Abstract
Mycobacterium avium subsp. paratuberculosis causes Johne's disease in ruminants, a chronic enteric disease responsible for severe economic losses in the dairy industry. Global gene regulators, including sigma factors are important in regulating mycobacterial virulence. However, the biological significance of such regulators in M. avium subsp. paratuberculosis rremains elusive. To better decipher the role of sigma factors in M. avium subsp. paratuberculosis pathogenesis, we targeted a key sigma factor gene, sigL, activated in mycobacterium-infected macrophages. We interrogated an M. avium subsp. paratuberculosis ΔsigL mutant against a selected list of stressors that mimic the host microenvironments. Our data showed that sigL was important in maintaining bacterial survival under such stress conditions. Survival levels further reflected the inability of the ΔsigL mutant to persist inside the macrophage microenvironments. Additionally, mouse infection studies suggested a substantial role for sigL in M. avium subsp. paratuberculosis virulence, as indicated by the significant attenuation of the ΔsigL-deficient mutant compared to the parental strain. More importantly, when the sigL mutant was tested for its vaccine potential, protective immunity was generated in a vaccine/challenge model of murine paratuberculosis. Overall, our study highlights critical role of sigL in the pathogenesis and immunity of M. avium subsp. paratuberculosis infection, a potential role that could be shared by similar proteins in other intracellular pathogens.
INTRODUCTION
Early infection of calves with Mycobacterium avium subsp. paratuberculosis leads to the development of Johne's disease (JD), also known as paratuberculosis, a chronic enteric infection responsible for significant economic losses throughout the world (1–3). A recent report suggested that more than 90% of the dairy operations in the United States include animals that are infected with M. avium subsp. paratuberculosis (4). The use of the current vaccine (Mycopar) to control JD is inadequate (5) and does not stop the severe economic losses estimated to be over $500 million annually to the U.S. dairy industry (6). Additionally, this inactivated vaccine cannot prevent the spread of the infection among animals (7, 8) and causes severe granuloma following accidental human injection (9). Following infection through the fecal-oral route, M. avium subsp. paratuberculosis is endocytosed by enterocytes and M cells in the Peyer's patches of the ileum (10) and subsequently phagocytosed by subepithelial and intraepithelial macrophages (11). Although the mechanism of intracellular survival of this pathogen within macrophages is not entirely understood, reports suggested the ability of M. avium subsp. paratuberculosis to block phagosome-lysosome maturation and interfere with macrophage activation (12, 13). Earlier reports indicated gene expression of alternative sigma factors encoded in the M. avium subsp. paratuberculosis genome following macrophage infection (14, 15). Additionally, among the 19 encoded alternative sigma factors (16), only the sigL transcript was induced at an early stage (2 h) of macrophage infection, indicating its importance for M. avium subsp. paratuberculosis during early adaptation. To better understand the genetic basis of paratuberculosis, we focused our efforts on the role played by sigL in M. avium subsp. paratuberculosis pathogenesis and examined whether a sigL mutant could constitute a candidate for a protective vaccine against paratuberculosis.
Earlier transcriptome analysis of M. avium subsp. paratuberculosis from JD-infected cattle identified novel virulence genes and pathogenic pathways that allow M. avium subsp. paratuberculosis to survive harsh environmental conditions (17). Interestingly, both M. avium subsp. paratuberculosis and Mycobacterium tuberculosis share several mechanisms, especially those involved in macrophage survival (15, 18). In M. tuberculosis, only 11 alternative sigma factors are encoded (19), where sigL acts as a stress response regulator and is implicated in cell membrane protein biosynthesis as well as virulence (20). In M. avium subsp. paratuberculosis, the role of sigL during intracellular survival and host infection remains elusive. To start, we examined the effect of sigL deletion on bacterial viability under various stress conditions. This analysis indicated the important role played by sigL in regulating the expression of a set of genes (similar to the sigL homologue in M. tuberculosis) needed for survival under the stress of cell wall-damaging agents as well as inside bovine macrophages. Further examination of the sigL mutant in a murine model of paratuberculosis suggested that sigL is an important virulence determinant for M. avium subsp. paratuberculosis. In addition, we evaluated the potential use of M. avium subsp. paratuberculosis ΔsigL as a live attenuated vaccine using the murine model. Interestingly, the sigL mutant elicited protective immune responses against paratuberculosis, validating the concept of using a global gene regulator (GGR) as a target for developing effective live attenuated vaccines.
MATERIALS AND METHODS
Bacterial strains and mutant construction.
M. avium subsp. paratuberculosis K10 and Mycobacterium smegmatis mc2155 strains were grown in Middlebrook 7H9 broth and on Middlebrook 7H10 plates as previously described (2, 15). For cloning, Escherichia coli DH5α and HB101 were used as host cells (15, 21).
To generate an isogenic sigL knockout mutant of M. avium subsp. paratuberculosis, the sigL (MAP4201) gene was deleted from the genome of M. avium subsp. paratuberculosis K10 (15). Primers (see Table S1 in the supplemental material) were designed to amplify ∼750-bp PCR fragments flanking each end of the sigL coding region and cloned into the pGEM-T Easy vector (Promega, Madison, WI). The fragments were digested with their flanking restriction enzyme sites (SpeI/HindIII and XbaI/Acc65I for upstream and downstream portions, respectively) and ligated into pYUB854 (15). After digestion by NotI and SpeI (Promega, Madison, WI), the linearized vector was ligated into pML19, a derivative of pPR27 (22) where a kanamycin resistance cassette has been inserted into the PstI site. The resulting vector, pML19_sigL, was electroporated into electrocompetent M. avium subsp. paratuberculosis using a Gene Pulser II machine (Bio-Rad, Hercules, CA), and cells were plated onto Middlebrook 7H10 medium containing 30 μg/ml kanamycin and 75 μg/ml hygromycin (Invitrogen, Carlsbad, CA, USA). Following 6 weeks of growth at 32°C, transformants were grown for 3 weeks with shaking at 32°C in Middlebrook 7H9 broth. These cultures were plated onto Middlebrook 7H10 medium containing 75 μg/ml hygromycin and 5% sucrose and incubated at 39°C for 6 weeks. Genotype of the hygromycin-resistant transformant M. avium subsp. paratuberculosis ΔsigL was confirmed by PCR and sequence analysis (15). To complement the M. avium subsp. paratuberculosis ΔsigL, an ∼4-kb fragment, encompassing sigL with its 5′ regulatory region and the distal genes (MAP4202 to MAP4205), was amplified by PCR (see Table S1 in the supplemental material for primers) and cloned into the HindIII/XbaI restriction site of the vector pMV306 (20). The M. avium subsp. paratuberculosis ΔsigL strain was transformed with this recombinant construct, and the genotype of the complemented strain (M. avium subsp. paratuberculosis ΔsigL::sigL) was identified by PCR analysis. A similar approach was applied to complement M. tuberculosis mutant strain lacking this alternative sigma factor (23).
Quantitative real-time PCR.
M. avium subsp. paratuberculosis strains were grown to mid-exponential phase (optical density at 600 nm [OD600] = 0.5), and the RNA was extracted by the TRIzol method as detailed earlier (15). Quantitative real-time PCR (qRT-PCR) was performed using a SYBR green-based reagent with (Bio-Rad, Hercules, CA). For qRT-PCR, cDNA was synthesized from 1 μg of total RNA using SuperScript III (Invitrogen), as directed by the manufacturer, in the presence 250 ng of mycobacterial genome-directed primers (15, 24). No genomic DNA was detected from the RNA samples for cDNA synthesis. A 100-ng cDNA was used as the template in a reaction in the presence of gene-specific primers (see Table S1 in the supplemental material) at a concentration of 200 nM using a StepOnePlus real-time PCR system (Applied Biosystems, Foster City, CA). Cycle conditions were 95°C for 5 min and 40 cycles of 95°C for 15 s and 59°C for 60 s. The threshold cycle (CT) of each gene was normalized to the CT of the sigA gene from the same cDNA sample. The changes in expression were calculated by comparing the normalized CT of wild-type or complemented M. avium subsp. paratuberculosis samples to the M. avium subsp. paratuberculosis ΔsigL sample as detailed earlier (15, 25).
