fetal bovine turbinate (FBT) cells, as described previously (19).
Protein quantification and identification.
The proteins present in the vaccine formulations were quantified by the Bradford assay, and the results were confirmed by amino acid analysis (Amino Acid Analysis Center, Uppsala University, Sweden) (28). The proteins were visualized by SDS-PAGE and colloidal Coomassie blue staining. The gel bands were analyzed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) at Uppsala University, Sweden, and at INRA, France, as described by Tran et al. (29). Furthermore, liquid chromatography tandem mass spectrometry (LC-MS/MS) was performed at the PAPPSO platform, INRA, France, as described previously (30), and queries of MS/MS data against a Macaca mulatta protein database (UniProtKB) and a BRSV database (strain A519086, UniProtKB, and strain 9402022, Denmark), together with an in-house contaminant database, were performed. The above analyses were carried out on a mixture of several ISCOM batches, which had been used for animal immunizations. For confirmation purposes, three of these ISCOM batches were run separately on SDS-PAGE gels, and selected bands were analyzed by MALDI-TOF MS.
Dot blot assays were performed to screen for BRSV proteins that did not form visible bands in the SDS-PAGE gels and to check for protein antigenicity. The proteins were visualized by using murine monoclonal antibodies (MAbs) specific to BRSV proteins (MAbs 19 [F], 16 [F], 44 [G], 57 [G], 6 [N], 88 [N], 8 [M2-1], 100 [M2-1], 12 [P], 106 [M], and 109 [M]) (31–33) and the results were confirmed by Western blot using MAbs against HRSV F (MCA490; Serotec), rabbit polyclonal antisera against recombinant HRSV M2-1, N, and P (34), and MAbs against BRSV G (MAb 44) and M (MAb 109). In addition, to investigate the recognition patterns of viral proteins in BRSV-ISCOMs by bovine antibodies acquired by BRSV infection, the proteins were blotted using serum from an unvaccinated experimentally BRSV-infected calf (strain 9402022, Denmark) and using hyperimmune serum (BRSV strain 2106, United Kingdom) from a calf immunized with the 391-2 strain of BRSV (35) from the BRSV genetic subgroup IV/antigenic subgroup A (27).
Immunization of calves.
Three- to 8-week-old conventional calves of Swedish red and white breed and Swedish Holstein breed were immunized subcutaneously (s.c.) twice at an interval of 3 weeks with either (i) BRSV-ISCOMs (containing 188 μg total protein in 2 ml PBS, n = 5), (ii) BRSV protein (188 μg total protein in 2 ml PBS, n = 5), (iii) 390 μg AbISCO-300 (in 2 ml PBS, n = 5), or (iv) 2 ml PBS (n = 5), as previously described in detail (19).
Bovine protein-specific antibody detection.
Serum samples collected 2 weeks after boost were used to verify the immunogenicity of the specific proteins in the vaccine preparations. For this purpose, enzyme-linked immunosorbent assays (ELISAs) using lysates of chicken embryo fibroblasts (CEF) infected with recombinant fowlpox viruses (FPV) expressing either BRSV G, F, M, P (cloned from the Snook strain [36]), or SH protein (cloned from strain 9402022, Denmark, and prepared by the Jenner Vector Core Facility, Oxford, United Kingdom) were used to detect protein-specific IgG. Lysates of CEF infected with wild-type FPV were used as a control antigen. The sera were additionally analyzed for IgG1 antibodies against the N protein by an ELISA based on purified recombinant BRSV N expressed by E. coli, as described earlier (37). The sera were serially diluted and endpoint titers calculated by linear regression. The endpoint titers were expressed as the reciprocals of the dilutions reaching a cutoff set to two times the mean corrected optical density (COD) of a negative-control serum sample. Moreover, antibodies that competed with MAb 19, which is specific for antigenic site IV on the F protein and neutralizes both HRSV and BRSV (38), were analyzed by competitive ELISA. In brief, 96-well plates were coated with BRSV protein and blocked with 2% horse milk prior to adding serially diluted samples, MAb 19, conjugate, 3,3′,5,5′-tetramethylbenzidine (TMB) substrate, and H2O2. The cutoff was determined to be the COD of a serum sample of a naturally BRSV-infected cow with a BRSV-specific IgG antibody titer of 25, as determined by indirect ELISA (Svanovir; Svanova Biotech AB).
Data analysis.
The data were analyzed using Minitab 16.1.1 statistical software using analysis of variance (ANOVA), followed by Dunnett's test. The statistical significances were set at 0.01 < P ≤ 0.05, 0.001 < P ≤ 0.01, and P ≤ 0.001, and the group variation was expressed as the standard deviation (SD).
