Abstract
There is a wealth of information on the genetic regulation and biochemical properties of bacterial C4-dicarboxylate transport systems. In sharp contrast, there are far fewer studies describing the transport and assimilation of C5-dicarboxylates among bacteria. In an effort to better our understanding on this subject, we identified the structural and regulatory genes necessary for the utilization of α-ketoglutarate (α-KG) in Pseudomonas aeruginosa PAO1. The PA5530 gene, encoding a putative dicarboxylate transporter, was found to be essential for the growth of P. aeruginosa PAO1 on both α-KG and glutarate (another C5-dicarboxylate). Metabolite analysis confirmed that the PA5530 gene was necessary for the uptake of extracellular α-KG. Like other substrate-inducible transporter genes, expression of the PA5530 gene was induced by extracellular C5-dicarboxylates. It was later found that the expression of the PA5530 gene was driven solely by a −24/−12 promoter recognized by the alternative sigma factor RpoN. Surprisingly, the enhancer binding protein MifR, which is known to have an essential role in biofilm development, was required for the expression of the PA5530 gene. The MifR protein is homologous to other transcriptional regulators involved in dicarboxylate assimilation, suggesting that MifR might interact with RpoN to activate the expression of the PA5530 gene in response to extracellular C5-dicarboxylates, especially α-KG. The results of this study provide a framework for exploring the assimilation of α-KG in other pseudomonads.
INTRODUCTION
The C5-dicarboxylate α-ketoglutarate (α-KG) is widely recognized for being an intermediate of both carbon and nitrogen metabolism. The other C5-dicarboxylate commonly encountered is glutarate, which is an intermediate of lysine and tryptophan metabolism. It has become increasingly apparent that α-KG plays a larger and more profound role in bacterial physiology than originally believed. For example, it is now known that α-KG directly regulates nitrogen metabolism via binding to and controlling the activity of the nitrogen regulatory protein PII (1–3). There is also compelling evidence that endogenous α-KG is a natural protectant against both oxidative damage (4, 5) and cyanide poisoning (6). α-KG-dependent hydroxylases are ubiquitous among bacteria and participate in various cellular functions (7), including the assimilation of key nutrients, e.g., sulfur and phosphate, and the biosynthesis of complex molecules such as antibiotics and lipid A. Interestingly, there have been a few recently published papers reporting that α-KG might be a preferred nutrient for some bacteria during the course of an infectious state (8–10). α-KG is a suitable growth substrate for most bacteria, but the genes involved in the transport and assimilation of α-KG have been investigated for only a small number of bacterial species, including Escherichia coli (10–12), Staphylococcus aureus (13), Bacillus licheniformis (14), Lactococcus lactis (15), Xanthomonas oryzae (9), and Rhizobium tropici (8). Missing from the literature are studies describing the genetic regulation of α-KG assimilation in Pseudomonas, which is surprising, because these bacteria are well known for their versatile metabolism, enabling them to consume a wide range of substrates.
The assimilation of α-KG has been documented in previous studies involving Pseudomonas (16–20). Campbell and Stokes observed that acetate-grown P. aeruginosa displayed a significant lag time when transferred into α-KG medium (19), hinting at the probable existence of an inducible α-KG transporter. Pseudomonas cultures grown under conditions of nitrogen limitation are well known to accumulate extracellular α-KG, which is readily depleted with the addition of exogenous ammonium. Probably the most complete study on α-KG transport in Pseudomonas was that by Edwards et al., which biochemically characterized C5-dicarboxylate transport in P. putida (21). Those authors observed that transport of α-KG in P. putida was induced by extracellular C5-dicarboxylates such as α-KG and glutarate (21). Despite these observations, the genes encoding the transport and regulatory proteins involved in α-KG uptake had not been identified for any given Pseudomonas species.
Because of the limited information available on α-KG assimilation in Pseudomonas, we decided to identify the genetic network surrounding α-KG transport in Pseudomonas aeruginosa PAO1. An initial survey of the sequenced genome of P. aeruginosa PAO1 revealed that the predicted PA5530 protein is homologous to the well-characterized α-KG transporter, KgtP, of E. coli K-12 (11). Subsequent analysis confirmed that the PA5530 gene was required for the uptake of extracellular α-KG for P. aeruginosa PAO1. Expression of the PA5530 gene was induced by extracellular C5-dicarboxylates, and this expression was dependent on the alternative sigma factor σ54 or RpoN. Unexpectedly, the expression of the PA5530 gene also required the transcriptional regulator MifR, which was found previously to be involved in biofilm formation (22). MifR shares significant homology (>60%) with other regulators involved in α-KG utilization, including KgtR of R. tropici CIAT899 (8) and KguR of E. coli CTF073 (10). The results presented here indicate that MifR is a homolog of the KgtR/KguR proteins and participates with RpoN to activate the expression of the putative α-KG transporter (PA5530) in response to extracellular α-KG.
MATERIALS AND METHODS
Bacteria, plasmids, and media.
Bacterial strains and plasmids used in the current study are given in Table 1. Bacteria were grown in Lennox broth (BD Difco), nutrient broth no. 2 (Oxoid), or modified M9 minimal medium (30), containing 5 μM FeSO4. Solid bacteriological medium was prepared with the addition of BD Bacto agar at 15 g liter−1. The following antibiotics were used for plasmid selection: kanamycin (Km) (50 μg ml−1 for E. coli), carbenicillin (Cb) (100 μg ml−1 for E. coli or 200 μg ml−1 for P. aeruginosa), and gentamicin (Gm) (10 μg ml−1 for E. coli or 30 μg ml−1 for P. aeruginosa).
TABLE 1.
