Abstract
Gold standard serological diagnostic methods focus on antigens that elicit a strong humoral immune response that is specific to a certain pathogen. In this study, we used bioinformatics approaches to identify linear B-cell epitopes that are conserved among Leishmania species but are divergent from the host species Homo sapiens and Canis familiaris and from Trypanosoma cruzi, the parasite that causes Chagas disease, to select potential targets for the immunodiagnosis of leishmaniasis. Using these criteria, we selected heat shock protein 83.1 of Leishmania braziliensis for this study. We predicted three linear B-cell epitopes in its sequence. These peptides and the recombinant heat shock protein 83.1 (rHSP83.1) were tested in enzyme-linked immunosorbent assays (ELISAs) against serum samples from patients with tegumentary leishmaniasis (TL) and visceral leishmaniasis (VL) and from dogs infected with Leishmania infantum (canine VL [CVL]). Our data show that rHSP83.1 is a promising target in the diagnosis of TL. We also identified specific epitopes derived from HSP83.1 that can be used in the diagnosis of human TL (peptide 3), both human and canine VL (peptides 1 and 3), and all TL, VL, and CVL clinical manifestations (peptide 3). Receiver operating characteristic (ROC) curves confirmed the superior performance of rHSP83.1 and peptides 1 and 3 compared to that of the soluble L. braziliensis antigen and the reference test kit for the diagnosis of CVL in Brazil (EIE-LVC kit; Bio-Manguinhos, Fiocruz). Our study thus provides proof-of-principle evidence of the feasibility of using bioinformatics to identify novel targets for the immunodiagnosis of parasitic diseases using proteins that are highly conserved throughout evolution.
INTRODUCTION
Leishmaniasis is a neglected vector-borne tropical disease that is caused by parasites of the Leishmania genus. Currently, it has a major impact on human health in tropical regions and affects approximately 12 million people worldwide (1). Two million new cases are reported annually, with an incidence of 1 to 1.5 million cases of tegumentary leishmaniasis (TL) and 500,000 cases of visceral leishmaniasis (VL) (1). The clinical forms of leishmaniasis range from self-healing cutaneous lesions to fatal visceral infections. In TL, a wide variety of skin manifestations, such as cutaneous leishmaniasis (CL) and mucocutaneous leishmaniasis (ML), have been described (2). The CL form is characterized by one or more painless ulcers with raised borders and a bed of granulation tissue that appear near the area of the sand fly bite, while ML is characterized by the progressive destruction of the nasopharyngeal mucosa (3, 4). Although the determining factors involved in the development of each clinical form have not been elucidated, it is likely that host and parasite genetics are involved (2, 5).
In Brazil, TL is distributed throughout the country, and among the various Leishmania species that can cause the disease, Leishmania braziliensis is responsible for the majority of the cases. L. braziliensis infection results in higher clinical severity due to larger ulcerated areas and a higher proportion of patients with mucosal involvement in the upper airway (6, 7). VL is caused by the Leishmania infantum parasite and is a zoonotic disease that has shown significant changes in transmission with progressive urbanization and geographic expansion; it now affects regions in which it was previously quite rare (2). Dogs are the main urban reservoirs and represent the major source of infection for the vector due to the high prevalence of canine infections and intense cutaneous parasitism that may contribute to urban spread of the disease (8, 9).
The major prophylactic practice recommended by the World Health Organization to control the human disease and canine visceral leishmaniasis (CVL) (8) involves early accurate diagnosis, systematic treatment of human cases, vector control with insecticide, and the elimination of seropositive dogs (10). At present, there is no gold standard serological test for diagnosing leishmaniasis, and a combination of different techniques is frequently necessary to obtain precise results. Therefore, the development of a new serological technique with higher sensitivity and specificity than the available commercial tests and that is able to discriminate postvaccination reactivity from active infections would represent an important innovation in the serological diagnosis of leishmaniasis (11). Additionally, due to the high conservation of proteins among the various species of Leishmania, the selection of an antigen that is able to simultaneously diagnose the various clinical forms of the disease would represent an interesting strategy for the technological development and large-scale production of tests for diagnosis (12, 13).
In this context, we performed genome-wide screening to identify linear B-cell epitopes in the predicted proteome of L. braziliensis in an attempt to identify conserved targets within the Leishmania genus for the serodiagnosis of the tegumentary and visceral forms of leishmaniasis. The protein selected in this study was heat shock protein 83.1 (HSP83.1), a highly conserved molecule in prokaryotes and eukaryotes that plays important roles in protein folding, assembly of protein complexes, and the translocation of proteins across cellular compartments (14). We mapped three B-cell linear epitopes in this protein whose sequences are divergent from its orthologs in Homo sapiens and Canis familiaris and in Trypanosoma cruzi, the etiologic agent of Chagas disease (CD), which is frequently associated with cross-reactivity with leishmaniasis (15, 16). The reactivities of recombinant HSP83.1 (rHSP83.1) and three derived B-cell epitopes with serum samples from TL and VL patients and from Leishmania-infected dogs were evaluated in enzyme-linked immunosorbent assays (ELISAs). rHSP83.1 demonstrated excellent performance for diagnosing TL. Additionally, we identified specific epitopes derived from HSP83.1 that may be useful in the diagnosis of human TL (peptide 3), both human and canine VL (peptides 1 and 3), and all TL, VL, and CVL clinical manifestations (peptide 3). Receiver operating characteristic (ROC) curves confirmed the superior performance of rHSP83.1 and peptides 1 and 3 compared to that of the soluble L. braziliensis antigen and the reference test for diagnosing CVL in Brazil (EIE-LVC kit; Bio-Manguinhos, Fiocruz) (17).
MATERIALS AND METHODS
Ethics statement and human and dog serum samples.
All samples that were used were anonymous and were obtained from the serum bank of the Laboratory of Immunology and Genomics of Parasites, Federal University of Minas Gerais. Approval to use the samples was obtained from the Human Research Ethics Committee (protocol CAAE – 00842112.2.0000.5149) and the Committee on Ethics of Animal Experimentation from the Federal University of Minas Gerais (protocol 44/2012).
The human serum panel consisted of 65 samples from tegumentary leishmaniasis patients infected with L. braziliensis that presented cutaneous (CL) (n = 45) or mucosal (ML) (n = 20) clinical manifestations obtained from the Centro de Referência em Leishmaniose (Januária, Minas Gerais, Brazil) and 55 samples from visceral leishmaniasis patients infected with L. infantum obtained from the University Hospital (Montes Claros, Minas Gerais, Brazil). The infection was confirmed by microscopic analyses of biopsy specimens from cutaneous lesions (for TL) or bone marrow aspirate samples (for VL), followed by specific PCR assays for the kinetoplastid DNA (kDNA) of Leishmania parasites (18). These individuals were known to be noninfected with T. cruzi. Information regarding the clinical evaluation and PCR results were obtained from the medical records of the patients. To evaluate the cross-reactivity with Chagas disease, serum samples from patients with chronic Chagas disease (CD) (n = 20) with infections confirmed by hemoculture or the Chagatest recombinant ELISA version 3.0 kit and the Chagatest hemagglutination inhibition (HAI) were used in this study. Serum samples from healthy Brazilian individuals (controls [CT]) (n = 50) from areas where TL and CD are not endemic (Belo Horizonte, Minas Gerais, Brazil) were used as negative controls in these assays. The resulting optical density (OD) values were compared with those obtained with the panel of TL and VL samples.
Male and female beagle dogs from an area where VL is not endemic that had Leishmania-negative tissue smears (bone marrow) were considered to be noninfected and were used as the control group (CT, n = 30). Serum samples from Leishmania-infected dogs (CVL) (n = 30) were obtained from the area where CVL is endemic in Minas Gerais in southeastern Brazil. The main inclusion criterion for the CVL serum samples used in this study was parasitological positivity for L. infantum confirmed by microscopic analysis of bone marrow aspirate samples. Samples from dogs that were experimentally infected with T. cruzi (CD, n = 15) but that were parasitologically negative for Leishmania were included in this study to evaluate possible cross-reactivity.
In silico prediction of linear B-cell epitopes.
Linear B-cell epitopes were predicted for the HSP83.1 protein of L. braziliensis (TriTrypDB identification [ID] [19] LbrM.33.0340) using the BepiPred 1.0 program with a cutoff of 1.3 (20). A BLASTp (21) search was performed against GeneDB (http://www.genedb.org/) to retrieve the HSP83.1 sequences of T. cruzi, H. sapiens, and C. familiaris using L. braziliensis HSP83.1 as a query. Multiple alignments of HSP83.1 were performed using the Clustal X 2.0 program (22), with the default parameters (23).