Stress phenotype of M. avium subsp. paratuberculosis.
M. avium subsp. paratuberculosis cultures were grown to log phase (OD600 = 0.5 to 1.0), and 200 μl was spread on 7H10 plates. For the disk diffusion assay (DDA), paper discs (6 mm; Whatman, Piscataway, NJ) containing 20 μl of 0.5, 1, or 1.5 M diamide (oxidative stressor) and 1, 2, or 3% sodium dodecyl sulfate (SDS; cell wall stressor) were placed on each of the spread plates. Plates were incubated at 37°C until a thick confluent lawn developed. To determine sustained effect of stressor on the viability of bacilli, after washing with phosphate-buffered saline (PBS), M. avium subsp. paratuberculosis cultures were exposed to the acidified 7H9 broth (pH 5.5 obtained by adding HCl) containing 0.3% bovine bile (cell wall stressor), and aliquots were collected at 0, 4, 15, 24, 48, and 72 h to monitor their viability by CFU counting (15).
Cell culture and infection.
The mouse macrophages (J774A.1) were regularly maintained as described elsewhere (15). To activate macrophages, cells were pretreated overnight (18 h) with 100 U/ml recombinant murine gamma interferon (IFN-γ) (Peprotech, Rocky Hill, NJ) before infection with M. avium subsp. paratuberculosis strains (15). For cell infection studies, wild-type and mutant strains were added to macrophage monolayers (multiplicity of infection [MOI], 20:1). Following incubation at 37°C in 5% CO2 for 3 h, macrophage monolayers were washed twice with warm PBS to remove extracellular bacteria, and RPMI 1640 medium containing 5% fetal bovine serum was added. Cells were lysed at 1 and 8 days postinfection for bacterial CFU counts. To examine M. avium subsp. paratuberculosis ΔsigL survival in bovine monocyte-derived macrophages (MDM), MDM were isolated from the peripheral blood of three cows, and cell infection studies were performed as described in detail elsewhere (15).
Mouse infections.
All animal experiments used in this study were performed according to the protocols approved by the Institutional Animal Care and Use Committee, University of Wisconsin—Madison. For the virulence study, two groups (15 per group) of female BALB/c mice (Harlan Laboratories, Indianapolis, IN, USA) were challenged intraperitoneally (i.p.) with the wild-type and mutant strains. Infection inocula (∼2 × 108 CFU/mouse) of the two strains were similar, as determined by plate count on the day of infection. Mouse groups (n = 5) were sacrificed at 3, 6, and 12 weeks postinfection (WPI), and organ samples were collected for bacterial CFU enumeration and histopathological examinations as described before (15).
For the immunization studies, female C57BL/6 mice (Taconic, Hudson, NY) were used. A mock-infected group (n = 12) was immunized with PBS buffer, while the M. avium subsp. paratuberculosis ΔsigL group (n = 14) received ∼2 × 106 CFU in 0.2 ml PBS subcutaneously (s.c.) into the neck scruff twice, 2 weeks apart. Four weeks following the booster dose, mice were challenged i.p. with ∼7 × 108 CFU wild-type M. avium subsp. paratuberculosis strain as determined by plate count on the day of infection. Mouse groups (n = 4 to 6) were sacrificed at 6 weeks postimmunization and 6 and 12 weeks postchallenge (WPC), and organ samples were collected for bacterial CFU counts, histopathological examinations, and immune responses. The liver and spleen tissue homogenates were serially diluted in PBS following plating on 7H10 medium. For intestines, 7H10 plates were supplemented with a mixture of 5 mg/ml vancomycin, 30 mg/ml amphotericin B, and 10 mg/ml nalidixic acid to reduce nonmycobacterial and fungal contamination. Organ homogenates were plated on 7H10 medium containing 50 μg/ml of hygromycin to determine the organ burden of the vaccine strain, M. avium subsp. paratuberculosis ΔsigL, and whenever necessary, vaccine strain CFU counts were subtracted from the total M. avium subsp. paratuberculosis organ loads to determine the actual level of organ colonization for the challenge strain, M. avium subsp. paratuberculosis K10.
Evaluation of immune responses.
Mouse spleens were collected aseptically and homogenized by gentle mechanical disruption using a stainless steel screen. Following isolation of mononuclear spleen cells, the red blood cells were lysed using 0.83% ammonium chloride. Spleen cells were washed and suspended in RPMI 1640 (Thermo Fisher) supplemented with 1× nonessential amino acids (Invitrogen), 1× l-glutamine, 1× penicillin-streptomycin, and 10% fetal bovine serum (Atlanta Biologicals). Following trypan blue dye enumeration for cell viability, splenocytes were cultured in duplicate in round-bottom 96-well tissue culture plates with 1 × 106 cells/well containing 100 U/ml human interleukin 2 (IL-2) (BD Biosciences) (26). Cells were restimulated in vitro with medium alone or medium supplemented with 10 μg/ml Johnin purified protein derivative (PPDj) (USDA–National Veterinary Services Laboratory, Ames, IA) for 48 and 72 h at 37°C with 5% CO2 (27). Cell supernatants were collected and levels of secreted cytokines were analyzed by mouse cytokine enzyme-linked immunosorbent assay (ELISA) kit according to the manufacturer's instructions (BioLegend, San Diego, CA). The level of cytokine secretion in the medium stimulation was subtracted from the PPDj-stimulated spleen cells. To determine the humoral immune response, sera were prepared from mouse whole blood, and M. avium subsp. paratuberculosis-specific antibody (anti-PPDj antibodies) was detected by ELISA using horseradish peroxidase-conjugated rabbit anti-mouse antibody (Pierce, Rockford, IL) (28, 29).
Statistical analysis.
Student's t test was performed to compare differences in mouse immune responses and bacterial CFU counts obtained from the in vitro stress studies. The Mann-Whitney U test was used to compare bacterial loads in mouse organs harvested from virulence and vaccine-challenge experiments. A probability value of <0.05 was considered significant for all tests.
RESULTS
Construction of sigL knockout mutant and effect of sigL mutation on gene expression.
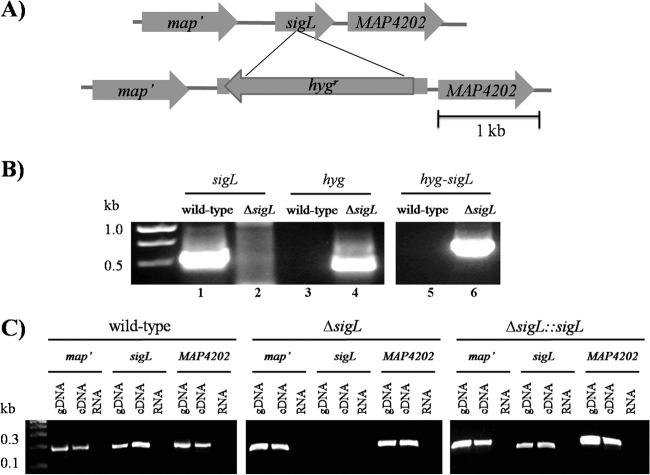
Recent analysis of the M. avium subsp. paratuberculosis transcriptome during macrophage infection suggested that sigL could be an important factor for M. avium subsp. paratuberculosis survival inside host macrophages (15). To test this hypothesis, we generated a sigL deletion mutant, M. avium subsp. paratuberculosis ΔsigL (Fig. 1A and B) and examined survival of this mutant under different stress conditions. Because sigL and its anti-sigma factor (MAP4202) are likely encoded in an operon (20), we examined M. avium subsp. paratuberculosis ΔsigL for possible polarity on the downstream gene MAP4202. By employing reverse transcriptase PCR analysis, the presence of the MAP4202 transcript was confirmed in the ΔsigL mutant (Fig. 1C). Additionally, we examined expression levels for neighboring genes around sigL in both the wild-type and ΔsigL::sigL complemented strains and compared their expression levels to those in the ΔsigL mutant. The data suggested that the expression levels of these genes in the complemented strain had a pattern similar to that found in the wild-type strain (see Fig. S1 in the supplemental material). Finally, a growth experiment of the complemented strain, the ΔsigL::sigL strain, in liquid medium (7H9 broth) revealed a growth pattern similar to that of the wild-type strain (see Fig. S2 in the supplemental material). However, the ΔsigL mutant showed slightly elevated bacterial growth, as measured by OD600, compared to the complemented and the wild-type strain.
