Abstract
Staphylococcal enterotoxin B (SEB) causes food poisoning in humans. It is considered a biological weapon, and inhalation can trigger lung injury and sometimes respiratory failure. Being a superantigen, SEB initiates an exaggerated inflammatory response. While the role of microRNAs (miRNAs) in immune cell activation is getting increasing recognition, their role in the regulation of inflammatory disease induced by SEB has not been studied. In this investigation, we demonstrate that exposure to SEB by inhalation results in acute inflammatory lung injury accompanied by an altered miRNA expression profile in lung-infiltrating cells. Among the miRNAs that were significantly elevated, miR-155 was the most overexpressed. Interestingly, miR-155−/− mice were protected from SEB-mediated inflammation and lung injury. Further studies revealed a functional link between SEB-induced miR-155 and proinflammatory cytokine gamma interferon (IFN-γ). Through the use of bioinformatics tools, suppressor of cytokine signaling 1 (SOCS1), a negative regulator of IFN-γ, was identified as a potential target of miR-155. While miR-155−/− mice displayed increased expression of Socs1, the overexpression of miR-155 led to its suppression, thereby enhancing IFN-γ levels. Additionally, the inhibition of miR-155 resulted in restored Socs1expression. Together, our data demonstrate an important role for miR-155 in promoting SEB-mediated inflammation in the lungs through Socs1 suppression and suggest that miR-155 may be an important target in preventing SEB-mediated inflammation and tissue injury.
INTRODUCTION
Staphylococcal enterotoxin B (SEB), a superantigen produced by Staphylococcus aureus, has deleterious effects in humans, such as food poisoning (1) and toxic shock (2). Because it can be easily aerosolized, SEB is classified as a category B agent by the Centers for Disease Control and Prevention (3). Upon inhalation exposure, SEB can trigger acute inflammatory lung injury characterized by immune cell infiltration, excessive cytokine production, tissue damage, and pulmonary edema (4, 5).
Due to the distinct manner in which SEB binds to the nonpolymorphic regions of major histocompatibility complex class II (MHC-II) on antigen-presenting cells and the specific Vβ regions of the T-cell receptor (TCR), such as murine Vβ8 (6), SEB exposure leads to the activation and proliferation of a large population (5 to 30%) of T lymphocytes (7). Activation of such a substantial number of T lymphocytes results in the robust production of inflammatory cytokines, such as interleukin 2 (IL-2), tumor necrosis factor alpha (TNF-α), and gamma interferon (IFN-γ) (8, 9). In most cases, IFN-γ is the main culprit in mediating the damaging and often lethal effects seen upon SEB exposure. For example, transgenic mice deficient in IFN-γ were protected from SEB-mediated toxic shock syndrome (TSS) and subsequent mortality (10). Additionally, the neutralization of IFN-γ, after SEB exposure was shown to prevent lethal systemic inflammation (11), further suggesting the importance of SEB-mediated IFN-γ production. While the interaction between SEB and TCR, along with the subsequent T-cell proliferation and cytokine secretion, has been extensively studied (12–14), the role of microRNA (miRNA) in mediating SEB-induced inflammation has not yet been elucidated.
MicroRNAs are ∼21 to 23 nt long, single-stranded noncoding RNA molecules that can translationally repress or target mRNA for degradation, thereby acting as primary modulators of gene expression (15). Several studies have demonstrated a role for miRNA in modulating immune responses under various inflammatory conditions (16). For example, while miR-125b is highly expressed in naive CD4+ T cells, it becomes significantly downregulated upon T-cell activation (17). Similarly, studies have demonstrated that overexpression of the (miR-17–92) cluster in T cells leads to lymphoproliferative disorders due to the repression of the proapoptotic molecule, BIM (18). Furthermore, mice deficient in miR-155 are resistant to developing experimental autoimmune encephalomyelitis (EAE), a mouse model of multiple sclerosis (19), while the overexpression of miR-155 exacerbates the symptoms associated with the disease. Taken together, these studies strongly suggest that miRNAs play a major role in modulating immune cell activation, particularly T cells, as well as promoting proinflammatory responses.
In the current study of SEB-induced acute inflammatory lung injury, we applied microarray analysis and quantitative real-time PCR (qRT-PCR) to establish important miRNAs that are dysregulated in response to SEB. Further, our data identified miR-155 as a major contributor to SEB-mediated lung inflammation. While it is known that SEB exposure leads to inflammation and the production of copious amounts of IFN-γ, we provide mechanistic insight through gain- and loss-of-function experiments into the role of miR-155 in this process. Our results may present an opportunity to further therapeutically target miR-155 in the treatment of SEB-mediated acute inflammatory lung injury.
MATERIALS AND METHODS
Mice.
Female C57BL/6 mice (6 to 8 weeks old) were purchased from the National Cancer Institute (NCI). miR-155−/− (B6.Cg-Mir155 tm1.1 Rsky/J) mice were purchased from The Jackson Laboratory. All mice were housed under pathogen-free conditions at the Animal Resource Facility (ARF), University of South Carolina (USC) School of Medicine. The use of vertebrate animals in the experiments performed was preapproved by the Institutional Animal Care and Use Committee (IACUC) at USC. This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Research Council (20).
Induction of SEB-induced acute lung injury (ALI).
SEB was obtained from Toxin Technologies (Sarasota, FL). SEB dissolved in sterile phosphate-buffered saline (PBS) (2 mg/ml) was administered by the intranasal (i.n.) route in a volume of 25 μl for a dose of 50 μg per mouse, as described previously (4, 21, 22). Mice were euthanized 48 h after SEB exposure.
Lung histopathological analysis.
At the time of euthanasia, lungs were obtained and fixed in 10% formalin. The tissue was then paraffin embedded and serial sections (5 μm) were made. The sections were subsequently deparaffinized by being dissolved with xylene, followed by rehydration in several changes of alcohol (100%, 95%, and 90%). The slides were then stained with hematoxylin and eosin (H&E) and evaluated with a Nikon E600 light microscopy system.
Antibodies.
Fluorescein isothiocyanate-conjugated anti-CD8 (clone 53.6.7) and phycoerythrin-conjugated anti-CD4 (clone GK1.5) antibodies (Abs) were purchased from BioLegend (San Diego, CA).
Preparation of lung-infiltrating cells and flow cytometry.
