Abstract
The N-terminal domain (winged-helix domain, or WH1) of the Pseudomonas pPS10 plasmid DNA replication protein RepA can assemble into amyloid fibers in vitro and, when expressed in Escherichia coli, leads to a unique intracellular amyloid proteinopathy by hampering bacterial proliferation. RepA-WH1 amyloidosis propagates along generations through the transmission of aggregated particles across the progeny, but it is unable to propagate horizontally as an infectious agent and is thus the first synthetic bacterial prionoid. RepA-WH1 amyloidosis is promoted by binding to double-stranded DNA (dsDNA) in vitro, and it is modulated by the Hsp70 chaperone DnaK in vivo. Different mutations in the repA-WH1 gene result in variants of the protein with distinct amyloidogenic properties. Here, we report that intracellular aggregates of the hyperamyloidogenic RepA with an A31V change in WH1 [RepA-WH1(A31V)] are able to induce and enhance the growth in vivo of new amyloid particles from molecules of wild-type RepA-WH1 [RepA-WH1(WT)], which otherwise would remain soluble in the cytoplasm. In contrast, RepA-WH1(ΔN37), a variant lacking a clear amyloidogenic sequence stretch that aggregates as conventional inclusion bodies (IBs), can drive the aggregation of the soluble protein into IBs only if expressed at high molar ratios over RepA-WH1(WT). The cytotoxic bacterial intracellular prionoid RepA-WH1 thus exhibits a hallmark feature of amyloids, as characterized in eukaryotes: cross-aggregation between variants of the same protein.
INTRODUCTION
Amyloid proteinopathies are neurodegenerative and systemic diseases with increasing incidence in human population and are currently one of the main burdens associated with aging. Although protein amyloidoses have broad clinical manifestations, they share an etiology: the accumulation of insoluble aggregates of a particular protein, either misfolded or posttranslationally processed, which are made of crossed β-sheet assemblies, the signature three-dimensional structure of amyloids (1). Oligomers of an amyloidogenic protein, in their process toward assembling as fibers, seem to be the most toxic amyloid species, acting either directly by targeting membranes or depleting essential cell factors through coaggregation or, indirectly, by contributing to generate reactive oxygen species (2). Apart from studies carried out with human cells in culture and with mice, which have uncovered many pathways possibly linked to disease, there is a need for simpler model systems that provide further insight into the essentials of amyloidosis. These include Drosophila and Caenorhabditis animal models and, among microorganisms, yeast. Fungal prions have indeed contributed to our knowledge about the molecular basis for amyloid assembly and propagation (3), but they are not optimal models for disease because they behave as selectable epigenetic determinants of non-Mendelian inheritance, conferring beneficial phenotypic traits to their carriers (4). Similarly, a number of natural amyloids have been recently identified in bacteria and found to be involved in scaffolding extracellular biofilms or as inert, mobilizable intracellular deposits of toxic peptides, such as microcins (5). The unsurpassed potential of Escherichia coli as a model system in biology has also been exploited to study the heterologous aggregation of amyloidogenic proteins, such as the Saccharomyces cerevisiae prion Sup35p ([PSI+]) or Alzheimer's Aβ peptides, which aggregate as intracellular inclusion bodies (IBs) (6, 7).
We have recently found that when the N-terminal winged-helix domain (WH1) of RepA, a plasmid DNA replication initiator/transcriptional repressor, includes a mutation (A31V) that has repeatedly been found to naturally enhance plasmid replication in vivo (8), it assembles into amyloid fibers in vitro upon allosteric binding to small double-stranded DNA (dsDNA) effector molecules (9, 10). Furthermore, fusions of RepA-WH1(A31V) to a fluorescent protein tag allowed tracking of the aggregation of the protein in the cytoplasm of E. coli, revealing a drastic reduction in cell proliferation upon protein aggregation (11). In addition to such toxicity, RepA-WH1(A31V) aggregates differ from conventional IBs in having a higher degree of staining with a specific amyloidotropic fluorophore and in exhibiting dynamic interconversion between distinct amyloid species, which resemble prion strains (12). The latter process is modulated by the Hsp70 chaperone DnaK that generates oligomeric RepA-WH1(A31V) particles that are readily transferred to the progeny during bacterial division (12). Being noninfectious but vertically transmissible from mother to daughter cells, RepA-WH1 qualifies as the first entirely bacterial prionoid (13).
In this work, by means of the pairwise coexpression of repA-WH1 alleles coding for proteins with diverse amyloidogenic properties, which were tracked through epitope tags and fusions to fluorescent proteins with distinct colors, we have addressed how they mutually influence their aggregation in vivo. We have found that the interplay between their intrinsic aggregation tendencies, expression levels, and intermolecular contacts determines the solubility versus aggregation balance for each RepA-WH1 variant.
MATERIALS AND METHODS
Bacterial strain and plasmid constructs.
