Abstract
Homologous recombination is a crucial mechanism that repairs a wide range of DNA lesions, including the most deleterious ones, double-strand breaks (DSBs). This multistep process is initiated by the resection of the broken DNA ends by a multisubunit helicase-nuclease complex exemplified by Escherichia coli RecBCD, Bacillus subtilis AddAB, and newly discovered Mycobacterium tuberculosis AdnAB. Here we show that in Streptomyces, neither recBCD nor addAB homologues could be detected. The only putative helicase-nuclease-encoding genes identified were homologous to M. tuberculosis adnAB genes. These genes are conserved as a single copy in all sequenced genomes of Streptomyces. The disruption of adnAB in Streptomyces ambofaciens and Streptomyces coelicolor could not be achieved unless an ectopic copy was provided, indicating that adnAB is essential for growth. Both adnA and adnB genes were shown to be inducible in response to DNA damage (mitomycin C) and to be independently transcribed. Introduction of S. ambofaciens adnAB genes in an E. coli recB mutant restored viability and resistance to UV light, suggesting that Streptomyces AdnAB could be a functional homologue of RecBCD and be involved in DNA damage resistance.
INTRODUCTION
Cells are under constant genotoxic pressure from both endogenous and exogenous sources. DNA damage needs to be repaired to avoid the formation of deleterious mutations, abortion of replication, and lethal chromosomal breakage. Homologous recombination (HR) is a crucial mechanism that repairs a variety of DNA lesions, including DNA double-strand breaks (DSBs), single-strand DNA gaps, and interstrand cross-links.
DSBs are probably the most deleterious DNA damage that a cell can encounter. They are induced in cells by physical agents such as ionizing radiation or UV light, chemical agents, and natural products such as mitomycin C (MMC) or bleomycin. DSBs also result from replication fork collapse during chromosomal replication (1). The failure to repair DSBs can lead to cell death and, in the case of disrepair, can trigger large-scale chromosome rearrangements, favoring the generation of genetic diversity.
In bacteria, DSBs are for the most part processed through HR, which requires a homologous DNA template to carry out faithful repair of the damaged DNA duplex. In intensively replicating cells (vegetative growth phase) or immediately after the passing of the replication fork, the sister chromatid can be used as an intact template. In nonreplicating phases, such as late stationary phase or in spores (which contain a single copy of the chromosome), DSBs are more likely repaired by an illegitimate repair pathway. Indeed, illegitimate recombination (IR) does not require an intact homologous sequence but as a consequence has reduced fidelity.
The involvement of DSB repair by HR in a range of fundamental cellular processes (e.g., chromosome integrity, replication, segregation, etc.) reveals that HR is conserved in all living organisms. The initiating step of the repair mechanism consists of the resection of the DSB end to generate a 3′ single-stranded tail onto which a synaptic protein (e.g., RecA in bacteria, RAD51 in humans) is loaded to form a nucleoprotein filament. The nucleoprotein filament will be used to perform homology search and strand invasion within a homologous DNA sequence. Subsequently, resolvase activity releases two intact DNA duplexes.
The end resection step is performed by a multisubunit helicase-nuclease complex, such as RecBCD in Escherichia coli (2, 3) and AddAB in Bacillus subtilis (4–6). In E. coli, defects in recB or recC impair HR, reduce cell viability, and increase sensitivity to DNA-damaging agents (7–9). In B. subtilis, a defect in AddAB moderately decreases DSB repair efficiency (10). RecBCD and AddAB are functional homologues, given that heterologous expression of B. subtilis AddAB restores cell viability, the ability to repair UV-damaged DNA, and recombination in the E. coli recBCD null mutant (11). Examination of the phylogenetic distribution of RecBCD and AddAB revealed that one or both of the complexes are encoded in the majority of sequenced bacterial genomes (12). Using a proteomic approach, Sinha et al. (13) uncovered a new heterodimeric DNA motor-driven nuclease complex called AdnAB (ATP-dependent nuclease) in Mycobacterium smegmatis. This is the third helicase-nuclease complex identified that has been shown to be involved in DSB repair, as an M. smegmatis adnAB-null mutant is hypersensitive to ionizing radiation (14).
Although all three of these helicase-nuclease enzymes appear to be involved in DSB repair in their respective context, they differ in distribution of the functional domains (helicase motor and nuclease domains within the different subunits of the complex). RecBCD is composed of two helicase motor domains carried by RecB and RecD (15, 16) and one nuclease domain located at the C terminus of RecB. For its part, AddAB possesses a single helicase motor at the N terminus of the AddA subunit, plus two distinct RecB-like nuclease domains located at the C terminus of AddA and AddB (6). The AdnA and AdnB subunits are each composed of an N-terminal UvrD-like motor domain and a C-terminal RecB-like nuclease domain (13).
In E. coli and B. subtilis, an alternative pathway (the RecFOR pathway) can substitute for RecBCD and AddAB, respectively, in the end resection step. In RecBCD-deficient E. coli cells, the RecFOR pathway, known to be involved in single-strand-break repair, can compensate for the RecBCD defect and promote recombinational DSB repair (8, 17–19). This pathway combines the single-strand DNA exonuclease RecJ, the RecQ helicase, and the RecF, RecO, and RecR proteins that act together to promote loading of RecA onto single-stranded DNA. In B. subtilis, genetic studies revealed that addAB or ΔrecJ mutations moderately impact DSB repair, whereas the addAB ΔrecJ double mutation has a synergistic negative effect (10).
In mycobacteria, both AdnAB and RecBCD complexes cohabit: while AdnAB is required for HR (14), the RecBCD complex is involved in a RecA-independent illegitimate recombination pathway called single-strand annealing (SSA) (20). Moreover, the RecFOR pathway is absent, since neither RecJ nor RecQ has been identified in mycobacterial genomes thus far (21).
Streptomyces are actinobacteria that are well known for their high genetic plasticity (22, 23), yet their recombination pathways remain poorly understood. Streptomyces are Gram-positive soil bacteria possessing a large linear chromosome (24) showing a remarkable genetic organization. While the central region is highly syntenic at the intraspecific level and gathers the essential genes, the terminal parts are variable and prone to chromosomal rearrangements affecting several hundreds of kilobases under laboratory conditions. Interestingly, comparative genomics reveals that synteny in the terminal regions does not abruptly stop but rather fades progressively toward the terminal regions. This degenerated synteny consists of the accumulation over evolutionary times of insertions and deletions (indels) that follows an increasing gradient toward the chromosomal ends (25). This phenomenon can be put into perspective with the recent report of high intraspecies and interspecies HR levels within Streptomyces revealed by multilocus sequence analysis (MLSA) (26). Further, the intraspecies recombination rate exceeded the interspecies rate by 2 orders of magnitude, supporting the possibility that gene exchange and recombination may have shaped the genome of streptomycetes. These genomic characteristics prompted us to investigate HR in Streptomyces and to search for helicase-nuclease-encoding genes and their involvement in survival, recombination, and DNA repair pathways.
MATERIALS AND METHODS
Bacterial strains and media.