Strains, plasmids, and oligonucleotides used in the current study
| Strain, plasmid, or oligonucleotide | Relevant characteristic(s) or sequence | Reference or source |
|---|---|---|
| Strains | ||
| Pseudomonas aeruginosa | ||
| PAO1 | Wild type | 23 |
| PW9687 | PA5166-F04::ISphoA/hah derivative of PAO1 | 24 |
| PW10371 | PA5530-C01::ISphoA/hah derivative of PAO1 | 24 |
| PAO6359 | rpoN::Ω-Km derivative of PAO1 | 25 |
| ΔmifR PAO1 | ΔmifR derivative of PAO1 | This study |
| ΔmifR PW9687 | ΔmifR derivative of PW9687 | This study |
| PA14 | Wild type | 26 |
| ΔPA5530 PA14 | ΔPA5530 derivative of PA14 | This study |
| PAK | Wild type | 27 |
| ΔPA5530 PAK | ΔPA5530 derivative of PAK | This study |
| Escherichia coli Top10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 nupG recA1 araD139 Δ(ara-leu)7697 galE15 galK16 rpsL (Strr) endA1 λ− | Invitrogen |
| Plasmids | ||
| pCR-Blunt | Cloning plasmid; Kmr | Invitrogen |
| pTrc99a | Expression plasmid; Cbr | Pharmacia |
| pBBR1MCS-5 | Broad-host-range plasmid; Gmr | 28 |
| pEX18ApGW | Plasmid for gene deletions in P. aeruingosa; Cbr Gmr | 29 |
| ΔPlac-pBBR1MCS-5 | pBBR1MCS-5 minus lac promoter; Gmr | This study |
| pBRL474 | PA5530 gene in pCR-Blunt; Kmr | This study |
| pBRL479 | PA5530 gene in pBBR1MCS-5; Gmr | This study |
| pBRL476 | PA5530-lacZ in pCR-Blunt; Kmr | This study |
| pBRL485 | PA5530-lacZ in ΔPlac-pBBR1MCS-5; Gmr | This study |
| pBRL487 | PA5530-lacZ with mutated RpoN promoter; Gmr | This study |
| pBRL511 | mifR::Gmr in pEX18ApGW; Cbr Gmr | This study |
| pBRL538 | PA5530::Gmr in pEXT18ApGW; Cbr Gmr | This study |
| pBRL549 | mifSR operon in pCR-Blunt; Kmr | This study |
| pBRL561 | mifS-ΔmifR operon in pCR-Blunt; Kmr | This study |
| pBRL566 | mifSR operon (forward orientation) in pTrc99a; Cbr | This study |
| pBRL567 | mifSR operon (backward orientation) in pTrc99a; Cbr | This study |
| pBRL570 | mifS-ΔmifR operon (forward orientation) in pTrc99a;Cbr | This study |
| pJRH07 | mifR gene in pCR-Blunt; Kmr | This study |
| pJRH08 | mifR gene in pBBR1MCS-5; Gmr | This study |
| Oligonucleotides | ||
| BL342.f | ATGACCATGATTACGGATTCACTG | |
| BL342.r | GCAGTTATTTTTGACACCAGACCAACTGGTA | |
| BL434.f | GCATCTAGACTGATCAGCACATCCAAGACAAC | |
| BL434.r | GCAGAGCTCTCAATCGGTCGTGATCTTCGA | |
| BL435.f | GCAGATCGGCGAGTTCTCC | |
| BL435.r | GAATCCGTAATCATGGTCATCGTGTTTCCTCTTTTCGTTGTTG | |
| BL438.f | GCTTTCCCGCGTTAACACGGCACCTGCTATC | |
| BL438.r | GATAGCAGGTGCCGTGTTAACGCGGGAAAGC | |
| BL466.f | TACAAAAAAGCAGGCTGACCAGGTGATCTTCGTCGAC | |
| BL466.r | TCAGAGCGCTTTTGAAGCTAATTCGGAAGGGTTTCTCGATGAAGTCG | |
| BL467.f | AGGAACTTCAAGATCCCCAATTCGCGAACATTTCGCCAGCGTTG | |
| BL467.r | TACAAGAAAGCTGGGTCTCTTCGCCGACGAAATCGC | |
| BL552.f | TACAAAAAAGCAGGCTGAAAGCGCCAACGCCATTTC | |
| BL552.r | TCAGAGCGCTTTTGAAGCTAATTCGGATGATCAGCGAGCCAAAGC | |
| BL553.f | AGGAACTTCAAGATCCCCAATTCGCTTCTACACCTACACCACCTAC | |
| BL553.r | TACAAGAAAGCTGGGTCAATCGGTCGTGATCTTCGAG | |
| BL554.f | GCATCTAGACGTTTCGATCCGCCGATGTC | |
| BL554.r | GCAGAGCTCAGTTGGCGAAGGATCTCTGAC | |
| JRH04.f | GACTCTAGATAAGAAGGAGATATACCATGAGCGACCAGGTGATCTTC | |
| JRH04.r | GACCTCGAGAGTTGGCGAAGGATCTCTGAC |
Molecular biology methods.
DNA was purified by using Promega nucleic acid purification kits. Restriction enzymes and ligases were products of New England BioLabs. PrimeStar polymerase (TaKaRa Biosciences) was used for all PCRs, which were done according to the recommended protocols for PrimeStar polymerase. Oligonucleotides used for PCR are provided in Table 1. Genomic DNA from P. aeruginosa PAO1 was used for all PCR applications. PCR products were cloned into pCR-Blunt by using the Zero Blunt PCR cloning kit (Invitrogen). Cloned DNA was verified by sequencing (Genewiz).
Construction of ΔPlac-pBBR1MCS-5.
A promoterless plasmid suitable for the housing and analysis of reporter genes in P. aeruginosa was constructed. The lac promoter positioned upstream of the multiple-cloning site of plasmid pBBR1MCS-5 was removed via SphI-KpnI double digestion. The SphI-KpnI-digested pBBR1MCS-5 fragment was blunted by using Pfu DNA polymerase (Agilent Technologies) and subsequently religated to yield ΔPlac-pBBR1MCS-5.
Cloning of the PA5530, mifR (PA5511), and mifSR (PA5512-PA5511) genes.
The PA5530, mifR, and mifSR genes were PCR amplified with primer pairs BL434.f/BL434.r, JRH04.f/JRH04.r, and BL554.f/BL554.r, respectively. The PA5530, mifR, and mifSR PCR products were cloned into pCR-Blunt to generate plasmids pBRL474, pJRH07, and pBRL549, respectively. The PA5530 and mifR genes were individually subcloned into the XbaI/SacI sites of pBBR1MCS-5 to yield pBRL479 and pJRH08, respectively.
The XbaI/SpeI mifSR fragment from pBRL549 was cloned into the XbaI site of pTrc99a with either a forward or backward orientation to give plasmid pBRL566 or pBRL567, respectively. Plasmid pBRL549 was digested with SalI to excise an internal portion (nucleotides 27 to 830) of the mifR gene, thereby yielding plasmid pBR561. The resulting XbaI/SpeI mifS-ΔmifR fragment was cloned from pBRL561 into the XbaI site of pTrc99a with a forward orientation, generating pBRL570.
Cloning of the PA5530-lacZ fusion.
The 1,069-bp 5′ region upstream of the predicted PA5530 open reading frame (ORF) was PCR amplified with primer pair BL435.f/BL435.r, and the β-galactosidase (lacZ) ORF of E. coli K-12 MG1655 was PCR amplified with primer pair BL342.f/BL342.r. The PA5530 promoter and lacZ PCR products were assembled into a single fragment via fusion PCR (31), using primer pair BL435.f/BL342.r. The PA5530-lacZ fusion was cloned into pCR-Blunt to give pBRL476. The XbaI/SpeI PA5530-lacZ fragment of pBRL482 was subcloned into the XbaI site of ΔPlac-pBBR1MCS-5 to yield pBRL485. The “GG” dinucleotide positioned 105 bp upstream of the lacZ ORF in the PA5530-lacZ reporter (in pBRL485) was changed to “AA” by using the QuikChange kit (Agilent Technologies) and primer pair BL438.f/BL438.r. The resulting plasmid, pBRL488, was sequenced to verify that the desired substitution was present.
Deletion of the PA5530 and mifR (PA5511) genes in P. aeruginosa.
The PA5530 and mifR genes were deleted in P. aeruginosa according to the method of Choi and Schweizer (29). Procedures for plasmid construction, electroporation, screening, selection, and marker removal were conducted exactly as described previously (29). Briefly, the PA5530::Gm and mifR::Gm cassettes were cloned into plasmid pEX18ApGW to give pBRL511 and pBRL538, respectively. The PA5530 gene was deleted in P. aeruginosa PA14 and PAK by using pBRL511, whereas the mifR gene was deleted in P. aeruginosa PAO1 and PW9687 by using pBRL538. The Gm markers were removed by using plasmid pFLP2. The ΔPA5530 and ΔmifR mutations were verified by PCR.
Growth experiments on C5-dicarboxylates.
All P. aeruginosa strains were grown in triplicate. For each replicate, 50 ml of M9 minimal medium supplemented with 5 μM FeSO4 and 20 mM C5-dicarboxylate (α-KG or glutarate) (in a 500-ml baffle shake flask) was inoculated to an initial optical density at 600 nm (OD600) of ∼0.1. Cultures were grown at 37°C at 200 rpm, and the absorbance at 600 nm (OD600) was measured at 0, 1.5, 3.0, 4.5, 6.0, 7.5, 9.0, and 24 h postinoculation. Note that M9 minimal medium did not have to be supplemented with gentamicin to select for recombinant P. aeruginosa strains harboring plasmid pBRL479 or pJRH08; the presence of C5-dicarboxylate as a sole carbon source was sufficient for selection.
Measurement of extracellular α-KG from P. aeruginosa cultures.