All experiments were carried out in the E. coli K-12 strain MDS42 recA, whose reduced genome has been depleted of mobile genetic elements (14) to avoid inactivation of the repA-WH1 gene by transposon insertion (11). Plasmids (Table 1) of the red series expressing either RepA-WH1(A31V) or RepA-WH1(ΔN37), a deletion mutant lacking its amyloidogenic peptide stretch, are derivatives of the RK2-based, low-copy-number pSEVA121 vector (http://seva.cnb.csic.es/SEVA) (15) and were described elsewhere (12), while construction of the wild-type (WT) variant was performed by PCR (Pfu DNApol) using the oligonucleotides described in Table 2. Plasmids of the yellow series were constructed by consecutive ligation of PCR-amplified fragments carrying the appropriate Ptac-repA-WH1-YFP (where YFP is yellow fluorescent protein) expression module from pWH1s (9) but including a cMyc-encoding tag in the 5′ primer (Table 2), a downstream lacIq repressor cassette, and the p15A replicon plus the Cm resistance gene (from the pACYC184 vector). Expression was carried out at 37°C in LB medium, supplemented with either ampicillin (100 μg · ml−1; for the red series) or chloramphenicol (30 μg · ml−1; yellow series), at an optical density at 600 nm (OD600) of 0.2 by means of the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to 0.5 mM.
TABLE 1.
Plasmids used in this study
| Plasmid | Relevant genotype | Reference or source |
|---|---|---|
| pACYC184 | Cmr Tcr | New England Biolabs |
| pSEVA121 | Apr | 15 |
| p3YFP | Apr, YFP | A. Lindner (CRI-INSERM) |
| pRK2-WH1(A31V)-mCherry | repA-WH1(A31V)-mCherry opsp18 lacIq+, Apr | 12 |
| pRK2-WH1(ΔN37)-mCherry | repA-WH1(ΔN37)-mCherry opsp18 lacIq+, Apr | 12 |
| pRK2-WH1(WT)-mCherry | repA-WH1(WT)-mCherry opsp18 lacIq+, Apr | This work |
| pRK2-mCherry | mCherry opsp18 lacIq+, Apr | This work |
| p15A-WH1(WT)-YFP | repA-WH1(WT)-YFPA206K opsp18 lacIq+, Cmr | This work |
| p15A-WH1(A31V)-YFP | repA-WH1(A31V)-YFPA206K opsp18 lacIq+, Cmr | This work |
| p15A-WH1(ΔN37)-YFP | repA-WH1(ΔN37)-YFPA206K opsp18 lacIq+, Cmr | This work |
| p15A-YFP | YFPA206K opsp18 lacIq+, Cmr | This work |
TABLE 2.
Oligonucleotides used in plasmid construction
| Oligonucleotidea | Restriction enzyme | Comment |
|---|---|---|
| F, 5′-GCTTCCGGAATGGTGAGCAAGGGCGAGG | BspEI | YFP cloning (PCR) |
| R, 5′-GCTGGATCCTTACTTGTACAGCTCGTCCATG | BamHI | |
| 5′-CTTTGCCCGTTATCCCGATCATATGAAACGG | YFP A206K mutantb | |
| 5′-CCGTTTCATATGATCGGGATAACGGGCAAAG | ||
| F, 5′-GCTACTAGTTGACAATTAATCATCGGCTCG | SpeI | p15A/Ptac cloning (PCR), cMyc-YFP and RepA-WH1 WT, A31V, or ΔN37 variant with YFP tag |
| R, 5′-GCTGGATCCTTACTTGTACAGCTCGTCCATG | BamHI | |
| F, 5′-GCTACTAGTTGACAATTAATCATCGGCTCG | SpeI | pRK2/Ptac cloning (PCR), His6-RepA-WH1 WT, A31V, or ΔN37 variant with mCherry tag |
| R, 5′-AGTGAATTCGAGCTCGGTAC | EcoRI | |
| F, 5′-GCTCGGTACCTCGATCCTCTACGCCGGAC | KpnI | lacIq cloning (PCR), pRK2/p15A |
| R, 5′-CGAGGAATTCTCATGCCCCGCGCCCAC | EcoRI | lacIq cloning (PCR), pRK2 |
| R, 5′-CGAGGAGCTCTCATGCCCCGCGCCCAC | SacI | lacIq cloning (PCR), p15A |
Restriction sites are underlined; mutations are in boldface. F, forward; R, reverse.
QuikChange kit (Stratagene).
Epifluorescence microscopy.
Bacteria carrying the fluorescent mCherry/YFP fusion proteins were observed, as described previously (12), with a Nikon Eclipse 90i microscope equipped with a CFI Plan APO VC ×100 (numerical aperture [NA], 1.40) oil immersion objective and a Hamamatsu ORCA-R2 charge-coupled-device (CCD) camera. The following excitation (EX) and emission (EM) filters and exposure times were used: for mCherry, EX at 543/22 nm, EM at 593/40 nm, and exposure of 600 ms; for YFP, EX at 500/24 nm, EM at 542/27 nm, and exposure of 700 ms. In dual-color expression experiments, exposure times were fixed to 1 s. A neutral 1/8 filter was interposed in the optical path in most cases, with the exception of cotransformants involving the monomeric red fluorescent protein (mRFP) or YFP controls, in which case a 1/32 filter was used due to the higher levels of expression. Differential interference contrast (DIC) images were also captured (100 to 300 ms). Image analysis was carried out with the NIS AR, version 3.1, software (Nikon).
Protein detection and quantitation.
