Abstract
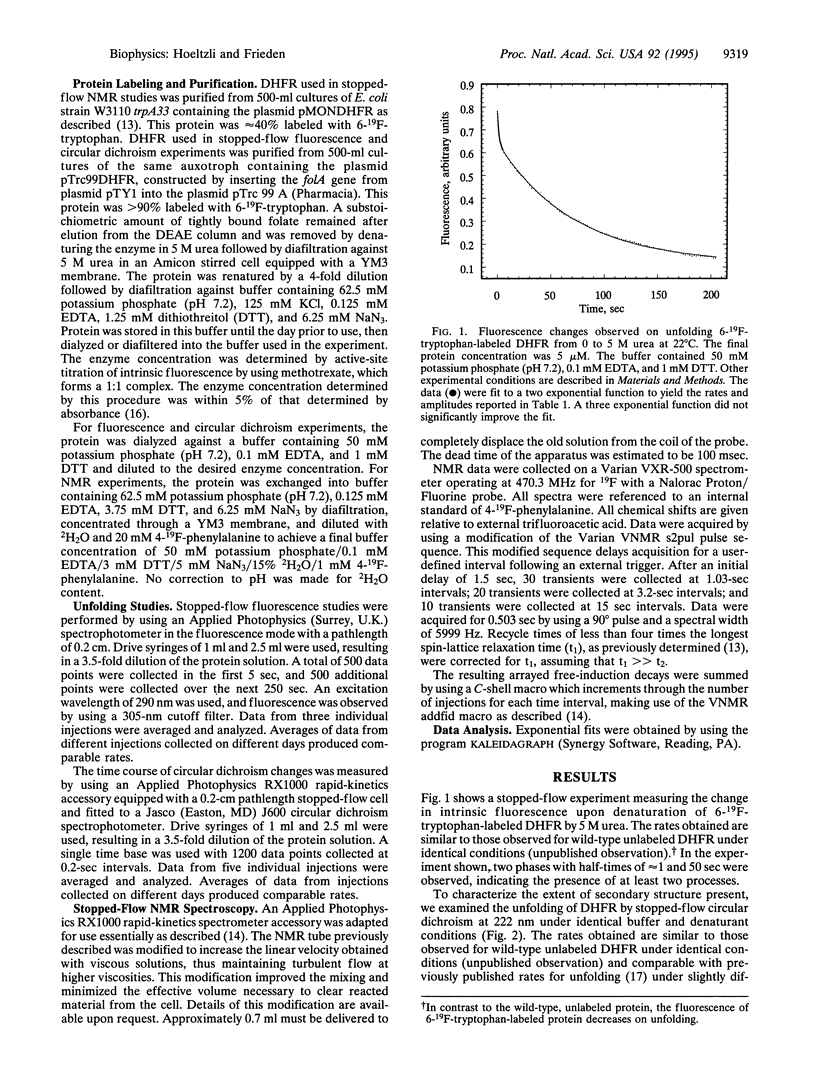
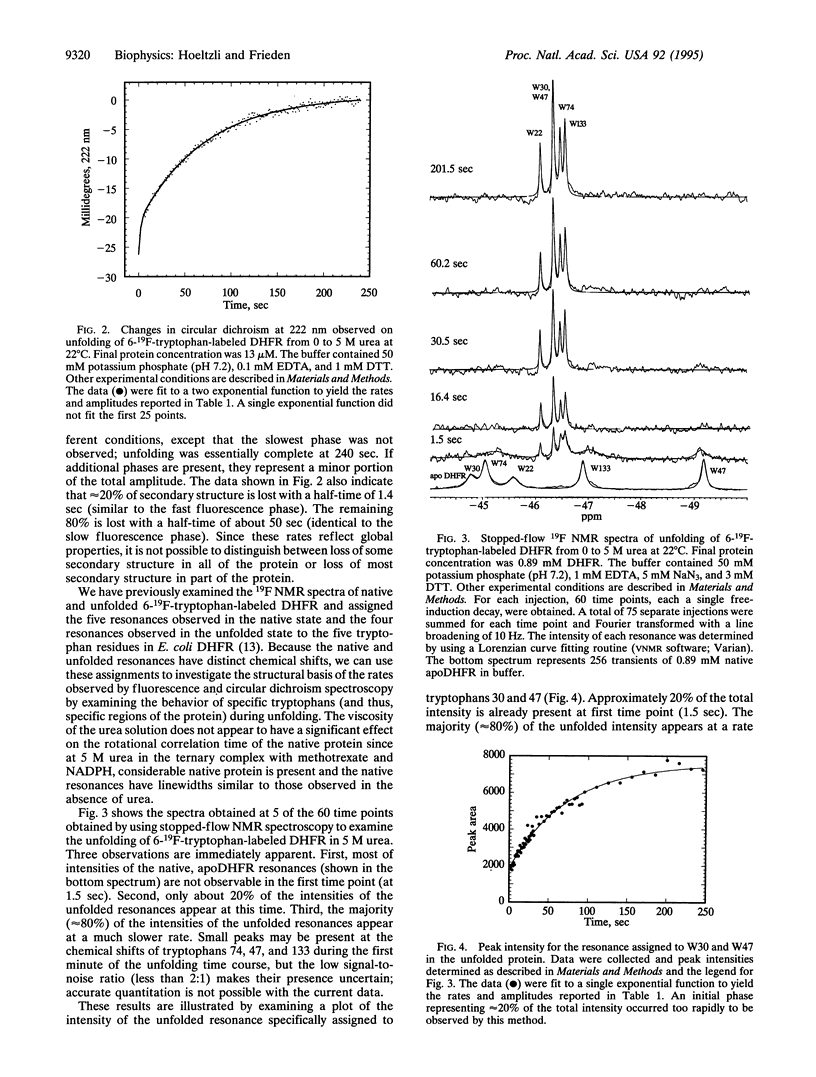
Escherichia coli dihydrofolate reductase (DHFR; EC 1.5.1.3) contains five tryptophan residues that have been replaced with 6-19F-tryptophan. The 19F NMR assignments are known in the native, unliganded form and the unfolded form. We have used these assignments with stopped-flow 19F NMR spectroscopy to investigate the behavior of specific regions of the protein in real time during urea-induced unfolding. The NMR data show that within 1.5 sec most of the intensities of the native 19F resonances of the protein are lost but only a fraction (approximately 20%) of the intensities of the unfolded resonances appears. We postulate that the early disappearance of the native resonances indicates that most of the protein rapidly forms an intermediate in which the side chains have considerable mobility. Stopped-flow far-UV circular dichroism measurements indicate that this intermediate retains native-like secondary structure. Eighty percent of the intensities of the NMR resonances assigned to the individual tryptophans in the unfolded state appear with similar rate constants (k approximately 0.14 sec-1), consistent with the major phase of unfolding observed by stopped-flow circular dichroism (representing 80% of total amplitude). These data imply that after formation of the intermediate, which appears to represent an expanded structural form, all regions of the protein unfold at the same rate. Stopped-flow measurements of the fluorescence and circular dichroism changes associated with the urea-induced unfolding show a fast phase (half-time of about 1 sec) representing 20% of the total amplitude in addition to the slow phase mentioned above. The NMR data show that approximately 20% of the total intensity for each of the unfolded tryptophan resonances is present at 1.5 sec, indicating that these two phases may represent the complete unfolding of the two different populations of the native protein.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baccanari D., Phillips A., Smith S., Sinski D., Burchall J. Purification and properties of Escherichia coli dihydrofolate reductase. Biochemistry. 1975 Dec 2;14(24):5267–5273. doi: 10.1021/bi00695a006. [DOI] [PubMed] [Google Scholar]
- Barshop B. A., Wrenn R. F., Frieden C. Analysis of numerical methods for computer simulation of kinetic processes: development of KINSIM--a flexible, portable system. Anal Biochem. 1983 Apr 1;130(1):134–145. doi: 10.1016/0003-2697(83)90660-7. [DOI] [PubMed] [Google Scholar]
- Bolin J. T., Filman D. J., Matthews D. A., Hamlin R. C., Kraut J. Crystal structures of Escherichia coli and Lactobacillus casei dihydrofolate reductase refined at 1.7 A resolution. I. General features and binding of methotrexate. J Biol Chem. 1982 Nov 25;257(22):13650–13662. [PubMed] [Google Scholar]