Experiments were done in triplicate. P. aeruginosa PAO1 and the PA5530 transposon mutant (strain PW10317) were grown in 2.0 ml of nutrient broth no. 2 (Oxoid) at 37°C at 200 rpm for 18 h. Afterwards, 50 ml of nutrient broth no. 2 (Oxoid) (in a 500-ml baffle shake flask) was inoculated with 0.5% of a seed culture, and the culture was grown at 37°C at 200 rpm. At OD600 values of 0.2, 0.4, 0.8, and 1.6, an aliquot of culture (1.0 ml) was passed through a 0.22-μm membrane (Millipore) via vacuum filtration. The filtrate (0.5 ml) was added to an equal volume (0.5 ml) of 0.8 M perchloric acid, and the solution was vortexed vigorously for 1 min.
Each prepared sample (20 μl) was combined with an equal volume (20 μl) of 1,2-diamino-4,5-methylenedioxybenzene (DMB) derivatization reagent (32), and the mixture was incubated in the dark at 50°C for 2.5 h. If needed, samples were diluted prior to the DMB derivatization so that the measured concentration of the DMB–α-KG derivative was between 20 and 200 μM (dynamic range of the method). The DMB-derivatized α-KG was analyzed in each sample by electrospray ionization-liquid chromatography mass spectrometry (ESI-LC-MS), which consisted of an API2000 LC tandem mass spectrometry (LC/MS/MS) system equipped with a turbo-ion spray source interfaced with a Prominence UFLC. The column used for separation was a C18 reversed-phase Hypersil (100-mm- by 2.1-mm internal diameter [ID], 3-μm particle size, and 120-Å pore size). Mobile phase A (H2O with 0.05% formic acid) and mobile phase B (95% acetonitrile–5% H2O with 0.05% formic acid) were used for all separations. The elution procedure consisted of an isocratic profile of 15 min of 20% mobile phase B in mobile phase A and a linear gradient from 20 to 100% mobile phase B in mobile phase A over 1 min, followed by an isocratic profile of 100% mobile phase B for 4 min with a flow rate of 0.30 ml min−1. Electrospray ionization in the positive mode was performed by using the turbo-ion spray source with an ion spray voltage of 4,500 V, a desolvation temperature of 300°C, and gas 1 and gas 2 set at 20 and 30, respectively. Mass spectra were collected over the range 200 to 300 m/z with a declustering potential of 22 V, a focusing potential of 400 V, and an entrance potential of 5 V. The DMB–α-KG derivative was detected by ion extraction of the [M + H]+ m/z of 262.9 to 263.5. All traces were smoothed by using a Savitzky-Golay filter, and the area under the peak was determined by using Analyst classic parameters from the Applied Biosystems software (version 1.5). Concentrations reported for α-KG represent mean values (± standard deviations [SD]).
PA5530-lacZ expression experiments.
Each treatment was tested in triplicate, and galactosidase (LacZ) activity was measured by using the Miller assay as described previously (23). Recombinant P. aeruginosa and E. coli strains harboring pBRL485 were grown in nutrient broth no. 2 (Oxoid) at 37°C at 200 rpm. When the cells reached an OD600 of ∼0.3, exogenous substrates were added to a final concentration of 0.02 to 20 mM, and LacZ activity was assayed at either 30 or 90 min postinduction.
Microarray analysis.
Microarray experiments were done in triplicate. P. aeruginosa PAO1 was grown in nutrient broth no. 2 (Oxoid) at 37°C at 200 rpm to an OD600 of 0.3. α-KG was then added to the cultures at a final concentration of either 0 or 20 mM. At 30 min postinduction, an aliquot of culture (1.0 ml) was mixed with 0.5 ml RNAprotect Bacteria reagent (Qiagen). The bacteria in the treated samples were lysed by using an enzymatic proteinase K digestion approach, as detailed in the RNAprotect Bacteria reagent handbook. The RNA was purified by using the RNeasy minikit (Qiagen) with an on-column DNase digestion step. PCR and a Bioanalyzer (Agilent Technologies) were used to check the purified RNA samples for DNA contamination and overall quality. Microarray studies were performed by the Microarray Core Facility (Upstate Medical University, Syracuse, NY) by using P. aeruginosa PAO1 Affymetrix GeneChip arrays. Methods for conducting the microarray experiments, including data processing and statistical analysis, were done exactly as previously described (23).
Microarray data accession number.
Data from the microarray experiment were deposited in the Gene Expression Omnibus (GEO) database (33) under accession no. GSE54032.
RESULTS
The PA5530 gene is required for growth of P. aeruginosa on C5-dicarboxylates.
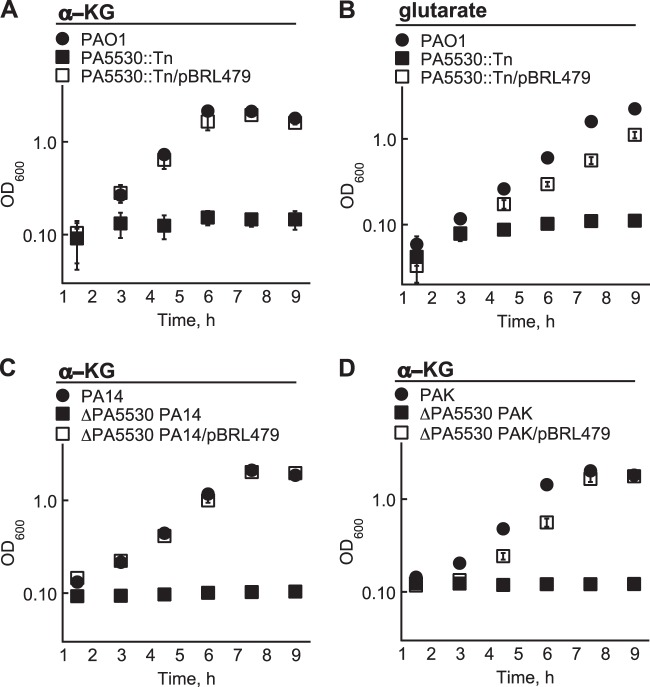
The protein responsible for α-KG transport in P. aeruginosa PAO1 was expected to possess significant homology to the α-KG:H+ symporter (KgtP) of E. coli K-12 MG1655. After performing a BLASTP search against the protein database of P. aeruginosa PAO1, the putative PA5530 protein was found to have 70% homology (55% identity) to KgtP of E. coli K-12. Consistent with the idea that the PA5530 protein functions as an α-KG transporter, a P. aeruginosa PAO1 strain harboring a transposon insertion within the PA5530 gene could not utilize α-KG as a carbon source (Fig. 1A). Additionally, the PA5530 mutant did not grow on glutarate-minimal medium (Fig. 1B), showing that the assimilation of C5-dicarboxylates converges on the PA5530 gene. Growth on C5-dicarboxylates was restored in the PA5530 mutant when the PA5530 gene was expressed from the lac promoter on the broad-host-range plasmid pBBR1MCS-5.
FIG 1.
The PA5530 gene is required for the growth of P. aeruginosa on C5-dicarboxylates. (A and B) A PA5530 transposon mutant (PA5530::Tn) of P. aeruginosa PAO1 did not grow in M9 minimal medium when the sole carbon source was 20 mM α-KG (A) or glutarate (B). (C and D) α-KG was also not a viable carbon source for ΔPA5530 derivatives of P. aeruginosa PA14 (C) and PAK (D). Growth on C5-dicarboxylates was recovered for all strains when the PA5530 gene was expressed from the lac promoter of pBBR1MCS-5 (pBRL479). Data points represent mean values ± SD (n = 3).
The PA5530 gene was also required for the growth of P. aeruginosa strains PA14 and PAK on α-KG. The PA5530 gene was deleted in P. aeruginosa strains PA14 and PAK, and the resulting ΔPA5530 P. aeruginosa mutants were assayed for growth on α-KG. As expected, neither ΔPA5530 mutant grew on α-KG unless the PA5530 gene was heterologously expressed from pBBR1MCS-5 (Fig. 1). The necessity of the PA5530 gene for α-KG assimilation appears to be a shared trait of Pseudomonas aeruginosa strains PAO1, PA14, and PAK.