The strains and plasmids used in this study are listed in Table 1. The DH5α E. coli strain was used as a cloning host for plasmid construction. E. coli ET12567/pUZ8002 and S17-1 are the nonmethylating plasmid donor strains used for intergeneric conjugation with Streptomyces ambofaciens and Streptomyces coelicolor. The E. coli strains JJC40 and JJC315 used for nuclease activity, cell viability, and UV sensitivity assays were kindly provided by Bénédicte Michel (CNRS, Gif-sur-Yvette, France). All E. coli strains were grown in Luria-Bertani (LB) broth at 37°C, with the exception of strain DY330, used for PCR targeting, which was grown at 30°C and transferred to 42°C for 17 min before transformation with linear DNA (27). Soya flour mannitol (SFM) agar medium was used for Streptomyces sporulation, and liquid Hickey-Tresner (HT) medium was used for growth of mycelia for RNA extraction (28). When appropriate, antibiotics were added to the media at concentrations of 50 μg · ml−1 kanamycin, 50 μg · ml−1 apramycin, 100 μg · ml−1 puromycin, 25 μg · ml−1 nalidixic acid, and 12 μg · ml−1 tetracycline.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Streptomyces | ||
| S. ambofaciens ATCC 23877 | Used as wild-type strain | 51 |
| S. coelicolor A3(2) | Used as wild-type strain | 52 |
| E. coli | ||
| DH5α | F− φdlacZΔM15 endA1 supE44 thi-1 recA1 relA1 gyrA96 deoR nupG Δ(lacZYA-argF)U169 λ− hsdR17(rK− mK+) phoA relA1 | 53 |
| DY330 | W3110 ΔlacU169 gal490 λcI857 Δ(cro-bioA) | 27 |
| ET12567/pUZ8002 | dam-13::Tn9 dcm cat tet hsdM hsdR zjj-201::Tn10 tra neo RP4 | 54, 55 |
| S17-1 | recA pro hsdR RP4–2-Tc::Mu Km::Tn7 | 56 |
| JJC40 | leu6 his4 argE3 lacY1 galK2 ara14 xyl5 mtl1 tsx33 rpsL31 supE44 hsdR | Provided by B. Michel (CNRS) |
| JJC315 | JJC40 derivative recB268::Tn10(Tet) | |
| BACs and plasmids | ||
| BAB15E8 | Resource of adnAB locus for construction of pSET152-adnAB | 25 |
| pSET152 | Integrative vector for actinomycetes; oriT(RK2), int and attP(φC31), acc(3)IV | 57 |
| pSET153 | pSET152 derivative containing neo instead of acc(3)IV | This work |
| pSET152-adnAB | pSET152 derivative containing the adnAB locus | This work |
| pSET153-adnAB | pSET152-adnAB derivative containing neo instead of acc(3)IV | This work |
| pIJ6902-LCN | pIJ6902 derivative | Unpublished data |
| pIJ6902-LCN-adnAB | pIJ6902-LCN derivative containing adnAB | This work |
Plasmid constructions.
The pNSV005 plasmid was designed for the deletion of S. ambofaciens adnAB: the adnAB flanking regions were amplified by PCR. The 5′ and 3′ regions were amplified from S. ambofaciens ATCC 23877 genomic DNA by PCR. The two PCR fragments were cloned into the pGEM-T Easy vector and then released by HindIII/EcoRV or EcoRV/NheI digestions. The excisable cassette att3-aac conferring apramycin resistance was excised from pOSV234 (29) by EcoRV digestion. The above-mentioned three fragments were simultaneously ligated with the HindIII/NheI-digested vector pWED2 (30), which confers puromycin resistance to generate an adnAB deletion vector.
Derivatives of the 2St3B6 cosmid carrying a Tn5062 insertion (31) in either the adnA or adnB gene were purchased from Paul Dyson, University of Swansea (http://strepdb.streptomyces.org.uk). These cosmids were used for adnA and adnB inactivation attempts in S. coelicolor.
For Streptomyces complementation experiments, the S. ambofaciens adnAB locus, including its own promoter, was isolated as a 11.4-kb NcoI fragment from a recombinant bacterial artificial chromosome (BAC) and cloned in pSL1180 (32). The locus was then excised as an XbaI/EcoRI fragment to be inserted into pSET152 and to yield pSET152-adnAB. A kanamycin-resistant version of pSET152 called pSET153 was constructed by replacement of the acc(3)IV gene by the neo gene from supercos1 (Agilent Technologies).
For E. coli complementation experiments, pIJ6902-LCN (unpublished), a low-copy-number plasmid derived from pIJ6902 (33), was used as a cloning vector. In plasmid pIJ6902-LCN, the high-copy-number pMB1 origin was replaced by the low-copy-number replication origin of the F factor (derived from the pBeloBAC11 vector). In order to avoid amplification of the long GC-rich fragment, including the adnAB genes (7.6 kb), we opted for the replacement of the sequence of pSET153 by pIJ6902-LCN in the pSET153-adnAB construct by using a PCR targeting procedure. By this approach, the adnAB genes were placed immediately downstream of the pTipA promoter, which was shown to be expressed in E. coli in our previous work (unpublished data).
Genetic procedures.
Plasmids, BACs, or cosmids were introduced into S. ambofaciens and S. coelicolor strains by intergeneric conjugation with E. coli ET12567/pUZ8002 or S17-1 (28). Single-crossover exconjugants leading to integration of the deletion construct into the chromosome were selected on plates containing apramycin. Double-crossover exconjugants leading to gene replacement and concomitant loss of the cloning vector were then screened for puromycin or kanamycin sensitivity according to the vector used (pNSV005 or 2St3B6 derivatives). Replacement events were confirmed by PCR. The frequencies of a second crossover were calculated from three independent experiments. P values were calculated by using the chi-square test.
Complemented E. coli strains were obtained by transformation of JJC315 with pIJ6902-LCN-adnAB.
RNA isolation and quantitative RT-PCR.
Pregerminated spores (28) of S. ambofaciens ATCC 23877 were grown until an optical density at 600 nm (OD600) of ∼0.2 was reached in HT liquid medium. RNA was harvested from cell aliquots treated with 1 μg · ml−1 mitomycin C (MMC) for 30 min. Total RNA was isolated from the harvested cells using the Aurum total RNA minikit (Bio-Rad). Reverse transcription (RT) reactions were performed using 1 μg of RNA and an iScript advanced cDNA synthesis kit from Bio-Rad according to the manufacturer's instructions. After quantitative RT-PCR was carried out on cDNA from each condition using iQ SYBR green supermix from Bio-Rad for each PCR, normalization of gene expression was achieved using hrdB as a reference. Primers and their positions in the target sequences are given in Table S1 in the supplemental material.
UV sensitivity and viability assays.