FIG 1.
Construction of M. avium subsp. paratuberculosis ΔsigL using wild-type M. avium subsp. paratuberculosis strain. (A) Physical map displaying the deletion of sigL (MAP4201) gene with homologous recombination via pYUB854 cosmid shuttle cloning vector, which resulted in the deletion of an ∼750-bp coding region and the insertion of an ∼2-kb region carrying a hygromycin resistance cassette. (B) The M. avium subsp. paratuberculosis ΔsigL mutant was confirmed with PCR and sequence verification using genomic DNA from the wild-type and the mutant strains. Primer pairs were designed for the sigL region, the hygromycin resistance gene (hyg), or the recombinant region after allelic exchange. A 1.5% agarose gel showed amplicons from the sigL region only when wild-type genomic DNA was used (lane 1), whereas hyg was amplified only from the M. avium subsp. paratuberculosis ΔsigL mutant genomic DNA (lane 4). Lane 6 showed amplicon from the recombinant region only when M. avium subsp. paratuberculosis ΔsigL mutant genomic DNA was used. (C) The polarity of the M. avium subsp. paratuberculosis ΔsigL mutant was assessed using reverse transcriptase PCR analysis to check for transcription of its neighboring genes. In the wild-type (left) and complemented (right) strains, positive bands show that map′, sigL, and the downstream gene MAP4202 are present in the genome and transcribed (amplified from cDNA), with no amplification from RNA used as a negative control. In the M. avium subsp. paratuberculosis ΔsigL mutant (middle), the sigL coding region is absent in the genome and as cDNA, but transcripts for the neighboring genes are present.
To determine possible effectors controlled by sigL in M. avium subsp. paratuberculosis, we employed qRT-PCR analysis to examine 8 genes (Table 1) that were under the control of a sigL homologue (85% similarity at protein level) in M. tuberculosis (20, 30). Overall, a significant induction (≥±1.5-fold induction when the fold change was greater than the standard deviation) in a list of 6 genes was observed in the sigL regulon when transcripts of wild-type and isogenic mutants were compared. Similar to the sigL regulon in M. tuberculosis, there was induction of pks10, while transcripts for the downstream gene pks7 were only slightly increased (30). In addition, modest induction of mce gene family members (e.g., lprL) was observed, suggesting their regulation by sigL. Interestingly, the mpt53 transcripts were elevated the most among the genes examined in this study, another agreement with the M. tuberculosis regulon (31). In contrast, no change was observed in the transcript levels of MAP3220c, which had the lowest level of protein similarity (55.1%) to M. tuberculosis orthologs. Generally, limited analysis of transcripts of the potential sigL regulon in M. avium subsp. paratuberculosis suggested their similarity to those present in M. tuberculosis.
TABLE 1.
Genes differentially expressed in wild-type M. avium subsp. paratuberculosis and M. avium subsp. paratuberculosis ΔsigL during growth in Middlebrook 7H9 broth at an OD600 of 0.5
| Gene | Gene product; possible function | M. tuberculosis ortholog | % protein similarity to M. tuberculosis | Fold changea |
|---|---|---|---|---|
| pks10 | Polyketide synthase; possible involvement in mycobacterial cell wall maintenance | Rv1660 | 82.2 | 3.5 ± 1.2 |
| pks7 | Polyketide synthase; possible involvement in mycobacterial cell wall maintenance | Rv1661 | 70.0 | 1.4 ± 0.1 |
| lprL | Mce family protein; involved in host cell invasion | Rv0593 | 88.7 | 1.9 ± 0.2 |
| MAP4089 | Mce family protein; involved in host cell invasion | Rv0594 | 82.6 | 2.1 ± 0.7 |
| MAP2635c | MMPL family protein; may be involved in lipid transport | Rv1145 | 89.1 | 2.4 ± 0.3 |
| MAP3220c | Hypothetical; possible transmembrane protein | Rv3166c | 55.1 | 1.2 ± 0.2 |
| mpt53 | Secreted protein; homologous to DsbE (involved in protein folding) | Rv2878c | 89.8 | 26.2 ± 0.9 |
| MAP2941c | Homologous to cytochrome c biogenesis protein; could be involved in membrane transport | Rv2877c | 55.9 | 1.6 ± 0.3 |
Values are means ± standard deviations and are representative of two independent experiments. A change was considered significant for genes with a ≥±1.5-fold change and a level that was >2× the standard deviation.
Role of sigL in viability of M. avium subsp. paratuberculosis under stress.
To examine a potential role for sigL in the response to unfavorable stress conditions, we analyzed the survival of M. avium subsp. paratuberculosis cultures under both oxidative (diamide) and cell wall (SDS and bovine bile) stresses. Growth inhibition zones in disk diffusion assays indicated that M. avium subsp. paratuberculosis ΔsigL was susceptible to diamide oxidation (Fig. 2A). Such phenotypic differences also indicated the inability of ΔsigL mutant to survive under SDS stress compared to the wild-type and complemented strains (Fig. 2B). Bile tolerance was also evaluated by culturing of the M. avium subsp. paratuberculosis strains in the presence of 0.3% bovine bile (oxgall). This concentration of bile is likely encountered by the bacteria within the intestinal contents following oral infection (32). In addition, because of the ability of M. avium subsp. paratuberculosis to resist killing by acidic conditions (33), we made culture broths slightly acidic (pH 5.5) to partially mimic the physiological conditions that M. avium subsp. paratuberculosis would encounter following infection in the gastrointestinal tract (e.g., the abomasum of a cow). Survival levels showed a significant drop in the viability of the ΔsigL mutant at 4 h after exposure to bovine bile compared to the wild-type and complemented strains (Fig. 2C). This difference in bacterial survival for M. avium subsp. paratuberculosis ΔsigL was increased more than 20-fold at 24 h, and the viability of the mutant continued to decline at later times, suggesting that sigL is important in establishing resistance when bacteria encounter initial bactericidal barriers in the host. Because complementation of M. avium subsp. paratuberculosis ΔsigL restored the wild-type phenotype under these stress conditions, the complemented strain was not included in further experiments.
FIG 2.
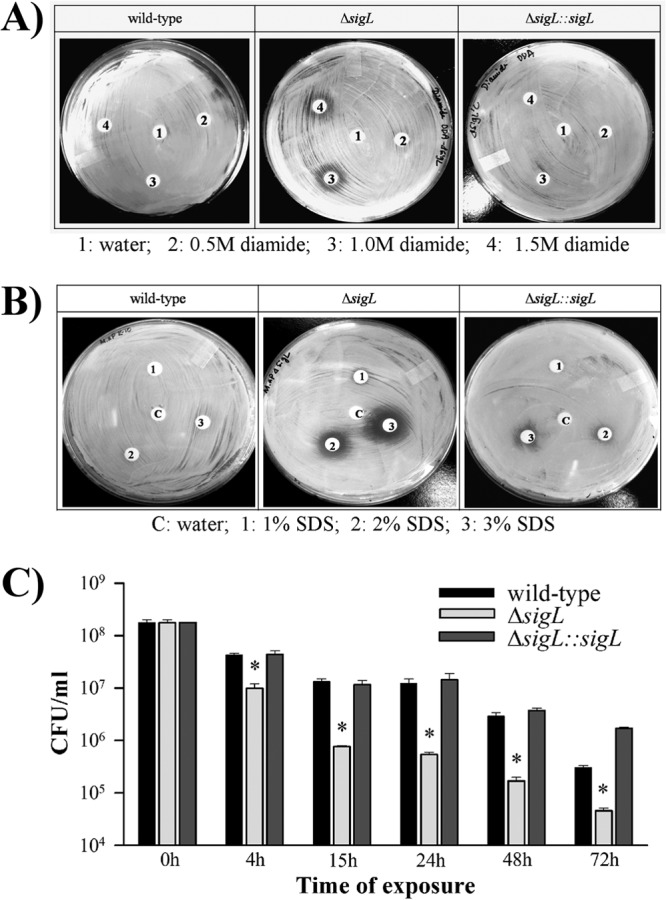
Phenotypic differences of M. avium subsp. paratuberculosis strains following exposure to stress environments. Disc diffusion assays were carried out with M. avium subsp. paratuberculosis, M. avium subsp. paratuberculosis ΔsigL, and M. avium subsp. paratuberculosis ΔsigL::sigL strains with various concentrations of diamide (A) and SDS (B). Halos indicate that M. avium subsp. paratuberculosis ΔsigL is susceptible to both the thiol-specific oxidant diamide and cell envelope stress. Images are representative of two biological replicates. (C) Sustained effect of exposure to acidified bovine bile on M. avium subsp. paratuberculosis survival. The wild type, M. avium subsp. paratuberculosis ΔsigL, and complemented strains were cultured in the presence of 0.3% acidified bile, and CFU counts were determined at 0, 4, 15, 24, 48, and 72 h postexposure via plating on 7H10 medium. The growth difference between M. avium subsp. paratuberculosis ΔsigL mutant and the wild-type strain or complemented strain was statistically significant (*, P < 0.05) at all time points following exposure to acidified bile. Error bars represent the standard deviations.