Soluble L. braziliensis antigen.
Soluble L. braziliensis antigen (SLbA) was prepared from L. (V.) braziliensis (MHOM/BR/75/M2904) stationary-phase promastigotes maintained in Schneider's insect medium (Sigma-Aldrich) supplemented with 10% inactivated fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin (Gibco). The parasites were initially subjected to three cycles of freezing (with liquid nitrogen) and thawing (42°C), followed by ultrasonication (Ultrasonic processor model GEX-600) with 10 alternating cycles of 30 s on/off sonication in an ice water bath at 35 MHz. The lysate was then centrifuged at 6,000 × g at 4°C for 15 min. The supernatant containing SLbA was collected, and the protein content was quantified using the Pierce BCA protein assay (Thermo Scientific).
Cloning, protein expression, and purification.
The primers used to amplify the HSP83.1 gene from the Leishmania genomic DNA were HSP83.1-Forward (5′-GCTAGCATGACGGAGACGTTCGCGTT) and HSP83.1-Reverse (5′-AAGCTTTCAGTCCACCTGCTCCATG). The sites for restriction enzymes (NheI and HindIII) that were added to the forward and reverse primers, respectively, to facilitate cloning, are shown in bold type. The DNA fragments obtained were excised from the gel, purified, digested with the restriction enzymes, and cloned into the pET28a-TEV vector (24). Electrocompetent Escherichia coli BL21 Arctic Express (DE3) cells (Agilent Technologies, USA) were transformed with the recombinant plasmid pET28a-TEV-HSP83.1 by electroporation using a MicroPulser electroporation apparatus (Bio-Rad Laboratories, USA). Gene insertion was confirmed by colony PCR and sequencing using T7 primers (Macrogen, South Korea).
Protein expression and purification were performed as previously described (25). Briefly, expression was induced by adding isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1.0 mM, and the bacterial culture was incubated for 24 h at 12°C with shaking at 200 rpm per min. The cells were then ruptured by sonication, the debris was removed by centrifugation, and the recombinant protein was purified using a HisTrap HP affinity column connected to an ÄKTAprime chromatography system (GE Healthcare, USA). The eluted fractions containing rHSP83.1 (705 amino acids, 80.6 kDa) were concentrated using the Amicon Ultra 15 centrifugal filters 10,000 nominal molecular weight limit (NMWL) (Millipore, Germany) and further purified on a Superdex 200 gel filtration column (GE Healthcare Life Sciences, USA).
Soluble peptide synthesis.
The soluble peptides were manually synthesized by 9-fluorenylmethoxy (Fmoc) chemistry in solid phase on a 30-μmol scale (26). Briefly, Fmoc-amino acids were activated with a 1:2 solution of Oxyme to N,N′-diisopropylcarbodiimide (DIC). The active amino acids were then incorporated into Rink amide resin with a substitution degree of 0.61. Fmoc deprotection was then performed using 25% 4-methylpiperidine. These steps were repeated until the synthesis of each peptide was completed. The peptides were then deprotected and released from the resin by treatment with a solution of 95% trifluoroacetic acid, 2.5% water, and 2.5% triisopropylsilane and precipitated with cold diisopropyl ether. The peptides were obtained with ≥90% purity, as confirmed by matrix-assisted laser desorption ionization–tandem time of flight mass spectrometry (MALDI-TOF-TOF MS) Autoflex III equipment (Bruker Daltonics, USA). Instrument calibration was achieved using peptide calibration standard II (Bruker Daltonics, USA) as a reference, and α-cyano-4-hydroxycinnamic acid was used as the matrix.
Serological assay and depletion ELISA.
rHSP83.1 and SLbA were coated onto 96-well microplates (Nalge Nunc International, USA) overnight at 2 to 8°C at a concentration of 250 ng/well for rHSP83.1 and 50 ng/well for SLbA. The peptides were coated onto flat-bottom plates (CoStar, USA) overnight at 37°C at a concentration of 10 μg/well. The plates were blocked with 150 μl of phosphate-buffered saline (PBS) containing 5% bovine serum albumin (BSA) for 1 h at 37°C and then treated with 1:100 dilutions of human or canine serum samples for 1 h at 37°C. Peroxidase-conjugated antibodies specific for human or dog IgG (Sigma-Aldrich, USA) were diluted at 1:5,000 and added to the plates for 1 h at 37°C. The wells were washed, and the 3,3′,5,5′-tetramethylbenzidine (TMB) substrate (Sigma-Aldrich, USA) in citrate buffer containing hydrogen peroxide was added to the plate. The plates were incubated for 30 min in the dark at 37°C. The reaction was halted by adding 4 N H2SO4, and the absorbance at 450 nm was read with an automatic microplate reader (VersaMax; Molecular Devices, USA). Each serum sample was assayed in duplicate. The results of the ELISA using rHSP83.1 as an antigen were compared with those using SLbA. The results of the ELISA using rHSP83.1 as an antigen for the canine serum samples were compared with those with the EIE-LVC kit immunoassay (Bio-Manguinhos, Fiocruz, Brazil), which is the test currently recommended by the Brazilian Ministry of Health for the screening of seroreactive animals (17). The assays were carried out according to the manufacturer's instructions.
Depletion ELISAs were performed as previously described (27, 28). Briefly, flat-bottom plates (CoStar, USA) were coated overnight at 37°C with 10 μg/well of peptide 1 (EEDESKKKSCGDEGEPKVE), peptide 2 (VTEGGEDKKK), and peptide 3 (EVAEAPPAEAAPA) and then washed and blocked as described above. All serum samples from individuals in the TL, VL, and CVL groups that were parasitologically positive for Leishmania and reactive for the tested peptide were used in the assay. Pools of sera were added to the plates at a 1:100 dilution and incubated overnight at 2 to 8°C. The depleted and undepleted sera were transferred to plates coated overnight with rHSP83.1 (50 ng/well) after appropriate washing and blocking, and the ELISAs were performed as described above.
Statistical analysis.
The lower limits of positivity (cutoff) for rHSP83.1, SLbA, and peptides 1, 2, and 3 were established for optimal sensitivity and specificity using the receiver operating characteristic (ROC) curve. The cutoff was chosen based on the point that provides the maximum of the sum of the sensitivity and specificity (29). The EIE-LVC cutoff was obtained according to the manufacturer (twice the average of the negative control provided by the kit). The performance of each test was evaluated according to the sensitivity (Se), specificity (Sp), positive predictive value (PPV), negative predictive value (NPV), area under the curve (AUC), and accuracy (AC). The degree of agreement between the ELISAs using rHSP83.1, SLbA, or the EIE-LVC kit with the parasitological test (biopsy specimen, aspirate sample, or PCR) was determined by the Kappa index (k) values with 95% confidence intervals and interpreted according to the following Fleiss scale: 0.00 to 0.20, poor; 0.21 to 0.40, fair; 0.41 to 0.60, moderate; 0.61 to 0.80, good; 0.81 to 0.99, very good; and 1.00, perfect (30). The one-sample Kolmogorov-Smirnov test was used to determine whether a variable was normally distributed. For the depletion assays, significant differences were detected using a two-way analysis of variance (ANOVA). The differences were considered statistically significant at a P value of <0.05. All of the statistical analyses were performed using GraphPad Prism (version 5.0) and the GraphPad QuickCalcs software.
RESULTS
Sequence divergence and prediction of B-cell linear epitopes in L. braziliensis HSP83.1 protein and its orthologs.