- Bystroff C., Kraut J. Crystal structure of unliganded Escherichia coli dihydrofolate reductase. Ligand-induced conformational changes and cooperativity in binding. Biochemistry. 1991 Feb 26;30(8):2227–2239. doi: 10.1021/bi00222a028. [DOI] [PubMed] [Google Scholar]
- Bystroff C., Oatley S. J., Kraut J. Crystal structures of Escherichia coli dihydrofolate reductase: the NADP+ holoenzyme and the folate.NADP+ ternary complex. Substrate binding and a model for the transition state. Biochemistry. 1990 Apr 3;29(13):3263–3277. doi: 10.1021/bi00465a018. [DOI] [PubMed] [Google Scholar]
- Cayley P. J., Dunn S. M., King R. W. Kinetics of substrate, coenzyme, and inhibitor binding to Escherichia coli dihydrofolate reductase. Biochemistry. 1981 Feb 17;20(4):874–879. doi: 10.1021/bi00507a034. [DOI] [PubMed] [Google Scholar]
- Chyan C. L., Wormald C., Dobson C. M., Evans P. A., Baum J. Structure and stability of the molten globule state of guinea-pig alpha-lactalbumin: a hydrogen exchange study. Biochemistry. 1993 Jun 1;32(21):5681–5691. doi: 10.1021/bi00072a025. [DOI] [PubMed] [Google Scholar]
- Englander S. W., Mayne L. Protein folding studied using hydrogen-exchange labeling and two-dimensional NMR. Annu Rev Biophys Biomol Struct. 1992;21:243–265. doi: 10.1146/annurev.bb.21.060192.001331. [DOI] [PubMed] [Google Scholar]
- Evans P. A., Topping K. D., Woolfson D. N., Dobson C. M. Hydrophobic clustering in nonnative states of a protein: interpretation of chemical shifts in NMR spectra of denatured states of lysozyme. Proteins. 1991;9(4):248–266. doi: 10.1002/prot.340090404. [DOI] [PubMed] [Google Scholar]
- Fierke C. A., Johnson K. A., Benkovic S. J. Construction and evaluation of the kinetic scheme associated with dihydrofolate reductase from Escherichia coli. Biochemistry. 1987 Jun 30;26(13):4085–4092. doi: 10.1021/bi00387a052. [DOI] [PubMed] [Google Scholar]
- Fink A. L. Compact intermediate states in protein folding. Annu Rev Biophys Biomol Struct. 1995;24:495–522. doi: 10.1146/annurev.bb.24.060195.002431. [DOI] [PubMed] [Google Scholar]
- Frieden C. Refolding of Escherichia coli dihydrofolate reductase: sequential formation of substrate binding sites. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4413–4416. doi: 10.1073/pnas.87.12.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeltzli S. D., Frieden C. 19F NMR spectroscopy of [6-19F]tryptophan-labeled Escherichia coli dihydrofolate reductase: equilibrium folding and ligand binding studies. Biochemistry. 1994 May 10;33(18):5502–5509. doi: 10.1021/bi00184a019. [DOI] [PubMed] [Google Scholar]
- Jennings P. A., Finn B. E., Jones B. E., Matthews C. R. A reexamination of the folding mechanism of dihydrofolate reductase from Escherichia coli: verification and refinement of a four-channel model. Biochemistry. 1993 Apr 13;32(14):3783–3789. doi: 10.1021/bi00065a034. [DOI] [PubMed] [Google Scholar]