Intracellular survival within macrophages.
Because sigL was upregulated inside activated murine macrophages (15), we examined intracellular survival of M. avium subsp. paratuberculosis ΔsigL in the IFN-γ pretreated murine macrophages. Our analysis showed an increase in the number of wild-type M. avium subsp. paratuberculosis at 8 days relative to the numbers obtained at day 1 postinfection, whereas viability of the ΔsigL mutant was significantly reduced at this time (see Fig. S3 in the supplemental material). To use a more relevant model for M. avium subsp. paratuberculosis infection, we evaluated the persistence of the M. avium subsp. paratuberculosis ΔsigL in both resting and IFN-γ-activated bovine monocyte-derived macrophages (MDM), the natural host cell for M. avium subsp. paratuberculosis. MDM monolayers in the culture wells were checked at regular intervals for cell confluence (>80%) under an inverted light microscope. There was no inhibitory effect on the survival of wild-type bacteria at 4 days postinfection, and at 8 days, the numbers of wild-type bacilli increased over 2-fold compared to the numbers obtained at day 1 in the resting MDM (Fig. 3A). In contrast, survival of the ΔsigL mutant was not supported inside naive macrophages, and at 8 days postinfection, the viability was significantly reduced, almost by half, indicating a potential function for sigL in defending M. avium subsp. paratuberculosis against intracellular stress. A similar survival trend for the M. avium subsp. paratuberculosis ΔsigL was seen inside IFN-γ-pretreated MDM, whereas this activation status did not result in a more inhibitory effect on the survival of wild-type bacilli (Fig. 3B). Collectively, survival assays indicated that deletion of sigL affected M. avium subsp. paratuberculosis viability following exposure to stress conditions, suggesting a significant function for sigL in defending M. avium subsp. paratuberculosis against intracellular insults.
FIG 3.
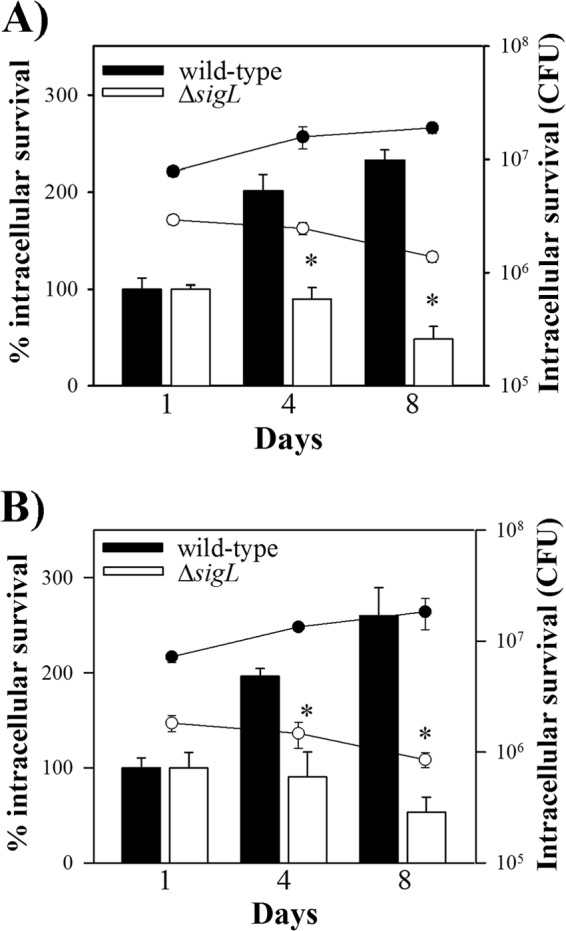
Survival of M. avium subsp. paratuberculosis ΔsigL in bovine macrophages. Naive (A) and IFN-γ-pretreated (B) MDM were infected with M. avium subsp. paratuberculosis ΔsigL and wild-type M. avium subsp. paratuberculosis. Cells were lysed at 1, 4, and 8 days postinfection, and numbers of viable bacilli were determined by serial dilutions for CFU plating. Survival was determined as the viable counts at 4 and 8 days relative to the viable counts at day 1. Data are the averages for macrophage infections in three different donor animals; significance levels were determined with Student's t test (*, P < 0.05). Error bars represent standard deviations.
Virulence analysis of M. avium subsp. paratuberculosis ΔsigL strain.
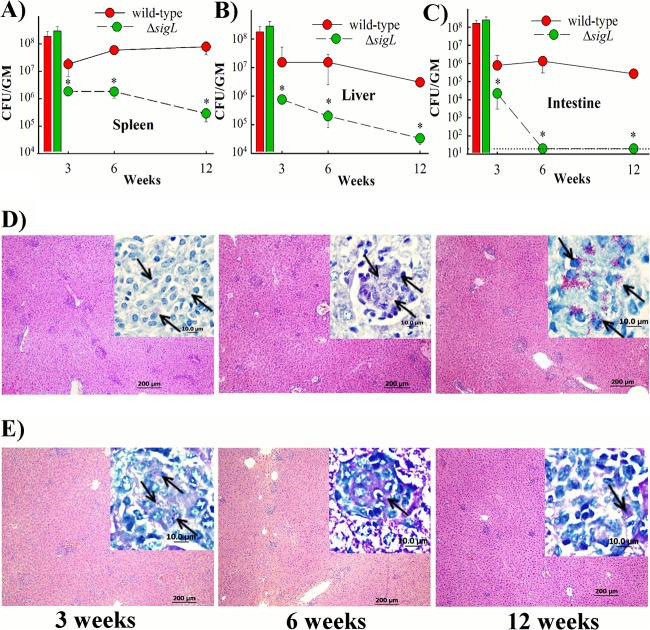
To evaluate the contribution of SigL to M. avium subsp. paratuberculosis virulence, we examined persistence of the isogenic ΔsigL mutant using the murine model of paratuberculosis. The survival pattern indicated significant attenuation of M. avium subsp. paratuberculosis ΔsigL as early as 3 WPI in all of the organs examined (Fig. 4A to C). In the spleen, the survival of ΔsigL mutant was reduced more than 40- and 400-fold relative to the wild-type strain at 6 and 12 WPI, respectively. Similarly, colonization levels of the ΔsigL mutant strain in the liver were significantly lower than those of the parental strain at all time points. Interestingly, the ΔsigL mutant did not persist in the intestine, as suggested by our inability to detect any bacteria (limit of detection 20 CFU) at 6 and 12 WPI in this organ.
FIG 4.