We next confirmed that the PA5530 gene is required for α-KG uptake in P. aeruginosa PAO1. P. aeruginosa PAO1 and the PA5530 mutant were grown in nutrient broth no. 2 (Oxoid), and the extracellular α-KG concentrations were measured at cell densities (OD600) of 0.2, 0.4, 0.8, and 1.6. At an OD600 of 0.2, P. aeruginosa PAO1 and the PA5530 mutant had comparable extracellular α-KG concentrations of 240 (±38) μM and 384 (±32) μM, respectively. By the next doubling (OD600 of 0.4), extracellular α-KG concentrations decreased to <20 μM (lower detection limit) for P. aeruginosa PAO1 but increased to 898 (±122) μM for the PA5530 mutant. Interestingly, P. aeruginosa PAO1 at an OD600 of 0.8 accumulated extracellular α-KG to a concentration of 88 (±24) μM, which decreased to <20 μM at an OD600 of 1.6. In contrast, the PA5530 mutant exhibited extracellular α-KG concentrations of 868 (±72) μM and 1,134 (±48) μM at OD600 values of 0.8 and 1.6, respectively. The elevated extracellular α-KG concentration observed for the PA5530 mutant is a characteristic also found in a ΔkgtP E. coli strain (12) and shows that the disruption of the PA5530 gene hinders the uptake of α-KG in P. aeruginosa PAO1. It should be noted that the α-KG content of nutrient broth no. 2 (Oxoid) was found to be <20 μM. Therefore, the measured extracellular α-KG concentration was a product of Pseudomonas metabolism and not an artifact of nutrient broth no. 2 (Oxoid).
Expression of the PA5530 gene is regulated by extracellular α-KG.
Previous biochemical studies have shown the existence of an inducible α-KG transporter in Pseudomonas (21). To determine if the expression of the PA5530 gene is induced by the presence of extracellular C5-dicarboxylates, the 1,069-bp 5′-regulatory region immediately upstream of the PA5530 start codon was fused to a DNA segment encoding the β-galactosidase (lacZ) ORF of E. coli K-12 MG1655. The resulting PA5530-lacZ fusion was cloned into a promoterless plasmid, ΔPlac-pBBR1MCS-5, and subsequently electroporated into P. aeruginosa PAO1. P. aeruginosa PAO1 harboring the PA5530-lacZ reporter was grown in nutrient broth no. 2 (Oxoid) to an OD600 of 0.3, at which time various organic acids were added to a final concentration of 20 mM. As shown in Fig. 2A, expression of PA5530-lacZ increased by 330% (±28%) within 90 min of the addition of α-KG. Glutarate also induced the expression of PA5530-lacZ by 224% (±8.0%). Of the remaining carbon sources tested, only succinate and citrate were found to have an inducing effect, increasing PA5530-lacZ expression levels by ∼50%. These data confirm that expression of the PA5530 gene is strongly induced in the presence of extracellular C5-dicarboxylates, with preference toward α-KG. For example, the PA5530-lacZ expression level increased by 48% (±22%) within 30 min after the addition of α-KG to a final concentration of 20 μM (Fig. 2B).
FIG 2.
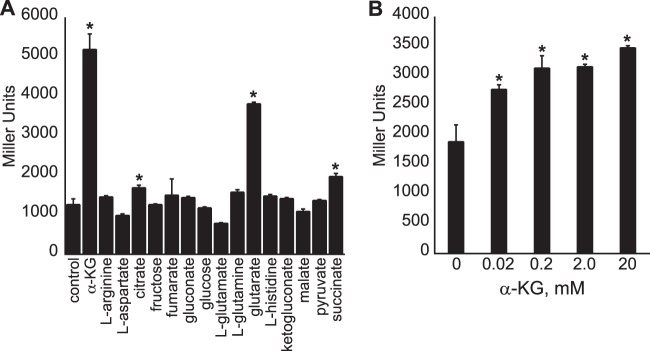
Expression of a PA5530-lacZ reporter is induced by extracellular C5-dicarboxylates. The 5′ 1,069-bp regulatory region upstream of the PA5530 ORF was fused to lacZ, and the resulting PA5530-lacZ reporter was introduced into P. aeruginosa PAO1. Recombinant cells were grown in nutrient broth no. 2 (Oxoid) to an OD600 of 0.3 and subsequently induced with various organic compounds provided at a final concentration of 20 mM. (A) At 90 min postinduction, LacZ activity was highest for cells treated with α-KG or glutarate. (B) The expression level of PA5530-lacZ increased within 30 min after the addition of α-KG to a final concentration of 0.02 to 20 mM. Data points represent mean values ± SD (n = 3). Analysis of variance was performed by using Dunnett's post hoc test (α-value of 0.05) to identify significant changes (P < 0.0001), which are marked (asterisks).
The data from the PA5530-lacZ reporter experiments showed that the expression of the PA5530 gene is induced by extracellular α-KG; however, the number and range of genes whose expression is regulated by α-KG were not known. We therefore determined the transcriptional response of P. aeruginosa PAO1 to extracellular α-KG using Affymetrix GeneChips. In triplicate, P. aeruginosa PAO1 cells were grown in nutrient broth no. 2 (Oxoid) to an OD600 of 0.3 and then treated with either 0 or 20 mM α-KG. At 30 min postinduction, cells were harvested, and total RNA was isolated. Following microarray and statistical analyses, a total of four genes displayed changes in transcript levels by >2-fold with the addition of exogenous α-KG.
The changes in transcript levels for three of the four genes were near the 2-fold cutoff: PA0865 (−2.8 ± 0.7), PA4131 (−2.5 ± 0.3), and PA5170 (3.7 ± 1.6). The PA4131 gene encodes a putative iron-sulfur protein and has no known function. The physiological roles of the PA0865 (hpd) and PA5170 (arcD) genes were previously analyzed (34, 35), and their known functions cannot be readily extrapolated to α-KG metabolism. Because of this uncertainty and the slight changes observed in their transcript levels, these genes are not considered to directly participate in α-KG assimilation. Expectedly, the transcript level of the PA5530 gene increased by 17-fold (±5.0-fold) with the addition of exogenous α-KG. The microarray results reinforce the findings of the PA5530-lacZ experiments, and collectively, they show that α-KG induces the expression of the PA5530 gene. The inducible expression of the PA5530 gene is a behavior reminiscent of other bacterial transporter genes and argues that the PA5530 gene most likely encodes the α-KG transporter in P. aeruginosa PAO1.
Expression of the PA5530 gene is dependent on RpoN.
The sigma factor RpoN recognizes a −24/−12 promoter with the consensus sequence 5′-TGGCACG-N4-TTGCW-3′ (W is A/T) (36). Located 92 bp upstream of the predicted start codon of the PA5530 ORF is a putative RpoN promoter, 5′-TGGCACG-N4-CTGCT-3′. To determine if this predicted RpoN promoter is functional, the highly conserved “GG” motif of the −24 element was changed to “AA” in the PA5530-lacZ reporter. This mutation in the RpoN promoter of the PA5530-lacZ reporter reduced its expression by >95% in P. aeruginosa PAO1 (Fig. 3); this finding is consistent with the “GG” motif being a key determinant of RpoN-mediated transcription. Furthermore, there was a >95% decrease in the expression level of the PA5530-lacZ reporter in an rpoN mutant compared to wild-type P. aeruginosa PAO1 (Fig. 3). The addition of exogenous α-KG did not change the expression of PA5530-lacZ in the rpoN mutant. These results support a model in which RpoN and its cognate −24/−12 promoter direct the transcription of the PA5530 gene in P. aeruginosa PAO1.