RESULTS
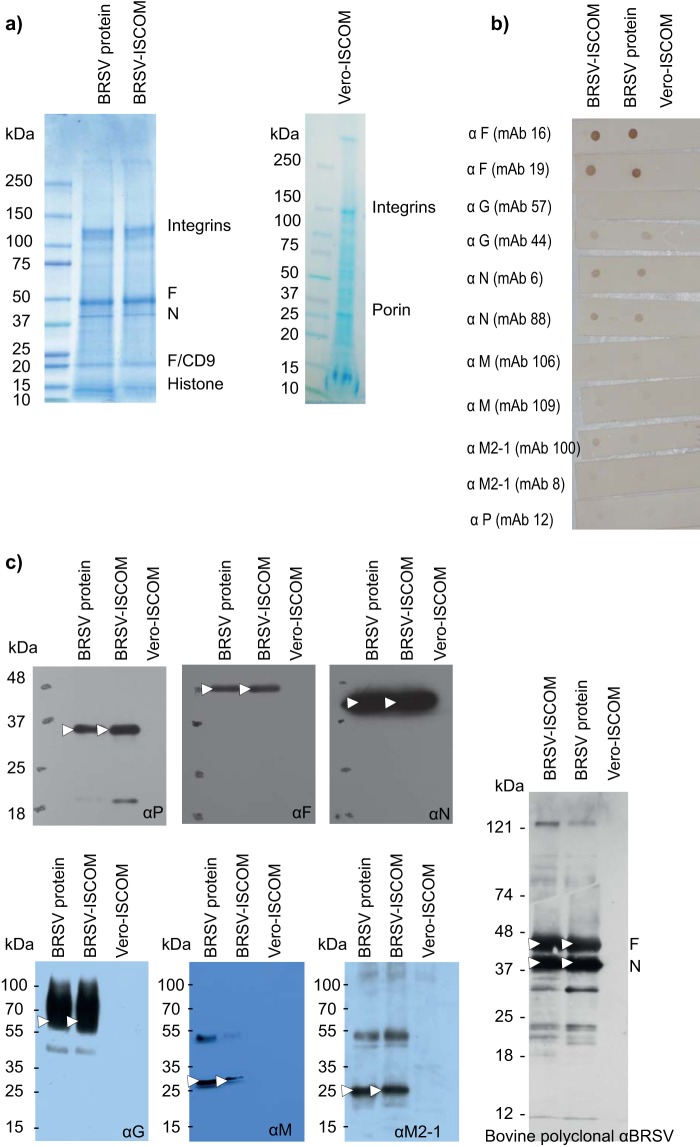
The BRSV-ISCOM preparation contained a heterogeneous mixture of pleomorphic ISCOM particles with a cage-like surface and less well-defined 10- to 20-nm structures (Fig. 1). A size distribution analysis showed that the majority of the ISCOM particles had a diameter of 40 to 90 nm, and the mean diameter of 1,644 detected particles was 56.4 nm, with a standard deviation of 11.1 nm. In order to characterize the proteins present in the BRSV-ISCOMs, the most intense bands revealed by Coomassie blue-stained SDS-PAGE gels were analyzed by MALDI-TOF MS. As shown in Fig. 2a and Table 1, the F and N proteins were clearly identified. Other bands were identified as cellular integrins, CD9 (colocalizing with F1), and histone H4. When analyzed separately, each of three separate ISCOM batches visually formed identical protein bands with comparable relative intensity on SDS-PAGE as those presented in Fig. 2a, and MALDI-TOF MS analysis of selected bands confirmed the presence of integrins and histones in every batch (data not shown). Using the ImageJ software (39), we estimated that the F and N proteins represented roughly half of the total proteins present in the extracts and that the F/N ratio was between 4 and 5. Consequently, in one vaccine dose for calves containing 188 μg total protein, the contents of F and N may be estimated to be approximately half of this quantity, e.g., 77 μg of F and 17 μg of N.
FIG 1.
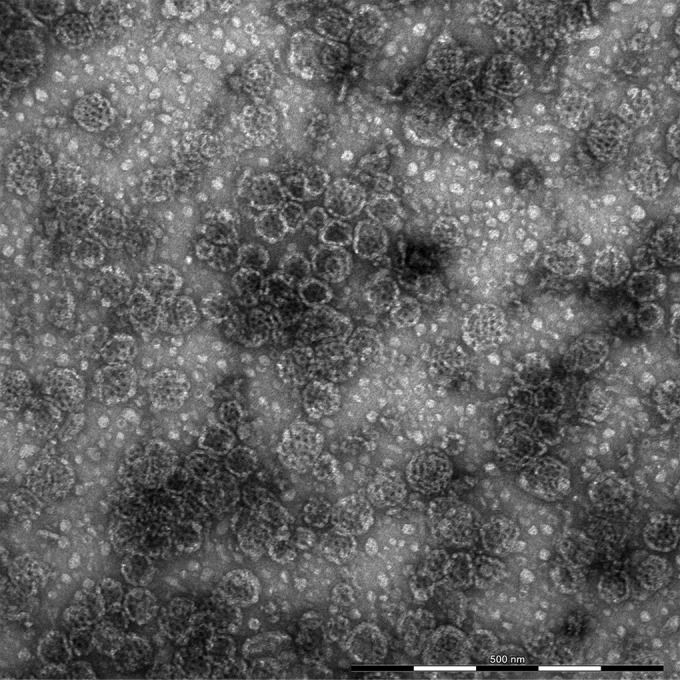
Negative-stain transmission electron microscopy image of BRSV-ISCOMs.
FIG 2.
BRSV protein content in BRSV-ISCOMs, BRSV proteins, and Vero-ISCOMs. (a) Coomassie blue-stained SDS-PAGE gel showing the bands analyzed by mass spectrometry and tandem mass spectrometry; (b) dot blots using monoclonal antibodies against BRSV proteins; (c) Western blots using monoclonal antibodies against HRSV F and BRSV G and M, rabbit polyclonal antiserum against recombinant HRSV M2-1, N, and P, and serum from an unvaccinated experimentally BRSV-infected calf. Arrows indicate BRSV proteins.