- Kiefhaber T., Baldwin R. L. Kinetics of hydrogen bond breakage in the process of unfolding of ribonuclease A measured by pulsed hydrogen exchange. Proc Natl Acad Sci U S A. 1995 Mar 28;92(7):2657–2661. doi: 10.1073/pnas.92.7.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefhaber T., Labhardt A. M., Baldwin R. L. Direct NMR evidence for an intermediate preceding the rate-limiting step in the unfolding of ribonuclease A. Nature. 1995 Jun 8;375(6531):513–515. doi: 10.1038/375513a0. [DOI] [PubMed] [Google Scholar]
- Kuwajima K., Garvey E. P., Finn B. E., Matthews C. R., Sugai S. Transient intermediates in the folding of dihydrofolate reductase as detected by far-ultraviolet circular dichroism spectroscopy. Biochemistry. 1991 Aug 6;30(31):7693–7703. doi: 10.1021/bi00245a005. [DOI] [PubMed] [Google Scholar]
- Kuwajima K., Mitani M., Sugai S. Characterization of the critical state in protein folding. Effects of guanidine hydrochloride and specific Ca2+ binding on the folding kinetics of alpha-lactalbumin. J Mol Biol. 1989 Apr 5;206(3):547–561. doi: 10.1016/0022-2836(89)90500-7. [DOI] [PubMed] [Google Scholar]
- Mücke M., Schmid F. X. A kinetic method to evaluate the two-state character of solvent-induced protein denaturation. Biochemistry. 1994 Nov 1;33(43):12930–12935. doi: 10.1021/bi00209a025. [DOI] [PubMed] [Google Scholar]
- Pace C. N. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 1986;131:266–280. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- Penner M. H., Frieden C. Kinetic analysis of the mechanism of Escherichia coli dihydrofolate reductase. J Biol Chem. 1987 Nov 25;262(33):15908–15914. [PubMed] [Google Scholar]
- Roder H. Structural characterization of protein folding intermediates by proton magnetic resonance and hydrogen exchange. Methods Enzymol. 1989;176:446–473. doi: 10.1016/0076-6879(89)76024-9. [DOI] [PubMed] [Google Scholar]
- Shakhnovich E. I., Finkelstein A. V. Theory of cooperative transitions in protein molecules. I. Why denaturation of globular protein is a first-order phase transition. Biopolymers. 1989 Oct;28(10):1667–1680. doi: 10.1002/bip.360281003. [DOI] [PubMed] [Google Scholar]
- Touchette N. A., Perry K. M., Matthews C. R. Folding of dihydrofolate reductase from Escherichia coli. Biochemistry. 1986 Sep 23;25(19):5445–5452. doi: 10.1021/bi00367a015. [DOI] [PubMed] [Google Scholar]
- Van Dael H., Haezebrouck P., Morozova L., Arico-Muendel C., Dobson C. M. Partially folded states of equine lysozyme. Structural characterization and significance for protein folding. Biochemistry. 1993 Nov 9;32(44):11886–11894. doi: 10.1021/bi00095a018. [DOI] [PubMed] [Google Scholar]