Mice were exposed to SEB as described above. Forty-eight hours after SEB exposure, lungs were harvested and homogenized using a Stomacher 80 Biomaster blender from Seward (Davie, FL) in 10 ml of sterile PBS. After being washed with sterile PBS, the cells were carefully layered on Ficoll Histopaque-1077 from Sigma-Aldrich (St. Louis, MO) and separated by density gradient centrifugation at 500 × g for 30 min at 24°C with the brake off. The mononuclear cell layer isolated was then enumerated using the Trypan blue exclusion method. To determine the subsets of immune cells infiltrating the lung, cells were stained with fluorescent-conjugated antibodies (anti-CD4, anti-CD8) and analyzed using the Beckman Coulter 500 flow cytometer (Indianapolis, IN).
Recovery of bronchoalveolar lavage fluid (BALF) and cytokine detection.
Forty-eight hours after SEB exposure, mice were euthanized, tracheae from vehicle- or SEB-treated mice were tied with a suture, and the lung was excised as an intact unit. With 1 ml sterile ice-cold PBS, the trachea was lavaged to collect the BALF fluid. Cytokine analysis for IFN-γ was carried out using BALF. All cytokines were measured using BioLegend (San Diego, CA) enzyme-linked immunosorbent assay (ELISA) MAX standard kits.
Total RNA isolation.
Total RNA (including small RNAs) was isolated from lung-infiltrating mononuclear cells or in vitro from lymph nodes (LN) or splenocytes using the miRNeasy kit from Qiagen (Valencia, CA) by following the manufacturer's instructions. The purity and concentration of the RNA was confirmed spectrophotometrically, while the integrity of miRNA was further assessed using an Agilent 2100 Bioanalyzer (Agilent Tech, Palo Alto, CA).
miRNA expression profiling and analysis.
To profile miRNA expression in the lung, the Affymetrix GeneChip miRNA 1.0 array platform was used. The array, which is composed of 609 mouse miRNA probes, makes use of the FlashTag biotin HSR hybridization technique and was carried out according to the manufacturer's instructions (Affymetrix, Santa Clara, CA). Fluorescent intensities obtained from hybridization were log transformed and visualized in the form of a heatmap. Hierarchical clustering was carried out using Ward's method, and similarity measurement was calculated using half square Euclidean distance. miRNA expression fold changes obtained from the microarray were then further analyzed using the commercially available analysis tool Ingenuity Systems Ingenuity Pathway analysis (IPA), (Mountain View, CA, USA.) In brief, the data set of 609 miRNAs was uploaded into IPA and only miRNAs that were 3-fold or higher were considered for analysis. Core analysis was carried out and a “top network” of miRNA and its associated molecules was generated.
Analysis of miRNA target genes.
IPA was also used to determine and collate the highly predicted, moderately predicted, and experimentally observed mRNA target genes of those miRNAs that were highly (≥3-fold) upregulated using IPA's miRNA target filter tool. These targets were further sorted based on their role in cytokine signaling, cellular growth and proliferation, and cellular immune response. Additionally, to assign immunological functions to our list of miRNA targets, gene ontology (GO) mapping of miRNA target genes was carried out using Cytoscape 3.0.1 equipped with ClueGO and CluePedia applications.
qRT-PCR.
Total RNA (miRNA and mRNA) was converted to cDNA using the miScript cDNA synthesis kit (Qiagen) according to the manufacturer's instructions. For miRNA validation, the miScript SYBR green PCR kit (Qiagen) was used, and fold change of miRNA was determined by using the 2−ΔΔCT method. Snord96a was used as the small RNA endogenous control. For mRNA validation, an SSO advanced SYBR green PCR kit from Bio-Rad (Hercules, CA) was used according to the manufacturer's instructions, and β-actin was used as the endogenous control. The following primers were used: β-actin forward (F) (5′-GGCTGTATTCCCCTCCAT G-3′) and reverse (R) (5′-CCAGTT GGTAACAATGCCATGT-3′); Socs1 F (5′-GGTTGTAGCAGCTTGTGTC-3′) and R (5′-AATGAAGCCAGAGACCCTC-3′); IFN-γ F (5′-GCGTCATTGAATCACACCTG-3′) and R (5′-GAGCTCATTGAATGCTTGGC-3′).
Transfection with miR-155 mimic and inhibitors.
Lymph nodes (axillary and inguinal) from naive C57BL/6 mice were harvested and cultured in 10 ml of complete medium at 37°C and 5% CO2. Complete medium was comprised of RPMI 1640 medium (Gibco Laboratories, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS), 10 mM l-glutamine, 10 mM HEPES, 50 μM β-mercaptoethanol, and 100 μg/ml penicillin. Cells were seeded at 2 × 105 cells in 24-well plates and transfected with either 40 nM synthetic mmu-miR-155-3p miScript miRNA mimic (CUCCUACCUGUUAGCAUUAAC) or AllStar negative-control small interfering RNA (siRNA). For inhibition of miR-155, cells were activated with SEB (1 μg/ml) and treated with 100 nM anti-mmu-miR-155-3p miScript miRNA inhibitor (CUCCUACCUGUUAGCAUUAAC) or miScript inhibitor negative control for 24 h using HiperFect transfection reagent (Qiagen) according to the manufacturer's instructions.
Luciferase assay.
The following plasmids were purchased from GeneCopoeia (Rockville, MD): 3′ untranslated region (3′UTR), Socs1 (MmiT028883), and control plasmid (CmiT000001-MT01). Chinese hamster ovary (CHO) cells were cotransfected with 100 ng of plasmid and 100 nM miRIDIAN microRNA mmu-miR-155-5p mimic or miRIDIAN microRNA mimic negative control using DharmaFECT DUO transfection reagent by following the manufacturer's instructions (Thermo Scientific, Pittsburgh, PA). Twenty-four hours following transfection, Luciferase activity was measured using LucPair miR-Duo Luciferase assay kit from GeneCopoeia.
Statistics.
All statistical analyses were carried out using GraphPad Prism Software (San Diego, CA). In all experiments, the number of mice used was 4 or 5 per group, unless otherwise specified. Results are expressed as means ± standard errors of the means (SEM). Student's t test was used to compare wild-type (WT) and miR-155−/− data, whereas multiple comparisons were made using one-way analysis of variance (ANOVA), followed by post hoc analysis using Tukey's method. A P value of <0.05 was considered statistically significant. Individual experiments were performed in triplicate, and each experiment was performed independently at least three times to test reproducibility of results.
Microarray accession number.
All microRNA microarray data were deposited the in ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-2379.
RESULTS
SEB exposure triggers inflammation in the lung.