TABLE 1.
MALDI-TOF MS analysis of proteins present in the BRSV-ISCOMs
| Molecular mass (kDa)a | BRSV-ISCOM (aa)b |
BRSV protein (aa)b |
Vero-ISCOMb |
||
|---|---|---|---|---|---|
| INRA | UU | INRA | UU | UU | |
| 120 | Integrin α3c | ||||
| 117 | Integrin αVc | Integrin αVc | |||
| 112 | Integrin αVc | ||||
| 93 | Integrin α3c,e | Integrin α3c | |||
| 92 | Integrin β1c | Integrin β1c | |||
| 90 | Integrin β3c | ||||
| 84 | Integrin β3c | Integrin α3c | Integrin Aiibb3c | ||
| 72 | 78-kDa glucose-related proteinc,e | ||||
| 64 | BRSV F protein (207–520, 157–336) | BRSV F protein (157–461)e | BRSV F protein (157–520) | BRSV F protein (157–520) | |
| 54 | BRSV F protein (62–331) | ||||
| 44 | BRSV N protein (8–308) | BRSV N protein (7–352) | BRSV N protein (47–358) | BRSV N protein (7–520) | |
| 31 | Porin 31 HM | ||||
| NAf | BRSV F protein/CD9c,g | BRSV F protein/CD9g | |||
| 11 | Histone H4 type VIIId | Histone H4 type VIIId | |||
Protein molecular mass as estimated by mass spectrometry.
Amino acid sequences (aa) covering the region of matched protein are in parentheses. UU, Uppsala University.
Matches an amino acid sequence of protein originating from Homo sapiens or Macaca mulatta (proteins probably originating from Vero cells).
Matches an amino acid sequence of protein originating from Gallus gallus (histones are almost identical between species).
Nonsignificant match (score is between 55 and 70) (P = 0.05).
NA, not analyzed.
Results were obtained by tandem mass spectrometry.
The presence of F and N was confirmed by dot blot (Fig. 2b) and Western blot (Fig. 2c) analyses, which also revealed the presence of G, M, M2-1, and P proteins in both the BRSV-ISCOMs and BRSV protein extracts. The G protein was recognized by MAb 44 but not 57 in both assays (Fig. 2b and data not shown). The F and N proteins were those best recognized in a Western blot when using sera from animals immunized with virus belonging to the genetic subgroup IV/antigenic subgroup A (Fig. 2c) and genetic subgroup II/antigenic subgroup AB (data not shown).
To further identify other viral proteins in the BRSV-ISCOMs, which are present in smaller amounts, an LC-MS/MS analysis of the BRSV-ISCOMs was conducted. Because the complete genome sequence of the BRSV strain used in this work (9402022, Denmark) is not available, we used a BRSV database consisting also of other BRSV sequences (see Materials and Methods), resulting in the acquisition of qualitative rather than quantitative data. As shown in Table 2, the F, M, M2-1, N, P, and SH proteins were clearly identified. As expected, the G protein was not identified because of the glycosylation of its peptides. Many cellular proteins, including integrins, were also identified using LC-MS/MS (data not shown). Contaminating virus was not detected in BRSV-ISCOM by either mass spectrometry or electron microscopy, and live BRSV was not detected by virus isolation, as determined by the absence of cytopathic effect during two passages in cell culture.
TABLE 2.
LC-MS/MS analysis of viral proteins present in the BRSV-ISCOMs
| Molecular mass (kDa) | Protein IDa | Name | % coverage | No. of spectrab | No. of unique sequencesc |
|---|---|---|---|---|---|
| 63.3 | P29791 | BRSV A51908 fusion glycoprotein | 36 | 55 | 25 |
| 43.3 | P22677 | BRSV A51908 nucleoprotein | 38 | 12 | 8 |
| 28.6 | P24615 | BRSV A51908 matrix protein | 49 | 12 | 9 |
| 27.2 | P33454 | BRSV A51908 phosphoprotein | 16 | 2 | 2 |
| 21.3 | P29792 | BRSV A51908 matrix M2-1 protein | 26 | 6 | 4 |
| 8.3 | P24616 | BRSV A51908 small hydrophobic protein | 14 | 4 | 2 |
ID, identifying number.
The number of MS/MS events attributed to the considered protein.
The number of unique peptidic sequences that matched with the considered protein.
Serum samples from the BRSV-ISCOM-immunized calves were analyzed for the presence of antibodies directed against BRSV proteins. High titers of antibodies directed against the F and N proteins were detected in calves immunized with BRSV-ISCOMs, and furthermore, antibody titers were high against the G and SH proteins (Fig. 3). The antibody titers against the M and P proteins were, in contrast, not significantly higher in immunized than in nonimmunized calves with MDA. The mean MAb 19-competitive antibody titers were 2.4 log10 (range, 1.7 to 2.6), 1.5 log10 (0 to 2.2), 0.3 log10 (0 to 1.0), and 1.2 log10 (0 to 1.5) in calves immunized with BRSV-ISCOM, BRSV proteins, AbISCO-300, and PBS, respectively.
FIG 3.