FIG 3.
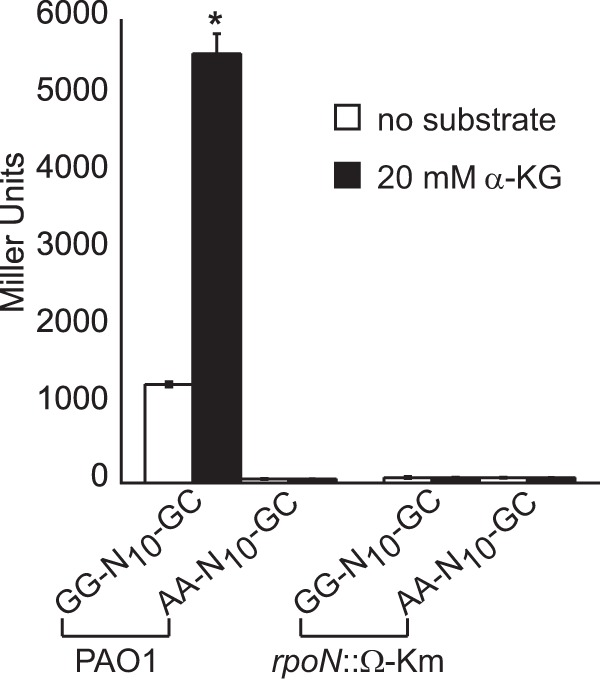
RpoN is essential for the expression of the PA5530-lacZ reporter. Positioned 93 bp upstream of the PA5530 ORF is a sequence resembling a −24/−12 promoter (GG-N10-GC). Replacement of the conserved “GG” dinucleotide of the −24 element with “AA” in the PA5530-lacZ reporter (denoted AA-N10-GC) reduced its expression by >95% in P. aeruginosa PAO1. Additionally, expression of the PA5530-lacZ reporter was null in an rpoN::Ω-Km derivative of P. aeruginosa PAO1. Note that all strains were grown in nutrient broth no. 2 (Oxoid) to an OD600 of 0.3 and then induced with 20 mM α-KG. LacZ activity was measured at 90 min postinduction. Data points represent mean values ± SD (n = 3). Analysis of variance was performed by using Dunnett's post hoc test (α-value of 0.05) to identify significant changes (P < 0.0001), which are marked (asterisk).
The mifR gene is required for expression of the PA5530 gene in P. aeruginosa PAO1.
RpoN requires additional regulators, known as enhancer binding proteins (EBPs), to activate the transcription of targeted genes (37). Having established that RpoN is completely necessary for the expression of the PA5530 gene, we next sought to determine the EBP that participates with RpoN in this process. To this end, we scanned the chromosomal region surrounding the PA5530 locus to identify genes encoding known or hypothetical EBPs. In the vicinity of the PA5530 locus is the mifR (PA5511) gene. The EBP MifR has been observed to be required for microcolony formation (22, 38), but it also possesses significant homology (>60%) to the DctD family of response regulators. DctD response regulators are EBPs that activate the transcription of genes whose products participate in the transport of dicarboxylates from the environment (39, 40). For P. aeruginosa PAO1, DctD (PA5166) regulates the transport of C4-dicarboxylates such as fumarate, malate, and succinate (41). We therefore wondered if MifR might be a DctD homolog that regulates the transport of C5-dicarboxylates in P. aeruginosa PAO1.
In support of MifR having a role in the utilization of C5-dicarboxylates, a ΔmifR mutant of P. aeruginosa PAO1 could not use α-KG or glutarate as a growth substrate (Fig. 4). Expression of the PA5530 gene from plasmid pBBR1MCS-5 rescued the growth of the ΔmifR mutant on both of these C5-dicarboxylates. This finding suggested that the diminished capacity of the ΔmifR mutant to grow on C5-dicarboxylates might be a result of insufficient expression of the PA5530 gene. This suspicion was confirmed when it was observed that the expression of the PA5530-lacZ reporter was reduced by >95% in the ΔmifR mutant compared to wild-type P. aeruginosa PAO1 (Fig. 5). We also found that the dctD gene had no impact on the expression of PA5530-lacZ; α-KG induced the expression of PA5530-lacZ in a dctD mutant to levels observed in wild-type cells (Fig. 5). As expected, deletion of the mifR gene in the dctD mutant abolished the expression of PA5530-lacZ. The inability of the ΔmifR mutant to grow on C5-dicarboxylates or express the PA5530-lacZ reporter gives credibility to the idea that the primary function of MifR is the regulation of C5-dicarboxylate transport in P. aeruginosa PAO1.
FIG 4.
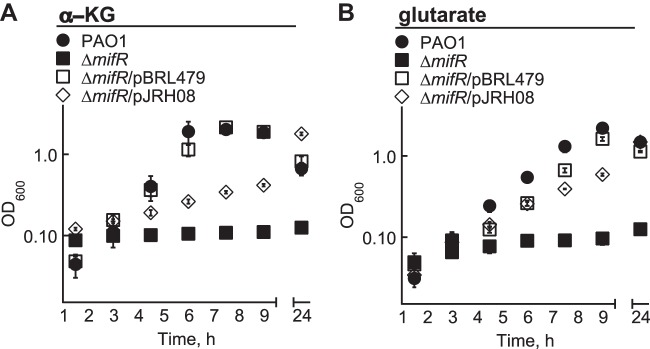
Heterologous expression of the PA5530 gene rescues the growth of a ΔmifR mutant of P. aeruginosa PAO1 on C5-dicarboxylates. A ΔmifR mutant of P. aeruginosa PAO1 could not grow in M9 minimal medium with either 20 mM α-KG (A) or glutarate (B) as the sole carbon source. Expression of the PA5530 gene from the lac promoter of pBBR1MCS-5 (pBRL479) restored the growth of the ΔmifR mutant on these C5-dicarboxylates. There was partial complementation of the ΔmifR mutant with pJRH08 (mifR gene in pBBR1MCS-5). Full complementation most likely requires optimizing the expression of the mifR gene in plasmid pJRH08 (for an example, see reference 34). Data points represent mean values ± SD (n = 3).
FIG 5.
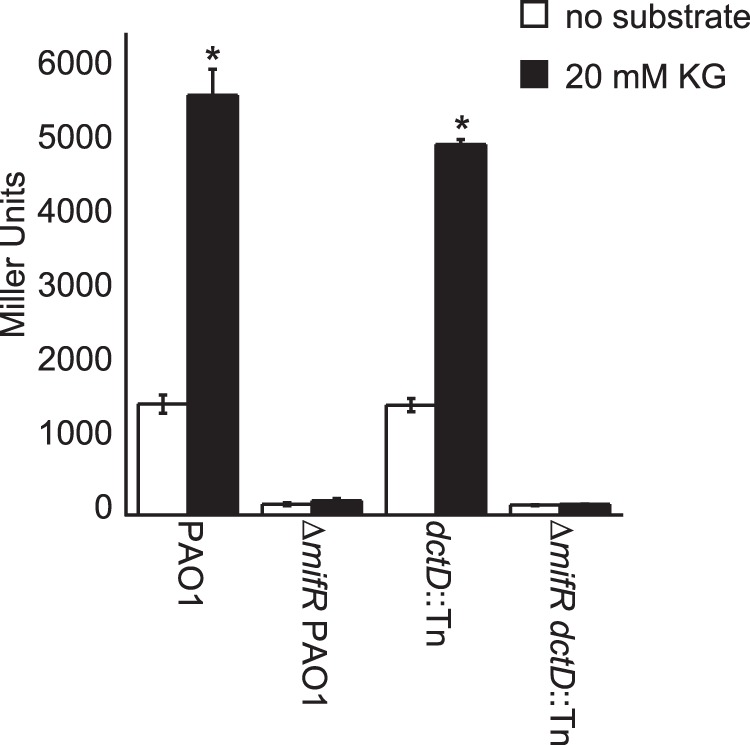
Expression of the PA5530-lacZ reporter is dependent on the mifR gene. Expression of the PA5530-lacZ reporter was reduced by >95% in a ΔmifR mutant of P. aeruginosa PAO1 compared to that observed in wild-type cells. Cells harboring a dctD transposon insertion (dctD::Tn) displayed wild-type levels of LacZ activity. However, deletion of the mifR gene in dctD::Tn cells eliminated the expression of PA5530-lacZ. These findings suggest that MifR regulates C5-dicarboxylate transport, whereas DctD is involved in the transport of C4-dicarboxylates. Note that all strains were grown in nutrient broth no. 2 (Oxoid) to an OD600 of 0.3 and then induced with 20 mM α-KG. LacZ activity was measured at 90 min postinduction. Data points represent mean values ± SD (n = 3). Analysis of variance was performed by using Dunnett's post hoc test (α-value of 0.05) to identify significant changes (P < 0.0001), which are marked (asterisks).