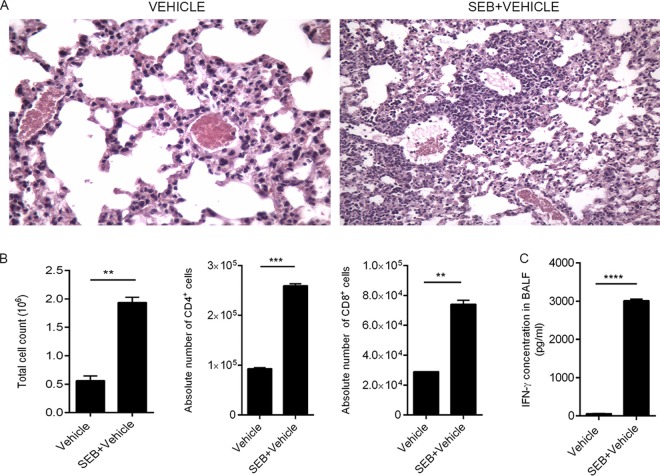
Previously, a single dose of SEB (50 μg) by intranasal delivery was found to induce cellular infiltration, increase cytokine production, and cause histopathological lesions and edema in C57BL/6 mice (4, 21, 22), mimicking the symptoms of acute inflammatory lung injury in humans (23). In this study, we first sought to investigate the inflammatory effect of SEB exposure in the lungs. Forty-eight hours after SEB exposure, H&E-stained sections of the lungs from SEB-exposed mice showed massive infiltration of cells and signs of edema as evidenced by fluid-filled bronchioles (Fig. 1A). Because SEB is a potent activator of T cells, we examined the effect of SEB on T-cell subsets within the lung. Immediately after euthanasia, the lungs were harvested and mononuclear cells were isolated from the lungs by density gradient centrifugation to determine the phenotypic characteristics of the cells. SEB exposure not only led to an overall increase in mononuclear cells but, specifically, an increase in CD4+ and CD8+ T cells (Fig. 1B) was seen. Because SEB exposure triggers an increase in IFN-γ, a major proinflammatory cytokine previously reported to orchestrate the inflammatory cascade and cause tissue damage (10, 24, 25), we analyzed the concentration of IFN-γ in the bronchoalveolar lavage fluid (BALF) and found a high concentration (up to 3,000 pg/ml) of IFN-γ in the lungs of SEB-exposed mice (Fig. 1C). These data suggested that SEB administration via the intranasal route triggers acute inflammation in the lungs.
FIG 1.
SEB induces lung inflammation. (A) Representative H&E images (×20) of cross sections of the lung from mice exposed to either vehicle or SEB. (B) Lung-infiltrating mononuclear cells obtained by density gradient centrifugation and total number of viable cells were counted using a hemocytometer. Cells were further stained with monoclonal antibody (MAb) to identify CD4+ and CD8+ cells and were analyzed on a flow cytometer. The percentage of the immune cell subsets was multiplied by the total number of cells found in the lung and divided by 100 to yield the absolute cell numbers shown. (C) The concentration of IFN-γ protein present in the BALF was determined using a standard ELISA kit. Data are means ± SEM (n = 5) and are representative of three independent experiments. Statistical significance compared to SEB-vehicle is indicated as follows: **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
SEB exposure modulates miRNA expression in the lungs.
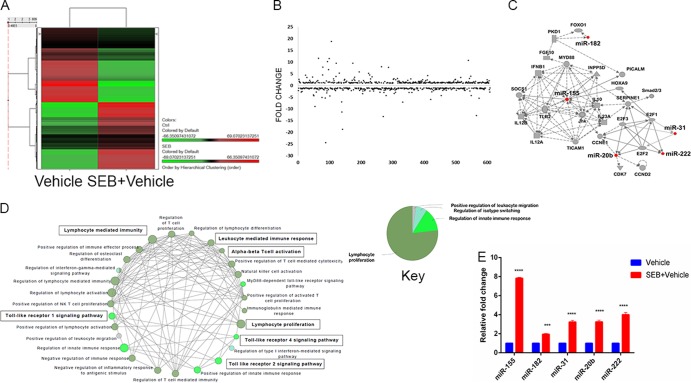
The dysregulation of specific miRNAs in response to SEB exposure has not been elucidated. Because miRNAs play a critical role in mediating inflammation, we examined the miRNA profile after SEB exposure. Total miRNA was isolated from lung-infiltrating mononuclear cells, and the relative abundance of miRNA in SEB-exposed and vehicle-treated mice was determined using microarray miRNA analysis. A heatmap was generated based on hierarchical clustering of miRNA, highlighting a stark difference between vehicle- and SEB-exposed mice (Fig. 2A). Further examination of miRNA expression revealed that of the 609 miRNAs assessed, most remained unchanged, but a few showed significant up- or downregulation, as seen in the fold change distribution plot (Fig. 2B). Ingenuity pathway analysis (IPA) generated a “top network” of miRNAs that was comprised of five upregulated miRNAs, including miR-155, miR-31, miR-182, miR-20b, and miR-222 and their associated molecules (Fig. 2C). This network was characterized by IPA as responses involving inflammation, cellular development, cellular growth, and proliferation. To assign significant immunological functions to the genes in the aforementioned top network, Cytoscape (ClueGO+CluePedia application) was employed. Gene ontology (GO) mapping revealed that the genes associated with the miRNA in the top network were functionally relevant to T-cell activation (GO 0042110) and proliferation (GO 0042098), IFN-γ signaling (GO 0060333), and Toll-like receptor signaling (GO 0002224) (Fig. 2D). Next, we validated the expression levels of these miRNAs in lung-infiltrating mononuclear cells by qRT-PCR, which corroborated the expression patterns seen using the microarray (Fig. 2E). Among the miRNAs we validated, miR-155 was the most highly expressed (∼8-fold) in the lungs upon SEB exposure. Based on this, the role of miR-155 in the development of SEB-induced ALI was further investigated.
FIG 2.
SEB exposure leads to dysregulation of miRNA. Forty-eight hours after vehicle or SEB administration, miRNA was isolated from lung-infiltrating mononuclear cells. (A) Heatmap depicting differential expression of miRNA in the lungs of mice exposed to SEB-vehicle compared to vehicle; (B) fold change distribution of the 609 miRNAs, indicating several upregulated and downregulated miRNAs; (C) ingenuity pathway analysis-generated “top network” with network function denoted as “inflammatory response, cellular development, cellular growth, and proliferation”; (D) Cytoscape-generated gene ontology (GO) network based on immunological processes for the molecules associated in the top network using the ClueGo 2.0.7 application. Analysis criteria consisted of a two-sided hypergeometric test with Benjamini Hochberg correction. Only results with a kappa score of 0.3 are displayed. (E) qRT-PCR validation of the IPA-generated top upregulated miRNA. Total RNA was isolated from lung-infiltrating mononuclear cells. Snord96a was used as the small RNA endogenous control, and the expression level of SEB-induced miRNA shown here was normalized to vehicle. Data are represented as means ± SEM from replicate samples (***, P < 0.05; ****, P < 0.01 compared to vehicle).
miR-155 is important for SEB-mediated inflammation.