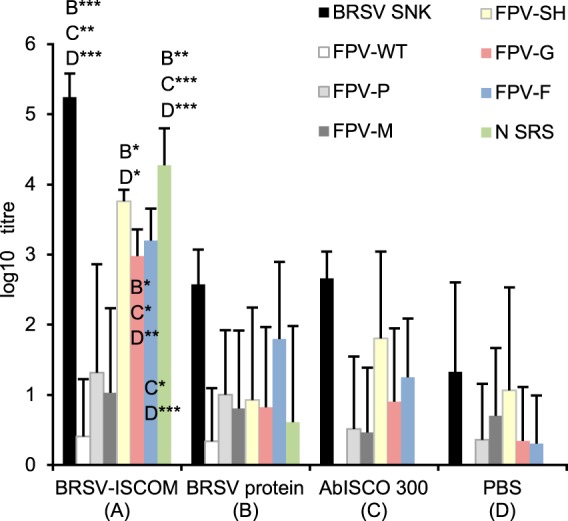
Reactivity of calf serum to BRSV proteins. Calves with various levels of BRSV-specific maternal antibodies were immunized twice subcutaneously with BRSV-ISCOMs containing 188 μg proteins (A), 188 μg BRSV proteins (B), 390 μg AbISCO-300 (C), or PBS (D), 33 and 12 days before blood sampling. The results are presented as a group mean of IgG antibody titers (log10) obtained by ELISAs using lysates of cells infected with BRSV Snook (BRSV SNK), wild-type fowlpox vector (FPV-WT), fowlpox vectors expressing BRSV G, F, M, P, or SH protein, or purified BRSV N protein expressed by E. coli (N SRS) as antigens. The titers presented against N are of the IgG1 isotype, whereas titers against G, F, M, P, or SH represent IgG. Standard deviations are shown by upward deflection lines. The asterisks indicate statistically significant differences between groups: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
DISCUSSION
Novel BRSV-DIVA vaccines need to be designed based on knowledge of protein-specific immune responses, including their protectiveness and safety. This paper describes the protein content of BRSV-ISCOMs by direct identification and through the antibody responses induced by this vaccine. As described previously for HRSV-ISCOMs similarly formulated by dialysis (22, 23), the present work confirmed that the main viral protein in BRSV-ISCOMs is the F protein, and that the G protein is present, probably in smaller amounts. Because the G protein migrates as a smear in SDS-PAGE, it cannot be quantified by Coomassie blue staining, but it was semiquantified by dot blot in this study. The G protein was recognized by only one out of two MAbs, which may be explained by the antigenic variation in this protein (33). In agreement with previous studies (25) in which BRSV-ISCOMs were prepared by ultracentrifugation, we also detected high quantities of the N protein. Furthermore, we extended these earlier observations by additionally identifying several other BRSV proteins, namely, the M, M2-1, P, and SH proteins, by direct or indirect immunological techniques and by using LC-MS/MS. These proteins were not abundant, since the corresponding bands could not be visualized on Coomassie blue-stained polyacrylamide gels. The polymerase (L), nonstructural (NS1 and NS2), and M2-2 proteins were not identified, indicating that they were absent or present only in very small amounts.
The presence of highly immunogenic F and N but also G and SH proteins was indirectly confirmed by antibody detection in sera from calves immunized with BRSV-ISCOMs. The IgG antibody titers were highest against the N and SH, followed by the F and G proteins. This apparent discrepancy in vaccine content according to signals observed in Western or dot blotting (F > N > G; SH not tested) or SDS-PAGE (F > N, G, or SH not detected) may be due to differences in the immunogenicity between proteins, a difference in the sensitivity or IgG isotype specificity (total IgG versus IgG1) of the ELISAs, or it might be a result of differences in the titers of MDA specific to the different proteins at the time of vaccination. The MDA detected in control calves injected with AbISCO-300 or PBS were nevertheless primarily directed against SH, and such antibodies in vaccinated calves did not seem to inhibit the SH-specific antibody response. Monoclonal Ab 19 neutralizes HRSV and BRSV, and MAb 19-competitive antibodies were induced by BRSV-ISCOMs, but the individual titers in calves did not correlate with the clinical or virological protection of these animals that was demonstrated earlier (reference 19 and data not shown). This is in line with previous observations of BRSV-neutralizing antibodies in BRSV-ISCOM-immunized calves (18) and suggests that other immune parameters, such as T-cell responses and mucosal IgA, are important for protection. It is likely that the F, G, N, and SH, as well as M, M2-1, and P proteins, were all contributing to such responses, and our data suggest that these proteins can safely be included in a BRSV-DIVA vaccine in the relative proportions detected by direct techniques. Whereas the protectiveness of the immune responses induced by the F, G, and N proteins are well established, the immune responses against SH are less well characterized, and SH is not needed for the induction of protection (40) (K. Blodörn, S. Hägglund, J. Fix, C. Dubuquoy, B. Makabi-Panzu, M. Thom, P. Karlsson, J. L. Roque, E. Karlstam, J. Pringle, J. F. Eléouët, S. Riffault, G. Taylor, and J. F. Valarcher, submitted for publication). The NS proteins, which are absent in the BRSV-ISCOMs, are other potential targets for DIVA. These were also not essential for the protection induced by gene-deleted live attenuated vaccines (41) and are already utilized as DIVA proteins for foot-and-mouth disease virus (42). The induction and duration of an anti-NS antibody response by BRSV infection should therefore be elucidated.