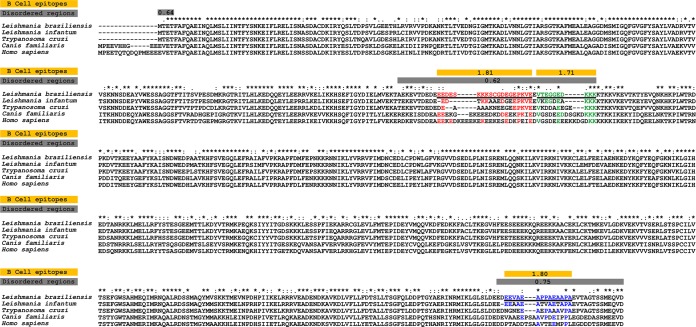
Gold standard serological diagnostic methods focus on markers that are able to elicit a strong antibody response specific to the pathogen. Although HSP83.1 is conserved throughout evolution, B-cell epitopes derived from Leishmania HSP83.1, with sequences that are divergent from those of the parasite hosts and other closely related parasites, may bind to specific antibodies and thus allow for the specific identification of samples from patients with leishmaniasis. To identify potential epitopes that can increase the accuracy of the currently available serological tests, we scanned the HSP83.1 sequence of L. braziliensis for B-cell linear epitopes that are divergent in the host orthologs and that cooccur with intrinsically unstructured regions (Fig. 1). The presence of these unstructured regions suggests that a given protein region is in an unfolded structure and therefore possibly accessible for antibody binding. As expected, the HSP83.1 ortholog of L. braziliensis, LbrM.33.0340, displayed a high level of identity to the orthologous proteins from L. infantum, T. cruzi, C. familiaris, and H. sapiens, with identities of >63.5%, as shown in Fig. 1 and Table 1. Despite this overall high level of similarity, we identified three potential linear B-cell epitopes (peptide 1, EEDESKKKSCGDEGEPKVE; peptide 2, VTEGGEDKKK; and peptide 3, EVAEAPPAEAAPA) in the LbrM.33.0340 protein that are highly divergent in T. cruzi as well as in dog and human orthologs, with identities ranging from 21.4 to 60.0% (Fig. 1 and Table 1). This result leads us to speculate that despite the high overall sequence similarity between HSP83 in Leishmania species and its orthologs in T. cruzi and in humans and dogs, this protein and the three predicted B-cell linear epitopes might be potential targets in the immunodiagnosis of leishmaniasis.
FIG 1.
Sequence divergence and prediction of B-cell linear epitopes and intrinsically unstructured/disordered regions in L. braziliensis HSP83.1 and its orthologs. Alignment between L. braziliensis HSP83.1 (TriTrypDB ID LbrM.33.0340) and orthologous proteins present in L. infantum (TriTrypDB ID Linj.33.0360), T. cruzi (TriTrypDB ID TcCLB.509105.140), H. sapiens (RefSeq ID NP_005339.3), and C. familiaris (RefSeq ID XP_532154.4). The yellow boxes mark predicted B-cell epitopes, and the gray boxes mark predicted disordered regions. The continuous black underlined amino acid sequences represent three potential B-cell epitopes predicted by BepiPred in the LbrM.33.0340 protein, and red, green, and blue highlight amino acid conservations in the T. cruzi, C. familiaris, and H. sapiens sequences in relation to the L. braziliensis sequence.
TABLE 1.
Sequence identity of the B-cell linear epitopes predicted in LbrM.33.0340 HSP83 protein of Leishmania braziliensis and its orthologs
| Species | % identity with LbrM.33.0340 | % identity with peptidea: |
||
|---|---|---|---|---|
| 1 | 2 | 3 | ||
| Leishmania infantum | 93.9 | 52.3 | 70.0 | 71.4 |
| Trypanosoma cruzi | 85.2 | 31.6 | 50.0 | 50.0 |
| Canis familiaris | 64.1 | 36.8 | 60.0 | 35.7 |
| Homo sapiens | 63.5 | 36.8 | 40.0 | 21.4 |
Peptide 1, EEDESKKKSCGDEGEPKVE; peptide 2, VTEGGEDKKK; peptide 3, EEVAEAPPAEAAPA.
Expression and purification of rHSP83.1 protein.
To verify whether the HSP83.1 protein is a candidate for the development of new diagnostic assays, we first expressed this protein as a His-tagged recombinant protein. The full-length coding region of the LbrM.33.0340 gene was amplified by PCR and cloned into the pET28a-TEV expression vector. rHSP83.1 was overexpressed in E. coli BL21 Arctic Express (DE3) cells as a soluble protein and purified by Ni2+ affinity chromatography, followed by gel filtration on a Superdex 200 column. The recombinant protein, with a predicted molecular mass of 80.6 kDa, was successfully expressed and obtained at a high level of purity (Fig. 2).
FIG 2.
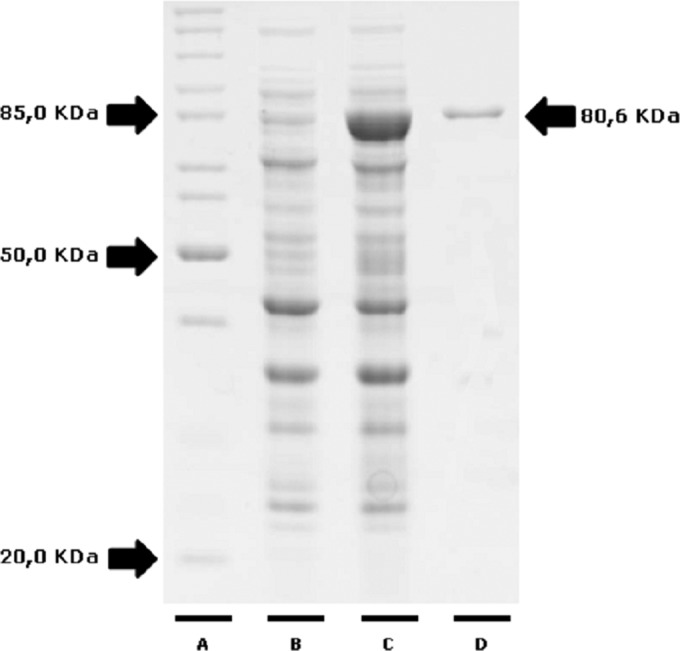
Expression and purification of recombinant HSP83.1 protein. Protein samples were separated by 12.5% SDS-PAGE. Lane A, molecular weight standard, lysate of culture before (lane B) and after (lane C) induction with IPTG and recombinant HSP83.1 protein (mass, 80.6 kDa) purified by gel filtration (lane D).
ELISA performance using rHSP83.1, peptides 1, 2, 3, and SLbA in TL diagnosis.
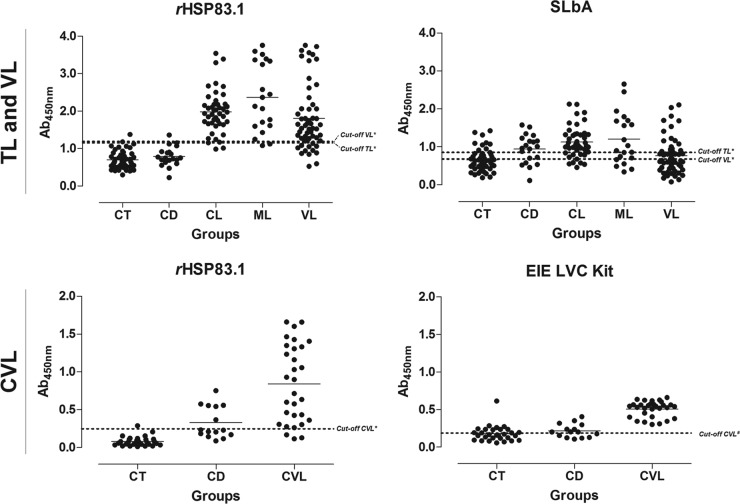
To evaluate the specific antibody responses against rHSP83.1 and the derived peptides 1, 2, and 3 in TL, ELISAs with these antigens were compared to SLbA ELISAs (Fig. 3 to 5 and Tables 2 and 3). When tested with serum samples from CL patients, the rHSP83.1 and peptide 1, 2, and 3 antigens showed sensitivities of 95.55%, 71.11%, 64.44%, and 95.55%, respectively. The serum samples from ML patients presented lower sensitivity values to all antigens tested (90.00%, 55.00%, 50.00%, and 75.00%, respectively). The antigens presented total sensitivity (CL + ML) for rHSP83.1 and sensitivities of 93.85% (95% confidence interval [CI], 84.99 to 98.30%), 63.08% (95% CI, 50.20 to 74.72), 63.08% (95% CI, 50.20 to 74.72), and 89.23% (95% CI, 79.06 to 95.56) for peptides 1, 2, and 3, respectively (Table 2). ELISAs using SLbA extract presented sensitivities of 75.55% and 60.00% for detecting the CL and ML clinical forms, respectively. The total sensitivity of SLbA in TL was 70.77% (95% CI, 58.17 to 81.40%), which was lower than that for rHSP83.1 and peptide 3 (Table 2). To evaluate specificity (Sp), serum samples from chagasic patients and negative-control individuals were also tested. rHSP83.1 and peptides 1, 2, and 3 showed total specificity values of 95.71% (95% CI, 87.98 to 99.11%), 94.29% (95% CI, 86.01 to 98.42), 90.00% (95% CI, 80.48 to 95.88%), and 91.43% (95% CI, 82.27 to 96.79%), respectively. The ELISA using SLbA presented a much lower specificity value of 68.57% (95% CI, 56.37 to 79.15%) (Table 2).