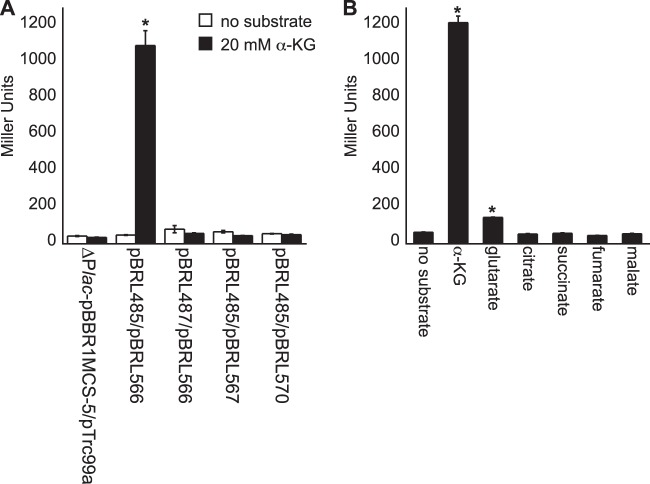
The response regulator gene mifR is in a putative operon with its predicted sensor kinase partner gene mifS. Sensing and responding to extracellular α-KG are most likely mediated by the MifSR two-component signal transduction system (TCS) in P. aeruginosa PAO1. To test this hypothesis, the mifSR genes were heterologously expressed in E. coli Top10 cells that also harbored the PA5530-lacZ reporter. As shown in Fig. 6, the basal expression of the mifSR operon from the trc promoter of plasmid pTrc99a allowed E. coli Top10 to express the PA5530-lacZ reporter when challenged with external α-KG. Specifically, LacZ activity increased by >1,000% within 120 min of the addition of 20 mM α-KG. This induction was dependent on the mifR gene, because E. coli Top10 harboring (i) a mifS-ΔmifR operon or (ii) a mifSR operon in the backward orientation of the trc promoter of pTrc99a failed to express the PA5530-lacZ reporter in the presence of extracellular α-KG. Glutarate induced the expression of the PA5530-lacZ reporter by 125% (±10%), whereas C4-dicarboxylates had no effect on PA5530-lacZ expression. These findings show that the mifSR genes are sufficient for activating the expression of the PA5530-lacZ reporter in nonnative E. coli and thus suggest that MifR is a direct regulator of the PA5530 gene.
FIG 6.
α-KG induces expression of the PA5530-lacZ reporter in nonnative E. coli only when the mifSR genes are heterologously expressed. (A) E. coli Top10 cells that basally expressed the mifSR operon from the trc promoter of pTrc99a (pBRL566) responded to extracellular α-KG via increasing the expression level of the PA5530-lacZ reporter by >1,000% compared to the level in noninduced cells. E. coli Top10 cells expressing nonfunctional mifSR alternatives, including a mifSR operon oriented in the opposite direction of the trc promoter (pBRL567) or a mifS-ΔmifR operon (pBRL570), did not express the PA5530-lacZ reporter. Furthermore, the mifSR operon did not cause a change in the expression of the PA5530-lacZ reporter in which the “GG” dinucleotide of the −24 element of the putative RpoN promoter was replaced with “AA” (pBRL487). (B) Lastly, expression of PA5530-lacZ in E. coli/pBRL566 was induced by extracellular C5-dicarboxylates. These results show that the mifSR genes can directly regulate the expression of the PA5530 gene in response to extracellular α-KG. For all experiments, cells were grown in nutrient broth no. 2 (Oxoid) to an OD600 of 0.3 and then induced with 20 mM substrate. LacZ activity was measured at 120 min postinduction. Data points represent mean values ± SD (n = 3). Analysis of variance was performed by using Dunnett's post hoc test (α-value of 0.05) to identify significant changes (P < 0.0001), which are marked (asterisks).
DISCUSSION
The main objective of this study was to determine the gene encoding the α-KG transporter in P. aeruginosa PAO1. Based on homology to the prototypic α-KG transporter KgtP, we identified two genes, PA0229 (pcaT) and PA5530, which might encode the α-KG transporter. PA0229 or PcaT is annotated as a putative α-KG transporter in the Pseudomonas Genome Database (42), but PcaT proteins function in the transport of β-ketoadipate (43), which is a C6-dicarboxylate. In P. aeruginosa PAO1, the pcaT gene is part of the predicted pcaTBCD operon, whose products participate in the breakdown of aromatic compounds, i.e., the protocatechuate branch of the β-ketoadipate pathway (44). Consequently, PcaT was considered to be a transporter for β-ketoadipate and not α-KG. This made the PA5530 gene a favorable candidate for encoding the α-KG transporter in P. aeruginosa PAO1.
A transposon insertion within the PA5530 gene in P. aeruginosa PAO1 prevented this bacterium from using C5-dicarboxylates as growth substrates. Follow-up experiments showed that the disruption of the PA5530 gene caused a significant accumulation of extracellular α-KG. For example, both wild-type P. aeruginosa PAO1 and a PA5530 mutant produced ∼200 μM extracellular α-KG within 90 min of being inoculated into a rich medium. Whereas wild-type cells were able to assimilate this extracellular α-KG, lowering its levels below 20 μM, the PA5530 mutant continued to accumulate extracellular α-KG, which eventually reached concentrations of >1.0 mM. This result is consistent with the PA5530 gene functioning in α-KG transport, but we did not anticipate that the levels of extracellular α-KG would be in the millimolar range. The generation and subsequent uptake of extracellular α-KG are well-known characteristics of P. aeruginosa cultures. However, the magnitude or significance of this activity when P. aeruginosa is grown in a rich medium has not been reported previously. The metabolic process responsible for the continuous production of extracellular α-KG is a topic worth exploring.
α-KG did induce the expression of the PA5530 gene, as determined by both microarray and lacZ reporter experiments. In fact, the exogenous addition of 20 μM α-KG was sufficient to stimulate the expression of the PA5530 gene. Expression of the PA5530 gene was also induced by extracellular glutarate, reaffirming that the PA5530 gene has all the elements associated with an inducible C5-dicarboxylate transporter. Namely, the PA5530 gene (i) encodes a putative KgtP homolog, (ii) was essential for the utilization of C5-dicarboxylates, (iii) was required for the uptake of extracellular α-KG, and (iv) was genetically regulated by extracellular C5-dicarboxylates.
Upstream of the PA5530 ORF is a −24/−12 promoter recognized by the sigma factor RpoN. We did not find any sequence resembling a σ70-type promoter within the vicinity of the predicted start codon of the PA5530 ORF. The changing of the highly conserved “GG” dinucleotide of the −24 element to “AA” completely eliminated the expression of the PA5530 gene. Additionally, the PA5530 gene was not expressed in an rpoN-deficient derivative of P. aeruginosa PAO1. Both of these results suggest that RpoN is the sigma factor responsible for the expression of the PA5530 gene. The genetic regulation of C5-dicarboxylate transport by RpoN might be a common mechanism employed by Pseudomonas, because the rpoN gene was also required for the growth of P. putida on α-KG (45).