As seen with other RSV-ISCOM preparations (23, 24), the BRSV-ISCOM particles were honeycombed through the assembly of subunit rings but were in general larger than reported previously (40 to 90 nm here versus 40 nm in Hu et al. [23] or 30 to 35 nm in Trudel et al. [24]) and pleomorphic instead of regular. There was also an unusual abundance of small 10- to 20-nm particles, which based on their morphology probably consisted of nonassembled or poorly assembled subunit rings. These differences were probably due to the different proportion of Quil A to lipids and proteins in this study compared to those in earlier publications (23).
The presence of high quantities of cellular proteins has, to our knowledge, not previously been reported for RSV-ISCOM formulations. However, major histocompatibility complex (MHC) class II was detected and depleted by immunosorbent chromatography from human immunodeficiency virus lysate used for ISCOM preparation (43). It was recently demonstrated that alloantibodies against MHC class I from a bovine cell line used to produce a bovine viral diarrhea virus (BVDV) vaccine for cattle likely caused neonatal pancytopenia in the calves of vaccinated dams (44). Since Vero cells used for BRSV-ISCOM production were derived from African green monkeys and not cattle, alloimmunity is unlikely to be induced by this vaccine. The repeated isolation of Vero cell proteins was probably favored by the laboratory protocols used. If, among these proteins, the primate integrins recognize the specific counterreceptors on bovine cells, they might have contributed to the adjuvant effect, antigen depot, and/or delivery, and this should be further investigated. Indeed, even calves immunized with BRSV protein extracts and no adjuvant exhibited weak but measurable immune responses (19). The integrins α3β1, αVβ1, and αVβ3 (the α and β chains of which were detected in the BRSV-ISCOMs) mediate cellular adhesion to the extracellular matrix, basement membranes, and endothelial cells (45–47). Furthermore, the integrin α3β1 interacts with tetraspanins (45), which are involved in endocytosis (48). An interaction with tetraspanin proteins, such as CD81 and CD9, could, theoretically, affect ISCOM recognition, uptake, and induction of alpha interferon in plasmacytoid dendritic cells, as previously demonstrated for hepatitis C virus-infected cells (49). We also detected CD9 in the BRSV-ISCOMs. The colocalization of this protein with the F1 protein on the SDS-PAGE gels may have been due to the fact that the sizes of these proteins in their glycosylated forms were similar, although the predicted mass of CD9 without glycosylation is 25 kDa and that of the cleaved F1 fragment is around 48 kDa. Another possibility that should be investigated further is that RSV uses CD9 as a coreceptor for the fusion of host cells, in addition to the recently identified receptor, nucleolin (50).
Before including any cellular protein in a recombinant subunit vaccine designed to resemble BRSV-ISCOMs, it must be determined whether such proteins can exert any of their biological functions across species (i.e., between primate integrins and bovine counterreceptors). Furthermore, although BRSV-ISCOMs appear to be safe so far, it must also be verified that such components do not induce any immunopathological effects.
In summary, BRSV-ISCOMs included viral proteins dominated by the F and N proteins, at an approximate ratio of 5:1, as well as cellular proteins dominated by integrins. The ISCOM-enclosed BRSV F, G, N, and SH proteins induced strong humoral responses in cattle, whereas the P, M, and M2-1 proteins, which were also present, might induce cellular responses not assessed in this work. The L, M2-2, NS1, and NS2 proteins, which were absent, do not appear to be essential for efficient vaccination. It remains to be investigated whether any of these proteins induces antibodies that could be used for DIVA assays.
ACKNOWLEDGMENTS
This work was supported by the Swedish Farmers' Foundation for Agricultural Research (grant H0750358) and the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (FORMAS), through EMIDA ERA-NET (FP-87), and by the Science for Life Mass Spectrometry Technology Platform in Uppsala, Sweden.
We thank the staff at SLU and SVA for rearing and maintaining the experimental animals, L. E. Larsen, DTU, Denmark, for generously sharing the BRSV isolate 9402022, B. Morein for advice, and C. Fossum for providing facilities. We also thank C. Vernersson, I. Dahlén, and L. Treiberg-Berndtsson, SVA, for FBT cells, Vero cells, and providing facilities for virus production, Å. Engström, A. Törnsten, and M. Ramström at Uppsala University, and J. Nilsson at Vironova AB (Stockholm, Sweden) for performing pertinent mass spectrometry, electron microscopy, and amino acid analyses.
Geraldine Taylor is a Jenner Investigator.
