Abstract
Sphingomonas sp. strain A1, a Gram-negative bacterium, directly incorporates alginate polysaccharide into the cytoplasm through a periplasmic alginate-binding protein-dependent ATP-binding cassette transporter. The polysaccharide is degraded to monosaccharides via the formation of oligosaccharides by endo- and exotype alginate lyases. The strain A1 proteins for alginate uptake and degradation are encoded in both strands of a genetic cluster in the bacterial genome and inducibly expressed in the presence of alginate. Here we show the function of the alginate-dependent transcription factor AlgO and its mode of action on the genetic cluster and alginate oligosaccharides. A putative gene within the genetic cluster seems to encode a transcription factor-like protein (AlgO). Mutant strain A1 (ΔAlgO mutant) cells with a disrupted algO gene constitutively produced alginate-related proteins. DNA microarray analysis indicated that wild-type cells inducibly transcribed the genetic cluster only in the presence of alginate, while ΔAlgO mutant cells constitutively expressed the genetic cluster. A gel mobility shift assay showed that AlgO binds to the specific intergenic region between algO and algS (algO-algS). Binding of AlgO to the algO-algS intergenic region diminished with increasing alginate oligosaccharides. These results demonstrated a novel alginate-dependent gene expression mechanism. In the absence of alginate, AlgO binds to the algO-algS intergenic region and represses the expression of both strands of the genetic cluster, while in the presence of alginate, AlgO dissociates from the algO-algS intergenic region via binding to alginate oligosaccharides produced through the lyase reaction and subsequently initiates transcription of the genetic cluster. This is the first report on the mechanism by which alginate regulates the expression of the gene cluster.
INTRODUCTION
Alginate is a linear heteropolysaccharide consisting of β-d-mannuronate (M) and the C5 epimer α-l-guluronate (G) (1). Three block regions, i.e., poly-β-d-mannuronate [poly(M)], poly-α-l-guluronate [poly(G)], and heteropolymer [poly(MG)], make up the alginate molecule. Brown seaweeds and certain bacteria are well-known producers of this polysaccharide. Pseudomonas aeruginosa produces extracellular alginate-containing biofilms that are involved in the expression of virulence factors during lung infections in cystic fibrosis patients (2, 3). In contrast, alginate produced by brown seaweeds is used as a gelling agent and thickener in food. Alginate is expected to become a potential marine biomass for biofuel production because, distinct from polysaccharides (e.g., starch and cellulose) obtained from terrestrial plants, the alginate abundant in brown seaweeds is readily extracted at a mild alkaline pH and causes no serious competing interests for foodstuffs (4). To achieve improvement of alginate gelling characteristics, removal of bacterial alginate biofilms, or saccharification of alginate for the preparation of a biofuel resource, a large number of alginate-assimilating microbes have been isolated from soil, sea, and wastewater (5).
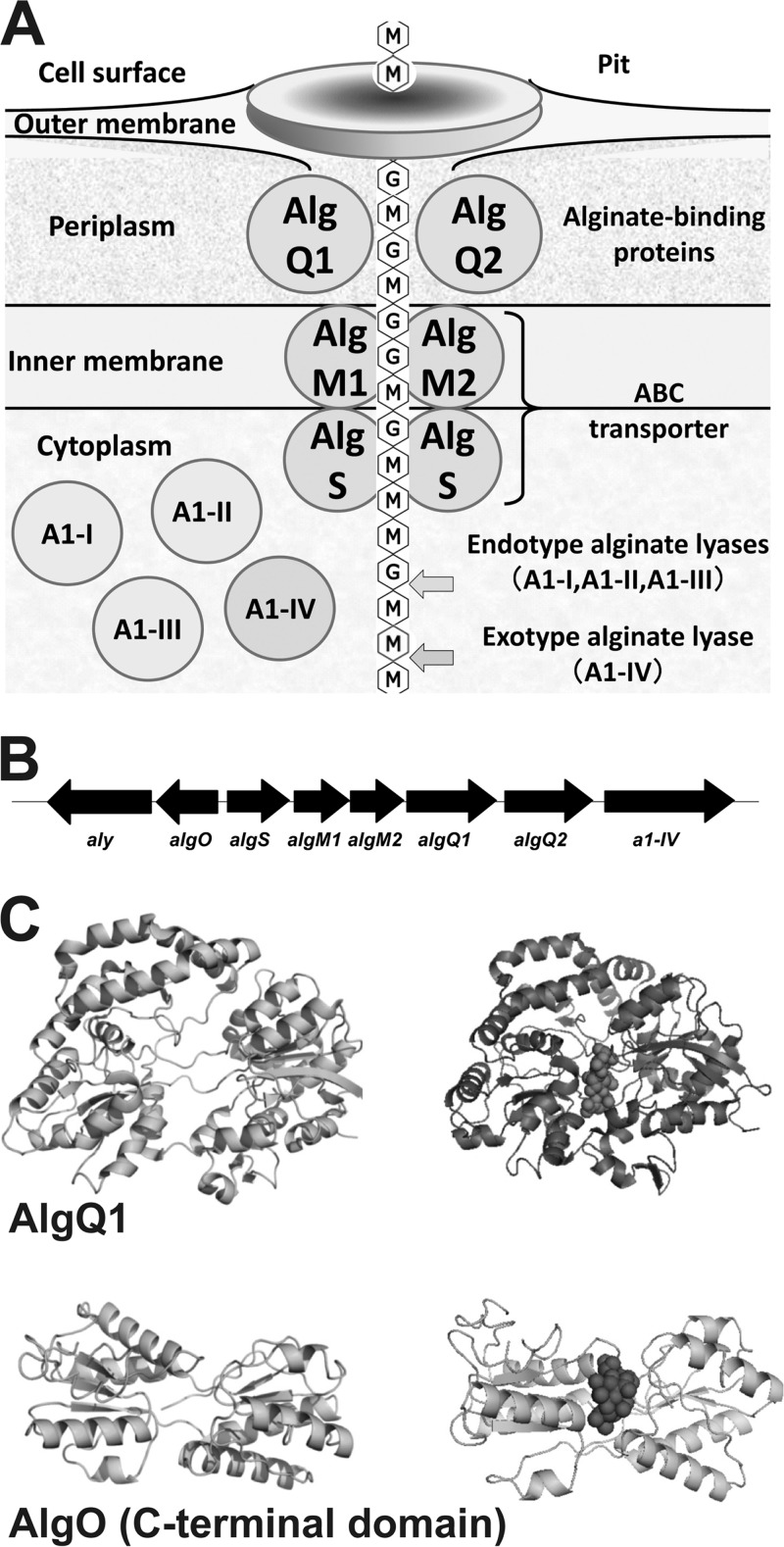
Sphingomonas sp. strain A1 is a Gram-negative, alginate-assimilating bacterium isolated from soil (6). The peculiar biosystem of strain A1 for macromolecule uptake, degradation, and metabolism has been well characterized, and the genes and proteins involved in the biosystem have been identified (Fig. 1A) (7). Strain A1 cells, when grown on alginate, form a mouth-like pit on the cell surface through the reorganization and fluidity of the pleat structures, resulting in the concentration of the external alginate in the pit (8). The concentrated polysaccharide is imported into the periplasm, where the alginate-binding protein AlgQ1 or AlgQ2 is expressed (9). AlgQ1 or AlgQ2 mediates the transfer of alginate from the outer membrane to the inner-membrane-bound ATP-binding cassette (ABC) transporter. A heterodimer of AlgM1 and AlgM2 as a membrane-spanning domain, as well as a homodimer of AlgS, as ATPase constitutes the ABC transporter for alginate uptake (10). The alginate polysaccharide incorporated into the cytoplasm is degraded into its constituent monosaccharides by endotype alginate lyases (A1-I, -II, and -III) and an exotype alginate lyase (A1-IV) (11, 12).
FIG 1.
Alginate-assimilating strain A1. (A) Alginate-uptake and degradation system in strain A1. (B) Alginate genetic cluster. (C) Structural comparison of AlgQ1 and the C-terminal domain of AlgO. Top, AlgQ1. Left, ligand-free AlgQ1; right, Δ3M-bound AlgQ1. Bottom, C-terminal domain of AlgO. Left, homology modeling-based C-terminal domain of AlgO; right, docking simulation of the C-terminal domain of AlgO and Δ3M. Balls represent Δ3M.
Strain A1 has a genetic cluster for alginate uptake and degradation in its genome (Fig. 1B). One (sense) strand of the alginate genetic cluster is composed of genes encoding the ABC transporter (AlgS, AlgM1, and AlgM2), alginate-binding proteins (AlgQ1 and AlgQ2), and exotype alginate lyase (A1-IV). The other (antisense) strand consists of the gene encoding endotype alginate lyase (Aly). aly encodes three types of lyases (A1-I, -II, and -III), and A1-I is autocatalytically processed to generate A1-II and A1-III (6, 13). All of these proteins and enzymes are inducibly expressed in the presence of alginate, and their expression levels are extremely low in the absence of alginate (9, 12). Such alginate-dependent gene expression has been well observed in most alginate-assimilating microbes, including strain A1 (5, 14).