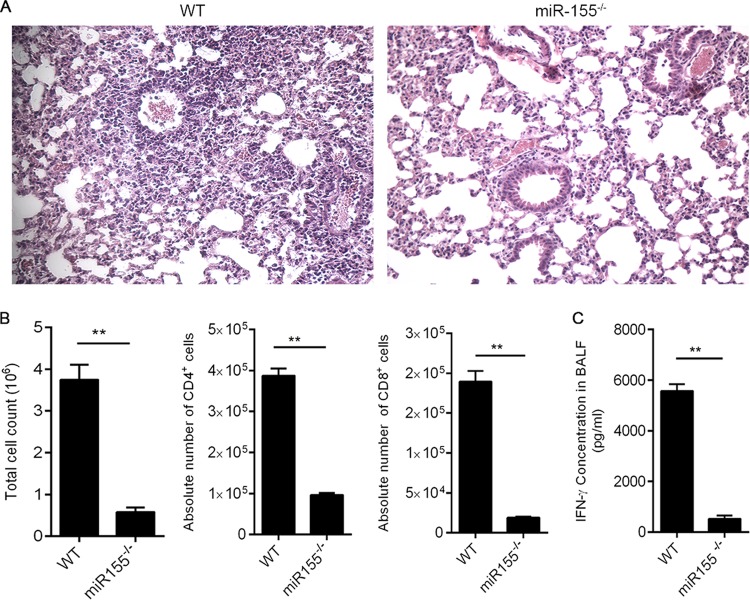
Because miR-155 was highly upregulated in response to SEB, we hypothesized that it might play a crucial role in facilitating the inflammation observed during the disease. To that end, WT and miR-155−/− mice were exposed to SEB to determine the effects on disease parameters. H&E-stained sections of the lung revealed that WT mice exposed to SEB exhibited numerous layers of infiltration interspersed with edema. Interestingly, miR-155−/− mice exposed to SEB presented with almost normal lung architecture (Fig. 3A). Additionally, the miR-155-deficient mice expressed significantly decreased total numbers of mononuclear cells within the lung upon SEB exposure compared to those of their WT counterparts. Upon closer examination of the mononuclear cell phenotype within the lungs, absolute numbers of CD4+ and CD8+ in the miR-155−/− mice were decreased compared to those of WT mice (Fig. 3B). Additionally, cytokine analysis of the bronchoalveolar lavage fluid (BALF) in the lungs of WT mice demonstrated high concentrations of proinflammatory cytokine IFN-γ. In contrast, IFN-γ levels were significantly diminished in miR-155−/− mice (Fig. 3C). Taken together, these results provided clear evidence that miR-155−/− mice were protected from SEB-mediated ALI, suggesting that miR-155 plays a critical role in SEB-induced inflammation.
FIG 3.
miR-155 plays a critical role in SEB-induced ALI. WT (C57BL/6) and miR-155−/− (B6.Cg-Mir155 tm1.1 Rsky/J) mice were exposed to SEB and euthanized 48 h later. (A) Representative H&E images (×20) of sections of lung indicating immune cell infiltration; (B) phenotypic characterization of cells infiltrating the lung was determined by staining of mononuclear cells with fluorescent-conjugated MAb against CD4 and CD8; (C) levels of IFN-γ cytokine in the BALF was determined by ELISA. Data are means ± SEM (n = 5) and are representative of two independent experiments. Statistical significance compared to WT is indicated as follows: **, P < 0.01.
miR-155 expression is critically linked to IFN-γ production.
Because SEB exposure leads to the release of copious amounts of IFN-γ and also results in increased miR-155 expression, we considered if there was a positive correlation between IFN-γ secretion and the expression of miR-155. To explore this possibility, we first assessed the expression of IFN-γ after transfection of LN T cells with a synthetic miR-155 mimic. Interestingly, we found a substantial increase in IFN-γ levels not only in WT mice (Fig. 4A) but also in miR-155-deficient mice that were transfected with the mimic (Fig. 4B). Additionally, inhibition of miR-155 with a synthetic inhibitor conversely resulted in the diminished expression of IFN-γ (Fig. 4C), suggesting a crucial link between miR-155 expression and that of IFN-γ after SEB exposure.
FIG 4.
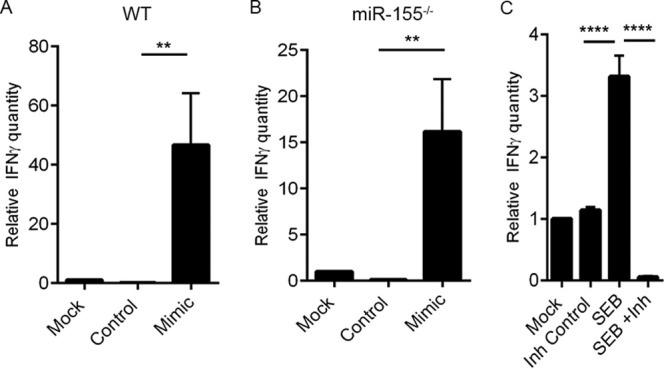
IFN-γ forms a critical link between SEB and subsequent miR-155 induction. (A) Lymph node (LN) T cells obtained from naive wild-type (WT) mice were transfected either with miR-155 mimic (Mimic) or mimic control (Control) for 24 h. IFN-γ levels were determined by RT-PCR. (B) LN T cells obtained from miR-155−/− mice were also transfected with 40 nM miR-155 mimic or mimic control as indicated for 24 h, and IFN-γ levels were assessed. (C) LN cells were activated with SEB (1 μg/ml) for 24 h. Cells were then transfected with 100 nM miR-155 inhibitor (Inh) or inhibitor control (Inh Control) for another 24 h. IFN-γ levels were determined via RT-PCR. Data are represented as means ± SEM from replicate samples. Statistical significance is indicated as follows: **, P < 0.01; ****, P < 0.0001.