Overnight cultures of E. coli were diluted at 1:100 into fresh LB medium with appropriate antibiotics and grown to an OD600 of ∼0.3. After dilution in 100 mM phosphate buffer (pH 7.0), 100 μl of the 10−3 dilution was spread on LB plates. Plates were irradiated with 0, 10, or 30 J · m−2 using a 254-nm UV light delivering 1 J · m2 · s−1 and then incubated for 24 h at 37°C in the dark to avoid photoreactivation. Survival was determined as the number of CFU after a given UV dose divided by the CFU of nonirradiated cells. Cell viability was defined, in the absence of stress, as the ratio of CFU per ml to the expected cell number per ml as estimated by OD600 measurement.
Cell extract preparation and nuclease activity assay.
E. coli bacteria were harvested from an overnight culture and resuspended in a lysis buffer containing 1 mM dithiothreitol (DTT), 1 mM EDTA, 0.05 M Tris (pH 8), and 1 mg · ml−1 lysozyme. Cell extracts were obtained after sonication. Nuclease activity assay mixtures contained 50 mM Tris-HCl (pH 8), 10 mM MgCl2, 50 μM ATP, 200 ng linear DNA (a 3,734-bp PCR product), and 1 μg cell extract, and the mixtures were incubated at 37°C for 0 h, 1.5 h, and 3 h. After electrophoretic separation and ethidium bromide staining, the intensity of each DNA band was estimated using ImageJ (http://rsb.info.nih.gov/ij/).
RESULTS AND DISCUSSION
AdnAB is predicted to be the only helicase-nuclease activity in Streptomyces.
Twenty complete Streptomyces genomes were searched for genes encoding helicase-nuclease activity. BLASTP searches confirmed a previous report (21) that no homologues for RecB, RecC, AddA, and AddB are present in Streptomyces. A weak homologue of RecD with partial amino acid identities was identified using Rv0629c (575 amino acids [aa]) encoding the RecD homologue of M. tuberculosis as the query sequence (36% aa identity over 73% of the protein length). This suggests the absence of a RecBCD-like and an AddAB-like helicase-nuclease complex in Streptomyces. Furthermore, searches for the RecQ-RecJ helicase-nuclease complex, known to carry out DNA end resection in the absence of RecBCD and AddAB in E. coli and B. subtilis, respectively (10, 34), resulted in the identification of a sole RecQ homologue in Streptomyces. In the absence of the RecJ component (exonuclease activity), this pathway is not expected to be functional.
In contrast, Streptomyces genomes showed the presence of genes encoding homologues of AdnA (Rv3202c) and AdnB (Rv3201c) from M. tuberculosis (complex called MtuAdnAB), with 35 to 40% amino acid identities (Table 2). Between Streptomyces species, AdnA-like and AdnB-like proteins are highly conserved (from 72% to 95% when using the S. ambofaciens gene products as references), and the amino acid identities tend to reflect the phylogenetic relationships. These two proteins will be here called AdnA and AdnB. The level of amino acid identity of Streptomyces AdnA and AdnB proteins with those of other actinomycetes (Saccharopolyspora erythraea, Rhodoccocus erythropolis, and Frankia sp.) falls below 50%. In all Streptomyces species, with the exception of Streptomyces cattleya and Streptomyces venezuelae, the genes encoding the AdnA and AdnB homologues are adjacent and show the same transcriptional orientation (Fig. 1), as in Mycobacterium species (13). In S. cattleya and S. venezuelae, a single putative gene of 702 and 834 nucleotides, respectively, is inserted between the adnA and adnB genes.
TABLE 2.
AdnAB in Streptomyces and other actinomycete sequenced genomesa
| Species and strain | Locus tag for sequence |
|
|---|---|---|
| AdnA | AdnB | |
| Streptomyces avermitilis MA-4680 | SAV_3077 | SAV_3076 |
| Streptomyces bingchenggensis BCW-1 | SBI_04047 | SBI_04046 |
| Streptomyces cattleya NRRL 8057 (=DSM 46488) | SCAT_4044 | SCAT_4047 |
| Streptomyces coelicolor A3(2) | SCO5183 | SCO5184 |
| Streptomyces flavogriseus ATCC 33331 | Sfla_2108 | Sfla_2107 |
| Streptomyces griseus subsp. griseus NBRC 13350 | SGR_2342 | SGR_2341 |
| Streptomyces scabiei 87.22 | SCAB_30771 | SCAB_30761 |
| Streptomyces sp. SirexAA-E | SACTE_4413 | SACTE_4414 |
| Streptomyces violaceusniger Tu 4113 | Strvi_1501 | Strvi_1502 |
| Streptomyces clavuligerus ATCC 27064 | SCLAV_4047 | SCLAV_4048 |
| Streptomyces lividans TK24 | SLI_5469 | SLI_5470 |
| Streptomyces davawensis JMC 4913 | BN159_3205 | BN159_3204 |
| Streptomyces collinus Tu 365 | B446_24410 | B446_24415 |
| Streptomyces hygroscopicus subsp. Jinggangensis TL01 | SHJGH_6050 | SHJGH_6052 |
| Streptomyces hygroscopicus subsp. Jinggangensis 5008 | SHJG_6289 | SHJG_6291 |
| Streptomyces fulvissimus DSM 40593 | SFUL_4994 | SFUL_4995 |
| Streptomyces albus J1074 | XNR_1620 | XNR_1619 |
| Streptomyces sp. PAMC26508 | F750_4709 | F750_4710 |
| Streptomyces venezuelae ATCC 10712 | SVEN_4833 | SVEN_4835b |
| Streptomyces rapamycinicus NRRL 5491 | M271_51085c | M271_18000 |
| Saccharopolyspora erythrea NRRL2338 | SACE_1077 | SACE_1078 |
| Rhodococcus erythropolis PR4 | RER_22180 | RER_22190 |
| Frankia sp. CcI3 | Francci3_3800 | Francci3_3799 |
Threshold e-value, <10−20 for AdnA and <10−40 for AdnB.
The aa identity with AdnB starts in SVEN_4835 and ends in SVEN_4836 (including the nuclease motif). The multiple homopolymers present in this region may contain a sequence error resulting in the predicted frameshift. Thus, these two coding sequences may form a single one.
M271_51085 of S. venezuelae includes a stretch of undetermined nucleotides interrupting the AdnA homologue.
FIG 1.
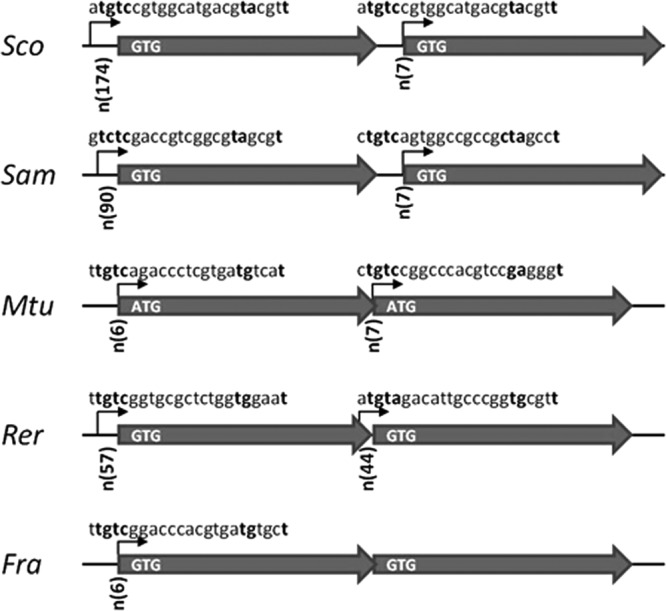
Organization of the adnAB locus in some actinomycetes. Putative promoter motifs are represented by arrows in front of both adnA and adnB genes of S. coelicolor (Sco), S. ambofaciens (Sam), M. tuberculosis (Mtu), Rhodococcus erythropolis (Rer), and Frankia sp. (Fra). The boldface nucleotides correspond to the strictly defined positions described by Gamulin et al. (5′-TGTC-12nt-TAnnnT-3′) (44). The distances between putative promoter motifs, indicated as n(number of nucleotides), and predicted start codons are indicated at the beginning of each arrow symbolizing the genes. The presence or absence of intergenic space is also indicated by a straight line between the arrows or by overlapping arrows, respectively.