Protein expression levels were determined by means of Western blotting, targeting with antibodies the N-terminal peptide tags His6 (mCherry fusions) and cMyc (fusions to YFP). Cells from 25 ml of culture at an OD600 of 2.0 were sedimented and lysed, and electrophoresis was run as described previously (12). Primary mouse antibodies (anti-His, 1:50,000; anti-cMyc, 1:10,000 [Sigma]) were incubated for 2 h, and a horseradish peroxidase (HRP)-conjugated secondary antibody (1:10,000) was incubated for 1 h. Chemiluminescence detection was performed with an ECL Prime kit (GE Healthcare), and band intensity analysis was carried out, with samples from three independent cultures, using Quantity One software (version 4.6.3; Bio-Rad).
SDD-AGE assay.
Semidenaturing detergent agarose gel electrophoresis (16) was carried out as previously described (12) on whole-cell lysates from 25-ml cultures that had been grown for 4 h after IPTG induction (see above). Three-microliter samples extracted from cells expressing RepA-WH1(WT)-mCherry or RepA-WH1(A31V)-YFP or 9-μl samples of the cotransformant expressing both proteins were diluted to 40 μl in 0.5× Tris-acetate EDTA (TAE), 5% glycerol, 1% Sarkosyl, 0.1 mg/ml bromophenol blue, and protease inhibitor cocktail (Roche). Electrophoresis was run at 50 V for 12 h at room temperature. The gel was electroblotted to a polyvinylidene difluoride (PVDF) membrane, and antibody detection was performed as described above, with the following primary antibody dilutions: anti-His at 1:50,000 and anti-cMyc at 1:5,000.
RESULTS AND DISCUSSION
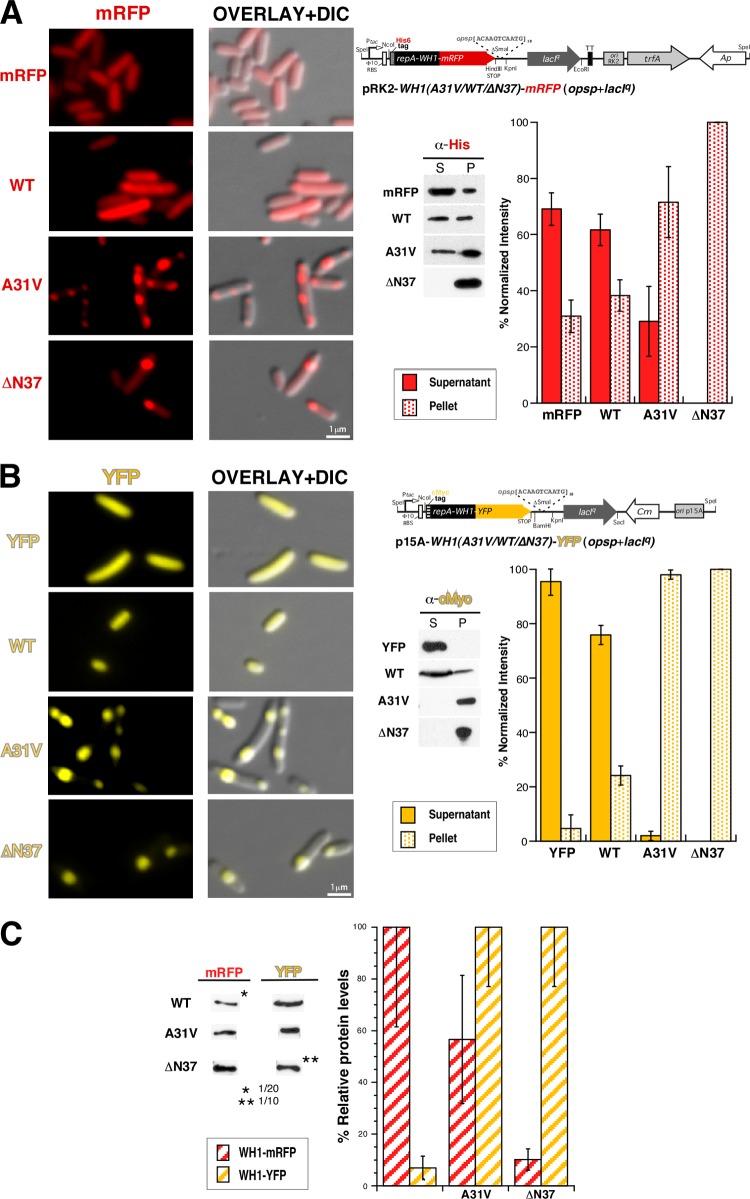
The soluble, although weakly amyloidogenic (9), wild-type (WT) RepA-WH1, its hyperamyloidogenic A31V mutant (9), or a deletion mutant lacking its amyloidogenic peptide stretch (ΔN37) (17) and thus aggregating as IBs (12), was cloned either into a low-copy-number RK2-based plasmid vector or into a compatible, medium-copy-number p15A replicon. These RepA-WH1 variants were fused, through a flexible linker placed at their C termini, to monomeric red (mCherry) or yellow (YFP) fluorescent protein as well as, at the N termini of the chimeras, to a His6 or cMyc tag, respectively (Fig. 1). A single point mutation (A206K) was introduced in YFP in order to get a mostly monomeric variant of the protein (18). Both series of fused genes were cloned under an inducible Ptac promoter and carried downstream tandem repeats of the specific amyloidogenic effector DNA sequence opsp18 (11, 12). When the genes were separately transformed into E. coli, IPTG-induced expression, followed for up to 4 h, led to the appearance of the characteristic red (Fig. 1A) or yellow (Fig. 1B) fluorescent label, which became clear after 30 min of induction. The fused RepA-WH1(WT) protein remained dispersed in the cytoplasm, showing diffused fluorescence, as did the mCherry or YFP protein when expressed alone as a control (Fig. 1A, mRFP/YFP). In contrast, the RepA-WH1(A31V) hyperamyloidogenic variant and ΔN37 protein appeared aggregated in the cytoplasm, albeit with distinct phenotypes: the former as multiple (mostly two or three) aggregates per cell and the latter mainly as monopolar IBs, as previously described (12). The relative protein levels in the soluble (supernatant) and aggregated (pellet) fractions were then quantified by Western blotting using monoclonal antibodies specific for the fused peptide tags. The expression of RepA-WH1(WT) led to higher relative levels of soluble protein when it was fused to YFP (Fig. 1B) than when it was fused to mCherry (Fig. 1A), in which a significant aggregated fraction was evident. This observation correlated with a higher total level of expression for RepA-WH1(WT), and thus an increased tendency toward aggregation, when it was cloned in the