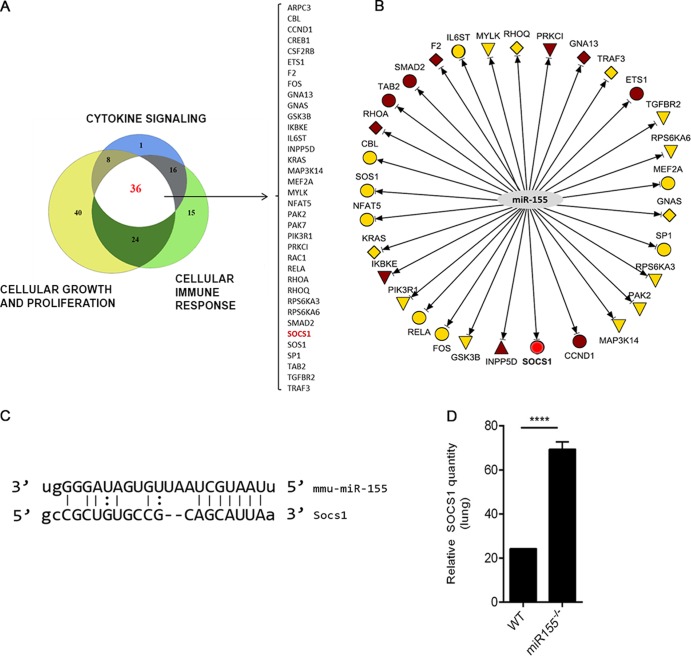
miR-155 targets Socs1, a negative regulator of IFN-γ.
miRNAs regulate the expression of genes by binding the 3′UTR of their respective target mRNA. To examine the link between miR-155 and its potential target genes, we undertook a bioinformatics-based approach. First, IPA miRNA target filter tools were used, selecting only those miR-155 target genes that were relevant to cytokine signaling, cellular immune response, and cellular growth and proliferation. Thirty six targets common to the filtering criteria applied were selected (Fig. 5A). Next, an IPA-generated network was used to sort the targets based on those that were highly predicted and experimentally observed (Fig. 5B). Conclusively, a gene known as suppressor of cytokine signaling 1 (Socs1) was found to be a prominent miR-155 target due to miR-155's ability to bind to the 3′UTR of the Socs1 mRNA (Fig. 5C). Because Socs1 is induced by IFN-γ and acts as a negative regulator of IFN-γ, the relationship between miR-155 and Socs1 in the context of IFN-γ production was examined. miR-155−/− mice that were exposed to SEB had significantly increased expression of Socs1 mRNA in lung-infiltrating mononuclear cells compared to that of the WT (Fig. 5D). Accordingly, we hypothesized that miR-155 may target Socs1 to promote IFN-γ-mediated inflammation during SEB-induced inflammatory ALI. To confirm Socs1 as an miR-155 target, we first measured relative luciferase activity after cotransfection of miR-155 mimic and plasmid containing the 3′ UTR of Socs1. Compared to the mimic control, miR-155 mimic led to a significant decrease in luciferase activity (Fig. 6A), validating Socs1 as a target for miR-155. Next, the impact of miR-155 mimic on Socs1 levels was explored. We found that both in WT (Fig. 6B) and miR-155-deficient cells (Fig. 6C) that were transfected with miR-155 mimic, Socs1 levels remained suppressed. On the other hand, whereas SEB activation continued to lead to a repression in Socs1, miR-155 inhibition of SEB-activated cells resulted in its derepression (Fig. 6D), confirming the role of miR-155 in suppressing Socs1 during SEB-mediated activation of immune cells.
FIG 5.
Identification of SEB-induced miR-155 targets. (A) miR-155 targets were filtered based on their role in cytokine signaling, cell growth and proliferation, and cellular immune response using IPA. A proportional Venn diagram indicating the miR-155 targets common to all three filtering criteria was generated. The list of targets is indicated within brackets, and Socs1, a highly predicted target, is in red. (B) IPA network was generated highlighting the highly predicted (yellow) and experimentally observed (brown) miR-155 targets, in addition to Socs1 (red), the target of interest. (C) Schematic illustration of the predicted target site for miR-155 within the 3′UTR of Socs1 mRNA. (D) Total mRNA was isolated from lung-infiltrating mononuclear cells of WT and miR-155−/− mice exposed to SEB. Relative expression of Socs1 mRNA was determined by qRT-PCR using β-actin as the endogenous control. Data are represented as means ± SEM from replicate samples. Statistical significance is indicated as follows: ****, P < 0.0001.
FIG 6.
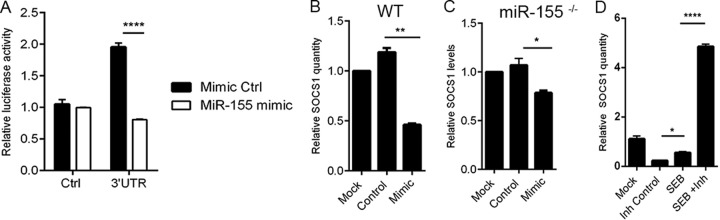
miR-155 targets Socs1. (A) Chinese hamster ovary (CHO) cells were cotransfected with miR-155 mimic or mimic control along with plasmid containing either the 3′UTR of Socs1 or control plasmid for 24 h. Relative luciferase activity (firefly normalized to renilla) was determined following transfection. (B) Lymph nodes (LN) cells obtained from naive wild-type (WT) mice were transfected either with 40 nM miR-155 mimic (Mimic) or mimic control (Control) for 24 h. Socs1 levels were determined by RT-PCR. (C) LN cells obtained from miR-155−/− were also transfected with 40 nM miR-155 mimic or mimic control as indicated for 24 h, and Socs1 levels were assessed. (D) LN cells were activated with SEB (1 μg/ml) for 24 h. Cells were then transfected with 100 nM miR-155 inhibitor (Inh) or inhibitor control (Inh Control) for another 24 h. Socs1 levels were determined via qRT-PCR. Data are represented as means ± SEM from replicate samples statistical significance is indicated as follows: *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
DISCUSSION
With the discovery of miRNA, a novel and exciting mechanism of gene regulation has arisen. miRNAs are small noncoding endogenous RNA molecules that bind 3′UTR of genes carrying complementary sites. A single miRNA usually targets several mRNAs, acting as a fine-tuner of gene regulation rather than an on-off system (26). In the context of inflammation, miRNAs have been found within a variety of immune cells often targeting genes involved in the regulation of inflammatory response (27). In the current study, we closely examined the miRNA profile after inhalation exposure to SEB. We observed that among the miRNAs that were dysregulated in response to SEB, miR-155 was one of the most significantly altered. It has been reported that naive CD4+ T cells initially display low levels of miR-155, which is increased after the engagement of TCR by an antigen (28, 29). This is consistent with our observation that miR-155 was upregulated following SEB activation.