Genetically engineered strain A1 cells produce bioethanol from alginate (15), and subsequently, recombinant ethanologenic Escherichia coli or Saccharomyces cerevisiae, with the addition of multiple genes involved in alginate uptake and assimilation, have been reported to convert brown microalgal sugars to bioethanol (16, 17). Deregulation of alginate-dependent gene expression probably contributes to the development of microbial strains constitutively expressing alginate-related genes. Analysis of the alginate-dependent gene expression mechanism is important for understanding the physiology of alginate-assimilating microbes and for its application in biofuel production from marine biomasses. However, the alginate-dependent gene expression mechanism has not been elucidated. On the other hand, the transcriptional regulation of alginate synthetic genes has been identified in P. aeruginosa (18). The three major transcription factors AlgR, AlgB, and AmrZ regulate the expression of genes involved in alginate biosynthesis by binding to the promoter region of algD (19). AlgR and AlgB are response regulators of a two-component system (20, 21), whereas AmrZ functions as an Arc-like DNA-binding domain (22). While many alginate-related transcription factors have been examined in P. aeruginosa, none have been shown to be directly regulated by the alginate polysaccharide.
This article focuses on the molecular identification of the strain A1 alginate-dependent transcription factor AlgO and its binding to DNA or alginates through gene disruption, DNA microarray, and the gel mobility shift assay.
MATERIALS AND METHODS
Materials.
Eisenia bicyclis sodium alginate, with an average molecular weight of 300,000 was purchased from Nacalai Tesque. Restriction endonucleases and DNA-modifying enzymes were from Toyobo. The oligonucleotides used in this study were synthesized by Hokkaido System Science; for their nucleotide sequences, see Table S1 in the supplemental material. Other analytical-grade chemicals were obtained from commercial sources.
Microorganisms and culture conditions.
To investigate the effect of algO disruption on bacterial gene expression, strain A1 (wild-type [WT] and algO-disruptant [ΔAlgO mutant]) cells were aerobically cultured at 30°C in alginate medium containing 0.1% (NH4)2SO4, 0.1% KH2PO4, 0.1% Na2HPO4, 0.01% MgSO4 · 7H2O, 0.01% yeast extract, and 0.5% sodium alginate (pH 7.2) or yeast extract medium containing 0.1% (NH4)2SO4, 0.1% KH2PO4, 0.1% Na2HPO4, 0.01% MgSO4 · 7H2O, and 0.5% yeast extract (pH 7.2). E. coli strains DH5α (Toyobo) and BL21-Gold(DE3)pLysS (Novagen) were used as hosts for plasmid amplification and recombinant AlgO expression, respectively, and were aerobically cultured at 30 or 37°C in Luria-Bertani (LB) medium consisting of 1% tryptone, 0.5% yeast extract, and 1% NaCl (23). Antibiotics were appropriately added at the following concentrations: sodium ampicillin (Amp), 100 μg/ml; kanamycin sulfate (Km), 50 μg/ml; chloramphenicol (Cm), 25 μg/ml.
DNA cloning.
DNA manipulations such as subcloning, transformation, and gel electrophoresis were performed as described previously (23). Nucleotide sequences of alginate-related genes amplified by PCR were determined by dideoxy chain termination with a 3730xl automated DNA sequencer (Applied Biosystems) (24). Plasmids for disruption of algO were constructed as follows. A DNA fragment containing algO was amplified by PCR with KOD (Toyobo) as DNA polymerase, genomic DNA of strain A1 as the template, and two synthetic oligonucleotides as primers (algO-F and algO-R). The fragment (algO) amplified by PCR was isolated and ligated with HincII-digested pUC119 (TaKaRa Bio). The resultant plasmid containing algO was designated pUC119-algO. A Km resistance-encoding gene cassette for aminoglycoside 3′-phosphotransferase (Kmr) was amplified by PCR with KOD as DNA polymerase, pUC4K (Amersham) as the template, and two synthetic oligonucleotides as primers (Km-F and Km-R). The resultant Km resistance-encoding gene cassette included its own promoter prior to the structural gene. The Kmr gene was introduced at the MscI site of algO in pUC119-algO. The resultant plasmid, with a disruption in algO, was designated pUC119-algO::Kmr. The AlgO::Kmr gene disrupted by insertion of the Kmr gene was amplified by PCR with KOD as DNA polymerase, pUC119-algO::Kmr as the template, and two synthetic oligonucleotides (M13_F and M13_R) with a PvuII site added to the 5′ regions. The resultant PCR product was digested with PvuII and ligated with PvuII-digested pKTY320 (25) containing the gene for β-lactamase (Ampr). The resultant plasmid was designated pKTY320-algO::Kmr and introduced into E. coli strain DH5α. The resultant strain, E. coli DH5α harboring pKTY320-algO::Kmr, was used to transconjugate strain A1 cells through triparental mating in the presence of E. coli strain HB101 harboring pRK2013 (26) as helper cells. The Km-resistant and Amp-sensitive ΔAlgO mutant was selected on 0.5% alginate medium solidified with 1.5% agar containing 0.5% yeast extract and 100 μg/ml Km. algO disruption in the ΔAlgO mutant was confirmed by the algO fragment size and DNA sequencing.
A plasmid for overexpression of AlgO with the addition of six histidine residues (His6) at its C terminus (AlgO-His6) was constructed as follows. To introduce algO into an expression vector, pET21b (Novagen) with the In-Fusion Advantage PCR cloning kit (Clontech), PCR was performed with KOD-Plus-Neo (Toyobo) as DNA polymerase, the genomic DNA of strain A1 as the template, and two synthetic oligonucleotides [algO(pET21b)-F and algO(pET21b)-R] as primers. These oligonucleotides overlap the pET21b cloning site by 15 bp in both 5′ regions. With the In-Fusion Advantage PCR cloning kit, the amplified algO fragment was cloned into the linear pET21b fragment, which was amplified by inverse PCR with pET21b-inv-F and pET21b-inv-R as primers. The resultant plasmid containing algO was designated pET21b-algO-His6 and introduced into BL21-Gold(DE3)pLysS.
Western blotting.
To investigate the expression of the proteins and enzymes involved in alginate uptake and degradation, strain A1 WT and ΔAlgO mutant cells were aerobically cultured at 30°C and 100 strokes/min for 2 days in 0.5% alginate medium or each of three alginate-free medium formulations, i.e., 0.5% yeast extract, 0.5% glucose, or 0.5% glucuronic acid medium, by using reciprocal shakers. Bacterial cells grown in each medium to an optical density at 600 nm (OD600) of 0.4 to 0.6 (i.e., exponential growth phase) were resuspended in 20 mM Tris-HCl (pH 7.5) and lysed with sodium lauryl sulfate (SDS). Resultant cell lysates were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) (27) and transferred to a polyvinylidene difluoride (PVDF) membrane at 1.5 mA/cm2 for 30 min. After blocking with Tris-glycine (pH 7.2) and 0.1% Tween 20 (TBS-T) containing 10% skim milk for 60 min at room temperature, the PVDF membrane was incubated with rabbit anti-AlgS, anti-AlgQ2, anti-AI-IV, or anti-AI-III antibody for 90 min at room temperature. After washes with TBS-T, the PVDF membrane was incubated with donkey anti-rabbit immunoglobulin antibody conjugated to horseradish peroxidase (HRP; GE Healthcare) for 40 min at room temperature. After washes with TBS-T, the PVDF membrane was incubated with the Immobilon Western Chemiluminescent HRP Substrate (Merck Millipore) to visualize immunoreactive proteins. The PVDF membrane was visualized with ImageQuant LAS 4010 (GE Healthcare).
Alginate lyase assay.
Strain A1 WT and ΔAlgO mutant cells grown in 0.5% alginate medium or 0.5% yeast extract medium were ultrasonically disrupted with an Insonator model 201 M (Kubota) at 0°C and 9 kHz for 10 min, and the clear solution obtained by centrifugation at 14,000 × g and 4°C for 15 min was used as a source of alginate lyase. Enzyme activity was measured by monitoring the increase in absorbance at 235 nm. One unit of enzyme activity was defined as the amount of enzyme required to produce an increase of 1.0 U of absorbance at 235 nm/min. Each assay datum represents the mean ± standard deviation of triplicate individual experiments.
DNA microarray.