FIG 3.
Comparison of the reactivity of ELISAs against rHSP83.1 and SLbA and the EIE-LVC kit against serum samples from TL and VL patients and from L. infantum-infected dogs. (Top) ELISAs were performed on samples from different groups of individuals (CT [control] group, n = 50; CD [Chagas disease] patients, n = 20; CL [cutaneous leishmaniasis], n = 45; ML [mucosal leishmaniasis], n = 20; VL [visceral leishmaniasis], n = 55). (Bottom) ELISAs were performed on samples from different groups of dogs (CT [control group], n = 30; CD [T. cruzi-infected dogs], n = 15; CVL [canine visceral leishmaniasis], n = 30). An asterisk indicates a cutoff value obtained by ROC curve; a pound sign indicates a cutoff value obtained according to the manufacturer. Dots represent individual absorbance values. The dotted line represents the cutoff value. The solid line corresponds to the mean values. Ab450nm, absorbance at 450 nm.
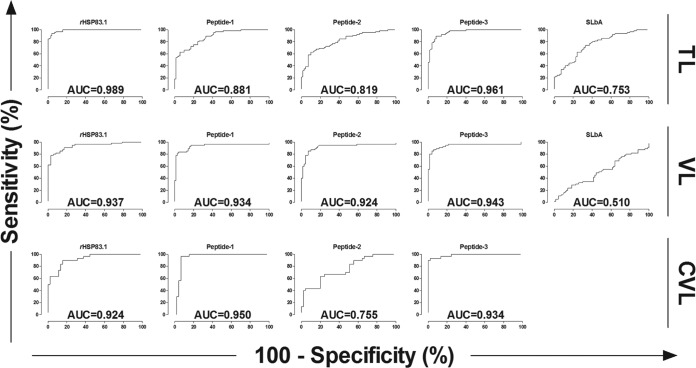
FIG 5.
Comparison of ROC curves obtained from rHSP83.1, peptides 1, 2, and 3, and SLbA. The ROC curves were used to determine the ELISA cutoff, sensitivity, specificity, and AUC. Ab450nm, absorbance at 450 nm.
TABLE 2.
Measure of diagnostic performance for rHSP83.1, peptides 1, 2, and 3, SLbA, and the EIE-LVC kit
| Testa | Disease | Cutoff value | Test performance parameter (% [95% CI])b |
||||
|---|---|---|---|---|---|---|---|
| TSe | TSp | PPV | NPV | AC | |||
| rHSP83.1 | TL | 1.1510 | 93.85 (84.99–98.30) | 95.71 (87.98–99.11) | 95.31 | 94.37 | 94.81 |
| Peptide 1 | TL | 0.5165 | 63.08 (50.20–74.72) | 94.29 (86.01–98.42) | 91.11 | 73.33 | 79.26 |
| Peptide 2 | TL | 0.4884 | 63.08 (50.20–74.72) | 90.00 (80.48–95.88) | 85.42 | 72.41 | 77.04 |
| Peptide 3 | TL | 0.2713 | 89.23 (79.06–95.56) | 91.43 (82.27–96.79) | 90.63 | 90.14 | 90.37 |
| SLbA | TL | 0.8530 | 70.77 (58.17–81.40) | 68.57 (56.37–79.15) | 67.65 | 71.64 | 69.63 |
| rHSP83.1 | VL | 1.1900 | 78.18 (64.99–88.19) | 97.14 (90.06–99.65) | 95.56 | 85.00 | 88.80 |
| Peptide 1 | VL | 0.5693 | 83.64 (71.20–92.23) | 95.71 (87.98–99.11) | 93.88 | 88.16 | 90.40 |
| Peptide 2 | VL | 0.5096 | 85.45 (73.34–93.50) | 92.86 (84.11–97.64) | 90.38 | 89.04 | 89.60 |
| Peptide 3 | VL | 0.2901 | 87.27 (75.52–94.73) | 94.29 (86.01–98.42) | 92.31 | 90.41 | 91.20 |
| SLbA | VL | 0.6780 | 52.73 (38.80–66.35) | 50.00 (37.80–62.20) | 45.31 | 57.38 | 51.20 |
| rHSP83.1 | CVL | 0.2473 | 90.00 (73.47–97.89) | 84.44 (70.54–93.51) | 79.41 | 92.68 | 86.67 |
| Peptide 1 | CVL | 0.1031 | 96.67 (82.78–99.92) | 93.33 (81.73–98.60) | 90.63 | 97.67 | 94.67 |
| Peptide 2 | CVL | 0.1669 | 63.33 (43.86–80.07) | 80.00 (65.40–90.42) | 67.86 | 76.60 | 73.33 |
| Peptide 3 | CVL | 0.0520 | 93.33 (77.93–99.18) | 97.78 (88.23–99.24) | 96.65 | 95.65 | 96.00 |
| EIE-LVC kit | CVL | 0.1894 | 100.00 (88.43–100.0) | 53.33 (37.87–68.34) | 58.82 | 100.00 | 72.00 |
Cutoff values for all tests obtained by ROC curve, except that for the EIE-LVC kit, which was obtained according to the manufacturer.
Parameters were calculated using all samples presented in this work for TL (CT + CD + CL + ML) (n = 135), VL (CT + CD + VL) (n = 125), and CVL (CT + CD + LM + CVL) (n = 75). Tse, total sensitivity; CI, confidence interval; TSp, total specificity; PPV, positive predictive value; NPV, negative predictive value; AC, accuracy.
TABLE 3.
Diagnostic performance for rHSP83.1, peptides 1, 2, and 3, SLbA, and the EIE-LVC kit using ROC curves, data validation, and agreement using kappa index
| Testa | Disease | AUCb (95% CIc) | No. with resultd: |
κe (95% CI) | Agreementf | |||
|---|---|---|---|---|---|---|---|---|
| TP | TN | FP | FN | |||||
| rHSP83.1 | TL | 0.989 (0.979–1.000) | 61 | 67 | 3 | 4 | 0.896 (0.821–0.971) | Very good |
| Peptide 1 | TL | 0.881 (0.826–0.936) | 41 | 66 | 4 | 24 | 0.580 (0.448–0.712) | Moderate |
| Peptide 2 | TL | 0.819 (0.748–0.889) | 41 | 63 | 7 | 24 | 0.536 (0.397–0.674) | Moderate |
| Peptide 3 | TL | 0.961 (0.933–0.989) | 58 | 64 | 6 | 7 | 0.807 (0.707–0.907) | Very good |
| SLbA | TL | 0.753 (0.673–0.834) | 48 | 46 | 22 | 19 | 0.393 (0.238–0.548) | Fair |
| rHSP83.1 | VL | 0.937 (0.894–0.981) | 43 | 68 | 2 | 12 | 0.768 (0.655–0.881) | Good |
| Peptide 1 | VL | 0.934 (0.880–0.988) | 46 | 67 | 3 | 9 | 0.803 (0.697–0.908) | Very good |
| Peptide 2 | VL | 0.924 (0.867–0.981) | 47 | 65 | 5 | 8 | 0.788 (0.679–0.897) | Good |
| Peptide 3 | VL | 0.943 (0.892–0.995) | 48 | 66 | 4 | 7 | 0.820 (0.719–0.922) | Very good |
| SLbA | VL | 0.510 (0.405–0.614) | 29 | 35 | 35 | 26 | 0.027 (−0.147–0.200) | Poor |
| rHSP83.1 | CVL | 0.924 (0.867–0.981) | 27 | 38 | 7 | 3 | 0.728 (0.572–0.884) | Good |
| Peptide 1 | CVL | 0.950 (0.894–1.005) | 29 | 42 | 3 | 1 | 0.890 (0.786–0.995) | Very good |
| Peptide 2 | CVL | 0.755 (0.644–0.866) | 19 | 36 | 9 | 11 | 0.438 (0.230–0.646) | Moderate |
| Peptide 3 | CVL | 0.987 (0.967–1.007) | 28 | 44 | 1 | 2 | 0.916 (0.823–1.000) | Very good |
| EIE-LVC kit | CVL | NA | 30 | 24 | 21 | 0 | 0.478 (0.316–0.639) | Moderate |
Cutoffs for all tests obtained by ROC curve, except that for the EIE-LVC kit, which was obtained according to the manufacturer.
AUC, area under the curve; NA, not applicable.
CI, confidence interval.
TP, true positive; TN, true negative; FP, false positive; FN, false negative.
Kappa index values were calculated using all samples presented in this work for TL (CT + CD + CL + ML) (n = 135), VL (CT + CD + VL) (n = 125), and CVL (CT + CD + CVL) (n = 75).