Virulence of wild-type and M. avium subsp. paratuberculosis ΔsigL strains of M. avium subsp. paratuberculosis. Mouse groups (n = 15) were inoculated with ∼2 × 108 CFU/mouse of wild-type M. avium subsp. paratuberculosis or M. avium subsp. paratuberculosis ΔsigL via intraperitoneal injection. Spleens (A), livers (B), and intestines (C) were collected at 3, 6, and 12 WPI (5 mice/group/time point) and cultured for bacterial counts. Colony counts for each group are represented by line plots, with error bars representing the standard deviations. Histograms show inoculum size for M. avium subsp. paratuberculosis strains. The limit of detection (20 CFU) is indicated by a dotted line. Organs with significant differences in bacterial load are indicated with asterisks (P < 0.05). The pathology of livers collected from mice infected with the wild-type strain (D) and its isogenic mutant M. avium subsp. paratuberculosis ΔsigL (E) was examined at 3, 6, and 12 WPI. Hematoxylin-and-eosin-stained sections (bar = 200 μm) are shown. Magnification, ×100. Inset images show the M. avium subsp. paratuberculosis bacilli in purple (arrows). Magnification, ×1,000. Bar, 10 μm.
The histological analysis revealed mild to moderate granulomatous inflammation in the liver tissues at both 3 and 6 WPI with each of the M. avium subsp. paratuberculosis strains, with higher lymphocytic infiltration in the mice infected with the ΔsigL mutant (Fig. 4D and E). At 12 WPI, ΔsigL mutant-infected animals showed less granulomatous inflammation, indicating a reduced ability of the mutant strain to establish paratuberculosis in animals. In accordance with the bacterial organ burden data, Ziehl-Neelsen staining showed higher numbers of acid-fast bacilli in livers from mice infected with the wild-type strain than those of mice infected with the ΔsigL mutant at all time points. A similar observation was noticed for the spleen and intestine tissues (data not shown). Both bacterial organ colonization and histological data analyses suggested that M. avium subsp. paratuberculosis ΔsigL was attenuated for survival, compared to the wild-type strain, in the mouse model of infection.
Immunization with M. avium subsp. paratuberculosis ΔsigL.
Because sigL encodes a mycobacterial GGR (30) and was critical for M. avium subsp. paratuberculosis survival in the present study, we investigated the vaccine potential of the ΔsigL mutant in a challenge model of murine paratuberculosis (Fig. 5A). To examine immunogenicity, groups of mice were immunized twice with M. avium subsp. paratuberculosis ΔsigL and 6 weeks postimmunization (WPI) mouse organs were analyzed for bacterial content. Two immunizations with this mutant resulted in low colonization (2 × 102 CFU) in the liver, whereas no bacteria were detected (limit of detection, 20 CFU) in the intestine or spleen (data not shown). To evaluate vaccine-induced immune responses before challenge, ELISA was used to estimate levels of key cytokines in stimulated spleen cells. Statistical analysis revealed a significantly (P < 0.05) higher level of IFN-γ secretion in the ΔsigL mutant-immunized mice than in naive animals (Fig. 5B). Because of the importance of T-helper 17 cells (34) for intracellular bacterial infection, we examined IL-17A production in the immunized animals. However, we did not find any detectable levels of IL-17A at 6 WPI. Additionally, the mouse group vaccinated with ΔsigL mutant had significantly (P < 0.05) higher anti-PPDj IgG levels (Fig. 5C). Thus, both IFN-γ and IgG data suggested an ability of the ΔsigL mutant strain to induce enhanced immune responses.
FIG 5.
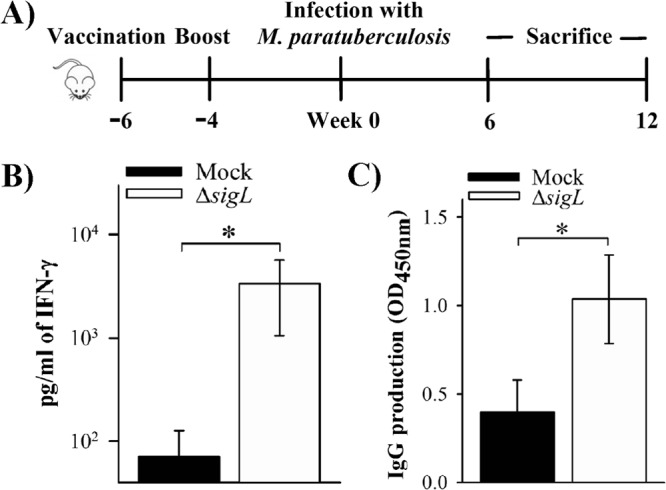
Analysis of immune responses in immunized mice before challenge. (A) Scheme illustrating the immunization study. C57BL/6 mice received a total of 2 doses, each containing ∼2 × 106 CFU of M. avium subsp. paratuberculosis ΔsigL, by subcutaneous injection. The mock group received PBS buffer. Following vaccination, both groups of mice were challenged with wild-type M. avium subsp. paratuberculosis as described above. After 6 weeks postimmunization (6 PWI; week 0 in the scheme), mice (n = 4 to 6) from each group were sacrificed for analysis of immune response. (B) Splenocytes (6 WPI) were isolated and restimulated in vitro with Johnin PPD to measure IFN-γ levels from culture supernatant by ELISA after 48 h. (C) M. avium subsp. paratuberculosis-specific antibody (anti-PPDj antibodies) in the mouse sera (6 WPI) was detected by ELISA (OD450) using horseradish peroxidase-conjugated rabbit anti-mouse antibody. *, P < 0.05.
Protection against challenge with M. avium subsp. paratuberculosis.
To examine the vaccine potential of sigL-based mutant, groups of mice were vaccinated with PBS (control) or the ΔsigL mutant as a live strain and challenged with the virulent M. avium subsp. paratuberculosis strain K10 at 6 WPI (Fig. 5A). At 6 weeks postchallenge (WPC), the ΔsigL-vaccinated mice displayed a significant reduction in the bacterial load in spleen and liver (>5-fold) compared to the PBS-vaccinated mice (Fig. 6A and B). More importantly, a greater reduction of the bacterial load (>10-fold) was observed in the intestine (Fig. 6C), an important organ for M. avium subsp. paratuberculosis infection. A similar colonization pattern was observed at 12 WPC, where the level of bacterial reduction was more than 5-, 10-, and 300-fold for spleen, intestine, and liver, respectively. It is noteworthy that overall colonization levels at 12 WPC were reduced compared to levels at 6 WPC in both immunized and mock-immunized groups, a phenotype that could result from the inherited resistance of the BL6 mice used in this study, as suggested before (35).
FIG 6.
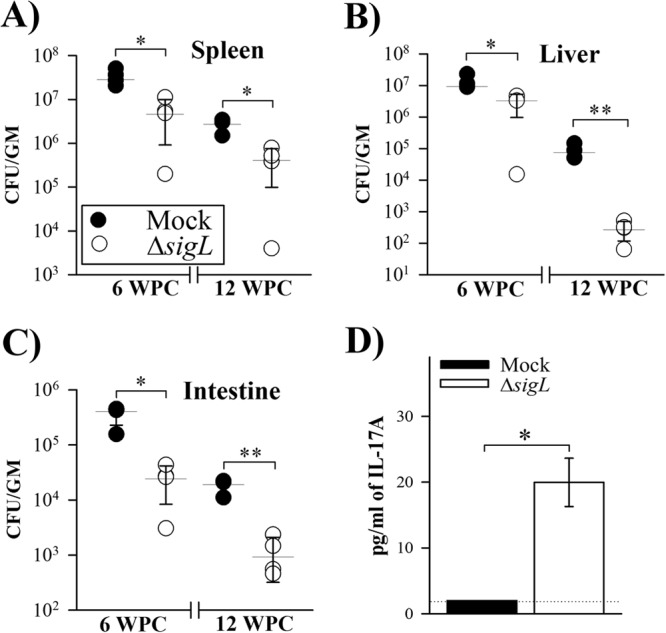
Protective efficacy of immunization. At 6 weeks following vaccination, mice received a challenge dose containing ∼7 × 108 CFU of wild-type M. avium subsp. paratuberculosis i.p. (Fig. 5A). Following challenge, mouse groups (n = 4) were sacrificed at 6 and 12 weeks, and bacterial burden was analyzed in spleen (A), liver (B), and intestine (C) organs. Horizontal lines indicate median values, with error bars representing standard deviations. Statistical analyses were done using the Mann-Whitney test to evaluate differences in bacterial organ load among mouse groups vaccinated with PBS (mock) or the M. avium subsp. paratuberculosis ΔsigL mutant. (D) Secretion of IL-17A (6 WPC) from the cell supernatant was measured by ELISA. Data are means and standard deviations. The dotted line shows the limit of detection (2 pg/ml). *, P < 0.05; **, P < 0.01.