Total RNA was extracted by the hot phenol method (23) from strain A1 WT and ΔAlgO mutant cells grown in 0.5% alginate medium or 0.5% yeast extract medium to an OD600 of 0.5 to 0.6 (i.e., exponential growth phase). The resultant RNA was isolated with the RNeasy minikit (Qiagen) and an RNase-Free DNase set (Qiagen) and was hybridized to the bacterial DNA microarray chip. The strain A1 DNA microarray chip designed by Roche NimbleGen includes 3,985 target genes fixed on a glass slide obtained by fixing two sets of nine unique probes comprising 60-mer synthetic oligonucleotides for each gene. Cyanine (Cy3) labeling, fragmentation, and hybridization were performed by Roche NimbleGen in accordance reference 28. Arrays were scanned with a NimbleGen MS200 microarray scanner at 532 nm and a resolution of 2 μm and analyzed by quantile normalization and robust multiarray averaging (29, 30). The about 18 raw expression data obtained per gene were subjected to statistical treatment. The normalized data were processed with the NANDEMO Analysis 1.0.0 software (Roche Diagnostics). Student's t test was used to analyze the mean log ratios of two samples, and subsequent Bonferroni adjustment for multiple testing (3,985 open reading frames on arrays) was used as a rigorous criterion for significant changes in signal intensity. Changes with P values of <0.05 were considered statistically significant.
Overexpression and purification of AlgO-His6.
After precultivation at 30°C, cells of E. coli BL21-Gold(DE3)pLysS harboring pET21b-algO-His6 were cultured at 16°C for 48 h in 1.5 liters of LB medium containing Amp and Cm (1.5 liters/flask) in the presence of 0.1 mM isopropyl-β-d-thiogalactopyranoside. The cells were collected by centrifugation at 8,000 ×g and 4°C for 15 min, washed with 20 mM Tris-HCl (pH 7.5), and resuspended in the same buffer. To avoid protein aggregation, arginine-HCl was added to the cell suspension at a final concentration of 1 M. The E. coli cells were ultrasonically disrupted with an Insonator model 201 M at 0°C and 9 kHz for 20 min, and the clear solution obtained after centrifugation at 14,000 ×g and 4°C for 15 min was used as the cell extract. The extract was subjected to chromatography with a TALON resin column (1.0 by 10 cm; Clontech) previously equilibrated with 20 mM Tris-HCl (pH 7.5) containing 0.5 M NaCl and 10 mM imidazole. The resin was washed with the same buffer, and absorbed proteins were eluted with a linear gradient of imidazole (10 to 200 mM) in 20 mM Tris-HCl (pH 7.5) containing 0.5 M NaCl. AlgO-His6 was detected by SDS-PAGE, followed by Coomassie brilliant blue staining. The fractions containing AlgO-His6 (approximately 40 kDa) were subjected to chromatography with a Sephacryl S-200 HR column (2.6 by 40 cm; GE Healthcare) previously equilibrated with 20 mM Tris-HCl (pH 7.5) containing 0.15 M NaCl and 1 M arginine-HCl. The proteins were eluted with the same buffer. The fractions containing AlgO-His6 were used as purified AlgO-His6. All purification procedures were performed at 0 to 4°C.
RT-PCR.
With the RNeasy minikit, total RNA was isolated from strain A1 WT cells grown in 0.5% alginate medium to an OD600 of 0.5 to 0.6. cDNA was synthesized with SuperScript III reverse transcriptase (RT; Invitrogen) from DNase I (Qiagen)-treated total RNA (1 μg). To confirm whether algO and aly located in the antisense strand of the alginate genetic cluster were organized as a single operon in the strain A1 genome, PCR was performed with KOD-Plus-Neo as DNA polymerase, cDNA as the template, and a set of two synthetic oligonucleotides as primers, i.e., a forward primer (algO-aly_F) designed from the internal region of algO and a reverse primer (algO-aly_R) designed from the internal region of aly. Similarly, algS, algM1, algM2, algQ1, algQ2, and a1-IV were confirmed to be assembled into a single operon in the strain A1 genome. For the primers used for RT-PCR, see Table S1 in the supplemental material.
Preparation of alginate derivatives.
Unsaturated alginate oligosaccharides were prepared from M- and G-rich saccharides (31) separated from sodium alginate. The M-rich saccharides (2%) were incubated at 30°C for 24 h with alginate lyase A1-I (16.1 U) (11) at a final concentration of 0.2 mg/ml. Enzymatically degraded saccharides were subjected to anion-exchange column chromatography (Q-Sepharose HP, 2.6 by 10 cm; GE Healthcare) and eluted with a linear gradient of ammonium bicarbonate (50 to 500 mM). Unsaturated trisaccharide (Δ3M) and tetrasaccharide (Δ4M) were purified to homogeneity. The homogeneity of the oligosaccharides was confirmed by thin-layer chromatography (11), and oligosaccharides were concentrated by freeze-drying. Similarly, the G-rich saccharides were used for the preparation of unsaturated trisaccharide (Δ3G), tetrasaccharide (Δ4G), and pentasaccharide (Δ5G).
DNA gel mobility shift assay.
To investigate the interaction between the recombinant AlgO (AlgO-His6) and target DNA, gel mobility shift assays were performed according to the protocols of the digoxigenin (DIG) gel shift kit, second generation (Roche). DNA probes 1, 2, 3, and 6 were synthesized by PCR with KOD-Plus-Neo as DNA polymerase, the genomic DNA of strain A1 as the template, and two synthetic oligonucleotides as primers. The following combinations of the two primers were used: 202bp_F and 202bp_R for probe 1, 146bp_F and 146bp_R for probe 2, 234bp_F and 234bp_R for probe 3, and 94bp_F and 94bp_R for probe 6. The PCR products were purified with the NucleoSpin Gel and PCR Cleanup kit (TaKaRa Bio) and confirmed by DNA sequencing. DNA probes 4, 5, 7, and 8 with molecular sizes of 72, 69, 35, and 34 bp, respectively, were prepared by annealing two mutually complementary oligonucleotides. The 3′ blunt end of probes were labeled by incorporating DIG-ddUTP with terminal transferase (Roche). The DIG-labeled probes were mixed at 20°C for 15 min with AlgO-His6 at a final concentration of 0.2 μM in 20 μl of binding reaction solution containing 1 μg of poly(dI-dC), 1 μg of poly-l-lysine, and 5× binding buffer. For the competition assay, an unlabeled DNA probe was added at a final concentration of 0.2 μM into the reaction mixture of the DIG-labeled probe with AlgO-His6. Mixtures of DNA and protein were subjected to 5% PAGE in 0.5× Tris-borate-EDTA buffer (89 mM Tris, 89 mM boric acid, 2 mM EDTA). After electrophoresis, gel contents were transferred to an equilibrated, positively charged nylon membrane (Hybond-N+; Amersham) by electroblotting at 1 mA/cm2 for 30 min. The luminescence signal was developed in disodium 3-(4-methoxyspiro{1,2-dioxetane-3,2′-(5′-chloro)tricyclo[3.3.1.13,7]decan}-4-yl)phenylphosphate working solution (Roche) at 37°C for 10 min after incubation of the nylon membrane with anti-DIG antibody (Roche). The nylon membrane was visualized with ImageQuant LAS 4010. The effect of alginate molecules on the interaction between AlgO-His6 and target DNA was analyzed by the gel mobility shift assay in the presence of alginate derivatives.
Modeling and docking simulation.
Homology modeling of the AlgO C-terminal domain was carried out with a SWISS-MODEL program (32–34). Interaction of the C-terminal domain model of AlgO with alginate trisaccharide was simulated with the AutoDock program package (35).
Microarray data accession number.
The microarray data from this study have been deposited in the Gene Expression Omnibus database at NCBI under accession number GSE54410.
RESULTS AND DISCUSSION
Primary structure analysis of AlgO.
The strain A1 protein AlgO (354 residues), with a theoretical molecular mass of 39.6 kDa, is encoded in the bacterial genome between aly and algS, which are involved in alginate uptake and degradation (Fig. 1B). Primary structure analysis with the Pfam database (36) indicates that AlgO is a member of the LacI family (37) and consists of N- and C-terminal regions. The N-terminal region (residues 18 to 59) shows significant similarity (E value, 2.4E-15) to a DNA-binding domain with a typical helix-turn-helix motif seen in LacI family transcriptional regulators. The C-terminal region (residues 107 to 328) shows similarity (E value, 1.2E-17) to the sugar-binding domain of the periplasmic binding protein. AlgO exhibits low homology with P. aeruginosa ribose operon repressor RbsR (E value, 8.0E-25) and a putative A. vinelandii transcriptional regulator (E value, 6.0E-23). None of the proteins deposited in the GenBank, UniProt, RefSeq, and PDBSTR databases exhibit significant identity (more than 40%, 3.0E-62) with AlgO. Therefore, AlgO is considered to be specific for strain A1.