Agreement was calculated using parasitological assays as a gold standard test.
The maximum PPV was achieved with rHSP83.1 (95.31%), followed by peptide 1 (91.11%), peptide 3 (90.63%), peptide 2 (85.42%), and SLbA (67.65%). High NPVs were also observed for rHSP83.1 (94.37%), followed by peptide 3 (90.14%), peptide 1 (73.33%), peptide 2 (72.41%), and SLbA (71.64%) (Table 2).
Based on the ELISA results, ROC curves were calculated to evaluate the capacity of the tested antigens to discriminate TL patients from healthy volunteers (Fig. 5 and Table 3). The area under the curve (AUC) and accuracy (AC) were used to compare the efficiencies of the different diagnostic antigens or tests (29) (Table 3). rHSP83.1 presented the highest AUC value (0.989; 95% CI, 0.979 to 1.000), followed by peptide 3 (0.961; 95% CI, 0.933 to 0.989), peptide 1 (0.881; 95% CI, 0.826 to 0.936), peptide 2 (0.819; 95% CI, 0.748 to 0.889), and SLbA (0.753; 95% CI, 0.673 to 0.834). The accuracy value for rHSP83.1 was also the highest (94.81%), followed by peptide 3 (90.37%), peptide 1 (79.26%), peptide 2 (77.04%), and SLbA (69.63%) (Table 2).
ELISA performance using rHSP83.1, peptides 1, 2, and 3, and SLbA in VL diagnosis.
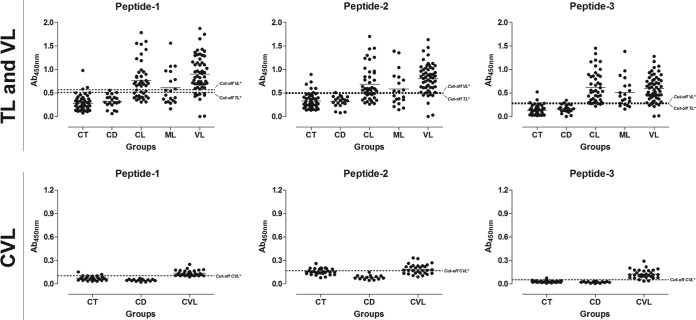
We also tested the performance of ELISAs using rHSP83.1 and the derived peptides 1, 2, and 3 in VL diagnosis and compared the results to those of the SLbA ELISA (Fig. 3 to 5 and Tables 2 and 3). rHSP83.1 and peptides 1, 2, and 3 presented total sensitivities of 78.18% (95% CI, 64.99 to 88.19%), 83.64% (95% CI, 71.20 to 92.23%), 85.45% (95% CI, 73.34 to 93.50%), and 87.27% (95% CI, 75.52 to 94.73%), respectively, for detecting VL (Table 2). The total sensitivity of SLbA for detecting VL was 52.73% (95% CI, 38.80 to 66.35%), which is much lower than those for the other tested antigens. rHSP83.1 and peptides 1, 2, and 3 presented specificity values of 97.14% (95% CI, 90.06 to 99.65%), 95.71% (95% CI, 87.98 to 99.11), 92.86% (95% CI, 84.11 to 97.64%), and 94.29% (95% CI, 86.01 to 98.42%), respectively; again, these values were much higher than that obtained for SLbA (50.00%; 95% CI, 37.80 to 62.20%) (Table 2).
The maximum PPV was achieved with rHSP83.1 (95.56%), followed by peptide 1 (93.88%), peptide 3 (92.31%), peptide 2 (90.38%), and SLbA (45.31%). The highest NPV was observed for peptide 3 (90.41%), followed by peptide 2 (89.04%), peptide 1 (88.16%), rHSP83.1 (85.00%), and SLbA (57.38%). Peptide 3 also presented the highest AUC value (0.943; 95% CI, 0.892 to 0.995), followed by rHSP83.1 (0.937; 95% CI, 0.894 to 0.981), peptide 1 (0.934; 95% CI, 0.880 to 0.988), peptide 2 (0.924; 95% CI, 0.867 to 0.981), and SLbA (0.510; 95% CI, 0.405 to 0.614) (Fig. 4 and Table 3). The accuracy value for peptide 3 was the highest (91.20%), followed by peptide 1 (90.40%), peptide 2 (89.60%), rHSP83.1 (88.80%), and SLbA (51.20%) (Table 2).
FIG 4.
Comparison of the reactivities of the ELISAs against peptides 1, 2, and 3 against serum samples from TL and VL patients and from L. infantum-infected dogs. (Top) ELISAs were performed on samples from the different groups of individuals (CT [control group], n = 50; CD [Chagas disease] patients, n = 20; CL [cutaneous leishmaniasis], n = 45; ML [mucosal leishmaniasis], n = 20; VL [visceral leishmaniasis], n = 55). (Bottom) ELISAs were performed on samples from the different groups of dogs (CT [control] group, n = 30; CD [T. cruzi-infected dogs], n = 15; CVL [canine visceral leishmaniasis], n = 30). An asterisk indicates a cutoff value obtained by ROC curve.
ELISA performance using rHSP83.1, peptides 1, 2, and 3, and the EIE-LVC kit in CVL diagnosis.
Next, we evaluated the performance of ELISAs using rHSP83.1 and the derived peptides 1, 2, and 3 in CVL diagnosis and compared the results to those obtained with the EIE-LVC kit (Fig. 3 to 5 and Tables 2 and 3). Serological assays using rHSP83.1 and peptides 1, 2, and 3 presented total sensitivities of 90.00% (95% CI, 73.47 to 97.89%), 96.67% (95% CI, 82.78 to 99.92%), 63.33% (95% CI, 43.86 to 80.07%), and 93.33% (95% CI, 77.93 to 99.18%), respectively, for detecting CVL (Table 2). All of the tested antigens showed sensitivity values for detecting CVL that were inferior to those obtained with the EIE-LVC kit (100.00% sensitivity; 95% CI, 88.43 to 100.00%). In contrast, ELISAs using the antigens tested in this study showed a better specificity value (84.44%; 95% CI, 70.54 to 93.51%) than that of the EIE-LVC kit (53.33%; 95% CI, 37.87 to 68.34%) (Table 2).
The maximum PPV was achieved with peptide 3 (96.65%), followed by peptide 1 (90.63%), rHSP83.1 (79.41%), peptide 2 (67.86%), and the EIE-LVC kit (58.82%). The highest NPV was observed for the EIE-LVC kit (100.00%), followed by peptide 1 (97.67%), peptide 3 (95.65%), rHSP83.1 (92.68%), and peptide 2 (76.60%) (Table 2). Peptide 3 presented the highest AUC value (0.987; 95% CI, 0.967 to 1.007), followed by peptide 1 (0.950; 95% CI, 0.894 to 1.005), rHSP83.1 (0.924; 95% CI, 0.867 to 0.981), and peptide 2 (0.755; 95% CI, 0.644 to 0.866). The AUC value was not calculated for the EIE-LVC kit because the cutoff value not was established using an ROC curve but was instead calculated as twice the average of the negative control provided by the kit, as recommended by the manufacturer. The accuracy value for peptide 3 was the highest (96.00%), followed by peptide 1 (94.67%), rHSP83.1 (86.67%), peptide 2 (73.33%), and the EIE-LVC kit (72.00%) (Table 2).
Agreement between rHSP83.1, peptides 1, 2, and 3, SLbA, and the EIE-LVC kit with parasitological assays.
The agreement (Kappa index) values between the serological tests using rHSP83.1, peptides 1, 2, and 3, SLbA, and the EIE-LVC kit with parasitological assays are shown in Table 3. Regarding TL diagnosis, the best agreement score was observed for rHSP83.1 (0.896, very good), followed by peptide 3 (0.807, very good), peptide 1 (0.580, moderate), peptide 2 (0.536, moderate), and SLbA (0.393, fair). For the diagnosis of human and canine VL, the best agreement scores were obtained for peptide 3 (0.820, very good for VL; 0.916, very good for CVL), followed by peptide 1 (0.803, very good for VL, and 0.890, very good for CVL).
Linear B-cell epitopes identified in this study contribute significantly to the overall antigenicity of rHSP83.1.