pRK2 vector than when it was expressed from p15A (Fig. 1C). However, the relative levels of expression observed were inverted for the RepA-WH1(A31V) and RepA-WH1(ΔN37) variants, which were mostly found aggregated, regardless of the vector or fluorescent protein fusion employed. It is noteworthy that, compared with the wild-type protein, RepA-WH1(A31V) is hyperamyloidogenic (9), and RepA-WH1(ΔN37) is a metastable mutant with marginal stability (17); therefore, both were expected to aggregate. These experiments indicated that the combination of RepA-WH1 variants fused to distinct fluorescent proteins, together with tuned expression levels from compatible vectors, would provide a suitable platform to perform cross-aggregation studies in vivo, as described in the next section.
FIG 1.
Expression in E. coli of single RepA-WH1 variants (WT, A31V, or ΔN37) fused to red (mCherry/mRFP) (A) or yellow (YFP) (B) fluorescent protein tags. Left panels show microscopy imaging (fluorescence and DIC) of representative cells, and right panels show linear schemes (not drawn to scale) of the plasmid vectors used for protein expression (Table 1). Gel insets correspond to Western blots of sedimentation analyses of whole-cell lysates from the same cultures, which were then quantitated (histograms). (C) A Western blot quantitation of the relative expression levels achieved for the distinct RepA-WH1 constructs when cloned in pRK2 (mRFP) or p15A (YFP). Samples were whole lysates from equivalent cell numbers. In all panels, histograms represent the mean intensities of the distinct protein bands, calculated from three independent experiments. Dilution factors were applied for gel loading of those samples in which high overexpression levels were achieved (asterisks) to allow for unsaturated detection. Dilutions were then taken into account for quantitation. Standard deviations are displayed on each histogram bar. RBS, ribosome binding site; α, anti.
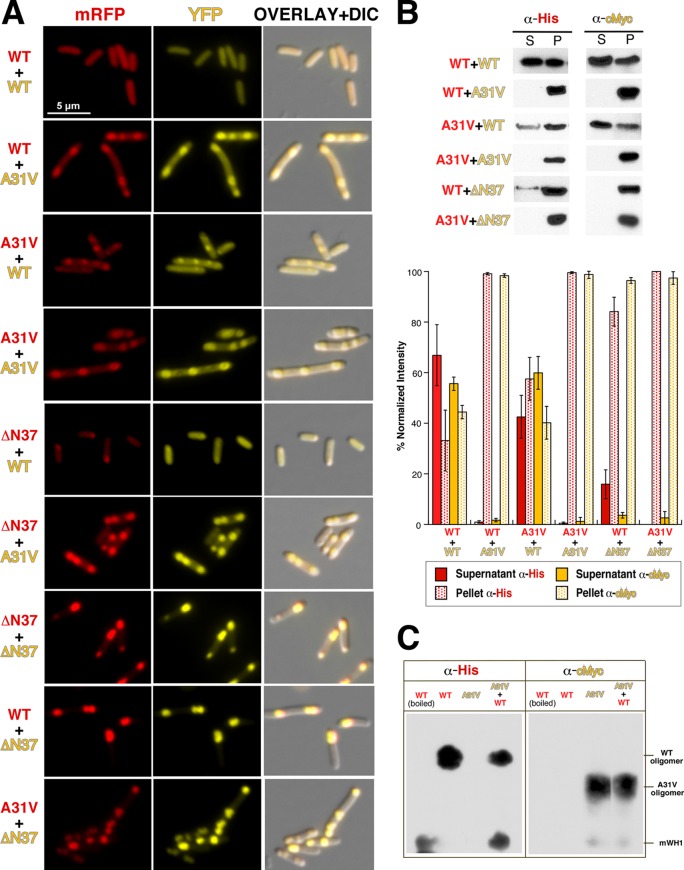
Coexpression of distinct RepA-WH1 pairs under conditions previously established for the individual proteins (12) led to the accumulation in bacteria of the corresponding red (mCherry) and yellow (YFP) fluorescence (Fig. 2A). Diffused fluorescence was observed only in cells where the soluble RepA-WH1(WT) variant was simultaneously expressed fused to both fluorescent tags. However, biochemical fractionation of the cells (Fig. 2B) suggests that the diffused fluorescence usually exhibited by RepA-WH1(WT) might correspond to oligomers susceptible to be sedimented rather than to soluble protein. This is consistent with the fact that the wild-type protein is mildly amyloidogenic (9). Otherwise, the aggregation-prone A31V and ΔN37 variants dominated the wild type, which became aggregated into foci that followed the tendency of the coexpressed partner, forming either multiple and dispersed (A31V) or single and monopolar (ΔN37) aggregates (Table 3); A31V was the strongest variant in spite of its level of expression. These results are compatible with previous experiments in vitro in which ex vivo-purified RepA-WH1(A31V) aggregates drove the formation of amyloid fibers by soluble molecules of the same variant and nucleated the cross-aggregation of its WT counterpart (11). In this sense, it is worth noting that recent nuclear magnetic resonance (NMR) studies have demonstrated that amyloid aggregates purified from biological sources actually do reproduce on soluble protein molecules the same amyloid conformations they originally adopted (19, 20). Using an amyloidotropic fluorophore (BTA-1), we have previously shown that the aggregates formed by RepA-WH1 inside bacterial cells indeed are amyloids and, furthermore, that a gradation in amyloidogenicity can be established between distinct