Periplasmic alginate-binding proteins (AlgQ1 and AlgQ2), a cytoplasmic-membrane-bound ABC transporter (AlgM1-AlgM2/AlS-AlgS), and cytoplasmic endo- and exotype alginate lyases (A1-I, -II, -III, and -IV) (Fig. 1A), all of which are encoded in the strain A1 alginate genetic cluster (Fig. 1B), are inducibly expressed in the presence of alginate (6). The primary structural characteristics of AlgO suggest that it could function as a regulator of this alginate genetic cluster.
Constitutive expression of alginate-related proteins in the ΔAlgO mutant.
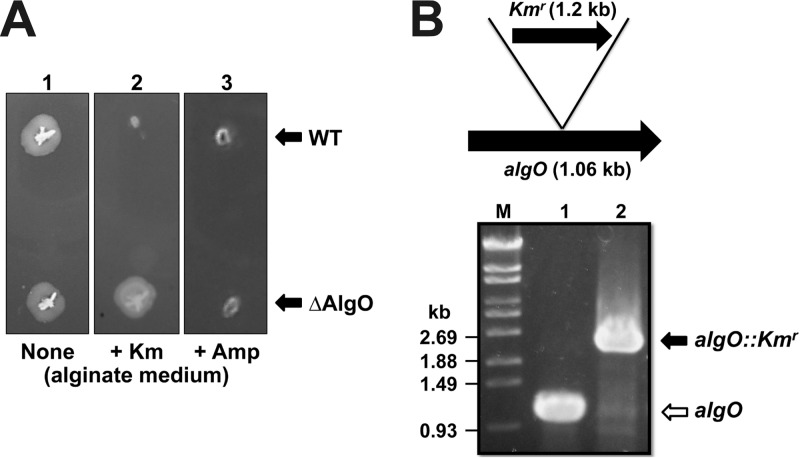
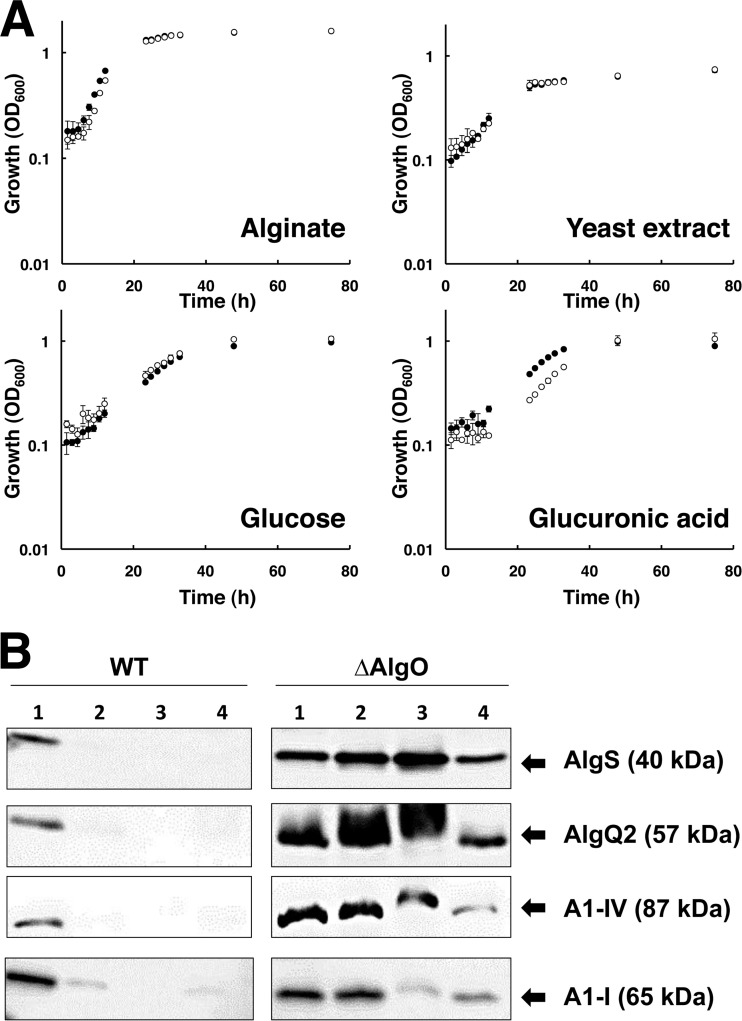
To investigate the physiological function of AlgO in strain A1 cells, the ΔAlgO mutant was constructed by inserting a Kmr-encoding gene cassette into the center of algO in the strain A1 genome. As expected, ΔAlgO mutant cells were Kmr but Amps (Fig. 2A). Gene disruption was confirmed by PCR amplification of a 1.2-kb-longer algO gene (algO::Kmr) from ΔAlgO mutant cells (Fig. 2B). The growth of ΔAlgO mutant cells in alginate medium or alginate-free (yeast extract, glucose, or glucuronic acid) medium was comparable to that of WT cells (Fig. 3A), indicating that algO disruption has no effect on strain A1 growth.
FIG 2.
algO disruption in strain A1. (A) Antibiotic sensitivity of strain A1 cells. Top, WT; bottom, ΔAlgO mutant. Lane 1, no antibiotics; lane 2, with Km; lane 3, with Amp. (B) algO disruption by insertion of the Kmr-encoding gene (1.2 kb). algO from WT or ΔAlgO mutant cells was amplified by PCR. Lane M, molecular size markers; lane 1, WT; and lane 2, ΔAlgO mutant.
FIG 3.
Characterization of the ΔAlgO mutant. (A) Growth profile of strain A1 WT and ΔAlgO mutant cells in alginate, yeast extract, glucose, or glucuronic acid medium. Closed circles, WT; open circles, ΔAlgO mutant. Growth experiments were performed in triplicate. Error bars represent the standard deviations of the means of the three experiments. (B) Western blotting. Left, WT cell extracts from each medium; right, ΔAlgO mutant cell extracts from each medium. Lane 1, alginate; lane 2, yeast extract; lane 3, glucose; lane 4, glucuronic acid.
Expression of proteins encoded in the sense strand of the alginate genetic cluster in strain A1 WT and ΔAlgO mutant cells was analyzed by Western blotting with anti-AlgS, anti-AlgQ2, and anti-A1-IV antibodies. Proteins with a molecular mass of 40, 57, or 87 kDa corresponding to AlgS, AlgQ2, or A1-IV, respectively, were detected in WT cells grown in alginate medium, while few protein bands were observed in WT cells grown in alginate-free medium (Fig. 3B, left). However, AlgS, AlgQ2, and A1-IV were expressed in ΔAlgO mutant cells grown in all of the media tested, even in alginate-free medium (Fig. 3B, right). These results indicate that AlgS, AlgQ2, and A1-IV encoded in the sense strand of the alginate genetic cluster are constitutively expressed in the ΔAlgO mutant cells independently of alginate. Similarly, expression of the A1-I protein encoded in the antisense strand of the alginate genetic cluster was analyzed in WT and ΔAlgO mutant cells by Western blotting with anti-A1-III antibody. A protein with a molecular mass of 65 kDa corresponding to A1-I was produced in WT cells specifically grown in alginate medium, while A1-I was expressed in ΔAlgO mutant cells grown in all of the media tested (Fig. 3B), demonstrating that A1-I encoded in the antisense strand of the alginate genetic cluster is constitutively expressed in ΔAlgO mutant cells, independently of alginate.
To compare protein expression levels, the alginate lyase activities of WT and ΔAlgO mutant cells grown on alginate or yeast extract were determined. WT cells grown on yeast extract exhibited little enzyme activity (21.9 ± 7.1 U/g of protein), whereas ΔAlgO mutant cells grown on yeast extract exhibited an activity of 453 ± 87 U/g of protein. This value corresponds to about 70% of the enzyme activity of ΔAlgO mutant cells grown on alginate (624 ± 213 U/g of protein).
These results indicate that, in the absence of alginate, strain A1 cells significantly repress the expression of alginate-related proteins encoded in both the sense and antisense strands of the alginate genetic cluster through the action of AlgO.
Comprehensive transcriptional analysis of alginate-related genes in the ΔAlgO mutant.