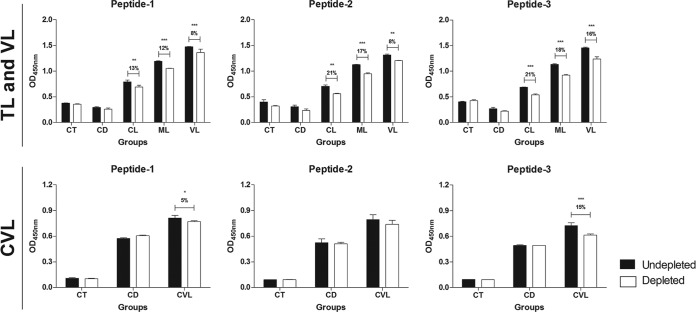
To determine the contribution of the linear B-cell epitopes identified in this study to the antigenicity of rHSP83.1, ELISA depletion assays were performed using the synthetic peptides EEDESKKKSCGDEGEPKVE, VTEGGEDKKK, and EVAEAPPAEAAPA, which correspond to the predicted linear B-cell epitopes present in the protein (Fig. 6). In this assay, the IgG reactivity against rHSP83.1 after the depletion of antibodies against peptide 1 was reduced by 13% for the CL (P < 0.01), 12% for the ML (P < 0.001), 8% for the VL (P < 0.001), and 5% for the CVL (P < 0.05) group. For peptide 2, we observed a reduction of 21% for the CL (P < 0.01), 17% for the ML (P < 0.001), 8% for the VL (P < 0.001), and no reduction for the CVL group. The depletion of antibodies against peptide 3 resulted in a reduction of 21% for the CL (P < 0.001), 18% for the ML (P < 0.001), 16% for the VL (P < 0.001), and 15% for the CVL (P < 0.001) group in the reactivity against rHSP83.1. No significant differences in the reduction of reactivity after depletion were observed in the human and canine CT and CD groups for any of the peptides evaluated. Our results suggest that specific antibodies against peptides 1 and 3 contribute significantly to the overall antigenicity of rHSP83.1 in all Leishmania-infected groups; however, peptide 2 contributes to the antigenicity of this protein in the TL and VL groups only.
FIG 6.
Immunodepletion assay showing specific IgG antibody recognition of the synthetic peptides with known reactivity to HSP83.1. Pools of serum (n = 10) from the different groups were depleted with peptide 1, 2, or 3 (CT [control] group; CD [Chagas disease]; CL [cutaneous leishmaniasis]; ML [mucosal leishmaniasis]; VL [visceral leishmaniasis]; CVL [canine visceral leishmaniasis]). The mean antibody OD values are shown on the y axis, and the error bars indicate the standard deviation. Significant differences are indicated on the graphs with P values (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
DISCUSSION
Genome-wide epitope predictions for pathogenic microorganisms have broadened the range of target epitopes and have provided clues to enhance peptide immunogenicity. Among the methods of selection, epitope mapping is a very useful procedure that has vast applications in the areas of antibody production, immunodiagnostics, epitope-based vaccine design, selective deimmunization of therapeutic proteins, and autoimmunity. The identification of these targets was facilitated by the publication of several parasite genomes that together with the availability of algorithms for epitope prediction allow for the use of large-scale approaches for the identification of new antigens (31–33). As an attempt to identify new targets for diagnosing multiple forms of leishmaniasis, we screened the L. braziliensis proteome to predict linear B-cell epitopes that are conserved in Leishmania spp. and divergent in H. sapiens, C. familiaris, and T. cruzi, the etiologic agent of Chagas disease, which often presents cross-reactivity with leishmaniasis due to sharing multiple common epitopes within the trypanosomatid species (34, 35).
Heat shock proteins (HSPs) are highly conserved molecules in prokaryotes and eukaryotes that play important roles in protein folding, assembly of protein complexes, and the translocation of proteins across cellular compartments (14). Despite the fact that their sequences are highly conserved throughout evolution, it has been proposed that the recognition of epitopes shared by HSPs from different pathogens may provide the immune system with a universal signal of infection (36). In this sense, HSP70 and HSP83 of L. infantum have been described as strong mitogens for B lymphocytes in a murine model (37). This phenomenon occurs even in the absence of T lymphocytes and adherent cells, suggesting a direct effect of Leishmania HSPs on B cells, and that Leishmania may serve to divert the immune response into the nonspecific activation of immune cells (37).
In our study, we identified three linear B-cell epitopes in HSP83.1 of L. braziliensis that are conserved within Leishmania species but are divergent from the orthologous proteins from human and canine hosts and from T. cruzi, the etiological agent of Chagas disease that has been responsible for several reports of cross-reactivity in the serological diagnosis of leishmaniasis. Previous studies have described that heat shock proteins, such as HSP83 and other chaperones, are among the most abundant proteins detected in Leishmania antigenic extracts and have obtained promising results that can be applied to the development of diagnostic kits (38–44). Here, the antigenicity of recombinant HSP83.1 (rHSP83.1) and three epitopes predicted in this protein were tested with sera from the TL, VL, and CVL groups.
rHSP83.1 showed excellent performance in the diagnosis of TL. Not all of the epitopes mapped in this study were recognized by sera from all clinical forms of leishmaniasis. Nevertheless, our results allowed for the identification of epitopes that presented high performance depending upon the species of the parasite. Interestingly, it was possible to identify specific epitopes derived from HSP83.1 that presented high performance for the diagnosis of human TL (peptide 3), both human and canine VL (peptides 1 and 3), and all TL, VL, and CVL clinical manifestations (peptide 3).
In recent years, synthetic peptides have shown high sensitivity and specificity when used as antigens in diagnostic tests (23, 45). These findings have also been associated with several advantages over chemically purified or recombinant antigens, because their production does not involve the manipulation of living organisms, and the peptides can be obtained with a high level of purity. Thus, it is appealing to use these peptides to develop devices to diagnose one or multiple types of leishmaniasis (46).
Comparing conventional and new techniques proposed to be innovative is essential to justify the implementation of the novel methods. In this sense, several authors have described ELISAs using soluble antigens from promastigotes for serological diagnosis that showed high sensitivity for detecting Leishmania-infected individuals (34, 35, 47, 48). Despite these findings, the use of these antigens still represents a relevant obstacle because it is common to observe a large number of false-positive reactions in individuals infected with other trypanosomatids due to the sharing of multiple common epitopes used in immunofluorescence assays or ELISAs sensitized with crude antigens (34, 35, 49, 50). As expected, using crude antigens, such as the soluble L. braziliensis antigen or the EIE-LVC kit, which uses antigen prepared from Leishmania major-like promastigotes, leads to a pronounced number of false positives compared to with rHSP83.1. This is the main limitation to their use in epidemiological surveys in areas where leishmaniasis is endemic (47). Furthermore, different species and strains of Leishmania can be used for the production of crude antigen, thus limiting the standardization and reproducibility of test results (51, 52). Therefore, there is a need to standardize an antigen for use in different regions of the world (47).
The serologic enzyme-linked immunosorbent assay is an important tool in the diagnosis of leishmaniasis compared to methods currently used to directly reveal parasites in tissue smears that are invasive, are risky, require considerable expertise, and are inappropriate for use in epidemiological surveillance (53, 54). Given the above, the rational development of a new serological technique featuring high sensitivity and specificity that is also able to discriminate cross-reactivity with other human and canine infections, at the individual level, represents an important innovation in the serological diagnosis of CVL (11).
The present study demonstrates that rHSP83.1 and the epitopes mapped on its sequence might be among the target molecules that could be used in an immunodiagnostic test. This was confirmed by the high sensitivity for diagnosing Leishmania infections, the high specificity for discriminating between other human and canine infections, and the high degree of agreement with parasitological techniques for the diagnosis of leishmaniasis. Further prospective studies using large cohorts of negative and positive individuals from areas where leishmaniasis is endemic are necessary to better characterize this approach as a possible marker in the diagnosis of TL, VL, and CVL and for monitoring posttherapeutic cures of TL and VL.
ACKNOWLEDGMENTS
C.M.C., E.A.F.C., R.T.F., and D.C.B. thank the Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazil, for their fellowships. We thank Michele Silva de Matos for her technical support.