Streptomyces AdnAB proteins share structural similarities with MtuAdnAB (13) and with other actinomycete AdnAB homologues (see Fig. S1 in the supplemental material): all Streptomyces AdnA and AdnB proteins possess the 8 helicase motifs (I, Ia, II to VI, and Q) (35, 36) in the N-terminal domain as well as the 3 motifs of the RecB nuclease active site in the C-terminal domain (37, 38). These data strongly suggest that Streptomyces AdnA and AdnB both contain an N-terminal UvrD-like motor domain and a C-terminal RecB-like nuclease domain, as in MtuAdnAB (13). Hence, in the absence of all other reported complexes (i.e., RecBCD, AddAB, and RecQ-RecJ), AdnAB may be the only DNA end-processing helicase-nuclease in Streptomyces.
The adnAB locus is essential in Streptomyces.
DNA end resection deficiency has a significant impact on bacterial cell viability and growth: E. coli recBC recJ (39, 40) and B. subtilis addAB ΔrecJ double mutants (10) are deeply affected, and the Acinetobacter baylyi ΔrecBCD ΔrecJ double mutant is not viable (41).
Hence, a strategy of deletion of the adnAB locus of S. ambofaciens was developed to assess the consequence of AdnAB deficiency on growth and survival against DNA-damaging agents. The aac(3)IV gene conferring apramycin resistance that is bordered by the 5′-end-flanking region (region U [upstream], 1,501 nt) and 3′-end-flanking region (region D [downstream], 1,790 nt) of the adnAB locus was inserted in the suicide plasmid pWED2 (carrying the pac gene, conferring resistance to puromycin). Apramycin- and puromycin-resistant (Aprar Purr) clones resulting from integration via a single crossover could be easily selected, and PCR analysis showed that this crossover occurred evenly in U and D regions. Therefore, there was no obstacle to a crossover event in either of the two regions, and the length of the regions was fully sufficient to support HR. In contrast, deletion of the adnAB locus by the formation of a second crossover (in the U region when the deletion cassette was initially inserted via the D region or vice versa) giving rise to Aprar Purs clones could not be obtained. Formally, no Aprar Purs clones could be found among 386 and 366 derivatives of 3 independent clones harboring the deletion construct in the U and D region, respectively.
In order to determine whether the failure to obtain adnAB gene replacement in S. ambofaciens results from the lethality of the adnAB deletion, an integrative plasmid (pSET153, conferring resistance to kanamycin) carrying an intact copy of the adnAB locus (as an 11.4-kb DNA fragment; see Materials and Methods) was inserted into the φC31 phage integration site of the chromosome. This insertion was carried out in strains harboring the adnAB deletion construct inserted by single crossover in the U or D region (3 strains for each). In the presence of this ectopic copy of adnAB, double crossovers resulting in Aprar Purs Kanr clones were selected at high frequencies. The loss of the puromycin resistance gene corresponding to the formation of the second crossover was observed at similar frequencies from strains with U (420 of 1,314 colonies tested, 32%) and D (464 of 1,472 colonies tested, 31.5%) region insertions and was confirmed by PCR analyses. The observation that the double crossover could be selected only in the presence of an ectopic extra copy of the adnAB genes indicates that the adnAB locus is essential in S. ambofaciens.
Similar experiments were carried out in the closely related strain S. coelicolor A3 (2). For this purpose, recombinant cosmids, including the adnAB locus interrupted by the Tn5062 transposon (in either adnA or adnB), were used in attempts to replace the adnA and adnB genes independently. Again, while single-crossover events resulting in Aprar Kanr clones were scored at high frequencies, no gene replacement by double crossover could be selected (Aprar Kans). In these experiments, the homologous upstream and downstream regions were at least 15 kb long and were thus obviously sufficient to support HR. From these data, we can conclude that both adnA and adnB are essential for viability, since this strategy targeted either open reading frame independently. The possibility that adnA mutation could have polar effects on adnB gene expression was excluded, since these two genes are independently transcribed (see below).
This essential nature of adnAB in Streptomyces is in contrast to all the other bacterial species investigated so far. Hence, the E. coli recBCD mutant, B. subtilis addAB mutant, and A. baylyi ΔrecBCD mutant are viable, probably as a result of the presence of alternative helicase-nuclease activities (e.g., RecJ/Q). E. coli recBC recJ and B. subtilis addAB ΔrecJ mutants are still viable although drastically impaired in growth, which suggests that they contain additional back-up systems. This is consistent with the lethality of E. coli recD recJ mutants in which additional mutations in the exonuclease I-encoding gene (xonA) and the exonuclease VII-encoding gene (xseA) are introduced (42). While an xseA homologue is found in S. ambofaciens and S. coelicolor genomes, it clearly does not provide the same back-up activity.
adnAB expression is induced in response to genotoxic agents.
The adnAB genes of M. tuberculosis were shown to be induced (9-fold) in response to MMC, a DNA-damaging agent causing genotoxic stress (43). In order to test whether adnAB genes are induced by DNA damage in Streptomyces, the level of expression of adnAB was measured by quantitative RT-PCR in response to MMC. The recA gene involved in the positive regulation of the SOS system in bacteria was chosen as a positive control in these experiments. The hrdB gene was used for normalization of gene expression levels. As expected, induction of recA was observed after exposure to 1 μg · ml−1 MMC (about 20-fold). Under the same conditions, a significant induction of about 4-fold was observed for both adnA and adnB genes (Fig. 2). In order to establish whether adnA and adnB constitute an operon, the transcription of the intergenic sequence (250 bp) was assessed in the presence or absence of MMC. No transcript was observed, indicating that both genes were independently expressed.
FIG 2.