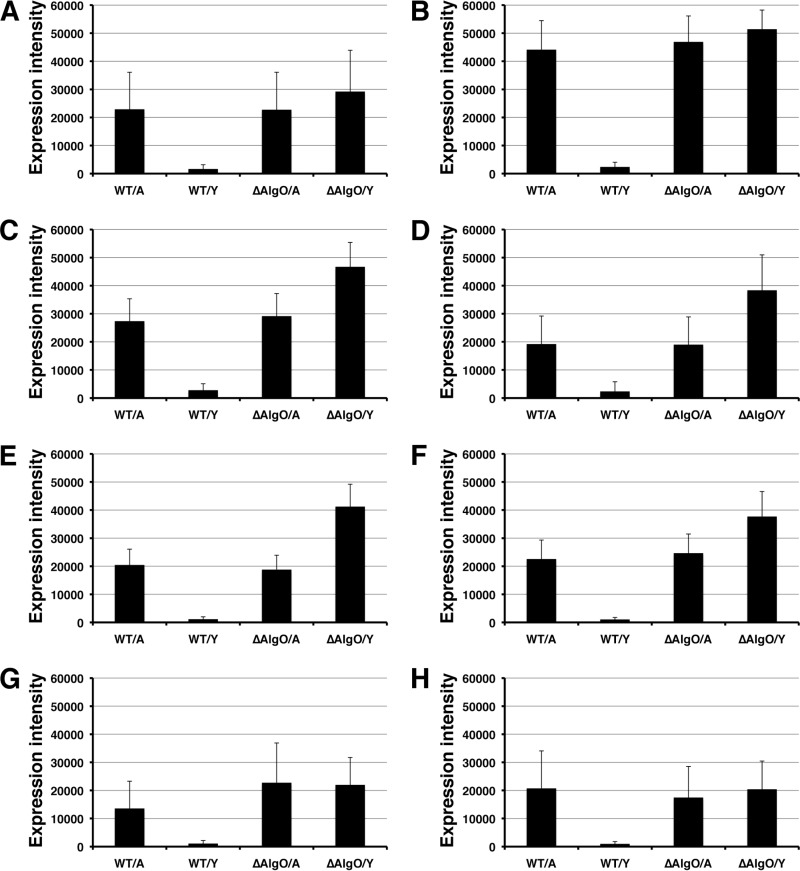
To identify the regulation step (i.e., transcriptional and/or translational stages) by AlgO, a DNA microarray was used to analyze WT and ΔAlgO mutant cells grown on alginate or yeast extract. The transcriptional levels of the alginate genetic cluster in WT and ΔAlgO mutant cells grown in 0.5% alginate medium or 0.5% yeast extract medium were compared (Fig. 4). All of the genes (aly, algO, algS, algM1, algM2, algQ1, algQ2, and a1-IV) in the alginate genetic cluster were inducibly transcribed in the WT cells grown on alginate, while their transcription was significantly repressed in WT cells grown on yeast extract. However, these alginate-related genes were transcribed at a high level in ΔAlgO mutant cells grown on either alginate or yeast extract. The transcription of the alginate-related genes was significantly upregulated (15- to 60-fold) in ΔAlgO mutant cells grown on yeast extract compared to that in WT cells grown on yeast extract. These results demonstrate that AlgO functions as a transcriptional regulator in the presence of alginate and represses the transcriptional stage of the alginate-related genes in the absence of alginate.
FIG 4.
mRNA transcript level of alginate genetic cluster in strain A1 cells. (A) a1-IV. (B) algQ2. (C) algQ1. (D) algM2. (E) algM1. (F) algS. (G) algO. (H) aly. WT/A, WT cells grown on alginate; WT/Y, WT cells grown on yeast extract; ΔAlgO/A, ΔAlgO mutant cells grown on alginate; ΔAlgO/Y, ΔAlgO mutant cells grown on yeast extract. Error bars represent the standard deviations of the means of about 18 raw expression data.
In all of the WT and ΔAlgO mutant cells tested, the transcriptional level of the alginate-related genes in the sense strand of the genetic cluster was higher than that in the antisense strand (Fig. 4), suggesting that the promoter activity in the sense strand is stronger. In fact, typical sequences (algS promoter) homologous to E. coli consensus −35 and −10 regions were found to be located upstream of algS (at around 150 bp from its initiation codon) in the sense strand with a program of GenetyxMac (Software Development), whereas the antisense strand also included a possible algO promoter in the complementary strand around the algS promoter. Although AlgO itself regulated its expression, AlgO molecules highly expressed in the presence of alginate seem to be inactivated by the alginate oligosaccharides described below. Strain A1 cells may also regulate gene expression on the basis of the promoter activity.
Alginate monosaccharides formed from both M- and G-rich saccharides through a reaction of A1-IV are nonenzymatically converted to 4-deoxy-l-erythro-5-hexoseulose uronic acid (DEH) (12). DEH is further converted to 2-keto-3-deoxy-d-gluconic acid by DEH reductase A1-R (38). Similar to the alginate genetic cluster, A1-R is inducibly expressed in the presence of alginate, although the gene for A1-R is located far from the alginate genetic cluster in the strain A1 genome. Moreover, ΔAlgO mutant cells grown on yeast extract showed repressed A1-R gene expression, while those grown on alginate inducibly expressed the gene. This suggests that a transcription factor other than AlgO regulates A1-R gene expression.
DNA sequence targeted by AlgO.
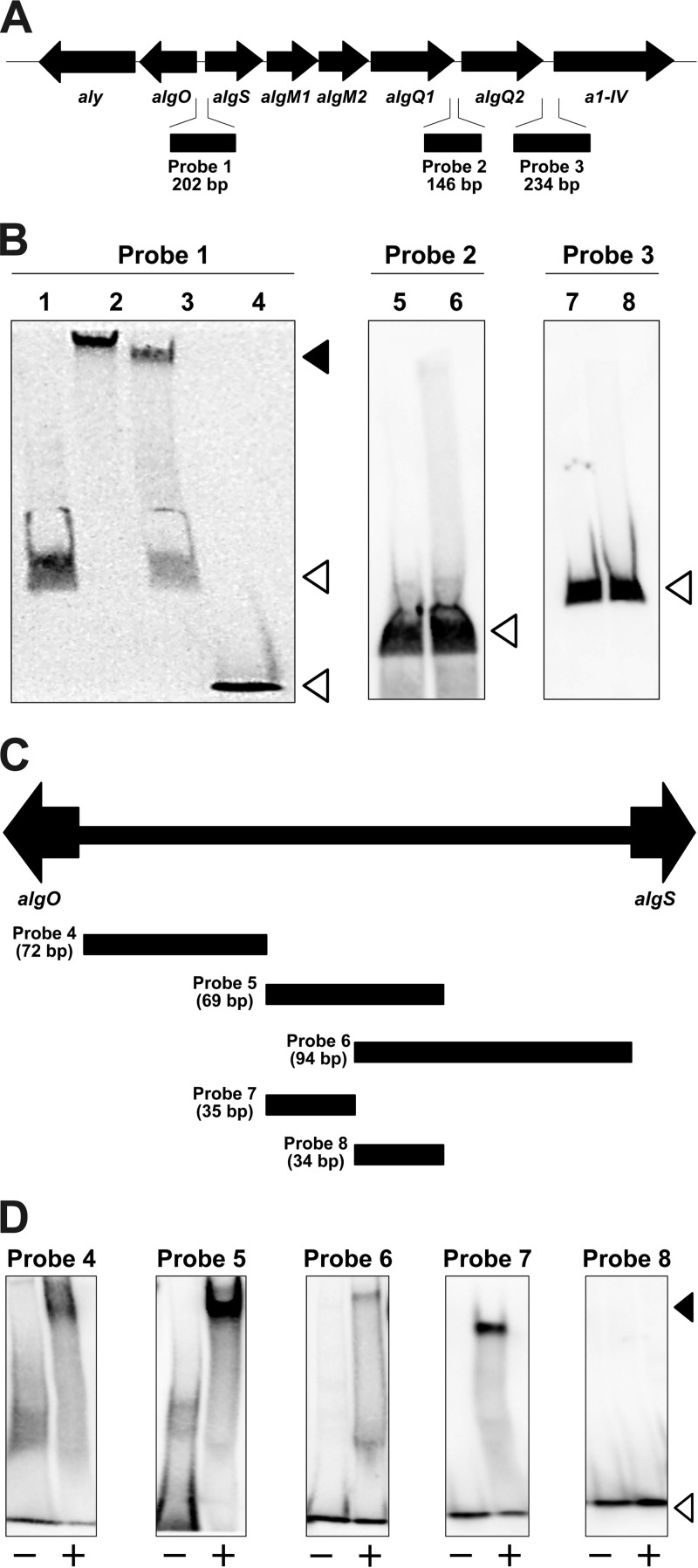
AlgO-binding sites in the intergenic regions of the alginate genetic cluster were examined by gel mobility shift assay with the purified AlgO-His6 protein (Fig. 5). Three DNA probes were designed as follows: probe 1, algO-algS intergenic region; probe 2, algQ1-algQ2 intergenic region; and probe 3, algQ2–a1-IV intergenic region (Fig. 5A). These sequences were selected as potential regulatory regions for the expression of the alginate-related genes on the basis of intergenic sequence length and/or the possible promoter location.