Footnotes
Published ahead of print 7 May 2014
REFERENCES
- 1.Desjeux P. 2001. Worldwide increasing risk factors for leishmaniasis. Med. Microbiol. Immunol. 190:77–79. 10.1007/s004300100085 [DOI] [PubMed] [Google Scholar]
- 2.Desjeux P. 2004. Leishmaniasis: current situation and new perspectives. Comp. Immunol. Microbiol. Infect. Dis. 27:305–318. 10.1016/j.cimid.2004.03.004 [DOI] [PubMed] [Google Scholar]
- 3.Gontijo B, de Carvalho Mde L. 2003. American cutaneous leishmaniasis. Rev. Soc. Bras. Med. Trop. 36:71–80 (In Portuguese.) 10.1590/S0037-86822003000100011 [DOI] [PubMed] [Google Scholar]
- 4.Martins AL, Barreto JA, Lauris JR, Martins AC. 2014. American tegumentary leishmaniasis: correlations among immunological, histopathological and clinical parameters. An. Bras. Dermatol. 89:52–58. 10.1590/abd1806-4841.20142226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costa JM, Marsden PD, Llanos-Cuentas EA, Netto EM, Carvalho EM, Barral A, Rosa AC, Cuba CC, Magalhães AV, Barreto AC. 1986. Disseminated cutaneous leishmaniasis in a field clinic in Bahia, Brazil: a report of eight cases. J. Trop. Med. Hyg. 89:319–323 [PubMed] [Google Scholar]
- 6.Marsden PD. 1985. Clinical presentations of Leishmania braziliensis braziliensis. Parasitol. Today 1:129–133. 10.1016/0169-4758(85)90057-2 [DOI] [PubMed] [Google Scholar]
- 7.Grimaldi G, Jr, Tesh RB. 1993. Leishmaniases of the New World: current concepts and implications for future research. Clin. Microbiol. Rev. 6:230–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tesh RB. 1995. Control of zoonotic visceral leishmaniasis: is it time to change strategies? Am. J. Trop. Med. Hyg. 52:287–292 [DOI] [PubMed] [Google Scholar]
- 9.Molina R, Amela C, Nieto J, San-Andrés M, González F, Castillo JA, Lucientes J, Alvar J. 1994. Infectivity of dogs naturally infected with Leishmania infantum to colonized Phlebotomus perniciosus. Trans. R. Soc. Trop. Med. Hyg. 88:491–493. 10.1016/0035-9203(94)90446-4 [DOI] [PubMed] [Google Scholar]
- 10.Chappuis F, Sundar S, Hailu A, Ghalib H, Rijal S, Peeling RW, Alvar J, Boelaert M. 2007. Visceral leishmaniasis: what are the needs for diagnosis, treatment and control? Nat. Rev. Microbiol. 5:S7–S16. 10.1038/nrmicro1748 [DOI] [PubMed] [Google Scholar]
- 11.de Andrade RA, Reis AB, Gontijo CM, Braga LB, Rocha RD, Araújo MS, Vianna LR, Martins-Filho OA. 2007. Clinical value of anti-Leishmania (Leishmania) chagasi IgG titers detected by flow cytometry to distinguish infected from vaccinated dogs. Vet. Immunol. Immunopathol. 116:85–97. 10.1016/j.vetimm.2007.01.002 [DOI] [PubMed] [Google Scholar]
- 12.de Oliveira CI, Nascimento IP, Barral A, Soto M, Barral-Netto M. 2009. Challenges and perspectives in vaccination against leishmaniasis. Parasitol. Int. 58:319–324. 10.1016/j.parint.2009.07.013 [DOI] [PubMed] [Google Scholar]
- 13.Souza AP, Soto M, Costa JM, Boaventura VS, de Oliveira CI, Cristal JR, Barral-Netto M, Barral A. 2013. Towards a more precise serological diagnosis of human tegumentary leishmaniasis using Leishmania recombinant proteins. PLoS One 8:e66110. 10.1371/journal.pone.0066110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henderson B. 2010. Integrating the cell stress response: a new view of molecular chaperones as immunological and physiological homeostatic regulators. Cell Biochem. Funct. 28:1–14. 10.1002/cbf.1609 [DOI] [PubMed] [Google Scholar]
- 15.Vexenat Ade C, Santana JM, Teixeira AR. 1996. Cross-reactivity of antibodies in human infections by the kinetoplastid protozoa Trypanosoma cruzi, Leishmania chagasi and Leishmania (Viannia) braziliensis. Rev. Inst. Med. Trop. Sao Paulo 38:177–185 [DOI] [PubMed] [Google Scholar]
- 16.Mancianti F, Pedonese F, Poli A. 1996. Evaluation of dot enzyme-linked immunosorbent assay (dot-ELISA) for the serodiagnosis of canine leishmaniosis as compared with indirect immunofluorescence assay. Vet. Parasitol. 65:1–9. 10.1016/0304-4017(96)00946-6 [DOI] [PubMed] [Google Scholar]
- 17.Alves WA, Bevilacqua PD. 2004. Quality of diagnosis of canine visceral leishmaniasis in epidemiological surveys: an epidemic in Belo Horizonte, Minas Gerais, Brazil, 1993–1997. Cad. Saude Publica. 20:259–265 (In Portuguese.) 10.1590/S0102-311X2004000100043 [DOI] [PubMed] [Google Scholar]
- 18.de Bruijn MH, Barker DC. 1992. Diagnosis of New World leishmaniasis: specific detection of species of the Leishmania braziliensis complex by amplification of kinetoplast DNA. Acta Trop. 52:45–58. 10.1016/0001-706X(92)90006-J [DOI] [PubMed] [Google Scholar]
- 19.Aslett M, Aurrecoechea C, Berriman M, Brestelli J, Brunk BP, Carrington M, Depledge DP, Fischer S, Gajria B, Gao X, Gardner MJ, Gingle A, Grant G, Harb OS, Heiges M, Hertz-Fowler C, Houston R, Innamorato F, Iodice J, Kissinger JC, Kraemer E, Li W, Logan FJ, Miller JA, Mitra S, Myler PJ, Nayak V, Pennington C, Phan I, Pinney DF, Ramasamy G, Rogers MB, Roos DS, Ross C, Sivam D, Smith DF, Srinivasamoorthy G, Stoeckert CJ, Jr, Subramanian S, Thibodeau R, Tivey A, Treatman C, Velarde G, Wang H. 2010. TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 38:D457–D462. 10.1093/nar/gkp851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larsen JE, Lund O, Nielsen M. 2006. Improved method for predicting linear B-cell epitopes. Immunome Res. 2:2. 10.1186/1745-7580-2-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. 10.1006/jmbi.1990.9999 [DOI] [PubMed] [Google Scholar]
- 22.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 23.Mendes TA, Reis Cunha JL, de Almeida Lourdes R, Rodriguez Luiz GF, Lemos LD, dos Santos AR, da Câmara AC, Galvão LM, Bern C, Gilman RH, Fujiwara RT, Gazzinelli RT, Bartholomeu DC. 2013. Identification of strain-specific B-cell epitopes in Trypanosoma cruzi using genome-scale epitope prediction and high-throughput immunoscreening with peptide arrays. PLoS Negl. Trop. Dis. 7:e2524. 10.1371/journal.pntd.0002524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carneiro FR, Silva TC, Alves AC, Haline-Vaz T, Gozzo FC, Zanchin NI. 2006. Spectroscopic characterization of the tumor antigen NY-REN-21 and identification of heterodimer formation with SCAND1. Biochem. Biophys. Res. Commun. 343:260–268. 10.1016/j.bbrc.2006.02.140 [DOI] [PubMed] [Google Scholar]
- 25.Coitinho JB, Costa DM, Guimarães SL, de Góes AM, Nagem RA. 2012. Expression, purification and preliminary crystallographic studies of NahF, a salicylaldehyde dehydrogenase from Pseudomonas putida G7 involved in naphthalene degradation. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 68:93–97. 10.1107/S174430911105038X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wellings DA, Atherton E. 1997. Standard Fmoc protocols. Methods Enzymol. 289:44–67. 10.1016/S0076-6879(97)89043-X [DOI] [PubMed] [Google Scholar]
- 27.Santiago HC, Bennuru S, Boyd A, Eberhard M, Nutman TB. 2011. Structural and immunologic cross-reactivity among filarial and mite tropomyosin: implications for the hygiene hypothesis. J. Allergy Clin. Immunol. 127:479–486. 10.1016/j.jaci.2010.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bueno LL, Lobo FP, Morais CG, Mourão LC, Machado de Avila RA, Soares IS, Fontes CJ, Lacerda MV, Chavez Olórtegui C, Bartholomeu DC, Fujiwara RT, Braga EM. 2011. Identification of a highly antigenic linear B cell epitope within Plasmodium vivax apical membrane antigen 1 (AMA-1). PLoS One 6:e21289. 10.1371/journal.pone.0021289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linnet K, Bossuyt PM, Moons KG, Reitsma JB. 2012. Quantifying the accuracy of a diagnostic test or marker. Clin. Chem. 58:1292–1301. 10.1373/clinchem.2012.182543 [DOI] [PubMed] [Google Scholar]