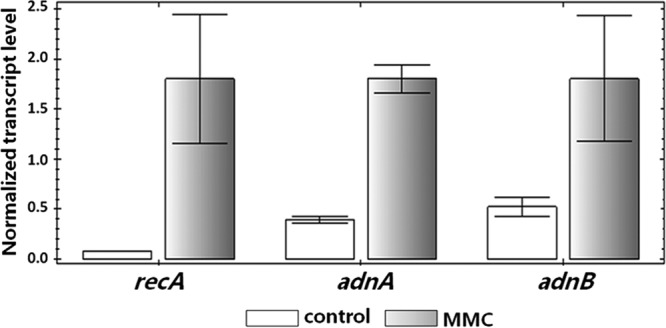
Induction of adnAB by exposure to mitomycin C. Quantitative RT-PCR experiments were carried out to test the induction of adnA and adnB in response to MMC. S. ambofaciens cultures were exposed to MMC (1 μg · ml−1) or not exposed for 30 min. recA served as a positive control. hrdB was used for normalization as a reference gene. This experiment was performed in triplicate.
Consistent with these data, we could identify putative motifs upstream of both genes that strongly resemble the consensus promoter sequence 5′-tTGTCRgtg-8nt-TAnnnT-3′ (where upper- and lowercase letters correspond to conserved and variable nucleotide positions, respectively) described by Gamulin et al. (44) in M. tuberculosis. This promoter motif was involved in the RecA-independent inducible response to DNA-damaging agents (MMC) (43, 44). In our analysis, a sequence approaching this consensus can be found just upstream of the translation start codon of both adnA and adnB (6 and 7 nucleotides, respectively) (Fig. 1). This promoter motif was also detected upstream of adnA in Frankia sp. and of adnA and adnB in Rhodococcus erythropolis, both actinomycetes. Interestingly, the promoter motifs are positioned only 7 nucleotides upstream from the predicted translational start codon of adnB in both S. coelicolor and S. ambofaciens, suggesting that these genes may be leaderless genes that are widespread in actinobacteria (45).
Phenotypic characterization of a Streptomyces mutant carrying two adnAB copies.
S. ambofaciens and S. coelicolor strains containing two copies of the adnAB locus were obtained during this work and were exploited to assess the impact of adnAB overexpression. S. ambofaciens harboring two copies of adnAB exhibited a colonial phenotype typical of genetic instability, i.e., with a high frequency of unpigmented papillae on bacterial colonies (Fig. 3A) (46). Colonies of S. coelicolor strains harboring two copies of the adnAB locus showed a marked impairment of their development (Fig. 3B). Furthermore, a small but reproducible increase in sensitivity to MMC was noticed in S. ambofaciens clones harboring two adnAB loci (not shown). Quantitative RT-PCR analysis showed that the presence of two copies of adnAB was associated with 6- and 3-fold increases in transcript quantity for adnA and andB, respectively, compared to the wild-type (WT) context. Therefore, the observed phenotypes may result from the modulation of the homeostasis of helicase-nuclease cellular activity. The excess of nuclease activity may be harmful and lead to an unexpected degradation of DNA during chromosomal replication, affecting cell growth. In the same way, genome instability and sensitivity to genotoxic agents may result from impairment of the HR process. A parallel could be drawn with E. coli, where overexpression of recBCD reduces resistance to UV exposure (47).
FIG 3.
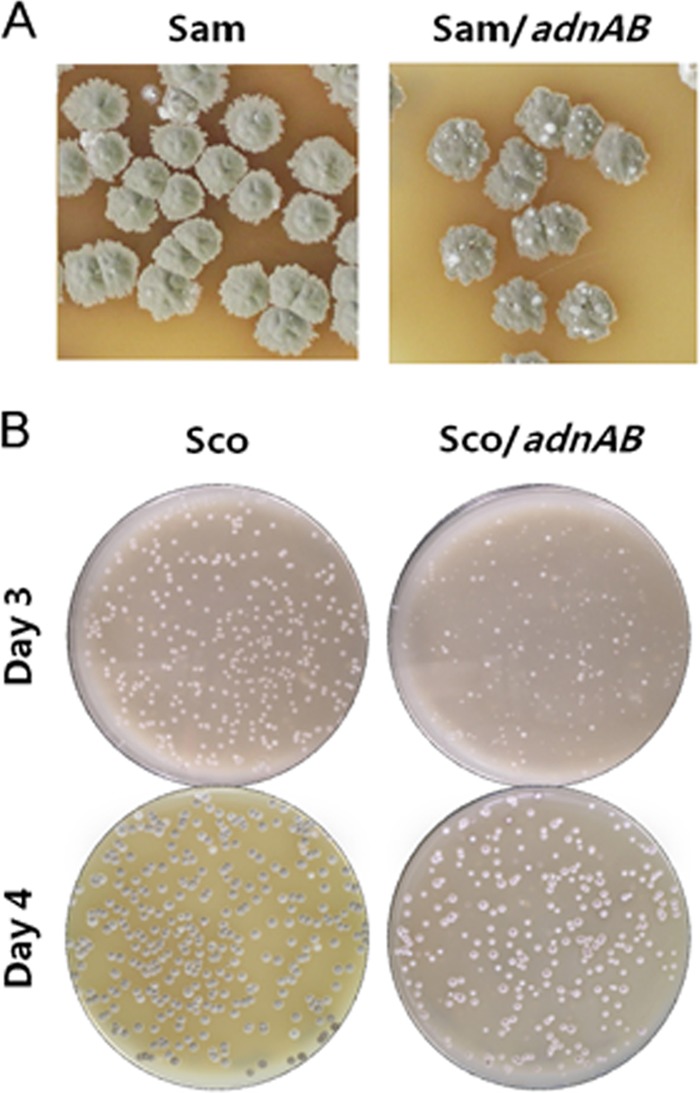
Phenotypes of Streptomyces harboring two copies of the adnAB locus. The S. ambofaciens (A) and S. coelicolor (B) strains harboring an empty plasmid pSET153 (Sam and Sco, respectively) as a control or harboring plasmid pSET153-adnAB (Sam/adnAB and Sco/adnAB, respectively) were grown on SFM agar. Genetic instability (represented by papillae) in S. ambofaciens was observed after 12 days of growth. The growth defect of S. coelicolor harboring two adnAB loci was visible from the third day of growth.
Streptomyces adnAB restores survival and UV light resistance to E. coli recB.
In E. coli, RecBCD (exonuclease V) is crucial for the initiation of HR at DNA double-strand ends, and its absence leads to a drastic decrease in survival compared to that of the WT strain in response to genotoxic agents (for a review, see reference 48). In order to test whether Streptomyces AdnAB works as a functional homologue of E. coli RecBCD, complementation of an E. coli recB mutant (JJC315) was examined. The recB mutant (allele recB268) is suggested to be deficient for the B and D subunits of the RecBCD complex due to a polar effect on the adjacent recD gene (49).
Plasmid pIJ6902-LCN either empty or containing the S. ambofaciens adnAB locus was used to transform E. coli JJC40 (WT) and JJC315 (recB) strains. First, crude extracts of E. coli clones harboring these plasmids were assayed for exonuclease activity. For that purpose, 300 ng of linear DNA was exposed to crude extracts. After 3 h of exposure, the degradation was more visible with extracts from cells containing the adnAB locus (Fig. 4). The degradation provided by the adnAB-complemented extract was roughly estimated to be 40 to 60% higher than that of the noncomplemented strain. This result strongly suggested that Streptomyces adnAB can be expressed in E. coli and exhibits an exonuclease activity.