amyloid variants (12). To explore if any additional difference can be found between the aggregated fractions of RepA-WH1(WT) and RepA-WH1(A31V), whole-cell lysates bearing these proteins, either separately or coexpressed, were analyzed by means of SDD-AGE, a technique widely used to separate distinct aggregated species of yeast prions (16) and one that we have previously adapted for RepA-WH1(A31V) aggregates in E. coli (12). Interestingly, SDD-AGE shows (Fig. 2C) that, in addition to the already reported RepA-WH1(A31V) oligomers (12), RepA-WH1(WT) assembles a distinct oligomeric aggregate with a lower electrophoretic mobility than the hyperamyloidogenic species. This observation actually is similar to what has been reported using SDD-AGE in yeast prions: phenotypically weak prion strains tend to form large amyloid assemblies, whereas strong strains assemble amyloid species with more discrete sizes (21, 22). However, it must be noted that although it is clear from the results shown in this work that coexpression of distinct RepA-WH1 variants mutually enhances aggregation, our SDD-AGE analysis does not provide evidence for their coexistence in a given type of oligomeric aggregate. This could reflect a limitation of a technique designed for the Q/N-rich (polar) amyloids assembled by yeast prions, whereas hydrophobic residues are the main drivers of amyloid aggregation in the case of RepA-WH1 (9).
FIG 2.
Study of the pairwise coexpression, from compatible pRK2 and p15A vectors (Fig. 1), of the indicated RepA-WH1 variants in E. coli cells. (A) Two-channel fluorescence microscopy plus DIC imaging. (B) Protein solubility analyses were carried out by sedimentation plus Western blotting with the indicated tag-specific antibodies. Below is a histogram representation of the intensities of the bands, averaged from three independent expression and blotting experiments. The intracellular distribution of fluorescent foci and the aggregation/solubility balance point to the following series in the strength/dominance of amyloid aggregation for the RepA-WH1 prionoid: A31V > ΔN37 (IBs) > WT. (C) SDD-AGE analysis of intracellular aggregation of the RepA-WH1(WT)-mRFP and RepA-WH1(A31V)-YFP amyloid variants. In addition to protein monomers, which were more abundant for the WT variant, two distinct oligomers were evident, with those assembled by RepA-WH1(A31V) being smaller than those assembled by the WT protein.
TABLE 3.
Quantification of aggregate numbers and cell lengths in microscopy fields captured after 4 h of coexpression of the specified RepA-WH1 pairsa
| RepA-WH1 pair | % of cells with the indicated no. of aggregates |
Avg cell length (μm) | ||||
|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | ≥4 | ||
| WT-mRFP + WT-YFP | 77.5 | 16 | 6.5 | 0 | 0 | 2.67 ± 0.79 |
| WT-mRFP + A31V-YFP | 0 | 7 | 58.5 | 19.5 | 15.5 | 5.07 ± 1.46 |
| WT-mRFP + YFP | 100 | 0 | 0 | 0 | 0 | 3.87 ± 1.13 |
| WT-mRFP + ΔN37-YFP | 0 | 43 | 39 | 13.5 | 4.5 | 3.91 ± 1.30 |
| A31V-mRFP + WT-YFP | 0 | 5.5 | 41.5 | 33.5 | 19.5 | 3.89 ± 1.32 |
| A31V-mRFP + A31V-YFP | 0 | 5 | 56 | 20.5 | 18.5 | 5.00 ± 1.66 |
| A31V-mRFP + YFP | 11 | 6 | 37.5 | 17.5 | 28 | 5.25 ± 1.54 |
| A31V-mRFP + ΔN37-YFP | 0 | 9 | 39.5 | 27 | 24.5 | 5.27 ± 1.58 |
| mRFP + WT-YFP | 100 | 0 | 0 | 0 | 0 | 2.33 ± 0.49 |
| mRFP + A31V-YFP | 0 | 10 | 64 | 21 | 5 | 4.91 ± 1.26 |
| mRFP + YFP | 100 | 0 | 0 | 0 | 0 | 4.23 ± 1.24 |
| mRFP + ΔN37-YFP | 0 | 90 | 10 | 0 | 0 | 3.21 ± 0.83 |
| ΔN37-mRFP + WT-YFP | 45 | 52.5 | 2.5 | 0 | 0 | 2.04 ± 0.59 |
| ΔN37-mRFP + A31V-YFP | 0 | 5.5 | 63.5 | 18.5 | 12.5 | 5.00 ± 1.49 |
| ΔN37-mRFP + YFP | 86.5 | 11.5 | 2 | 0 | 0 | 3.79 ± 1.15 |
| ΔN37-mRFP + ΔN37-YFP | 0 | 68 | 27.5 | 3 | 1.5 | 3.33 ± 0.99 |
A total of 200 cells of each class were used for these measurements.