Recent studies have reported that miR-155−/− mice are resistant to EAE, demonstrating its importance in mediating disease development (30). In other studies, miR-155−/− mice failed to control Helicobacter pylori infection due to defective Th1 signaling (31). Moreover, in a mouse model of collagen-induced arthritis (CIA), the deficiency of miR-155 led to a decrease in pathogenic T cells. In humans, it has also been reported that soldiers undergoing a battlefield-like stress program demonstrated an increase in hsa-miR-155 levels in leukocytes exposed to SEB ex vivo (32), indicating that stress-related inflammation could also potentially lead to the increase in miR-155. The current study further suggests that the acute inflammatory response in the lungs to a bacterial superantigen is also regulated by miR-155, inasmuch as SEB-exposed miR-155−/− mice having fewer infiltrating T cells than their WT counterparts and almost normal lung histopathology.
Usually, an appropriate regulation of IFN-γ is necessary for mediating Th1 responses and blunting infection (33). SEB exposure, however, causes an excessive release of IFN-γ. T cells exposed to IFN-γ proliferate further, thus perpetuating a cycle of inflammation (34, 35). Studies carried out with SEB activation of splenocytes have demonstrated that early cytokines released include IL-2 and TNF-α, followed by the massive production of IFN-γ by T-helper cells (36). Additionally, in vitro SEB activation of rat splenocytes results in the release of IFN-γ for up to 48 h postactivation and promotes the proliferation of CD4+ T cells, similar to that seen in mice and humans (37). We have noted in the current model (data not shown) that the peak of acute inflammatory lung injury occurs at 48 h after SEB exposure. In line with the typical kinetics of cytokine release seen with SEB activation, we did not detect early cytokines, TNF-α and IL-2, in the BALF at this time point (data not shown). However, our data indicated substantial release of SEB-induced IFN-γ in WT mice, suggesting that this particular cytokine may significantly contribute to SEB-induced inflammation. Moreover, the correlation between decreased IFN-γ in the BALF of miR-155−/− mice exposed to SEB and lack of significant inflammation in the lungs is suggestive of a major role for IFN-γ in our model.
The relationship between miR-155 and IFN-γ has been briefly explored in previous studies. For example, when miR-155 is overexpressed in human NK cells, the subsequent downregulation of a target, SHIP-1, promotes IFN-γ expression (38). During collagen-induced arthritis, miR-155−/− mice display significantly lower numbers of IFN-γ-producing cells than WT mice (39). Likewise, our data demonstrated that while transfection with a synthetic miR-155 mimic leads to the induction of IFN-γ, the blockade of miR-155 diminishes IFN-γ production. Our results clearly indicate that the SEB-mediated induction of IFN-γ can be explained, at least in part, by the induction of miR-155.
To uncover the relationship between miR-155 and IFN-γ in response to SEB exposure, we employed bioinformatics tools and conducted an extensive literature search. These efforts suggested suppressor of cytokine signaling 1 (SOCS1) as a possible link between miR-155 and IFN-γ. SOCS1 belongs to a family of eight proteins (SOCS1 to SOCS7 and CIS) that regulate the production of several cytokines (40). In particular, SOCS1 is induced by IFN-γ for autoregulation of the IFN-γ proinflammatory response by inhibiting the JAK/STAT1 signaling pathway (41). Recent experiments in numerous cell types have revealed that miR-155 targets SOCS1 (42–44). For example, macrophages infected with an RNA virus demonstrated enhanced type I interferon production due to miR-155 targeting of Socs1 (45). In the current study, we noted that the miR-155−/− mice that are exposed to SEB showed increased expression in Socs1 mRNA levels compared to those of SEB-exposed WT mice. The expression of Socs1 correlated inversely with IFN-γ production in the BALF. In addition, our gain- and loss-of-function studies with miR-155 mimic and inhibitor clearly demonstrated that miR-155-mediated targeting of Socs1 regulates IFN-γ production.
The results of the present study highlight the role of miR-155 in SEB-induced acute inflammatory lung injury. Specifically, we demonstrate that the high levels of IFN-γ production associated with SEB exposure can be attributed to the miR-155-mediated repression of Socs1, a critical regulator of IFN-γ (Fig. 7). Furthermore, the importance of miR-155 is made particularly evident, as miR-155-deficient mice were found to be protected from SEB-mediated inflammation and acute lung injury, thereby suggesting that therapeutic targeting of miR-155 may be useful in the treatment of SEB-triggered acute inflammatory lung injury.
FIG 7.
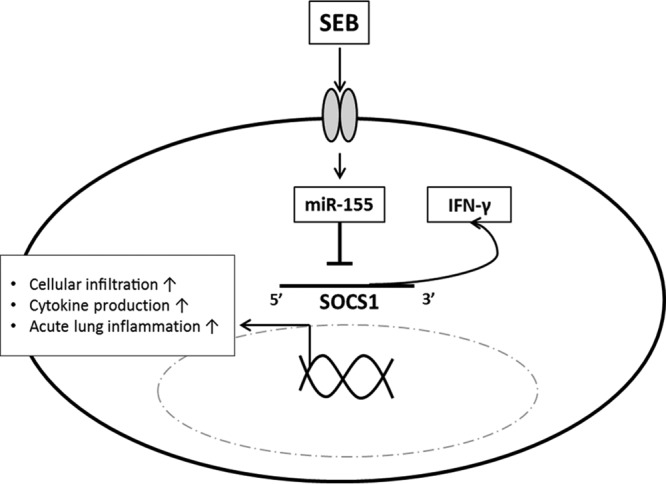
Schematic of SEB-mediated downregulation of Socs1 via miR-155. SEB exposure leads to the release of IFN-γ and subsequent expression of miR-155. miR-155-mediated suppression of Socs1 prevents appropriate control of IFN-γ, leading to cell proliferation and sustained cytokine signaling and damage to the lung.
ACKNOWLEDGMENTS
This work was supported by NIH grants P01AT003961, R01AT006888, R01ES019313, R01MH094755, and P20RR032684 and VA merit award BX001357.