One of the most interesting findings of this study was the requirement of the mifR gene for the assimilation of C5-dicarboxylates in P. aeruginosa PAO1. The response regulator MifR (PA5511) and its partner sensor kinase MifS (PA5512) comprise a TCS that was initially found to be essential for the development of mature biofilms (22). Later studies showed that in the absence of the mifR gene, pyruvate fermentation was suboptimal and thus unable to support the formation of microcolonies (38). In our study, a ΔmifR mutant could not utilize C5-dicarboxylates due to a significant reduction (>95%) in the expression of the PA5530 gene.
A reasonable explanation as to why the mifR gene was required for the expression of the PA5530 gene was found when we closely examined the amino acid sequence of the MifR protein. Like all other enhancer binding proteins, MifR has the conserved RpoN interaction domain, but more importantly, MifR is 70% homologous (54% identical) to the response regulator DctD (PA5166) of P. aeruginosa PAO1. The presence of dual DctD regulators (commonly annotated DctD1 and DctD2) has been reported for the sequenced genomes of several Pseudomonas-related bacteria, including Ralstonia solanacearum, Azotobacter vinelandii, Pseudomonas protegens, and Burkholderia mallei (NCBI Genome Database). The DctBD TCS of P. aeruginosa PAO1 is involved in the assimilation of C4-dicarboxylates (41), and we found that the dctD gene had no effect on the expression of the PA5530 gene. It is plausible that MifS and MifR function as a DctBD-type TCS that responds to extracellular C5-dicarboxylates via activating the expression of the PA5530 gene. Some bacteria are known to use a TCS, e.g., KguSR (10) and KgtSR (8), to regulate α-KG assimilation, and we found that the heterologous expression of the mifSR genes in E. coli was sufficient to activate the expression of the PA5530-lacZ reporter in response to extracellular α-KG. Biochemical evidence is needed to verify that MifR binds to and regulates the expression of the PA5530 gene.
Lastly, it will be valuable to determine if the PA5530 gene or α-KG has any role in biofilm development in P. aeruginosa PAO1. Cells within the interior of matured biofilms are considered to be in an anaerobic environment (46, 47), and recently, pathogenic E. coli strain CTF073 was reported to preferentially assimilate α-KG as a carbon source under anaerobic growth conditions (10). It is possible that MifR is required for the anaerobic metabolism of α-KG in P. aeruginosa PAO1. A recent transcriptomic study found that the transcript levels of several genes whose products are involved in anaerobic metabolism were deregulated in a ΔmifR mutant of P. aeruginosa PAO1 (38). We have now just begun to analyze α-KG utilization for anaerobically grown P. aeruginosa PAO1, including its ΔPA5530 and ΔmifR derivatives.
ACKNOWLEDGMENTS
We acknowledge NIH grant P30 DK089507 for funding the P. aeruginosa PAO1 transposon mutant used for our study. This study was made possible by NIH R15 GM104880-01A1 and NSF CBET 1263905 awards to C.T.N.
Footnotes
Published ahead of print 2 May 2014
REFERENCES
- 1.Kamberov ES, Atkinson MR, Ninfa AJ. 1995. The Escherichia coli PII signal transduction protein is activated upon binding 2-ketoglutarate and ATP. J. Biol. Chem. 270:17797–17807. 10.1074/jbc.270.30.17797 [DOI] [PubMed] [Google Scholar]
- 2.Jiang P, Peliska JA, Ninfa AJ. 1998. The regulation of Escherichia coli glutamine synthetase revisited: role of 2-ketoglutarate in the regulation of glutamine synthetase adenylylation state. Biochemistry 37:12802–12810. 10.1021/bi980666u [DOI] [PubMed] [Google Scholar]
- 3.Forchhammer K. 2004. Global carbon/nitrogen control by PII signal transduction in cyanobacteria: from signals to targets. FEMS Microbiol. Rev. 28:319–333. 10.1016/j.femsre.2003.11.001 [DOI] [PubMed] [Google Scholar]
- 4.Mailloux RJ, Singh R, Brewer G, Auger C, Lemire J, Appanna VD. 2009. Alpha-ketoglutarate dehydrogenase and glutamate dehydrogenase work in tandem to modulate the antioxidant alpha-ketoglutarate during oxidative stress in Pseudomonas fluorescens. J. Bacteriol. 191:3804–3810. 10.1128/JB.00046-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lemire J, Milandu Y, Auger C, Bignucolo A, Appanna VP, Appanna VD. 2010. Histidine is a source of the antioxidant, alpha-ketoglutarate, in Pseudomonas fluorescens challenged by oxidative stress. FEMS Microbiol. Lett. 309:170–177. 10.1111/j.1574-6968.2010.02034.x [DOI] [PubMed] [Google Scholar]
- 6.Kunz DA, Chen JL, Pan G. 1998. Accumulation of alpha-keto acids as essential components in cyanide assimilation by Pseudomonas fluorescens NCIMB 11764. Appl. Environ. Microbiol. 64:4452–4459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hausinger RP. 2004. FeII/alpha-ketoglutarate-dependent hydroxylases and related enzymes. Crit. Rev. Biochem. Mol. Biol. 39:21–68. 10.1080/10409230490440541 [DOI] [PubMed] [Google Scholar]
- 8.Batista S, Patriarca EJ, Tatè R, Martinez-Drets G, Gill PR. 2009. An alternative succinate (2-oxoglutarate) transport system in Rhizobium tropici is induced in nodules of Phaseolus vulgaris. J. Bacteriol. 191:5057–5067. 10.1128/JB.00252-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo W, Cai LL, Zou HS, Ma WX, Liu XL, Zou LF, Li YR, Chen XB, Chen GY. 2012. Ketoglutarate transport protein KgtP is secreted through the type III secretion system and contributes to virulence in Xanthomonas oryzae pv. oryzae. Appl. Environ. Microbiol. 78:5672–5681. 10.1128/AEM.07997-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai W, Wannemuehler Y, Dell'anna G, Nicholson B, Barbieri NL, Kariyawasam S, Feng Y, Logue CM, Nolan LK, Li G. 2013. A novel two-component signaling system facilitates uropathogenic Escherichia coli's ability to exploit abundant host metabolites. PLoS Pathog. 9:e1003428. 10.1371/journal.ppat.1003428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seol W, Shatkin AJ. 1991. Escherichia coli kgtP encodes an alpha-ketoglutarate transporter. Proc. Natl. Acad. Sci. U. S. A. 88:3802–3806. 10.1073/pnas.88.9.3802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan D, Lenz P, Hwa T. 2011. Overcoming fluctuation and leakage problems in the quantification of intracellular 2-oxoglutarate levels in Escherichia coli. Appl. Environ. Microbiol. 77:6763–6771. 10.1128/AEM.05257-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tynecka Z, Korona-Glowniak I, Loœ R. 2001. 2-Oxoglutarate transport system in Staphylococcus aureus. Arch. Microbiol. 176:143–150. 10.1007/s002030100306 [DOI] [PubMed] [Google Scholar]
- 14.Pajor AM, Sun NN, Leung A. 2013. Functional characterization of SdcF from Bacillus licheniformis, a homolog of the SLC13 Na(+)/dicarboxylate transporters. J. Membr. Biol. 246:705–715. 10.1007/s00232-013-9590-3 [DOI] [PubMed] [Google Scholar]
- 15.Pudlik AM, Lolkema JS. 2013. Uptake of alpha-ketoglutarate by citrate transporter CitP drives transamination in Lactococcus lactis. Appl. Environ. Microbiol. 79:1095–1101. 10.1128/AEM.02254-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Von Tigerstrom M, Campbell JJ. 1966. The accumulation of alpha-ketoglutarate by suspensions of Pseudomonas aeruginosa. Can. J. Microbiol. 12:1005–1013. 10.1139/m66-135 [DOI] [PubMed] [Google Scholar]