- 30.Fleiss JL, Spitzer RL, Endicott J, Cohen J. 1972. Quantification of agreement in multiple psychiatric diagnosis. Arch. Gen. Psychiatry 26:168–171. 10.1001/archpsyc.1972.01750200072015 [DOI] [PubMed] [Google Scholar]
- 31.Singh H, Ansari HR, Raghava GP. 2013. Improved method for linear B-cell epitope prediction using antigen's primary sequence. PLoS One 8:e62216. 10.1371/journal.pone.0062216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dudek NL, Perlmutter P, Aguilar MI, Croft NP, Purcell AW. 2010. Epitope discovery and their use in peptide based vaccines. Curr. Pharm. Des. 16:3149–3157. 10.2174/138161210793292447 [DOI] [PubMed] [Google Scholar]
- 33.Bryson CJ, Jones TD, Baker MP. 2010. Prediction of immunogenicity of therapeutic proteins: validity of computational tools. BioDrugs 24:1–8. 10.2165/11318560-000000000-00000 [DOI] [PubMed] [Google Scholar]
- 34.Badaró R, Reed SG, Barral A, Orge G, Jones TC. 1986. Evaluation of the micro enzyme-linked immunosorbent assay (ELISA) for antibodies in American visceral leishmaniasis: antigen selection for detection of infection-specific responses. Am. J. Trop. Med. Hyg. 35:72–78 [DOI] [PubMed] [Google Scholar]
- 35.Barbosa-De-Deus R, dos Mares-Guia ML, Nunes AZ, Costa KM, Junqueira RG, Mayrink W, Genaro O, Tavares CAP. 2002. Leishmania major-like antigen for specific and sensitive serodiagnosis of human and canine visceral leishmaniasis. Clin. Diagn. Lab. Immunol. 9:1361–1366. 10.1128/CDLI.9.6.1361-1366.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaufmann SH. 1990. Heat shock proteins and the immune response. Immunol. Today 11:129–136. 10.1016/0167-5699(90)90050-J [DOI] [PubMed] [Google Scholar]
- 37.Rico AI, Gironès N, Fresno M, Alonso C, Requena JM. 2002. The heat shock proteins, Hsp70 and Hsp83, of Leishmania infantum are mitogens for mouse B cells. Cell Stress Chaperones 7:339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coelho VT, Oliveira JS, Valadares DG, Chávez-Fumagalli MA, Duarte MC, Lage PS, Soto M, Santoro MM, Tavares CA, Fernandes AP, Coelho EA. 2012. Identification of proteins in promastigote and amastigote-like Leishmania using an immunoproteomic approach. PLoS Negl. Trop. Dis. 6:e1430. 10.1371/journal.pntd.0001430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Costa MM, Andrade HM, Bartholomeu DC, Freitas LM, Pires SF, Chapeaurouge AD, Perales J, Ferreira AT, Giusta MS, Melo MN, Gazzinelli RT. 2011. Analysis of Leishmania chagasi by 2-D difference gel electrophoresis (2-D DIGE) and immunoproteomic: identification of novel candidate antigens for diagnostic tests and vaccine. J. Proteome Res. 10:2172–2184. 10.1021/pr101286y [DOI] [PubMed] [Google Scholar]
- 40.Skeiky YA, Benson DR, Costa JL, Badaro R, Reed SG. 1997. Association of Leishmania heat shock protein 83 antigen and immunoglobulin G4 antibody titers in Brazilian patients with diffuse cutaneous leishmaniasis. Infect. Immun. 65:5368–5370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rey-Ladino JA, Joshi PB, Singh B, Gupta R, Reiner NE. 1997. Leishmania major: molecular cloning, sequencing, and expression of the heat shock protein 60 gene reveals unique carboxy terminal peptide sequences. Exp. Parasitol. 85:249–263. 10.1006/expr.1996.4137 [DOI] [PubMed] [Google Scholar]
- 42.Quijada L, Requena JM, Soto M, Alonso C. 1998. Analysis of the antigenic properties of the L. infantum Hsp70: design of synthetic peptides for specific serodiagnosis of human leishmaniasis. Immunol. Lett. 63:169–174. 10.1016/S0165-2478(98)00071-6 [DOI] [PubMed] [Google Scholar]
- 43.Celeste BJ, Angel SO, Castro LG, Gidlund M, Goto H. 2004. Leishmania infantum heat shock protein 83 for the serodiagnosis of tegumentary leishmaniasis. Braz. J. Med. Biol. Res. 37:1591–1593. 10.1590/S0100-879X2004001100001 [DOI] [PubMed] [Google Scholar]
- 44.Kaur J, Kaur S. 2013. ELISA and Western blotting for the detection of Hsp70 and Hsp83 antigens of Leishmania donovani. J. Parasit. Dis. 37:68–73. 10.1007/s12639-012-0133-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carmona SJ, Sartor PA, Leguizamón MS, Campetella OE, Agüero F. 2012. Diagnostic peptide discovery: prioritization of pathogen diagnostic markers using multiple features. PLoS One 7:e50748. 10.1371/journal.pone.0050748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aguirre S, Silber AM, Brito ME, Ribone ME, Lagier CM, Marcipar IS. 2006. Design, construction, and evaluation of a specific chimeric antigen to diagnose chagasic infection. J. Clin. Microbiol. 44:3768–3774. 10.1128/JCM.01043-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosário EY, Genaro O, Franca-Silva JC, da Costa RT, Mayrink W, Reis AB, Carneiro M. 2005. Evaluation of enzyme-linked immunosorbent assay using crude Leishmania and recombinant antigens as a diagnostic marker for canine visceral leishmaniasis. Mem. Inst. Oswaldo Cruz 100:197–203. 10.1590/S0074-02762005000200015 [DOI] [PubMed] [Google Scholar]
- 48.Mettler M, Grimm F, Capelli G, Camp H, Deplazes P. 2005. Evaluation of enzyme-linked immunosorbent assays, an immunofluorescent-antibody test, and two rapid tests (immunochromatographic-dipstick and gel tests) for serological diagnosis of symptomatic and asymptomatic Leishmania infections in dogs. J. Clin. Microbiol. 43:5515–5519. 10.1128/JCM.43.11.5515-5519.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.da Costa CA, Genaro O, de Lana M, Magalhães PA, Dias M, Michalick MS, Melo MN, da Costa RT, Magalhães-Rocha NM, Mayrink W. 1991. Canine visceral leishmaniasis: evaluation of the serologic method used in epidemiologic studies. Rev. Soc. Bras. Med. Trop. 24:21–25. 10.1590/S0037-86821991000100004 [DOI] [PubMed] [Google Scholar]
- 50.Roffi J, Dedet JP, Desjeux P, Garré MT. 1980. Detection of circulating antibodies in cutaneous leishmaniasis by enzyme-linked immunosorbent assay (ELISA). Am. J. Trop. Med. Hyg. 29:183–189 [DOI] [PubMed] [Google Scholar]
- 51.Burns JM, Jr, Shreffler WG, Benson DR, Ghalib HW, Badaro R, Reed SG. 1993. Molecular characterization of a kinesin-related antigen of Leishmania chagasi that detects specific antibody in African and American visceral leishmaniasis. Proc. Natl. Acad. Sci. U. S. A. 90:775–779. 10.1073/pnas.90.2.775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martin SK, Thuita-Harun L, Adoyo-Adoyo M, Wasunna KM. 1998. A diagnostic ELISA for visceral leishmaniasis, based on antigen from media conditioned by Leishmania donovani promastigotes. Ann. Trop. Med. Parasitol. 92:571–577. 10.1080/00034989859267 [DOI] [PubMed] [Google Scholar]
- 53.Pereira VR, Reis Lde C, Souza Mde A, de Oliveira AP, de Brito ME, Lage PS, Andrade MC, Rocha RD, Martins-Filho OA. 2012. Evaluation of anti-lived and anti-fixed Leishmania (Viannia) braziliensis promastigote IgG antibodies detected by flow cytometry for diagnosis and post-therapeutic cure assessment in localized cutaneous leishmaniasis. Diagn. Microbiol. Infect. Dis. 74:292–298. 10.1016/j.diagmicrobio.2012.06.025 [DOI] [PubMed] [Google Scholar]
- 54.Kumar S, Kumar D, Chakravarty J, Rai M, Sundar S. 2012. Identification and characterization of a novel Leishmania donovani antigen for serodiagnosis of visceral leishmaniasis. Am. J. Trop. Med. Hyg. 86:601–605. 10.4269/ajtmh.2012.11-0261 [DOI] [PMC free article] [PubMed] [Google Scholar]