Footnotes
Published ahead of print 28 April 2014
REFERENCES
- 1.Pinchuk IV, Beswick EJ, Reyes VE. 2010. Staphylococcal enterotoxins. Toxins 2:2177–2197. 10.3390/toxins2082177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Savransky V, Rostapshov V, Pinelis D, Polotsky Y, Korolev S, Komisar J, Fegeding K. 2003. Murine lethal toxic shock caused by intranasal administration of staphylococcal enterotoxin B. Toxicol. Pathol. 31:373–378. 10.1080/01926230309725 [DOI] [PubMed] [Google Scholar]
- 3.Ulrich RG, Sidell S, Taylor TJ, Wilhelmsen CL, Franz DR. 2001. Textbook of military medicine: medical aspects of chemical and biological warfare. Office of The Surgeon General at TMM Publications, Borden Institute, Walter Reed Army Medical Center, Washington, DC [Google Scholar]
- 4.Saeed AI, Rieder SA, Price RL, Barker J, Nagarkatti P, Nagarkatti M. 2012. Acute lung injury induced by staphylococcal enterotoxin B: disruption of terminal vessels as a mechanism of induction of vascular leak. Microsc. Microanal. 18:445–452. 10.1017/S1431927612000190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neumann B, Engelhardt B, Wagner H, Holzmann B. 1997. Induction of acute inflammatory lung injury by staphylococcal enterotoxin B. J. Immunol. 158:1862–1871 [PubMed] [Google Scholar]
- 6.Bavari S, Ulrich RG. 1995. Staphylococcal enterotoxin A and toxic shock syndrome toxin compete with CD4 for human major histocompatibility complex class II binding. Infect. Immun. 63:423–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kozono H, Parker D, White J, Marrack P, Kappler J. 1995. Multiple binding sites for bacterial superantigens on soluble class II MHC molecules. Immunity 3:187–196. 10.1016/1074-7613(95)90088-8 [DOI] [PubMed] [Google Scholar]
- 8.Miethke T, Duschek K, Wahl C, Heeg K, Wagner H. 1993. Pathogenesis of the toxic shock syndrome: T cell mediated lethal shock caused by the superantigen TSST-1. Eur. J. Immunol. 23:1494–1500. 10.1002/eji.1830230715 [DOI] [PubMed] [Google Scholar]
- 9.Bette M, Schafer MK, van Rooijen N, Weihe E, Fleischer B. 1993. Distribution and kinetics of superantigen-induced cytokine gene expression in mouse spleen. J. Exp. Med. 178:1531–1539. 10.1084/jem.178.5.1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tilahun AY, Holz M, Wu TT, David CS, Rajagopalan G. 2011. Interferon gamma-dependent intestinal pathology contributes to the lethality in bacterial superantigen-induced toxic shock syndrome. PLoS One 6:e16764. 10.1371/journal.pone.0016764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Florquin S, Amraoui Z, Goldman M. 1995. T cells made deficient in interleukin-2 production by exposure to staphylococcal enterotoxin B in vivo are primed for interferon-gamma and interleukin-10 secretion. Eur. J. Immunol. 25:1148–1153. 10.1002/eji.1830250503 [DOI] [PubMed] [Google Scholar]
- 12.Fleischer B, Gerardy-Schahn R, Metzroth B, Carrel S, Gerlach D, Kohler W. 1991. An evolutionary conserved mechanism of T cell activation by microbial toxins. Evidence for different affinities of T cell receptor-toxin interaction. J. Immunol. 146:11–17 [PubMed] [Google Scholar]
- 13.Herman A, Kappler JW, Marrack P, Pullen AM. 1991. Superantigens: mechanism of T-cell stimulation and role in immune responses. Annu. Rev. Immunol. 9:745–772. 10.1146/annurev.immunol.9.1.745 [DOI] [PubMed] [Google Scholar]
- 14.Saeed AI, Rieder SA, Price RL, Barker J, Nagarkatti P, Nagarkatti M. 2012. Acute lung injury induced by staphylococcal enterotoxin B: disruption of terminal vessels as a mechanism of induction of vascular leak. Microsc. Microanal. 18:445–452. 10.1017/S1431927612000190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai Y, Yu X, Hu S, Yu J. 2009. A brief review on the mechanisms of miRNA regulation. Genomics Proteomics Bioinformatics 7:147–154. 10.1016/S1672-0229(08)60044-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Connell RM, Rao DS, Baltimore D. 2012. microRNA regulation of inflammatory responses. Annu. Rev. Immunol. 30:295–312. 10.1146/annurev-immunol-020711-075013 [DOI] [PubMed] [Google Scholar]
- 17.Rossi RL, Rossetti G, Wenandy L, Curti S, Ripamonti A, Bonnal RJ, Birolo RS, Moro M, Crosti MC, Gruarin P, Maglie S, Marabita F, Mascheroni D, Parente V, Comelli M, Trabucchi E, De Francesco R, Geginat J, Abrignani S, Pagani M. 2011. Distinct microRNA signatures in human lymphocyte subsets and enforcement of the naive state in CD4+ T cells by the microRNA miR-125b. Nat. Immunol. 12:796–803. 10.1038/ni.2057 [DOI] [PubMed] [Google Scholar]
- 18.Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K. 2008. Lymphoproliferative disease and autoimmunity in mice with increased miR-17–92 expression in lymphocytes. Nat. Immunol. 9:405–414. 10.1038/ni1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Connell RM, Kahn D, Gibson WS, Round JL, Scholz RL, Chaudhuri AA, Kahn ME, Rao DS, Baltimore D. 2010. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity 33:607–619. 10.1016/j.immuni.2010.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Committee for the Update of the Guide for the Care and Use of Laboratory Animals. 2011. Guide for the care and use of laboratory animals, 8th ed. The National Academies Press, Washington, DC [Google Scholar]
- 21.Rieder SA, Nagarkatti P, Nagarkatti M. 2012. Multiple anti-inflammatory pathways triggered by resveratrol lead to amelioration of staphylococcal enterotoxin B-induced lung injury. Br. J. Pharmacol. 167:1244–1258. 10.1111/j.1476-5381.2012.02063.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rieder SA, Nagarkatti P, Nagarkatti M. 2011. CD1d-independent activation of invariant natural killer T cells by staphylococcal enterotoxin B through major histocompatibility complex class II/T cell receptor interaction results in acute lung injury. Infect. Immun. 79:3141–3148. 10.1128/IAI.00177-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wheeler AP, Bernard GR. 2007. Acute lung injury and the acute respiratory distress syndrome: a clinical review. Lancet 369:1553–1564. 10.1016/S0140-6736(07)60604-7 [DOI] [PubMed] [Google Scholar]