FIG 5.
Gel mobility shift assay. (A) Alginate genetic cluster. Probe 1, algO-algS intergenic region; probe 2, algQ1-algQ2 intergenic region; probe 3, algQ2–a1-IV intergenic region. (B) Gel mobility shift assay with DIG-labeled DNA probes and AlgO-His6. Lane 1, probe 1; lane 2, probe 1 and AlgO-His6; lane 3, probe 1, nonlabeled probe 1, and AlgO-His6; lane 4, negative control DIG-labeled probe and AlgO-His6; lane 5, probe 2; lane 6, probe 2 and AlgO-His6; lane 7, probe 3; lane 8, probe 3 and AlgO-His6. (C) Probes 4 to 8 in the algO-algS intergenic region. (D) Gel mobility shift assay with probes 4 to 8. Symbols: −, each probe in the absence of AlgO-His6; +, each probe in the presence of AlgO-His6. The closed and open arrowheads indicate the positions of the DNA-protein complex and free DNA, respectively.
DIG-labeled probe 1 was shifted upward in the presence of AlgO-His6 (Fig. 5B, lane 2), suggesting that AlgO-His6 binds to probe 1. The complex of labeled probe 1 and AlgO-His6 decreased as nonlabeled probe 1 was added to the reaction mixture (Fig. 5B, lane 3). In contrast, no AlgO-His6 bound to probes 2 and 3. These results indicate that AlgO specifically binds to the algO-algS intergenic region to repress the transcription of alginate-related genes. The algQ1-algQ2 and algQ2–a1-IV intergenic regions are considered not to be target DNA sequences of AlgO.
To identify the AlgO-binding site in the algO-algS intergenic region, the gel mobility shift assay was further performed with five short DIG-labeled DNA probes in the algO-algS intergenic region (Fig. 5C). algO-sided probes 4, 5, and 7 clearly showed the DNA mobility shift in the presence of AlgO-His6 (Fig. 5D), although few shifted DNA bands were observed in the mixture of AlgO-His6 and algS-sided probe 6 or 8. This gel mobility shift assay demonstrates that AlgO binds to the algO-sided sequence between algO and algS.
Operon structure of the alginate genetic cluster.
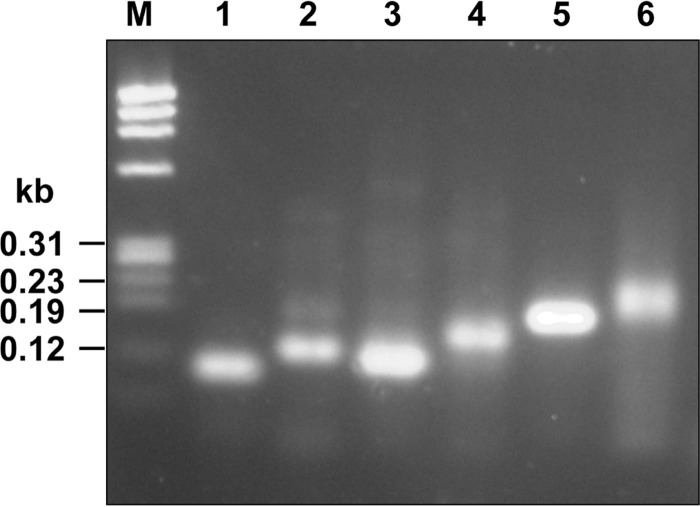
Through the above-mentioned gel mobility shift assay, AlgO was observed to bind to the algO-sided sequence of the algO-algS intergenic region and to regulate the expression of the alginate-related genes located in both the sense and antisense strands. This suggests that the alginate genetic cluster includes two operons, i.e., operon 1 (algOaly) and operon 2 (algSalgM1algM2algQ1algQ2a1-IV). To analyze the operon structure, cDNA synthesized from mRNA transcripts was examined by PCR (Fig. 6). A DNA fragment about 60 bp long was amplified from cDNA with internal primers of algO and aly (Fig. 6, lane 1). This indicates that at least algO and aly are transcribed to a single mRNA by the algO promoter. AlgO and Aly are therefore located as an operon in the antisense strand of the alginate genetic cluster. In the case of independent transcription of algO and aly, no PCR product could be detected with the primer set used. Similarly, each DNA fragment with a predicted molecular size was amplified from cDNA with an internal primer set of each gene, algS, algM1, algM2, algQ1, algQ2, or a1-IV (Fig. 6, lanes 2 to 6), demonstrating that the six genes are located as an operon in the sense strand of the alginate genetic cluster.
FIG 6.
Operon analysis. Lane M, molecular size markers; lane 1, alyalgO; lane 2, algSalgM1; lane 3, algM1algM2; lane 4, algM2algQ1; lane 5, algQ1algQ2; lane 6, algQ2a1-IV.
Inhibition of AlgO binding to DNA by alginate oligosaccharides.
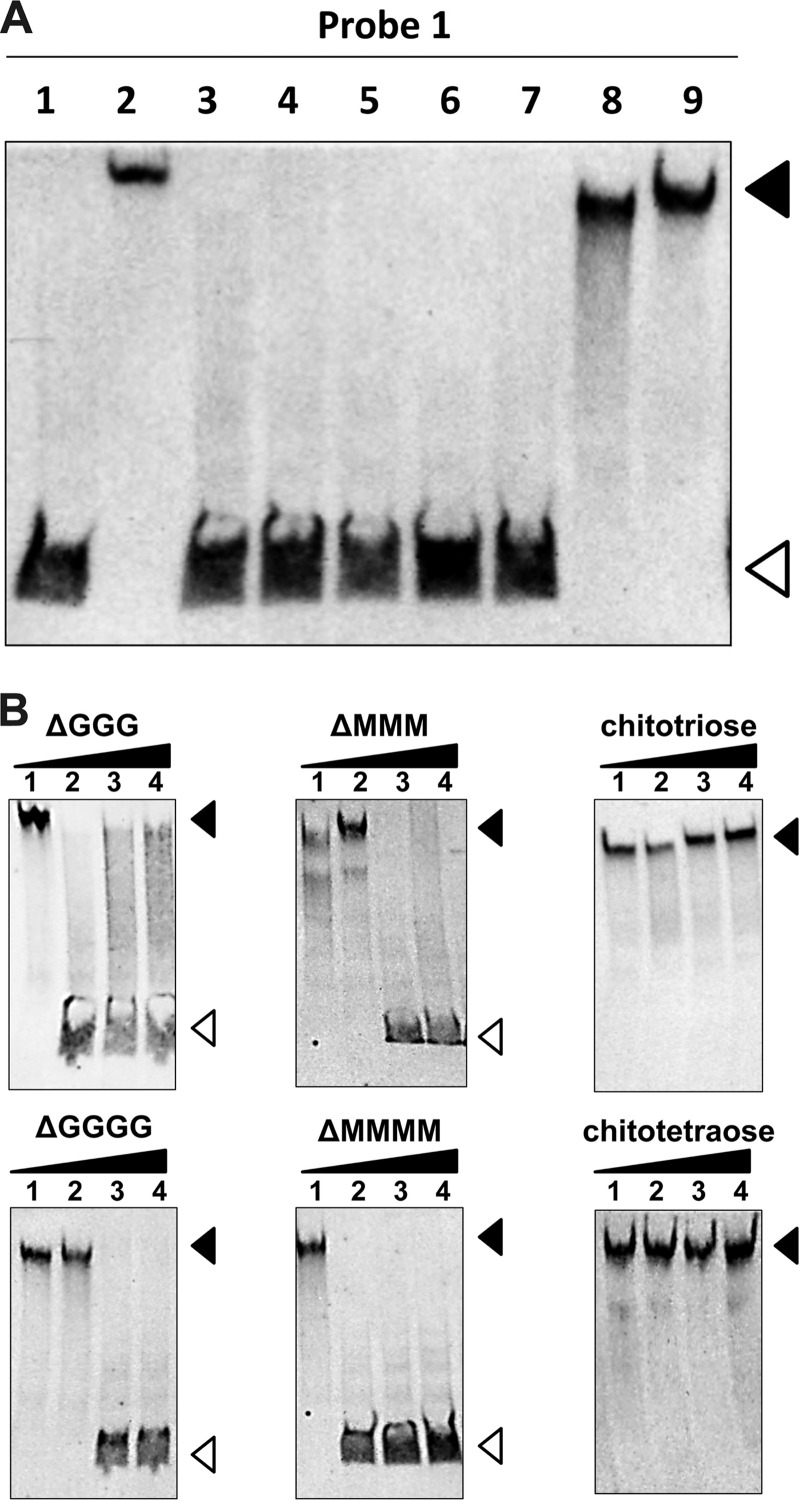
The effect of alginate derivatives on the interaction between AlgO and its target DNA (the algO-algS intergenic region) was analyzed by gel mobility shift assay in the presence of alginate oligosaccharides. In the presence of each unsaturated alginate oligosaccharide (Δ3G, Δ4G, Δ5G, Δ3M, or Δ4M), no formation of the AlgO-His6/DNA (DIG-labeled probe 1) complex was observed, and instead, protein-free probe 1 was detected (Fig. 7A, lanes 3 to 7). Both chitotriose and chitotetraose, used as nonalginate oligosaccharides, had no effect on complex formation, indicating that alginate oligosaccharides specifically inhibit the formation of the AlgO/DNA complex. These results demonstrate that unsaturated alginate oligosaccharides produced from alginate through reactions with alginate lyases function as an effector molecule for the dissociation of AlgO from target DNA (the algO-algS intergenic region), subsequently leading to the initiation of transcription. Quantitative dependence of alginate oligosaccharides on the inhibition of AlgO-His6 binding to DIG-labeled probe 1 was analyzed by gel mobility shift assay to determine the physiologically functional concentration. The amount of free probes increased as the concentration of each alginate oligosaccharide (Δ3G, Δ4G, Δ3M, or Δ4M) increased, while no such change in the amounts of probes was observed at any concentration of chitotriose or chitotetraose (Fig. 7B). The effective alginate oligosaccharide concentration may seem relatively high in this assay compared with the concentration of AlgO (AlgO/alginate oligosaccharide ratio, 1:800). This ratio is unremarkable because one molecule of alginate polysaccharide, with an average molecular weight of 300,000, gives rise to more than 500 molecules of alginate trisaccharides through reactions with alginate lyases.