- 17.Koepsell HJ, Stodola FH, Sharpe ES. 1952. Production of α-ketoglutarate in glucose oxidation by Pseudomonas fluorescens. J. Am. Chem. Soc. 74:5142–5144. 10.1021/ja01140a044 [DOI] [Google Scholar]
- 18.Duncan MG, Campbell JJ. 1962. Oxidative assimilation of glucose by Pseudomonas aeruginosa. J. Bacteriol. 84:784–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campbell JJ, Stokes FN. 1951. Tricarboxylic acid cycle in Pseudomonas aeruginosa. J. Biol. Chem. 190:853–858 [PubMed] [Google Scholar]
- 20.Clarke PH, Meadow PM. 1959. Evidence for the occurrence of permeases for tricarboxylic acid cycle intermediates in Pseudomonas aeruginosa. J. Gen. Microbiol. 20:144–155. 10.1099/00221287-20-1-144 [DOI] [PubMed] [Google Scholar]
- 21.Edwards WV, Sando JJ, Hartline RA. 1979. Transport of C5 dicarboxylate compounds by Pseudomonas putida. J. Bacteriol. 139:748–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petrova OE, Sauer K. 2009. A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. PLoS Pathog. 5:e1000668. 10.1371/journal.ppat.1000668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lundgren BR, Thornton W, Dornan MH, Villegas-Peñaranda LR, Boddy CN, Nomura CT. 2013. Gene PA2449 is essential for glycine metabolism and pyocyanin biosynthesis in Pseudomonas aeruginosa PAO1. J. Bacteriol. 195:2087–2100. 10.1128/JB.02205-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, Chun-Rong L, Guenthner D, Bovee D, Olson MV, Manoil C. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 100:14339–14344. 10.1073/pnas.2036282100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heurlier K, Dénervaud V, Pessi G, Reimmann C, Haas D. 2003. Negative control of quorum sensing by RpoN (sigma54) in Pseudomonas aeruginosa PAO1. J. Bacteriol. 185:2227–2235. 10.1128/JB.185.7.2227-2235.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price-Whelan A, Dietrich LE, Newman DK. 2007. Pyocyanin alters redox homeostasis and carbon flux through central metabolic pathways in Pseudomonas aeruginosa PA14. J. Bacteriol. 189:6372–6381. 10.1128/JB.00505-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sadovskaya I, Vinogradov E, Li J, Hachani A, Kowalska K, Filloux A. 2010. High-level antibiotic resistance in Pseudomonas aeruginosa biofilm: the ndvB gene is involved in the production of highly glycerol-phosphorylated beta-(1→3)-glucans, which bind aminoglycosides. Glycobiology 20:895–904. 10.1093/glycob/cwq047 [DOI] [PubMed] [Google Scholar]
- 28.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, II, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. 10.1016/0378-1119(95)00584-1 [DOI] [PubMed] [Google Scholar]
- 29.Choi KH, Schweizer HP. 2005. An improved method for rapid generation of unmarked Pseudomonas aeruginosa deletion mutants. BMC Microbiol. 5:30. 10.1186/1471-2180-5-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 31.Szewczyk E, Nayak T, Oakley CE, Edgerton H, Xiong Y, Taheri-Talesh N, Osmani SA, Oakley BR. 2007. Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 1:3111–3120. 10.1038/nprot.2006.405 [DOI] [PubMed] [Google Scholar]
- 32.Klein A, Diaz S, Ferreira I, Lamblin G, Roussel P, Manzi AE. 1997. New sialic acids from biological sources identified by a comprehensive and sensitive approach: liquid chromatography-electrospray ionization-mass spectrometry (LC-ESI-MS) of SIA quinoxalinones. Glycobiology 7:421–432. 10.1093/glycob/7.3.421 [DOI] [PubMed] [Google Scholar]
- 33.Edgar R, Domrachev M, Lash AE. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207–210. 10.1093/nar/30.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palmer GC, Palmer KL, Jorth PA, Whiteley M. 2010. Characterization of the Pseudomonas aeruginosa transcriptional response to phenylalanine and tyrosine. J. Bacteriol. 192:2722–2728. 10.1128/JB.00112-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Verhoogt HJ, Smit H, Abee T, Gamper M, Driessen AJ, Haas D, Konings WN. 1992. arcD, the first gene of the arc operon for anaerobic arginine catabolism in Pseudomonas aeruginosa, encodes an arginine-ornithine exchanger. J. Bacteriol. 174:1568–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barrios H, Valderrama B, Morett E. 1999. Compilation and analysis of sigma(54)-dependent promoter sequences. Nucleic Acids Res. 27:4305–4313. 10.1093/nar/27.22.4305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morett E, Segovia L. 1993. The sigma 54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J. Bacteriol. 175:6067–6074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petrova OE, Schurr JR, Schurr MJ, Sauer K. 2012. Microcolony formation by the opportunistic pathogen Pseudomonas aeruginosa requires pyruvate and pyruvate fermentation. Mol. Microbiol. 86:819–835. 10.1111/mmi.12018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Janausch IG, Zientz E, Tran QH, Kröger A, Unden G. 2002. C4-dicarboxylate carriers and sensors in bacteria. Biochim. Biophys. Acta 1553:39–56. 10.1016/S0005-2728(01)00233-X [DOI] [PubMed] [Google Scholar]
- 40.Yurgel SN, Kahn ML. 2004. Dicarboxylate transport by rhizobia. FEMS Microbiol. Rev. 28:489–501. 10.1016/j.femsre.2004.04.002 [DOI] [PubMed] [Google Scholar]
- 41.Valentini M, Storelli N, Lapouge K. 2011. Identification of C4-dicarboxylate transport systems in Pseudomonas aeruginosa PAO1. J. Bacteriol. 193:4307–4316. 10.1128/JB.05074-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winsor GL, Lam DK, Fleming L, Lo R, Whiteside MD, Yu NY, Hancock RE, Brinkman FS. 2011. Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 39:D596–D600. 10.1093/nar/gkq869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ondrako JM, Ornston LN. 1980. Biological distribution and physiological role of the beta-ketoadipate transport system. J. Gen. Microbiol. 120:199–209 [DOI] [PubMed] [Google Scholar]
- 44.Harwood CS, Parales RE. 1996. The beta-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 50:553–590. 10.1146/annurev.micro.50.1.553 [DOI] [PubMed] [Google Scholar]
- 45.Köhler T, Harayama S, Ramos JL, Timmis KN. 1989. Involvement of Pseudomonas putida RpoN sigma factor in regulation of various metabolic functions. J. Bacteriol. 171:4326–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hentzer M, Eberl L, Givskov M. 2005. Transcriptome analysis of Pseudomonas aeruginosa biofilm development: anaerobic respiration and iron limitation. Biofilms 2:37–61. 10.1017/S1479050505001699 [DOI] [Google Scholar]
- 47.Hassett DJ, Cuppoletti J, Trapnell B, Lymar SV, Rowe JJ, Yoon SS, Hilliard GM, Parvatiyar K, Kamani MC, Wozniak DJ, Hwang SH, McDermott TR, Ochsner UA. 2002. Anaerobic metabolism and quorum sensing by Pseudomonas aeruginosa biofilms in chronically infected cystic fibrosis airways: rethinking antibiotic treatment strategies and drug targets. Adv. Drug Deliv. Rev. 54:1425–1443. 10.1016/S0169-409X(02)00152-7 [DOI] [PubMed] [Google Scholar]