Measurements of the average cell length in bacterial populations expressing the distinct RepA-WH1 pairs (Table 3) showed that bacteria expressing the RepA-WH1(A31V) variant were consistently longer (by at least 1 μm) than those bearing combinations of the WT variant and the ΔN37 and mRFP/YFP controls. This is an indication of delayed cell division and thus of the toxicity of the RepA-WH1(A31V) aggregates (23, 24). The significant differences in the lengths of bacteria bearing RepA-WH1(A31V) reflect their tendency toward filamentation, as previously observed (11, 12).
It is noteworthy that in the case of the variant forming conventional IBs (ΔN37), the level of expression (Fig. 1C) determined its ability to enhance the aggregation of RepA-WH1(WT) (Fig. 2A); e.g., when expressed from the p15A/YFP vector, high expression of ΔN37 drove the aggregation of WT into IBs. Since ΔN37 lacks the main amyloidogenic stretch in RepA-WH1 (9), this presumably occurs through a different region in the protein with lower amyloidogenicity or through general hydrophobic interactions. In contrast, its lower relative expression from pRK2/mCherry resulted in smaller IBs with weak colocalization of RepA-WH1(WT) but with a vast excess of this yellow-tagged protein dispersed in the cytoplasm (Fig. 2A). Thus, RepA-WH1(ΔN37) can either drive the aggregation of a fraction of RepA-WH1(WT) toward IBs or become attached to the foci preestablished by RepA-WH1(A31V).
As controls, neither the individual mCherry nor YFP protein expressed under the same conditions coaggregated with RepA-WH1s but remained diffused across the cytoplasm (Fig. 3). It is interesting that in these controls, the regions occupied by the nucleoid are often not labeled, either by mCherry or YFP, suggesting that both fluorescent proteins might be forming oligomers large enough to have reduced diffusion rates inside the crowded nucleoid milieu (25).
FIG 3.
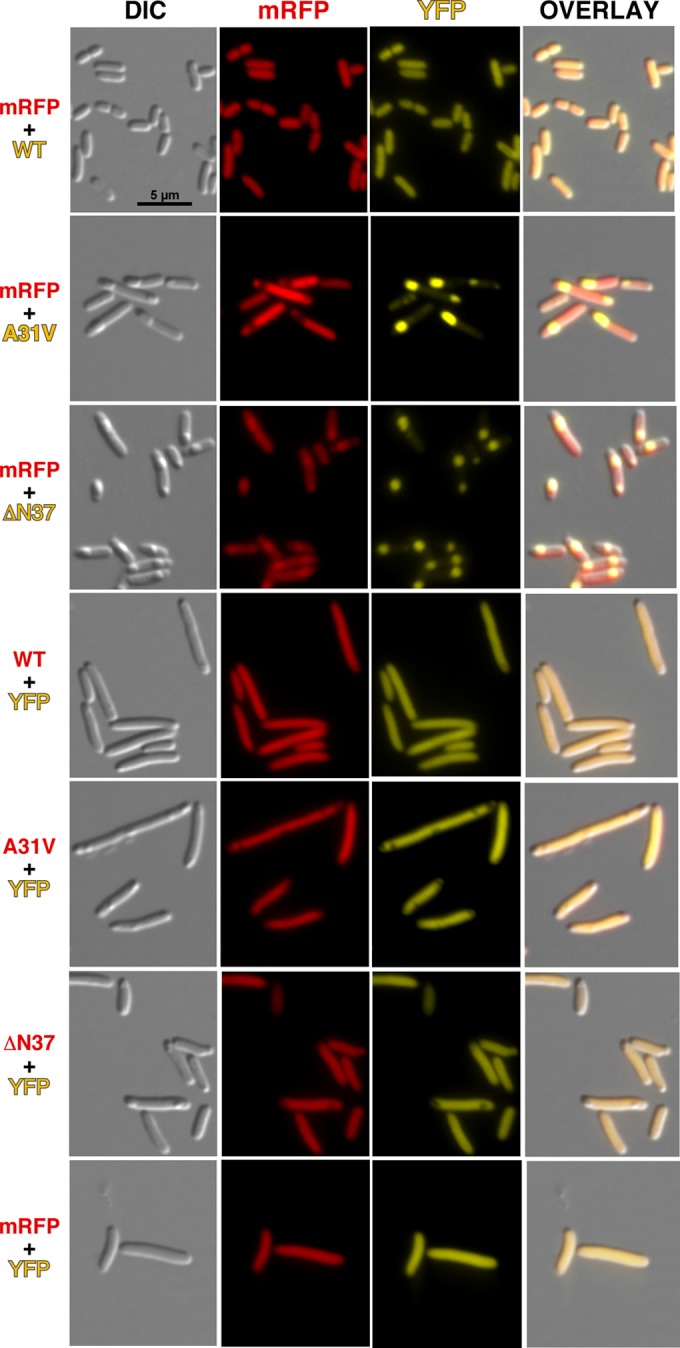
DIC and fluorescence microscopy imaging of bacteria coexpressing either mRFP (red) or YFP (yellow), as controls for the fluorescent protein reporters, together with the indicated RepA-WH1 partner (WT, A31V, or ΔN37 variant) fused to the complementary color.