FIG 7.
Effects of alginate derivatives on AlgO binding to DNA. (A) Gel mobility shift assay with DIG-labeled probe 1 in the presence of each alginate derivative. Lane 1, probe 1; lane 2, probe 1 and AlgO-His6; lanes 3 to 10, probe 1 and AlgO-His6 in the presence of 25 mM Δ3G (lane 3), Δ4G (lane 4), Δ5G (lane 5), Δ3M (lane 6), Δ4M (lane 7), chitotriose (lane 8), and chitotetraose (lane 9). (B) Concentration dependence of the inhibitory effect of AlgO binding to DIG-labeled probe 1. Lanes 1 to 4, probe 1 and AlgO-His6 in the presence of 0 mM (lane 1), 10 mM (lane 2), 25 mM (lane 3), and 50 mM (lane 4) of each oligosaccharide. The closed and open arrowheads indicate the positions of the DNA-protein complex and free DNA, respectively.
This study revealed that AlgO binds specifically to the algO-algS intergenic region in the alginate genetic cluster and alginate-degraded products function as an effector for the dissociation of AlgO from the algO-algS intergenic region. Primary structure analysis of AlgO and gel mobility shift assays suggest that the C-terminal domain of AlgO contains the structural fold common to periplasmic solute-binding proteins. The strain A1 alginate-binding proteins (AlgQ1 and AlgQ2) are typical periplasmic solute-binding proteins (9). To investigate the binding mode of AlgO to alginate oligosaccharides, the tertiary structure of the C-terminal domain of AlgO was built by homology modeling with a SWISS-MODEL program (Fig. 1C, bottom). The structural architecture shows that the C-terminal domain comprises two domains with an α/β fold. The two domains are connected by a long linker and separated by a cleft. These structural features are also observed in AlgQ1 and AlgQ2 (39, 40). X-ray crystallography of AlgQ1 complexed with the alginate oligosaccharide demonstrates (Protein Data Bank ID 3VLU) that AlgQ1 accommodates the oligosaccharide at the interface cleft between two domains through a conformational change (Fig. 1C, top). Common structural features (cleft between two domains) and functional characteristics (alginate binding) suggest that the C-terminal domain of AlgO accommodates the alginate oligosaccharide in the cleft between the two domains. In fact, docking simulation of the C-terminal domain model of AlgO and Δ3M with the AutoDock program package indicates that the alginate oligosaccharide is bound to the cleft between the two domains (Fig. 1C, bottom right). It is well known that ligand binding in the C-terminal domain of the LacI family proteins causes structural changes within the N-terminal DNA-binding domain and subsequently affects its DNA-binding affinity (41–43). Moreover, once the C-terminal domain of AlgO specifically recognizes and binds to the unsaturated alginate oligosaccharides, structural changes probably occur in the helix-turn-helix motif of the N-terminal domain and reduce its DNA-binding affinity.
DNA microarray data indicated that ΔAlgO mutant cells grown on yeast extract produced transcripts from all of the genes in the alginate genetic cluster at the highest level among the strain A1 cells tested (Fig. 4). This suggests that the expression of the alginate genetic cluster with an algO disruption is independent of alginate and is not affected by any other repression mechanism. To produce biofuel such as bioethanol from alginate, ethanologenic microbes are expected to become promising hosts for the introduction of the alginate assimilation ability. On the other hand, the strain A1 alginate genetic cluster with an algO disruption is a potential gene resource for the addition of alginate-uptake/degradation capability to ethanologenic hosts.
Other than alginate-related genes, some genes were more highly expressed (more than 20-fold) in yeast extract-grown ΔAlgO mutant cells than in WT cells. For example, these gene products show a homology with enzymes (synthase, lyase, and dehydrogenase) involved in the methylcitrate cycle. The expression of these genes is possibly regulated by AlgO, although the involvement of the methylcitrate cycle in alginate metabolism is unclear and no consensus sequence was observed between the algO-algS intergenic region and upstream region of methylcitrate-related genes.
In conclusion, the strain A1 AlgO-dependent regulation mechanism of gene expression of the alginate genetic cluster is postulated as follows (see Fig. S1 in the supplemental material). In the absence of alginate, AlgO binds to DNA (the algO-algS intergenic region) through a helix-turn-helix motif in the N-terminal region and represses the transcription of both sense and antisense strands of the alginate genetic cluster, whereas in the presence of alginate, the concentration of unsaturated alginate oligosaccharides degraded by alginate lyases reaches a sufficiently high level in strain A1 cells, and subsequently, alginate oligosaccharides act as an effecter molecule and bind to the C-terminal domain of AlgO. Sugar binding induces a conformational change in AlgO and the dissociation of AlgO from DNA (the algO-algS intergenic region), thereby initiating the transcription of all of the alginate-related genes in the cluster. This is the first report demonstrating the involvement of alginate molecules in bacterial gene expression.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by grants-in-aid from the Japan Society for the Promotion of Science (to K.M. and W.H.) and a research fellowship from the Japan Society for the Promotion of Science for Young Scientists (to R.T.).
Footnotes
Published ahead of print 9 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01666-14.