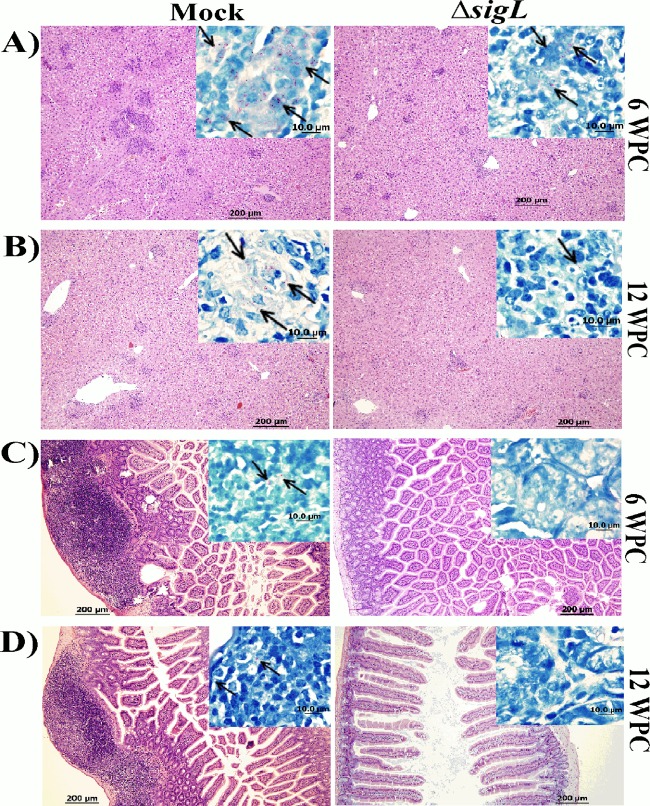
For histological examination, we focused our efforts on the liver because it is the most reflective organ for M. avium subsp. paratuberculosis infection (36). Liver sections from the ΔsigL mutant-immunized animals displayed lower granulomatous scores and smaller granulomas than the PBS control group at 6 and 12 WPC (Fig. 7A and B). In addition, low numbers of acid-fast bacilli were observed when liver sections were stained with Ziehl-Neelsen stain, another support for the colonization data discussed above. Interestingly, sections from the intestines of the ΔsigL mutant-immunized mice appeared normal compared to those from mock-infected mice, with no detectable acid-fast bacteria in Ziehl-Neelsen-stained sections at both at 6 and 12 WPC (Fig. 7C and D). Overall, the reduction in M. avium subsp. paratuberculosis colonization levels combined with histological scores indicated the ability of the ΔsigL mutant to control tissue damage by a challenge with the virulent strain of M. avium subsp. paratuberculosis.
FIG 7.
Pathological analysis of mouse organs following vaccination. Photographs shows hematoxylin and eosin staining of liver (A and B) and intestine (C and D) sections from mock- and M. avium subsp. paratuberculosis ΔsigL-vaccinated animals following challenge with wild-type M. avium subsp. paratuberculosis at 6 WPC and 12 WPC. Magnification, ×100; bar, 200 μm. (Insets) Ziehl-Neelsen staining of both liver and intestine displayed more acid-fast bacilli in the mock-vaccinated animals than in the ones that received M. avium subsp. paratuberculosis ΔsigL vaccination. Magnification, ×1,000. Bar, 10 μm.
Expansion of immune responses following challenge in immunized mice.
To evaluate expansion of the cellular immune response following challenge, splenocytes of immunized and challenged mice were analyzed for the production of key cytokines associated with protection against paratuberculosis (37, 38). The PPDj-stimulated splenocytes from ΔsigL mutant-immunized and challenged mice secreted higher levels of IFN-γ than those from mock-challenged animals at 6 WPC, indicating increased levels of T cell activity (T-helper 1 cells) in the animals that received the ΔsigL mutant (see Fig. S4 in the supplemental material). However, at 12 WPC there was no difference in IFN-γ response between these two groups of animals (see Fig. S4 in the supplemental material). Interestingly, our data also showed that the ΔsigL mutant-vaccinated animals had a better ability to induce PPDj-specific IL-17A secretion than the mock-challenged group at 6 WPC (Fig. 6D), suggesting an importance for T-helper 17 cells during vaccine-induced protection against paratuberculosis. Taken together, the colonization and histological and immune response levels suggest that M. avium subsp. paratuberculosis ΔsigL induced protective immunity against challenge with virulent M. avium subsp. paratuberculosis.
DISCUSSION
Infection with M. avium subsp. paratuberculosis represents a major threat to dairy cattle (4, 6) with the potential to spread to humans (39–43). Earlier reports indicated that M. avium subsp. paratuberculosis counts on a large number of sigma factors (n = 19) to establish the infection and survive diverse stress conditions that the bacteria face during infection (15, 21). In this study, we targeted sigL because of its activation during early macrophage infection, suggesting a role in controlling an important stage(s) of M. avium subsp. paratuberculosis pathogenesis (15). Moreover, the orthologous sigL deletion mutant of M. tuberculosis was attenuated in mice relative to the wild-type strain (30). Our analysis indicated that deletion of sigL affected the ability of M. avium subsp. paratuberculosis to survive exposure to intracellular stimuli, including oxidative stress and stresses damaging the mycobacterial cell wall (e.g., diamide and SDS) (44–46). This reduced survival of the mutant could be partially explained by the type and magnitude of regulated transcripts in mutant and wild-type strains of M. avium subsp. paratuberculosis. Limited analysis of the sigL mutant transcripts compared to those of the wild-type strain suggested that the sigL regulon includes genes encoding polyketide synthases (involved in mycobacterial cell envelope maintenance) (20), the mce gene family proteins (important during macrophage infection) (47), genes encoding proteins involved in lipid transport (48), and finally genes involved in proper folding of secreted proteins (49), suggesting similarity to its counterpart in M. tuberculosis. However, a detailed transcriptional profiling during infection is warranted to analyze the complete regulon of SigL and decipher the differences between both groups in M. avium subsp. paratuberculosis and M. tuberculosis.
Both macrophage and animal colonization levels indicated that the sigL mutant strain was unable to survive inside both macrophages and mouse tissue at levels similar to those of the wild-type strain, an indication of the significant attenuation of this mutant. Both histological and bacteriological analyses revealed reduced organ colonization of the M. avium subsp. paratuberculosis ΔsigL with low inflammatory scores compared to the parental strain. Interestingly, this result was different from the M. tuberculosis ΔsigL murine infection, suggesting a different role for sigL in the survival of M. avium subsp. paratuberculosis (20, 30). Such a disparity is expected with the difference in the pathogenesis of both tuberculosis and paratuberculosis and the animal model used for each (e.g., aerosol versus enteric infection, respectively).
Previously, alternative sigma factor (e.g., sigE) mutants were targeted for the vaccine development and found to provide protection against infection with pathogenic bacteria, including mycobacteria (50, 51). In our study, observations gained from both in vitro and murine model experiments encouraged us to investigate the vaccine potential of M. avium subsp. paratuberculosis ΔsigL as a live attenuated vaccine against murine paratuberculosis. In our hands, mice that received the ΔsigL mutant were very efficient in producing IFN-γ (e.g., at 6 WPI), an important cytokine involved in controlling mycobacterial infection (52). Importantly, culturing tissue samples from immunized animals indicated the ability of the vaccine candidate to persist in animals following immunization but to a low level, which could be a critical factor in inducing protective immune responses. We further evaluated the longevity of immune responses in the mouse groups following challenge (e.g., at 6 WPC), and these data suggest that ΔsigL mutant-immunized mice maintained strong T-cell responses with secretion of more IFN-γ and IL-17. These observations suggested an important role for both IFN-γ and IL-17A during M. avium subsp. paratuberculosis infection. However, we noticed a decrease in IFN-γ response in the ΔsigL mutant-immunized and challenged group at 12 WPC, which could be partially explained by the decrease in the mycobacterial burden in the mouse organs. Alternatively, it is possible that complete elimination of the ΔsigL mutant at later times may have resulted in a decreased IFN-γ response, and if this is the case, then a higher vaccine dose with or without adjuvants may be necessary to achieve long-lasting vaccine-induced protective immunity against M. avium subsp. paratuberculosis infection.