Footnotes
Published ahead of print 14 May 2014
REFERENCES
- 1.Nicholas RA, Ayling RD. 2003. Mycoplasma bovis: disease, diagnosis, and control. Res. Vet. Sci. 74:105–112. 10.1016/S0034-5288(02)00155-8 [DOI] [PubMed] [Google Scholar]
- 2.Assié S, Seegers H, Makoschey B, Désiré-Bousquié L, Bareille N. 2009. Exposure to pathogens and incidence of respiratory disease in young bulls on their arrival at fattening operations in France. Vet. Rec. 165:195–199. 10.1136/vr.165.7.195 [DOI] [PubMed] [Google Scholar]
- 3.Stott EJ, Thomas LH, Collins AP, Crouch S, Jebbett J, Smith GS, Luther PD, Caswell R. 1980. A survey of virus infections of the respiratory tract of cattle and their association with disease. J. Hyg. (Lond.) 85:257–270. 10.1017/S0022172400063294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kimman TG, Westenbrink F, Schreuder BE, Straver PJ. 1987. Local and systemic antibody response to bovine respiratory syncytial virus infection and reinfection in calves with and without maternal antibodies. J. Clin. Microbiol. 25:1097–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kimman TG, Westenbrink F, Straver PJ. 1989. Priming for local and systemic antibody memory responses to bovine respiratory syncytial virus: effect of amount of virus, virus replication, route of administration and maternal antibodies. Vet. Immunol. Immunopathol. 22:145–160. 10.1016/0165-2427(89)90057-3 [DOI] [PubMed] [Google Scholar]
- 6.Sandbulte MR, Roth JA. 2002. T-cell populations responsive to bovine respiratory syncytial virus in seronegative calves. Vet. Immunol. Immunopathol. 84:111–123. 10.1016/S0165-2427(01)00393-2 [DOI] [PubMed] [Google Scholar]
- 7.Getahun A, Heyman B. 2009. Studies on the mechanism by which antigen-specific IgG suppresses primary antibody responses: evidence for epitope masking and decreased localization of antigen in the spleen. Scand. J. Immunol. 70:277–287. 10.1111/j.1365-3083.2009.02298.x [DOI] [PubMed] [Google Scholar]
- 8.Tran TL, Castagné N, Bhella D, Varela PF, Bernard J, Chilmonczyk S, Berkenkamp S, Benhamo V, Grznarova K, Grosclaude J, Nespoulos C, Rey FA, Eléouët JF. 2007. The nine C-terminal amino acids of the respiratory syncytial virus protein P are necessary and sufficient for binding to ribonucleoprotein complexes in which six ribonucleotides are contacted per N protein protomer. J. Gen. Virol. 88:196–206. 10.1099/vir.0.82282-0 [DOI] [PubMed] [Google Scholar]
- 9.Valarcher JF, Taylor G. 2007. Bovine respiratory syncytial virus infection. Vet. Res. 38:153–180. 10.1051/vetres:2006053 [DOI] [PubMed] [Google Scholar]
- 10.Gaddum RM, Cook RS, Furze JM, Ellis SA, Taylor G. 2003. Recognition of bovine respiratory syncytial virus proteins by bovine CD8+ T lymphocytes. Immunology 108:220–229. 10.1046/j.1365-2567.2003.01566.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riffault S, Meyer G, Deplanche M, Dubuquoy C, Durand G, Soulestin M, Castagné N, Bernard J, Bernardet P, Dubosclard V, Bernex F, Petit-Camurdan A, Deville S, Schwartz-Cornil I, Eléouët JF. 2010. A new subunit vaccine based on nucleoprotein nanoparticles confers partial clinical and virological protection in calves against bovine respiratory syncytial virus. Vaccine 28:3722–3734. 10.1016/j.vaccine.2010.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schrijver RS, Langedijk JP, Keil GM, Middel WG, Maris-Veldhuis M, Van Oirschot JT, Rijsewijk FA. 1997. Immunization of cattle with a BHV1 vector vaccine or a DNA vaccine both coding for the G protein of BRSV. Vaccine 15:1908–1916. 10.1016/S0264-410X(97)00129-1 [DOI] [PubMed] [Google Scholar]
- 13.Taylor G, Bruce C, Barbet AF, Wyld SG, Thomas LH. 2005. DNA vaccination against respiratory syncytial virus in young calves. Vaccine 23:1242–1250. 10.1016/j.vaccine.2004.09.005 [DOI] [PubMed] [Google Scholar]
- 14.Taylor G, Rijsewijk FA, Thomas LH, Wyld SG, Gaddum RM, Cook RS, Morrison WI, Hensen E, van Oirschot JT, Keil G. 1998. Resistance to bovine respiratory syncytial virus (BRSV) induced in calves by a recombinant bovine herpesvirus-1 expressing the attachment glycoprotein of BRSV. J. Gen. Virol. 79:1759–1767 [DOI] [PubMed] [Google Scholar]
- 15.Taylor G, Thomas LH, Furze JM, Cook RS, Wyld SG, Lerch R, Hardy R, Wertz GW. 1997. Recombinant vaccinia viruses expressing the F, G or N, but not the M2, protein of bovine respiratory syncytial virus (BRSV) induce resistance to BRSV challenge in the calf and protect against the development of pneumonic lesions. J. Gen. Virol. 78:3195–3206 [DOI] [PubMed] [Google Scholar]
- 16.Thomas LH, Cook RS, Wyld SG, Furze JM, Taylor G. 1998. Passive protection of gnotobiotic calves using monoclonal antibodies directed at different epitopes on the fusion protein of bovine respiratory syncytial virus. J. Infect. Dis. 177:874–880. 10.1086/515234 [DOI] [PubMed] [Google Scholar]