REFERENCES
- 1.Gacesa P. 1988. Alginates. Carbohydr. Polym. 8:161–182. 10.1016/0144-8617(88)90001-X [DOI] [Google Scholar]
- 2.May TB, Chakrabarty AM. 1994. Pseudomonas aeruginosa: genes and enzymes of alginate synthesis. Trends Microbiol. 2:151–157. 10.1016/0966-842X(94)90664-5 [DOI] [PubMed] [Google Scholar]
- 3.Schwarzmann S, Boring JR. 1971. Antiphagocytic effect of slime from a mucoid strain of Pseudomonas aeruginosa. Infect. Immun. 3:762–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stokstad E. 2012. Biofuels. Engineered superbugs boost hopes of turning seaweed into fuel. Science 335:273. 10.1126/science.335.6066.273 [DOI] [PubMed] [Google Scholar]
- 5.Wong TY, Preston LA, Schiller NL. 2000. ALGINATE LYASE: review of major sources and enzyme characteristics, structure-function analysis, biological roles, and applications. Annu. Rev. Microbiol. 54:289–340. 10.1146/annurev.micro.54.1.289 [DOI] [PubMed] [Google Scholar]
- 6.Murata K, Kawai S, Mikami B, Hashimoto W. 2008. Superchannel of bacteria: biological significance and new horizons. Biosci. Biotechnol. Biochem. 72:265–277. 10.1271/bbb.70635 [DOI] [PubMed] [Google Scholar]
- 7.Hashimoto W, Kawai S, Murata K. 2010. Bacterial supersystem for alginate import/metabolism and its environmental and bioenergy applications. Bioeng. Bugs. 1:97–109. 10.4161/bbug.1.2.10322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hisano T, Kimura N, Hashimoto W, Murata K. 1996. Pit structure on bacterial cell surface. Biochem. Biophys. Res. Commun. 220:979–982. 10.1006/bbrc.1996.0526 [DOI] [PubMed] [Google Scholar]
- 9.Momma K, Mishima Y, Hashimoto W, Mikami B, Murata K. 2005. Direct evidence for Sphingomonas sp. A1 periplasmic proteins as macromolecule-binding proteins associated with the ABC transporter: molecular insights into alginate transport in the periplasm. Biochemistry 44:5053–5064. 10.1021/bi047781r [DOI] [PubMed] [Google Scholar]
- 10.Momma K, Okamoto M, Mishima Y, Mori S, Hashimoto W, Murata K. 2000. A novel bacterial ATP-binding cassette transporter system that allows uptake of macromolecules. J. Bacteriol. 182:3998–4004. 10.1128/JB.182.14.3998-4004.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoon H-J, Hashimoto W, Miyake O, Okamoto M, Mikami B, Murata K. 2000. Overexpression in Escherichia coli, purification, and characterization of Sphingomonas sp. A1 alginate lyases. Protein Expr. Purif. 19:84–90. 10.1006/prep.2000.1226 [DOI] [PubMed] [Google Scholar]
- 12.Hashimoto W, Miyake O, Momma K, Kawai S, Murata K. 2000. Molecular identification of oligoalginate lyase of Sphingomonas sp. strain A1 as one of the enzymes required for complete depolymerization of alginate. J. Bacteriol. 182:4572–4577. 10.1128/JB.182.16.4572-4577.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyake O, Ochiai A, Hashimoto W, Murata K. 2004. Origin and diversity of alginate lyases of families PL-5 and -7 in Sphingomonas sp. strain A1. J. Bacteriol. 186:2891–2896. 10.1128/JB.186.9.2891-2896.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ochiai A, Hashimoto W, Murata K. 2006. A biosystem for alginate metabolism in Agrobacterium tumefaciens strain C58: molecular identification of Atu3025 as an exotype family PL-15 alginate lyase. Res. Microbiol. 157:642–649. 10.1016/j.resmic.2006.02.006 [DOI] [PubMed] [Google Scholar]
- 15.Takeda H, Yoneyama F, Kawai S, Hashimoto W, Murata K. 2011. Bioethanol production from marine biomass alginate by metabolically engineered bacteria. Energy Environ. Sci. 4:2575–2581. 10.1039/c1ee01236c [DOI] [Google Scholar]
- 16.Wargacki AJ, Leonard E, Win MN, Regitsky DD, Santos CN, Kim PB, Cooper SR, Raisner RM, Herman A, Sivitz AB, Lakshmanaswamy A, Kashiyama Y, Baker D, Yoshikuni Y. 2012. An engineered microbial platform for direct biofuel production from brown macroalgae. Science 335:308–313. 10.1126/science.1214547 [DOI] [PubMed] [Google Scholar]
- 17.Enquist-Newman M, Faust AM, Bravo DD, Santos CN, Raisner RM, Hanel A, Sarvabhowman P, Le C, Regitsky DD, Cooper SR, Peereboom L, Clark A, Martinez Y, Goldsmith J, Cho MY, Donohoue PD, Luo L, Lamberson B, Tamrakar P, Kim EJ, Villari JL, Gill A, Tripathi SA, Karamchedu P, Paredes CJ, Rajgarhia V, Kotlar HK, Bailey RB, Miller DJ, Ohler NL, Swimmer C, Yoshikuni Y. 2014. Efficient ethanol production from brown macroalgae sugars by a synthetic yeast platform. Nature 505:239–243. 10.1038/nature12771 [DOI] [PubMed] [Google Scholar]
- 18.Remminghorst U, Rehm BH. 2006. Bacterial alginates: from biosynthesis to applications. Biotechnol. Lett. 28:1701–1712. 10.1007/s10529-006-9156-x [DOI] [PubMed] [Google Scholar]
- 19.Wozniak DJ, Ohman DE. 1994. Transcriptional analysis of the Pseudomonas aeruginosa genes algR, algB, and algD reveals a hierarchy of alginate gene expression which is modulated by algT. J. Bacteriol. 176:6007–6014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deretic V, Dikshit R, Konyecsni WM, Chakrabarty AM, Misra TK. 1989. The algR gene, which regulates mucoidy in Pseudomonas aeruginosa, belongs to a class of environmentally responsive genes. J. Bacteriol. 171:1278–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wozniak DJ, Ohman DE. 1991. Pseudomonas aeruginosa AlgB, a two-component response regulator of the NtrC family, is required for algD transcription. J. Bacteriol. 173:1406–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baynham PJ, Ramsey DM, Gvozdyev BV, Cordonnier EM, Wozniak DJ. 2006. The Pseudomonas aeruginosa ribbon-helix-helix DNA-binding protein AlgZ (AmrZ) controls twitching motility and biogenesis of type IV pili. J. Bacteriol. 188:132–140. 10.1128/JB.188.1.132-140.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 24.Sanger F, Nicklen S, Coulson AR. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U. S. A. 74:5463–5467. 10.1073/pnas.74.12.5463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimbara K, Hashimoto T, Fukuda M, Koana T, Takagi M, Oishi M, Yano K. 1989. Cloning and sequencing of two tandem genes involved in degradation of 2,3-dihydroxybiphenyl to benzoic acid in the polychlorinated biphenyl-degrading soil bacterium Pseudomonas sp. strain KKS102. J. Bacteriol. 171:2740–2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruvkun GB, Ausubel FM. 1981. A general method for site-directed mutagenesis in prokaryotes. Nature 289:85–88. 10.1038/289085a0 [DOI] [PubMed] [Google Scholar]
- 27.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- 28.Roche NimbleGen. 2010. NimbleGen Arrays user's guide: gene expression arrays version 5.1. Roche NimbleGen, Madison, WI [Google Scholar]
- 29.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31:e15. 10.1093/nar/gng015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 4:249–264. 10.1093/biostatistics/4.2.249 [DOI] [PubMed] [Google Scholar]
- 31.Haug A, Larsen B, Smidsrod O. 1966. A study of the constitution of alginic acid by partial acid hydrolysis. Acta Chem. Scand. 20:183–190. 10.3891/acta.chem.scand.20-0183 [DOI] [Google Scholar]
- 32.Arnold K, Bordoli L, Kopp J, Schwede T. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201. 10.1093/bioinformatics/bti770 [DOI] [PubMed] [Google Scholar]
- 33.Kiefer F, Arnold K, Künzli M, Bordoli L, Schwede T. 2009. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 37:D387–D392. 10.1093/nar/gkn750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peitsch MC. 1995. Protein modeling by e-mail. Biotechnology (NY) 13:658–660. 10.1038/nbt0795-658 [DOI] [Google Scholar]
- 35.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. 2009. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30:2785–2791. 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer ELL, Eddy SR, Bateman A, Finn RD. 2012. The Pfam protein families database. Nucleic Acids Res. 40:D290–D301. 10.1093/nar/gkr1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matthews KS, Nichols JC. 1998. Lactose repressor protein: functional properties and structure. Prog. Nucleic Acid Res. Mol. Biol. 58:127–164 [DOI] [PubMed] [Google Scholar]
- 38.Takase R, Ochiai A, Mikami B, Hashimoto W, Murata K. 2010. Molecular identification of unsaturated uronate reductase prerequisite for alginate metabolism in Sphingomonas sp. A1. Biochim. Biophys. Acta 1804:1925–1936. 10.1016/j.bbapap.2010.05.010 [DOI] [PubMed] [Google Scholar]
- 39.Mishima Y, Momma K, Hashimoto W, Mikami B, Murata K. 2003. Crystal structure of AlgQ2, a macromolecule (alginate)-binding protein of Sphingomonas sp. A1, complexed with an alginate tetrasaccharide at 1.6-Å resolution. J. Biol. Chem. 278:6552–6559. 10.1074/jbc.M209932200 [DOI] [PubMed] [Google Scholar]
- 40.Nishitani Y, Maruyama Y, Itoh T, Mikami B, Hashimoto W, Murata K. 2012. Recognition of heteropolysaccharide alginate by periplasmic solute-binding proteins of a bacterial ABC transporter. Biochemistry 51:3622–3633. 10.1021/bi300194f [DOI] [PubMed] [Google Scholar]
- 41.Falcon CM, Matthews KS. 1999. Glycine insertion in the hinge region of lactose repressor protein alters DNA binding. J. Biol. Chem. 274:30849–30857. 10.1074/jbc.274.43.30849 [DOI] [PubMed] [Google Scholar]
- 42.Wilson CJ, Zhan H, Swint-Kruse L, Matthews KS. 2007. The lactose repressor system: paradigms for regulation, allosteric behavior and protein folding. Cell. Mol. Life Sci. 64:3–16. 10.1007/s00018-006-6296-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weickert MJ, Adhya S. 1992. A family of bacterial regulators homologous to Gal and Lac repressors. J. Biol. Chem. 267:15869–15874 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.