The simple colocalization experiments reported here provide evidence for the ability of the RepA-WH1(A31V) prionoid to induce cross-aggregation in the cytoplasm of E. coli, i.e., to enhance the growth of amyloid particles by a distinct variant of the same protein with reduced amyloidogenicity. Such assays easily discriminate bona fide amyloid aggregation from formation of IBs, which, albeit they exhibit some amyloid-like character (26, 27), differ in cellular distribution and number, toxicity, and chaperone-modulated transmissibility (11, 12). In addition to the reports on heterologous expression of human amyloidogenic proteins in E. coli (7) and the pioneering work by Chapman et al. with the amyloid curli fibers (28), recently adapted by Hochschild and Sivanathan for the extracellular secretion of proteins (29, 30), the RepA-WH1 prionoid has been shown to constitute a fully bacterial system to test intracellular amyloid cross-seeding, a hot spot in current research on the etiology and pathogenesis of human amyloidosis (31).
Conclusions.
The results presented in our previous reports established that the synthetic bacterial prionoid RepA-WH1 recapitulates some of the universal features of protein amyloidosis: (i) it adopts two alternative conformations, soluble dimers and insoluble monomers, which assemble in vitro as amyloid (cross-β) sheets; (ii) it forms intracellular aggregates that hamper bacterial proliferation; (iii) amyloid variants with distinct toxicities and morphologies have been identified in vivo; and (iv) these intracellular aggregates can seed in vitro the growth of amyloid fibers from soluble RepA-WH1 molecules. Exploring the influence on amyloidosis of distinct protein fusion tags, expression levels, gene dosage, and allelic variants, we have reported here a fifth feature: the ability of a hyperamyloidogenic RepA-WH1 point mutant (A31V) to drive the soluble, mildly amyloidogenic, wild-type protein to aggregate as intracellular inclusions. Given the reduced toxicity, extracellular location, and relaxed cross-seeding specificity of other bacterial amyloids, RepA-WH1 stands out as a minimal model system for human amyloid proteinopathies.
ACKNOWLEDGMENTS
We thank the members of our lab for their encouragement and continuous support. We are grateful to V. de Lorenzo for the gift of the pSEVA121 vector and R. Tsien and A. Lindner for plasmid sources of the mCherry and the YFP markers, respectively.
This work has been supported by grants from MINECO of Spain (BIO2012-30852 and CSD2009-00088).
Footnotes
Published ahead of print 2 May 2014
REFERENCES
- 1.Chiti F, Dobson CM. 2006. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75:333–366. 10.1146/annurev.biochem.75.101304.123901 [DOI] [PubMed] [Google Scholar]
- 2.Blancas-Mejía LM, Ramírez-Alvarado M. 2013. Systemic amyloidosis. Annu. Rev. Biochem. 82:745–774. 10.1146/annurev-biochem-072611-130030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liebman SW, Chernoff YO. 2012. Prions in yeast. Genetics 191:1041–1072. 10.1534/genetics.111.137760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halfmann R, Jarosz DF, Jones SK, Chang A, Lancaster AK, Lindquist S. 2012. Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature 482:363–368. 10.1038/nature10875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DePas WH, Chapman MR. 2012. Microbial manipulation of the amyloid fold. Res. Microbiol. 163:592–606. 10.1016/j.resmic.2012.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garrity SJ, Sivanathan V, Dong J, Lindquist S, Hochschild A. 2010. Conversion of a yeast prion to an infectious form in bacteria. Proc. Natl. Acad. Sci. U. S. A. 107:10596–10601. 10.1073/pnas.0913280107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim W, Kim Y, Min J, Kim DJ, Chang YT, Hecht MH. 2006. A high-throughput screen for compounds that inhibit aggregation of the Alzheimer's peptide. ACS Chem. Biol. 1:461–469. 10.1021/cb600135w [DOI] [PubMed] [Google Scholar]
- 8.Gasset-Rosa F, Díaz-López T, Lurz R, Prieto A, Fernández-Tresguerres ME, Giraldo R. 2008. Negative regulation of pPS10 plasmid replication: Origin pairing by zipping-up DNA-bound RepA monomers. Mol. Microbiol. 68:560–572. 10.1111/j.1365-2958.2008.06166.x [DOI] [PubMed] [Google Scholar]
- 9.Giraldo R. 2007. Defined DNA sequences promote the assembly of a bacterial protein into distinct amyloid nanostructures. Proc. Natl. Acad. Sci. U. S. A. 104:17388–17393. 10.1073/pnas.0702006104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gasset-Rosa F, Maté MJ, Dávila-Fajardo C, Bravo J, Giraldo R. 2008. Binding of sulphonated indigo derivatives to RepA-WH1 inhibits DNA-induced protein amyloidogenesis. Nucleic Acids Res. 36:2249–2256. 10.1093/nar/gkn067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernández-Tresguerres ME, de la Espina SM, Gasset-Rosa F, Giraldo R. 2010. A DNA-promoted amyloid proteinopathy in Escherichia coli. Mol. Microbiol. 77:1456–1469. 10.1111/j.1365-2958.2010.07299.x [DOI] [PubMed] [Google Scholar]