- 17.Antonis AF, van der Most RG, Suezer Y, Stockhofe-Zurwieden N, Daus F, Sutter G, Schrijver RS. 2007. Vaccination with recombinant modified vaccinia virus Ankara expressing bovine respiratory syncytial virus (bRSV) proteins protects calves against RSV challenge. Vaccine 25:4818–4827. 10.1016/j.vaccine.2007.04.002 [DOI] [PubMed] [Google Scholar]
- 18.Hägglund S, Hu KF, Larsen LE, Hakhverdyan M, Valarcher JF, Morein B, Taylor G, Belák S, Alenius S. 2004. Bovine respiratory syncytial virus ISCOMs: protection in the presence of maternal antibodies. Vaccine 23:646–655. 10.1016/j.vaccine.2004.07.006 [DOI] [PubMed] [Google Scholar]
- 19.Hägglund S, Hu K, Vargmar K, Poré L, Olofson AS, Blodörn K, Anderson J, Ahooghalandari P, Pringle J, Taylor G, Valarcher JF. 2011. Bovine respiratory syncytial virus ISCOMs—immunity, protection and safety in young conventional calves. Vaccine 29:8719–8730. 10.1016/j.vaccine.2011.07.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morein B, Sundquist B, Höglund S, Dalsgaard K, Osterhaus A. 1984. Iscom, a novel structure for antigenic presentation of membrane proteins from enveloped viruses. Nature 308:457–460. 10.1038/308457a0 [DOI] [PubMed] [Google Scholar]
- 21.Hu KF, Ekström J, Merza M, Lövgren-Bengtsson K, Morein B. 1999. Induction of antibody responses in the common mucosal immune system by respiratory syncytical [sic] virus immunostimulating complexes. Med. Microbiol. Immunol. 187:191–198. 10.1007/s004300050092 [DOI] [PubMed] [Google Scholar]
- 22.Hu KF, Elvander M, Merza M, Akerblom L, Brandenburg A, Morein B. 1998. The immunostimulating complex (ISCOM) is an efficient mucosal delivery system for respiratory syncytial virus (RSV) envelope antigens inducing high local and systemic antibody responses. Clin. Exp. Immunol. 113:235–243. 10.1046/j.1365-2249.1998.00650.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu KF, Regner M, Siegrist CA, Lambert P, Chen M, Bengtsson KL, Morein B. 2005. The immunomodulating properties of human respiratory syncytial virus and immunostimulating complexes containing Quillaja saponin components QH-A, QH-C and ISCOPREP703. FEMS Immunol. Med. Microbiol. 43:269–276. 10.1016/j.femsim.2004.08.010 [DOI] [PubMed] [Google Scholar]
- 24.Trudel M, Nadon F, Séguin C, Brault S, Lusignan Y, Lemieux S. 1992. Initiation of cytotoxic T-cell response and protection of BALB/c mice by vaccination with an experimental ISCOMs respiratory syncytial virus subunit vaccine. Vaccine 10:107–112. 10.1016/0264-410X(92)90026-G [DOI] [PubMed] [Google Scholar]
- 25.Trudel M, Nadon F, Séguin C, Simard C, Lussier G. 1989. Experimental polyvalent ISCOMs subunit vaccine induces antibodies that neutralize human and bovine respiratory syncytial virus. Vaccine 7:12–16. 10.1016/0264-410X(89)90004-2 [DOI] [PubMed] [Google Scholar]
- 26.Viuff B, Uttenthal A, Tegtmeier C, Alexandersen S. 1996. Sites of replication of bovine respiratory syncytial virus in naturally infected calves as determined by in situ hybridization. Vet. Pathol. 33:383–390. 10.1177/030098589603300403 [DOI] [PubMed] [Google Scholar]
- 27.Valarcher JF, Schelcher F, Bourhy H. 2000. Evolution of bovine respiratory syncytial virus. J. Virol. 74:10714–10728. 10.1128/JVI.74.22.10714-10728.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spackman DH, Stein WH, Moore S. 1958. Automatic recording apparatus for use in the chromatography of amino acids. Anal. Chem. 30:1190–1206. 10.1021/ac60139a006 [DOI] [PubMed] [Google Scholar]
- 29.Tran TL, Castagné N, Dubosclard V, Noinville S, Koch E, Moudjou M, Henry C, Bernard J, Yeo RP, Eléouët JF. 2009. The respiratory syncytial virus M2-1 protein forms tetramers and interacts with RNA and P in a competitive manner. J. Virol. 83:6363–6374. 10.1128/JVI.00335-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arfi Y, Chevret D, Henrissat B, Berrin JG, Levasseur A, Record E. 2013. Characterization of salt-adapted secreted lignocellulolytic enzymes from the mangrove fungus Pestalotiopsis sp. Nat. Commun. 4:1810. 10.1038/ncomms2850 [DOI] [PubMed] [Google Scholar]
- 31.Taylor G, Stott EJ, Bew M, Fernie BF, Cote PJ, Collins AP, Hughes M, Jebbett J. 1984. Monoclonal antibodies protect against respiratory syncytial virus infection in mice. Immunology 52:137–142 [PMC free article] [PubMed] [Google Scholar]
- 32.Taylor G, Stott EJ, Furze J, Ford J, Sopp P. 1992. Protective epitopes on the fusion protein of respiratory syncytial virus recognized by murine and bovine monoclonal antibodies. J. Gen. Virol. 73 2217–2223 [DOI] [PubMed] [Google Scholar]