Earlier studies demonstrated the potential use of M. avium subsp. paratuberculosis mutants (e.g., WAg915 and M. avium subsp. paratuberculosis ΔleuD) as live attenuated vaccine candidates in the murine model of paratuberculosis (53, 54). The mutant strain WAg915 (M. avium subsp. paratuberculosis ΔppiA), defective in the peptidyl-prolyl cis-trans-isomerase, showed a mild attenuated phenotype relative to the wild-type strain and resulted in limited tissue colonization following challenge with parental M. avium subsp. paratuberculosis strain (53). Alternatively, the attenuated M. avium subsp. paratuberculosis ΔleuD strain, which was defective in leucine biosynthesis, exhibited better protection in a similar vaccine/challenge model (54). However, it will be difficult to compare the vaccine potential of the ΔsigL mutant to that of the ΔleuD mutant because tissue colonization levels in the leuD experiments were analyzed using only Ziehl-Neelsen staining (55, 56), a less sensitive assay than the counting of CFU, used in our study. Finally, it would be very helpful to compare the performance of the sigL mutant and other vaccine candidates against a licensed vaccine, such as Mycopar, once approvals from veterinary health authority are obtained for use in mice. In future experiments, it would be helpful to generate a vaccine candidate with a deletion of one of the genes in the sigL regulon in the sigL mutant genetic background to increase vaccine safety and avoid the risk of vaccine reversion.
Overall, an isogenic mutant of M. avium subsp. paratuberculosis lacking sigL had a limited ability to survive in macrophages and mice, most likely because of a defective bacterial cell wall. Such an attenuated strain of M. avium subsp. paratuberculosis (ΔsigL) persisted in murine tissues following subcutaneous immunization and generated a substantial immune response. The generated immune responses were sufficient to reduce tissue colonization and lesion scores in animals following a challenge with the wild-type strain of M. avium subsp. paratuberculosis. Further vaccine testing in natural hosts of Johne's disease, e.g., goats or calves, will demonstrate the viability of developing an effective control strategy against paratuberculosis in animals.
Supplementary Material
ACKNOWLEDGMENTS
We thank Matyas Sandor for providing murine IFN-γ and Gary Splitter for reading the manuscript. We also acknowledge the assistance of Kay Nelson and Meagan A. Cooney at the University of Wisconsin—Madison in MDM isolation.
This work was supported by grants NRI 2007-35204-18400, NIFA 2013-67015, and JDIP-Q6286224301 from the U.S. Department of Agriculture.
A.M.T. is the founder of Pan Genome Systems, Madison, WI, a company involved in developing vaccines against Johne's disease.
Footnotes
Published ahead of print 5 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00001-14.
REFERENCES
- 1.Nielsen S, Sr, Toft N. 2009. A review of prevalences of paratuberculosis in farmed animals in Europe. Prev. Vet. Med. 88:1–14. 10.1016/j.prevetmed.2008.07.003 [DOI] [PubMed] [Google Scholar]
- 2.Ghosh P, Hsu C, Alyamani EJ, Shehata MM, Al-Dubaib MA, Al-Naeem A, Hashad M, Mahmoud OM, Alharbi KB, Al-Busadah K, Al-Swailem AM, Talaat AM. 2012. Genome-wide analysis of the emerging infection with Mycobacterium avium subspecies paratuberculosis in the Arabian camels (Camelus dromedarius). PLoS One 7:e31947. 10.1371/journal.pone.0031947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumthekar S, Manning EJ, Ghosh P, Tiwari K, Sharma RN, Hariharan H. 2013. Mycobacterium avium subspecies paratuberculosis confirmed following serological surveillance of small ruminants in Grenada, West Indies. J. Vet. Diagn. Invest. 25:527–530. 10.1177/1040638713490688 [DOI] [PubMed] [Google Scholar]
- 4.Lombard JE, Gardner IA, Jafarzadeh SR, Fossler CP, Harris B, Capsel RT, Wagner BA, Johnson WO. 2013. Herd-level prevalence of Mycobacterium avium subsp. paratuberculosis infection in United States dairy herds in 2007. Prev. Vet. Med. 108:234–238. 10.1016/j.prevetmed.2012.08.006 [DOI] [PubMed] [Google Scholar]
- 5.Lei L, Plattner BL, Hostetter JM. 2008. Live Mycobacterium avium subsp. paratuberculosis and a killed-bacterium vaccine induce distinct subcutaneous granulomas, with unique cellular and cytokine profiles. Clin. Vaccine Immunol. 15:783–793. 10.1128/CVI.00480-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Losinger WC. 2005. Economic impact of reduced milk production associated with Johne's disease on dairy operations in the U. S. A. J. Dairy Res. 72:425–432. 10.1017/S0022029905001007 [DOI] [PubMed] [Google Scholar]
- 7.Uzonna JE, Chilton P, Whitlock RH, Habecker PL, Scott P, Sweeney RW. 2003. Efficacy of commercial and field-strain Mycobacterium paratuberculosis vaccinations with recombinant IL-12 in a bovine experimental infection model. Vaccine 21:3101–3109. 10.1016/S0264-410X(03)00261-5 [DOI] [PubMed] [Google Scholar]
- 8.Kalis CH, Hesselink JW, Barkema HW, Collins MT. 2001. Use of long-term vaccination with a killed vaccine to prevent fecal shedding of Mycobacterium avium subsp paratuberculosis in dairy herds. Am. J. Vet. Res. 62:270–274. 10.2460/ajvr.2001.62.270 [DOI] [PubMed] [Google Scholar]
- 9.Patterson CJ, LaVenture M, Hurley SS, Davis JP. 1988. Accidental self-inoculation with Mycobacterium paratuberculosis bacterin (Johne's bacterin) by veterinarians in Wisconsin. J. Am. Vet. Med. Assoc. 192:1197–1199 [PubMed] [Google Scholar]
- 10.Bermudez LE, Petrofsky M, Sommer S, Barletta RG. 2010. Peyer's patch-deficient mice demonstrate that Mycobacterium avium subsp. paratuberculosis translocates across the mucosal barrier via both M cells and enterocytes but has inefficient dissemination. Infect. Immun. 78:3570–3577. 10.1128/IAI.01411-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chacon O, Bermudez LE, Barletta RG. 2004. Johne's disease, inflammatory bowel disease, and Mycobacterium paratuberculosis. Annu. Rev. Microbiol. 58:329–363. 10.1146/annurev.micro.58.030603.123726 [DOI] [PubMed] [Google Scholar]
- 12.Hostetter J, Steadham E, Haynes J, Bailey T, Cheville N. 2003. Phagosomal maturation and intracellular survival of Mycobacterium avium subspecies paratuberculosis in J774 cells. Comp. Immunol. Microbiol. Infect. Dis. 26:269–283. 10.1016/S0147-9571(02)00070-X [DOI] [PubMed] [Google Scholar]
- 13.Arsenault RJ, Li Y, Bell K, Doig K, Potter A, Griebel PJ, Kusalik A, Napper S. 2012. Mycobacterium avium subsp. paratuberculosis inhibits gamma interferon-induced signaling in bovine monocytes: insights into the cellular mechanisms of Johne's disease. Infect. Immun. 80:3039-3048. 10.1128/IAI.00406-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cossu A, Sechi LA, Zanetti S, Rosu V. 2012. Gene expression profiling of Mycobacterium avium subsp. paratuberculosis in simulated multi-stress conditions and within THP-1 cells reveals a new kind of interactive intramacrophage behaviour. BMC Microbiol. 12:87. 10.1186/1471-2180-12-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghosh P, Wu CW, Talaat AM. 2013. Key role for the alternative sigma factor, SigH, in the intracellular life of Mycobacterium avium subsp. paratuberculosis during macrophage stress. Infect. Immun. 81:2242–2257. 10.1128/IAI.01273-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li L, Bannantine JP, Zhang Q, Amonsin A, May BJ, Alt D, Banerji N, Kanjilal S, Kapur V. 2005. The complete genome sequence of Mycobacterium avium subspecies paratuberculosis. Proc. Natl. Acad. Sci. U. S. A. 102:12344–12349. 10.1073/pnas.0505662102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janagama HK, Lamont EA, George S, Bannantine JP, Xu WW, Tu ZJ, Wells SJ, Schefers J, Sreevatsan S. 2010. Primary transcriptomes of Mycobacterium avium subsp. paratuberculosis reveal proprietary pathways in tissue and macrophages. BMC Genomics 11:561. 10.1186/1471-2164-11-561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schnappinger D, Ehrt S, Voskuil MI, Liu Y, Mangan JA, Monahan IM, Dolganov G, Efron B, Butcher PD, Nathan C, Schoolnik GK. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198:693–704. 10.1084/jem.20030846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, III, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544. 10.1038/31159 [DOI] [PubMed] [Google Scholar]