- 24.Miyata A, Natsuaki M, Yamanishi K. 2008. Staphylococcal enterotoxin B enhances a flare-up reaction of murine contact hypersensitivity through up-regulation of interferon-gamma. Exp. Dermatol. 17:843–848. 10.1111/j.1600-0625.2008.00714.x [DOI] [PubMed] [Google Scholar]
- 25.Plaza R, Rodriguez-Sanchez JL, Juarez C. 2007. Staphylococcal enterotoxin B in vivo modulates both gamma interferon receptor expression and ligand-induced activation of signal transducer and activator of transcription 1 in T cells. Infect. Immun. 75:306–313. 10.1128/IAI.01220-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sonkoly E, Pivarcsi A. 2009. microRNAs in inflammation. Int. Rev. Immunol. 28:535–561. 10.3109/08830180903208303 [DOI] [PubMed] [Google Scholar]
- 27.Lindsay MA. 2008. microRNAs and the immune response. Trends Immunol. 29:343–351. 10.1016/j.it.2008.04.004 [DOI] [PubMed] [Google Scholar]
- 28.Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, Schmidt-Supprian M, Rajewsky N, Yancopoulos G, Rao A, Rajewsky K. 2007. Regulation of the germinal center response by microRNA-155. Science 316:604–608. 10.1126/science.1141229 [DOI] [PubMed] [Google Scholar]
- 29.Stahl HF, Fauti T, Ullrich N, Bopp T, Kubach J, Rust W, Labhart P, Alexiadis V, Becker C, Hafner M, Weith A, Lenter MC, Jonuleit H, Schmitt E, Mennerich D. 2009. miR-155 inhibition sensitizes CD4+ Th cells for TREG mediated suppression. PLoS One 4:e7158. 10.1371/journal.pone.0007158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murugaiyan G, Beynon V, Mittal A, Joller N, Weiner HL. 2011. Silencing microRNA-155 ameliorates experimental autoimmune encephalomyelitis. J. Immunol. 187:2213–2221. 10.4049/jimmunol.1003952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oertli M, Engler DB, Kohler E, Koch M, Meyer TF, Muller A. 2011. MicroRNA-155 is essential for the T cell-mediated control of Helicobacter pylori infection and for the induction of chronic gastritis and colitis. J. Immunol. 187:3578–3586. 10.4049/jimmunol.1101772 [DOI] [PubMed] [Google Scholar]
- 32.Muhie S, Hammamieh R, Cummings C, Yang D, Jett M. 2013. Transcriptome characterization of immune suppression from battlefield-like stress. Genes Immun. 14:19–34. 10.1038/gene.2012.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schoenborn JR, Wilson CB. 2007. Regulation of interferon-gamma during innate and adaptive immune responses. Adv. Immunol. 96:41–101. 10.1016/S0065-2776(07)96002-2 [DOI] [PubMed] [Google Scholar]
- 34.Krakauer T. 2013. Update on staphylococcal superantigen-induced signaling pathways and therapeutic interventions. Toxins 5:1629–1654. 10.3390/toxins5091629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee CL, Lee SH, Jay FT, Rozee KR. 1990. Immunobiological study of interferon-gamma-producing cells after staphylococcal enterotoxin B stimulation. Immunology 70:94–99 [PMC free article] [PubMed] [Google Scholar]
- 36.Assenmacher M, Lohning M, Scheffold A, Manz RA, Schmitz J, Radbruch A. 1998. Sequential production of IL-2, IFN-gamma and IL-10 by individual staphylococcal enterotoxin B-activated T helper lymphocytes. Eur. J. Immunol. 28:1534–1543. [DOI] [PubMed] [Google Scholar]
- 37.Huang W, Koller LD. 1998. Superantigen activation and kinetics of cytokines in the Long-Evans rat. Immunology 95:331–338. 10.1046/j.1365-2567.1998.00613.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trotta R, Chen L, Ciarlariello D, Josyula S, Mao C, Costinean S, Yu L, Butchar JP, Tridandapani S, Croce CM, Caligiuri MA. 2012. miR-155 regulates IFN-gamma production in natural killer cells. Blood 119:3478–3485. 10.1182/blood-2011-12-398099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kurowska-Stolarska M, Alivernini S, Ballantine LE, Asquith DL, Millar NL, Gilchrist DS, Reilly J, Ierna M, Fraser AR, Stolarski B, McSharry C, Hueber AJ, Baxter D, Hunter J, Gay S, Liew FY, McInnes IB. 2011. MicroRNA-155 as a proinflammatory regulator in clinical and experimental arthritis. Proc. Natl. Acad. Sci. U. S. A. 108:11193–11198. 10.1073/pnas.1019536108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krebs DL, Hilton DJ. 2001. SOCS proteins: negative regulators of cytokine signaling. Stem Cells 19:378–387. 10.1634/stemcells.19-5-378 [DOI] [PubMed] [Google Scholar]
- 41.Tamiya T, Kashiwagi I, Takahashi R, Yasukawa H, Yoshimura A. 2011. Suppressors of cytokine signaling (SOCS) proteins and JAK/STAT pathways: regulation of T-cell inflammation by SOCS1 and SOCS3. Arterioscler. Thromb. Vasc. Biol. 31:980–985. 10.1161/ATVBAHA.110.207464 [DOI] [PubMed] [Google Scholar]
- 42.Cardoso AL, Guedes JR, Pereira de Almeida L, Pedroso de Lima MC. 2012. miR-155 modulates microglia-mediated immune response by down-regulating SOCS-1 and promoting cytokine and nitric oxide production. Immunology 135:73–88. 10.1111/j.1365-2567.2011.03514.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang M, Zhang Q, Liu F, Yin L, Yu B, Wu J. 2011. MicroRNA-155 may affect allograft survival by regulating the expression of suppressor of cytokine signaling 1. Med. Hypotheses 77:682–684. 10.1016/j.mehy.2011.07.016 [DOI] [PubMed] [Google Scholar]
- 44.Jiang S, Zhang HW, Lu MH, He XH, Li Y, Gu H, Liu MF, Wang ED. 2010. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res. 70:3119–3127. 10.1158/0008-5472.CAN-09-4250 [DOI] [PubMed] [Google Scholar]
- 45.Wang P, Hou J, Lin L, Wang C, Liu X, Li D, Ma F, Wang Z, Cao X. 2010. Inducible microRNA-155 feedback promotes type I IFN signaling in antiviral innate immunity by targeting suppressor of cytokine signaling 1. J. Immunol. 185:6226–6233. 10.4049/jimmunol.1000491 [DOI] [PubMed] [Google Scholar]