- 33.Furze J, Wertz G, Lerch R, Taylor G. 1994. Antigenic heterogeneity of the attachment protein of bovine respiratory syncytial virus. J. Gen. Virol. 75(Pt 2):363–370 [DOI] [PubMed] [Google Scholar]
- 34.Castagné N, Barbier A, Bernard J, Rezaei H, Huet JC, Henry C, Da Costa B, Eléouët JF. 2004. Biochemical characterization of the respiratory syncytial virus P-P and P-N protein complexes and localization of the P protein oligomerization domain. J. Gen. Virol. 85:1643–1653. 10.1099/vir.0.79830-0 [DOI] [PubMed] [Google Scholar]
- 35.Furze JM, Roberts SR, Wertz GW, Taylor G. 1997. Antigenically distinct G glycoproteins of BRSV strains share a high degree of genetic homogeneity. Virology 231:48–58. 10.1006/viro.1997.8490 [DOI] [PubMed] [Google Scholar]
- 36.Thomas LH, Gourlay RN, Stott EJ, Howard CJ, Bridger JC. 1982. A search for new microorganisms in calf pneumonia by the inoculation of gnotobiotic calves. Res. Vet. Sci. 33:170–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roux X, Dubuquoy C, Durand G, Tran-Tolla TL, Castagné N, Bernard J, Petit-Camurdan A, Eléouët JF, Riffault S. 2008. Sub-nucleocapsid nanoparticles: a nasal vaccine against respiratory syncytial virus. PLoS One 3:e1766. 10.1371/journal.pone.0001766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arbiza J, Taylor G, López JA, Furze J, Wyld S, Whyte P, Stott EJ, Wertz G, Sullender W, Trudel M, Melero J. 1992. Characterization of two antigenic sites recognized by neutralizing monoclonal antibodies directed against the fusion glycoprotein of human respiratory syncytial virus. J. Gen. Virol. 73:2225–2234 [DOI] [PubMed] [Google Scholar]
- 39.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9:671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karron RA, Wright PF, Belshe RB, Thumar B, Casey R, Newman F, Polack FP, Randolph VB, Deatly A, Hackell J, Gruber W, Murphy BR, Collins PL. 2005. Identification of a recombinant live attenuated respiratory syncytial virus vaccine candidate that is highly attenuated in infants. J. Infect. Dis. 191:1093–1104. 10.1086/427813 [DOI] [PubMed] [Google Scholar]
- 41.Valarcher JF, Furze J, Wyld S, Cook R, Conzelmann KK, Taylor G. 2003. Role of alpha/beta interferons in the attenuation and immunogenicity of recombinant bovine respiratory syncytial viruses lacking NS proteins. J. Virol. 77:8426–8439. 10.1128/JVI.77.15.8426-8439.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hema M, Nagendrakumar SB, Yamini R, Chandran D, Rajendra L, Thiagarajan D, Parida S, Paton DJ, Srinivasan VA. 2007. Chimeric tymovirus-like particles displaying foot-and-mouth disease virus non-structural protein epitopes and its use for detection of FMDV-NSP antibodies. Vaccine 25:4784–4794. 10.1016/j.vaccine.2007.04.023 [DOI] [PubMed] [Google Scholar]
- 43.Höglund S, Akerblom L, Ozel M, Villacres M, Eriksson M, Gelderblom HR, Arthur L, Morein B. 1990. Characterization of immunostimulating complexes (ISCOMs) of HIV-1. Viral Immunol. 3:195–206. 10.1089/vim.1990.3.195 [DOI] [PubMed] [Google Scholar]
- 44.Foucras G, Corbière F, Tasca C, Pichereaux C, Caubet C, Trumel C, Lacroux C, Franchi C, Burlet-Schiltz O, Schelcher F. 2011. Alloantibodies against MHC class I: a novel mechanism of neonatal pancytopenia linked to vaccination. J. Immunol. 187:6564–6570. 10.4049/jimmunol.1102533 [DOI] [PubMed] [Google Scholar]
- 45.Kreidberg JA. 2000. Functions of alpha3beta1 integrin. Curr. Opin. Cell Biol. 12:548–553. 10.1016/S0955-0674(00)00130-7 [DOI] [PubMed] [Google Scholar]
- 46.Morgan MR, Byron A, Humphries MJ, Bass MD. 2009. Giving off mixed signals: distinct functions of alpha5beta1 and alphavbeta3 integrins in regulating cell behaviour. IUBMB Life 61:731–738. 10.1002/iub.200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weerasinghe D, McHugh KP, Ross FP, Brown EJ, Gisler RH, Imhof BA. 1998. A role for the alphavbeta3 integrin in the transmigration of monocytes. J. Cell Biol. 142:595–607. 10.1083/jcb.142.2.595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scheffer KD, Gawlitza A, Spoden GA, Zhang XA, Lambert C, Berditchevski F, Florin L. 2013. Tetraspanin CD151 mediates papillomavirus type 16 endocytosis. J. Virol. 87:3435–3446. 10.1128/JVI.02906-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang S, Kodys K, Babcock GJ, Szabo G. 2012. CD81/CD9 tetraspanins aid plasmacytoid dendritic cells in recognition of hepatitis C virus-infected cells and induction of interferon-alpha. Hepatology 58:940–949. 10.1002/hep.25827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mastrangelo P, Hegele RG. 2012. The RSV fusion receptor: not what everyone expected it to be. Microbes Infect. 14:1205–1210. 10.1016/j.micinf.2012.07.015 [DOI] [PubMed] [Google Scholar]