FIG 4.
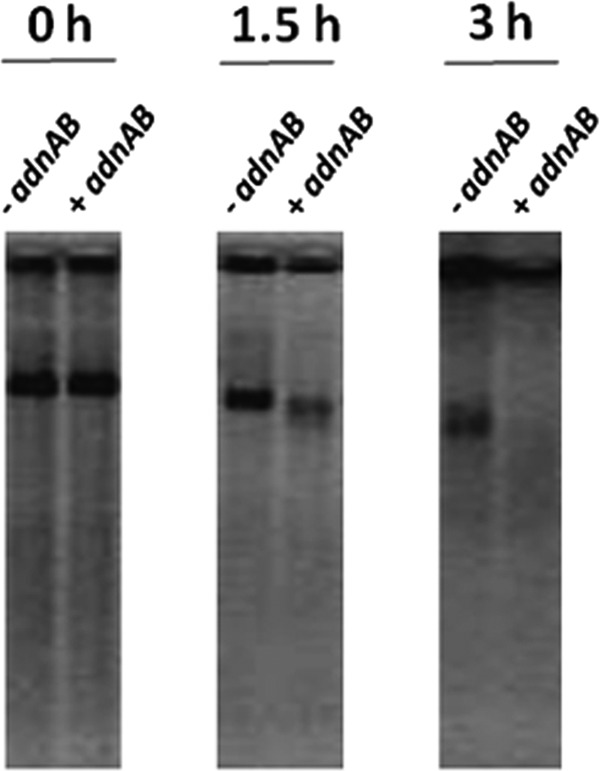
Streptomyces adnAB can restore the nuclease activity of an E. coli ΔrecB mutant. Cell extracts were prepared from an E. coli recB mutant strain carrying an empty plasmid (-adnAB) or a plasmid that included the locus adnAB from S. ambofaciens (+adnAB). Reaction mixtures were incubated for 0 h, 1.5 h, or 3 h at 37°C.
To examine whether the viability of an E. coli recB mutant could be restored by the Streptomyces adnAB genes, the plating efficiency of E. coli cultures grown to an OD600 of ∼0.3 was measured. Cell viability was expressed as the number of CFU · ml−1 per unit OD600. recB mutation in E. coli resulted in a 20-fold reduction of cell viability. In contrast, in the E. coli recB mutant carrying S. ambofaciens adnAB, the viability was partially restored, reaching 60% of the WT level of survival (Fig. 5A).
FIG 5.
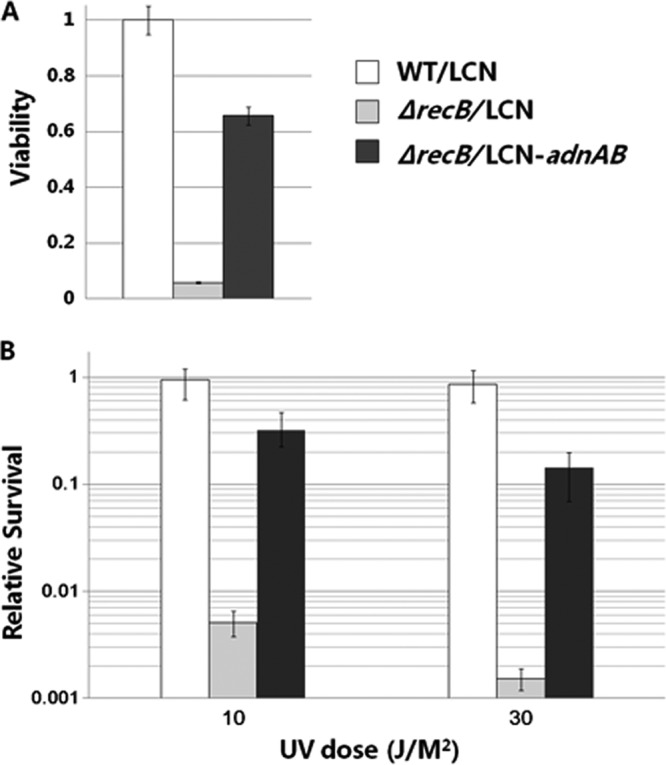
S. ambofaciens adnAB can restore cell viability and UV resistance of E. coli ΔrecB. The wild-type E. coli strain carrying pIJ6902-LCN (WT/LCN) and the E. coli ΔrecB strain carrying pIJ6902-LCN (ΔrecB/LCN) or pIJ6902-LCN-adnAB (ΔrecB/LCN-adnAB) were plated to measure cell viability (A) and exposed to UV at 10 J · m−2 and 30 J · m−2 to measure relative survival (B). This experiment was performed in triplicate.
The role of Streptomyces AdnAB in DNA repair was studied by examining whether S. ambofaciens adnAB genes could restore the impaired ability of the E. coli ΔrecB mutant to repair UV-induced DNA damage. Samples of cultures in exponential phase were irradiated with UV light, and cell survival was determined (Fig. 5B). In our experimental conditions, the E. coli ΔrecB mutant was highly sensitive to UV, as cell survival was reduced about 200-fold compared to that of the WT at 10 J · m−2. However, the strain complemented with S. ambofaciens adnAB was about 60-fold more resistant to UV than the E. coli ΔrecB mutant, which demonstrates that Streptomyces adnAB could restore the impaired ability of ΔrecB mutant to repair UV lesions. Altogether, these data suggest that S. ambofaciens AdnAB is a functional homologue of RecBCD and is likely to be involved in DNA repair.
In conclusion, the helicase-nuclease encoded by adnAB of Streptomyces is assumed to be involved in HR. Therefore, adnAB essentiality may be linked to some HR-dependent critical cell processes such as DNA replication, DNA repair, or chromosome segregation. It is also tempting to speculate that integrity and maintenance of the chromosomal linearity and bacterial telomeres could require a functional HR pathway. The terminal redundancies, characteristic of the Streptomyces chromosome, may result from HR or require HR for its maintenance. However, HR does not seem to be essential, since recA-null mutants can be generated in Streptomyces (50). Hence, the essentiality of AdnAB would result from the involvement of AdnAB in an unknown crucial cell process.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the French National Research Agency program Streptoflux (ANR Blanc 0096_01), by ANR through the Laboratory of Excellence ARBRE (ANR-12-LABXARBRE-01), and by the Région Lorraine.
We are grateful to Bénédicte Michel (CNRS, Gif-sur-Yvette, France) for the kind gift of E. coli strains and helpful advice and Janet Gibson and Susan Rosenberg (Baylor College of Medicine, Houston, TX) for helpful discussions. We also thank Paul Hoskisson (University of Strathclyde, Glasgow, United Kingdom) for critical reading of the manuscript.
Footnotes
Published ahead of print 16 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01513-14.