- 12.Gasset-Rosa F, Coquel AS, Moreno-del Álamo M, Chen P, Song X, Serrano AM, Fernández-Tresguerres ME, Moreno-Díaz de la Espina S, Lindner AB, Giraldo R. 2014. Direct assessment in bacteria of prionoid propagation and phenotype selection by Hsp70 chaperone. Mol. Microbiol. 91:1070–1087. 10.1111/mmi.12518 [DOI] [PubMed] [Google Scholar]
- 13.Giraldo R, Moreno-Díaz de la Espina S, Fernández-Tresguerres ME, Gasset-Rosa F. 2011. RepA prionoid: a synthetic amyloid proteinopathy in a minimalist host. Prion 5:60–64. 10.4161/pri.5.2.14913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pósfai G, Plunkett G, Fehér T, Frisch D, Keil GM, Umenhoffer K, Kolisnychenko V, Stahl B, Sharma SS, de Arruda M, Burland V, Harcum SW, Blattner FR. 2006. Emergent properties of reduced-genome Escherichia coli. Science 312:1044–1046. 10.1126/science.1126439 [DOI] [PubMed] [Google Scholar]
- 15.Silva-Rocha R, Martínez-García E, Calles B, Chavarría M, Arce-Rodríguez A, de las Heras A, Páez-Espino AD, Durante-Rodríguez G, Kim J, Nikel PI, Platero R, de Lorenzo V. 2013. The Standard European Vector Architecture (SEVA): a coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Res. 41:D666–D675. 10.1093/nar/gks1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bagriantsev SN, Kushnirov VV, Liebman SW. 2006. Analysis of amyloid aggregates using agarose gel electrophoresis. Methods Enzymol. 412:33–48. 10.1016/S0076-6879(06)12003-0 [DOI] [PubMed] [Google Scholar]
- 17.Giraldo R, Andreu JM, Díaz-Orejas R. 1998. Protein domains and conformational changes in the activation of RepA, a DNA replication initiator. EMBO J. 17:4511–4526. 10.1093/emboj/17.15.4511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zacharias DA, Violin JD, Newton AC, Tsien RY. 2002. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296:913–916. 10.1126/science.1068539 [DOI] [PubMed] [Google Scholar]
- 19.Lu JX, Qiang W, Yau WM, Schwieters CD, Meredith SC, Tycko R. 2013. Molecular structure of β-amyloid fibrils in Alzheimer's disease brain tissue. Cell 154:1257–1268. 10.1016/j.cell.2013.08.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frederick KK, Debelouchina GT, Kayatekin C, Dorminy T, Jacavone AC, Griffin RG, Lindquist S. 2014. Distinct prion strains are defined by amyloid core structure and chaperone binding site dynamics. Chem. Biol. 21:295–305. 10.1016/j.chembiol.2013.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka M, Collins SR, Toyama BH, Weissman JS. 2006. The physical basis of how prion conformations determine strain phenotypes. Nature 442:585–589. 10.1038/nature04922 [DOI] [PubMed] [Google Scholar]
- 22.Derdowski A, Sindi SS, Klaips CL, Disalvo S, Serio TR. 2010. A size threshold limits prion transmission and establishes phenotypic diversity. Science 330:680–683. 10.1126/science.1197785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stewart EJ, Madden R, Paul G, Taddei F. 2005. Aging and death in an organism that reproduces by morphologically symmetric division. PLoS Biol. 3:e45. 10.1371/journal.pbio.0030045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindner A, Madden R, Demarez A, Stewart EJ, Taddei F. 2008. Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proc. Natl. Acad. Sci. U. S. A. 105:3076–3081. 10.1073/pnas.0708931105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jun S, Wright A. 2010. Entropy as the driver of chromosome segregation. Nat. Rev. Microbiol. 8:600–607. 10.1038/nrmicro2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carrió M, González-Montalbán N, Vera A, Villaverde A, Ventura S. 2005. Amyloid-like properties of bacterial inclusion bodies. J. Mol. Biol. 347:1025–1037. 10.1016/j.jmb.2005.02.030 [DOI] [PubMed] [Google Scholar]
- 27.Wang L, Maji SK, Sawaya MR, Eisenberg D, Riek R. 2008. Bacterial inclusion bodies contain amyloid-like structure. PLoS Biol. 6:e195. 10.1371/journal.pbio.0060195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ. 2002. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295:851. 10.1126/science.1067484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sivanathan V, Hochschild A. 2012. Generating extracellular amyloid aggregates using E. coli cells. Genes Dev. 26:2659–2667. 10.1101/gad.205310.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sivanathan V, Hochschild A. 2013. A bacterial export system for generating extracellular amyloid aggregates. Nat. Protoc. 8:1381–1390. 10.1038/nprot.2013.081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morales R, Moreno-González I, Soto C. 2013. Cross-seeding of misfolded proteins: implications for etiology and pathogenesis of protein misfolding diseases. PLoS Pathog. 9:e1003537. 10.1371/journal.ppat.1003537 [DOI] [PMC free article] [PubMed] [Google Scholar]