REFERENCES
- 1.Michel B, Ehrlich SD, Uzest M. 1997. DNA double-strand breaks caused by replication arrest. EMBO J. 16:430–438. 10.1093/emboj/16.2.430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singleton MR, Dillingham MS, Gaudier M, Kowalczykowski SC, Wigley DB. 2004. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature 432:187–193. 10.1038/nature02988 [DOI] [PubMed] [Google Scholar]
- 3.Dillingham MS, Kowalczykowski SC. 2008. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 72:642–671. 10.1128/MMBR.00020-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haijema BJ, Meima R, Kooistra J, Venema G. 1996. Effects of lysine-to-glycine mutations in the ATP-binding consensus sequences in the AddA and AddB subunits on the Bacillus subtilis AddAB enzyme activities. J. Bacteriol. 178:5130–5137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haijema BJ, Venema G, Kooistra J. 1996. The C terminus of the AddA subunit of the Bacillus subtilis ATP-dependent DNase is required for the ATP-dependent exonuclease activity but not for the helicase activity. J. Bacteriol. 178:5086–5091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yeeles JTP, Dillingham MS. 2007. A dual-nuclease mechanism for DNA break processing by AddAB-type helicase-nucleases. J. Mol. Biol. 371:66–78. 10.1016/j.jmb.2007.05.053 [DOI] [PubMed] [Google Scholar]
- 7.Capaldo-Kimball F, Barbour SD. 1971. Involvement of recombination genes in growth and viability of Escherichia coli K-12. J. Bacteriol. 106:204–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horii Z-I, Clark AJ. 1973. Genetic analysis of the RecF pathway to genetic recombination in Escherichia coli K12: isolation and characterization of mutants. J. Mol. Biol. 80:327–344. 10.1016/0022-2836(73)90176-9 [DOI] [PubMed] [Google Scholar]
- 9.Kushner SR. 1974. In vivo studies of temperature-sensitive recB and recC mutants. J. Bacteriol. 120:1213–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanchez H, Kidane D, Castillo Cozar M, Graumann PL, Alonso JC. 2006. Recruitment of Bacillus subtilis RecN to DNA double-strand breaks in the absence of DNA end processing. J. Bacteriol. 188:353–360. 10.1128/JB.188.2.353-360.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kooistra J, Haijema BJ, Venema G. 1993. The Bacillus subtilis addAB genes are fully functional in Escherichia coli. Mol. Microbiol. 7:915–923. 10.1111/j.1365-2958.1993.tb01182.x [DOI] [PubMed] [Google Scholar]
- 12.Cromie GA. 2009. Phylogenetic ubiquity and shuffling of the bacterial RecBCD and AddAB recombination complexes. J. Bacteriol. 191:5076–5084. 10.1128/JB.00254-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sinha KM, Unciuleac M-C, Glickman MS, Shuman S. 2009. AdnAB: a new DSB-resecting motor-nuclease from mycobacteria. Genes Dev. 23:1423–1437. 10.1101/gad.1805709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Unciuleac M-C, Shuman S. 2010. Double strand break unwinding and resection by the mycobacterial helicase-nuclease AdnAB in the presence of single strand DNA-binding protein (SSB). J. Biol. Chem. 285:34319–34329. 10.1074/jbc.M110.162925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dillingham MS, Spies M, Kowalczykowski SC. 2003. RecBCD enzyme is a bipolar DNA helicase. Nature 423:893–897. 10.1038/nature01673 [DOI] [PubMed] [Google Scholar]
- 16.Taylor AF, Smith GR. 2003. RecBCD enzyme is a DNA helicase with fast and slow motors of opposite polarity. Nature 423:889–893. 10.1038/nature01674 [DOI] [PubMed] [Google Scholar]
- 17.Clark AJ, Sandler SJ, Willis DK, Chu CC, Blanar MA, Lovett ST. 1984. Genes of the RecE and RecF pathways of conjugational recombination in Escherichia coli. Cold Spring Harbor Symp. Quant. Biol. 49:453–462. 10.1101/SQB.1984.049.01.051 [DOI] [PubMed] [Google Scholar]
- 18.Nakayama H, Nakayama K, Nakayama R, Irino N, Nakayama Y, Hanawalt PC. 1984. Isolation and genetic characterization of a thymineless death-resistant mutant of Escherichia coli K12: identification of a new mutation (recQ1) that blocks the RecF recombination pathway. Mol. Gen. Genet. 195:474–480. 10.1007/BF00341449 [DOI] [PubMed] [Google Scholar]
- 19.Kolodner R, Fishel RA, Howard M. 1985. Genetic recombination of bacterial plasmid DNA: effect of RecF pathway mutations on plasmid recombination in Escherichia coli. J. Bacteriol. 163:1060–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta R, Barkan D, Redelman-Sidi G, Shuman S, Glickman MS. 2011. Mycobacteria exploit three genetically distinct DNA double-strand break repair pathways. Mol. Microbiol. 79:316–330. 10.1111/j.1365-2958.2010.07463.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rocha EPC, Cornet E, Michel B. 2005. Comparative and evolutionary analysis of the bacterial homologous recombination systems. PLoS Genet. 1:e15. 10.1371/journal.pgen.0010015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirby R. 2011. Chromosome diversity and similarity within the Actinomycetales. FEMS Microbiol. Lett. 319:1–10. 10.1111/j.1574-6968.2011.02242.x [DOI] [PubMed] [Google Scholar]
- 23.Thibessard A, Leblond P. 2014. Subtelomere plasticity in the bacterium Streptomyces, p 243–258 In Louis EJ, Becker MM. (ed), Subtelomeres. Springer, Berlin, Germany [Google Scholar]
- 24.Lin YS, Kieser HM, Hopwood DA, Chen CW. 1993. The chromosomal DNA of Streptomyces lividans 66 is linear. Mol. Microbiol. 10:923–933. 10.1111/j.1365-2958.1993.tb00964.x [DOI] [PubMed] [Google Scholar]
- 25.Choulet F, Aigle B, Gallois A, Mangenot S, Gerbaud C, Truong C, Francou F-X, Fourrier C, Guérineau M, Decaris B, Barbe V, Pernodet J-L, Leblond P. 2006. Evolution of the terminal regions of the Streptomyces linear chromosome. Mol. Biol. Evol. 23:2361–2369. 10.1093/molbev/msl108 [DOI] [PubMed] [Google Scholar]
- 26.Doroghazi JR, Buckley DH. 2010. Widespread homologous recombination within and between Streptomyces species. ISME J. 4:1136–1143. 10.1038/ismej.2010.45 [DOI] [PubMed] [Google Scholar]
- 27.Yu D, Ellis HM, Lee E-C, Jenkins NA, Copeland NG, Court DL. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 97:5978–5983. 10.1073/pnas.100127597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, United Kingdom [Google Scholar]
- 29.Nguyen HC, Karray F, Lautru S, Gagnat J, Lebrihi A, Huynh TDH, Pernodet J-L. 2010. Glycosylation steps during spiramycin biosynthesis in Streptomyces ambofaciens: involvement of three glycosyltransferases and their interplay with two auxiliary proteins. Antimicrob. Agents Chemother. 54:2830–2839. 10.1128/AAC.01602-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karray F, Darbon E, Oestreicher N, Dominguez H, Tuphile K, Gagnat J, Blondelet-Rouault M-H, Gerbaud C, Pernodet J-L. 2007. Organization of the biosynthetic gene cluster for the macrolide antibiotic spiramycin in Streptomyces ambofaciens. Microbiology 153:4111–4122. 10.1099/mic.0.2007/009746-0 [DOI] [PubMed] [Google Scholar]
- 31.Fernández-Martínez LT, Del Sol R, Evans MC, Fielding S, Herron PR, Chandra G, Dyson PJ. 2011. A transposon insertion single-gene knockout library and new ordered cosmid library for the model organism Streptomyces coelicolor A3(2). Antonie Van Leeuwenhoek 99:515–522. 10.1007/s10482-010-9518-1 [DOI] [PubMed] [Google Scholar]
- 32.Brosius J. 1989. Superpolylinkers in cloning and expression vectors. DNA 8:759–777. 10.1089/dna.1989.8.759 [DOI] [PubMed] [Google Scholar]
- 33.Huang J, Shi J, Molle V, Sohlberg B, Weaver D, Bibb MJ, Karoonuthaisiri N, Lih C-J, Kao CM, Buttner MJ, Cohen SN. 2005. Cross-regulation among disparate antibiotic biosynthetic pathways of Streptomyces coelicolor. Mol. Microbiol. 58:1276–1287. 10.1111/j.1365-2958.2005.04879.x [DOI] [PubMed] [Google Scholar]
- 34.Ivancić-Baće I, Peharec P, Moslavac S, Skrobot N, Salaj-Smic E, Brci-Kosti K. 2003. RecFOR function is required for DNA repair and recombination in a RecA loading-deficient recB mutant of Escherichia coli. Genetics 163:485–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gorbalenya AE, Koonin EV. 1993. Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 3:419–429. 10.1016/S0959-440X(05)80116-2 [DOI] [Google Scholar]
- 36.Tanner NK, Cordin O, Banroques J, Doère M, Linder P. 2003. The Q motif: a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol. Cell 11:127–138 [DOI] [PubMed] [Google Scholar]
- 37.Yu M, Souaya J, Julin DA. 1998. Identification of the nuclease active site in the multifunctional RecBCD enzyme by creation of a chimeric enzyme. J. Mol. Biol. 283:797–808. 10.1006/jmbi.1998.2127 [DOI] [PubMed] [Google Scholar]
- 38.Wang J, Chen R, Julin DA. 2000. A single nuclease active site of the Escherichia coli RecBCD enzyme catalyzes single-stranded DNA degradation in both directions. J. Biol. Chem. 275:507–513. 10.1074/jbc.275.1.507 [DOI] [PubMed] [Google Scholar]
- 39.Lloyd RG, Evans NP, Buckman C. 1987. Formation of recombinant lacZ+ DNA in conjugational crosses with a recB mutant of Escherichia coli K12 depends on recF, recJ, and recO. Mol. Gen. Genet. 209:135–141. 10.1007/BF00329848 [DOI] [PubMed] [Google Scholar]
- 40.Lovett ST, Clark AJ. 1984. Genetic analysis of the recJ gene of Escherichia coli K-12. J. Bacteriol. 157:190–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kickstein E, Harms K, Wackernagel W. 2007. Deletions of recBCD or recD influence genetic transformation differently and are lethal together with a recJ deletion in Acinetobacter baylyi. Microbiology 153:2259–2270. 10.1099/mic.0.2007/005256-0 [DOI] [PubMed] [Google Scholar]
- 42.Dermić D. 2006. Functions of multiple exonucleases are essential for cell viability, DNA repair and homologous recombination in recD mutants of Escherichia coli. Genetics 172:2057–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rand L, Hinds J, Springer B, Sander P, Buxton RS, Davis EO. 2003. The majority of inducible DNA repair genes in Mycobacterium tuberculosis are induced independently of RecA. Mol. Microbiol. 50:1031–1042. 10.1046/j.1365-2958.2003.03765.x [DOI] [PubMed] [Google Scholar]
- 44.Gamulin V, Cetkovic H, Ahel I. 2004. Identification of a promoter motif regulating the major DNA damage response mechanism of Mycobacterium tuberculosis. FEMS Microbiol. Lett. 238:57–63. 10.1111/j.1574-6968.2004.tb09737.x [DOI] [PubMed] [Google Scholar]
- 45.Zheng X, Hu G-Q, She Z-S, Zhu H. 2011. Leaderless genes in bacteria: clue to the evolution of translation initiation mechanisms in prokaryotes. BMC Genomics 12:361. 10.1186/1471-2164-12-361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin P, Dary A, Decaris B. 1998. Generation of a genetic polymorphism in clonal populations of the bacterium Streptomyces ambofaciens: characterization of different mutator states. Mutat. Res. 421:73–82. 10.1016/S0027-5107(98)00156-0 [DOI] [PubMed] [Google Scholar]
- 47.Dermić D, Halupecki E, Zahradka D, Petranović M. 2005. RecBCD enzyme overproduction impairs DNA repair and homologous recombination in Escherichia coli. Res. Microbiol. 156:304–311. 10.1016/j.resmic.2004.10.005 [DOI] [PubMed] [Google Scholar]
- 48.Kuzminov A. 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage λ. Microbiol. Mol. Biol. Rev. 63:751–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lloyd RG, Buckman C, Benson FE. 1987. Genetic analysis of conjugational recombination in Escherichia coli K12 strains deficient in RecBCD enzyme. J. Gen. Microbiol. 133:2531–2538 [DOI] [PubMed] [Google Scholar]
- 50.Huang T-W, Chen CW. 2006. A recA null mutation may be generated in Streptomyces coelicolor. J. Bacteriol. 188:6771–6779. 10.1128/JB.00951-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pinnert-Sindico S. 1954. Une nouvelle espèce de Streptomyces productrice d'antibiotiques: Streptomyces ambofaciens n. sp. caractères culturaux. Ann. Inst. Pasteur 87:702–707 [PubMed] [Google Scholar]
- 52.Hopwood DA. 1999. Forty years of genetics with Streptomyces: from in vivo through in vitro to in silico. Microbiology 145:2183–2202 [DOI] [PubMed] [Google Scholar]
- 53.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–580. 10.1016/S0022-2836(83)80284-8 [DOI] [PubMed] [Google Scholar]
- 54.MacNeil DJ, Gewain KM, Ruby CL, Dezeny G, Gibbons PH, MacNeil T. 1992. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 111:61–68. 10.1016/0378-1119(92)90603-M [DOI] [PubMed] [Google Scholar]
- 55.Paget MSB, Chamberlin L, Atrih A, Foster SJ, Buttner MJ. 1999. Evidence that the extracytoplasmic function sigma factor σE is required for normal cell wall structure in Streptomyces coelicolor A3(2). J. Bacteriol. 181:204–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1:784–791. 10.1038/nbt1183-784 [DOI] [Google Scholar]
- 57.Bierman M, Logan R, O'Brien K, Seno ET, Rao RN, Schoner BE. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43–49. 10.1016/0378-1119(92)90627-2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