- 20.Hahn MY, Raman S, Anaya M, Husson RN. 2005. The mycobacterium tuberculosis extracytoplasmic-function sigma factor SigL regulates polyketide synthases and secreted or membrane proteins and is required for virulence. J. Bacteriol. 187:7062–7071. 10.1128/JB.187.20.7062-7071.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu CW, Schmoller SK, Shin SJ, Talaat AM. 2007. Defining the stressome of Mycobacterium avium subsp paratuberculosis in vitro and in naturally infected cows. J. Bacteriol. 189:7877–7886. 10.1128/JB.00780-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pelicic V, Jackson M, Reyrat JM, Jacobs WR, Gicquel B, Guilhot C. 1997. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 94:10955–10960. 10.1073/pnas.94.20.10955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaushal D, Schroeder BG, Tyagi S, Yoshimatsu T, Scott C, Ko C, Carpenter L, Mehrotra J, Manabe YC, Fleischmann RD, Bishai WR. 2002. Reduced immunopathology and mortality despite tissue persistence in a Mycobacterium tuberculosis mutant lacking alternative sigma factor, SigH. Proc. Natl. Acad. Sci. U. S. A. 99:8330–8335. 10.1073/pnas.102055799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Talaat AM, Hunter P, Johnston SA. 2000. Genome-directed primers for selective labeling of bacterial transcripts for DNA microarray analysis. Nat. Biotechnol. 18:679–682. 10.1038/76543 [DOI] [PubMed] [Google Scholar]
- 25.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murali-Krishna K, Altman JD, Suresh M, Sourdive DJ, Zajac AJ, Miller JD, Slansky J, Ahmed R. 1998. Counting antigen-specific CD8 T cells: a reevaluation of bystander activation during viral infection. Immunity 8:177–187. 10.1016/S1074-7613(00)80470-7 [DOI] [PubMed] [Google Scholar]
- 27.Stabel JR, Barnhill A, Bannantine JP, Chang YF, Osman MA. 2012. Evaluation of protection in a mouse model after vaccination with Mycobacterium avium subsp. paratuberculosis protein cocktails. Vaccine 31:127–134. 10.1016/j.vaccine.2012.10.090 [DOI] [PubMed] [Google Scholar]
- 28.Frey A, Di CJ, Zurakowski D. 1998. A statistically defined endpoint titer determination method for immunoassays. J. Immunol. Methods 221:35–41. 10.1016/S0022-1759(98)00170-7 [DOI] [PubMed] [Google Scholar]
- 29.Settles EW, Kink JA, Talaat A. 2014. Attenuated strains of Mycobacterium avium subspecies paratuberculosis as vaccine candidates against Johne's disease. Vaccine 32:2062–2069. 10.1016/j.vaccine.2014.02.010 [DOI] [PubMed] [Google Scholar]
- 30.Dainese E, Rodrigue S, Delogu G, Provvedi R, Laflamme L, Brzezinski R, Fadda G, Smith I, Gaudreau L, Palu G, Manganelli R. 2006. Posttranslational regulation of Mycobacterium tuberculosis extracytoplasmic-function sigma factor sigma(L) and roles in virulence and in global regulation of gene expression. Infect. Immun. 74:2457–2461. 10.1128/IAI.74.4.2457-2461.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson S, Brusasca P, Lyashchenko K, Spencer JS, Wiker HG, Bifani P, Shashkina E, Kreiswirth B, Harboe M, Schluger N, Gomez M, Gennaro ML. 2001. Characterization of the secreted MPT53 antigen of Mycobacterium tuberculosis. Infect. Immun. 69:5936–5939. 10.1128/IAI.69.9.5936-5939.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gilliland SE, Staley TE, Bush LJ. 1984. Importance of bile tolerance of Lactobacillus acidophilus used as a dietary adjunct. J. Dairy Sci. 67:3045–3051. 10.3168/jds.S0022-0302(84)81670-7 [DOI] [PubMed] [Google Scholar]
- 33.Sung N, Collins MT. 2003. Variation in resistance of Mycobacterium paratuberculosis to acid environments as a function of culture medium. Appl. Environ. Microbiol. 69:6833–6840. 10.1128/AEM.69.11.6833-6840.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khader SA, Gopal R. 2010. IL-17 in protective immunity to intracellular pathogens. Virulence 1:423–427. 10.4161/viru.1.5.12862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roupie V, Rosseels V, Piersoel V, Zinniel DK, Barletta RG, Huygen K. 2008. Genetic resistance of mice to Mycobacterium paratuberculosis is influenced by Slc11a1 at the early but not at the late stage of infection. Infect. Immun. 76:2099–2105. 10.1128/IAI.01137-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shin SJ, Wu C-W, Steinberg H, Talaat AM. 2006. Identification of novel virulence determinants in Mycobacterium paratuberculosis by screening a library of insertional mutants. Infect. Immun. 74:3825–3833. 10.1128/IAI.01742-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Begg DJ, Griffin JF. 2005. Vaccination of sheep against M. paratuberculosis: immune parameters and protective efficacy. Vaccine 23:4999–5008. 10.1016/j.vaccine.2005.05.031 [DOI] [PubMed] [Google Scholar]
- 38.Stabel JR, Robbe-Austerman S. 2011. Early immune markers associated with Mycobacterium avium subsp. paratuberculosis infection in a neonatal calf model. Clin. Vaccine Immunol. 18:393–405. 10.1128/CVI.00359-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Naser SA, Ghobrial G, Romero C, Valentine JF. 2004. Culture of Mycobacterium avium subspecies paratuberculosis from the blood of patients with Crohn's disease. Lancet 364:1039–1044. 10.1016/S0140-6736(04)17058-X [DOI] [PubMed] [Google Scholar]
- 40.Bull TJ, McMinn EJ, Sidi-Boumedine K, Skull A, Durkin D, Neild P, Rhodes G, Pickup R, Hermon-Taylor J. 2003. Detection and verification of Mycobacterium avium subsp. paratuberculosis in fresh ileocolonic mucosal biopsy specimens from individuals with and without Crohn's disease. J. Clin. Microbiol. 41:2915–2923. 10.1128/JCM.41.7.2915-2923.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hermon-Taylor J. 2009. Mycobacterium avium subspecies paratuberculosis, Crohn's disease and the Doomsday scenario. Gut Pathog. 1:15. 10.1186/1757-4749-1-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Naser SA, Thanigachalam S, Dow CT, Collins MT. 2013. Exploring the role of Mycobacterium avium subspecies paratuberculosis in the pathogenesis of type 1 diabetes mellitus: a pilot study. Gut Pathog. 5:14. 10.1186/1757-4749-5-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Masala S, Paccagnini D, Cossu D, Brezar V, Pacifico A, Ahmed N, Mallone R, Sechi LA. 2011. Antibodies recognizing Mycobacterium avium paratuberculosis epitopes cross-react with the beta-cell antigen ZnT8 in Sardinian type 1 diabetic patients. PLoS One 6:e26931. 10.1371/journal.pone.0026931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gunn JS. 2000. Mechanisms of bacterial resistance and response to bile. Microbes Infect. 2:907–913. 10.1016/S1286-4579(00)00392-0 [DOI] [PubMed] [Google Scholar]
- 45.Prieto AI, Ramos-Morales F, Casadesus J. 2006. Repair of DNA damage induced by bile salts in Salmonella enterica. Genetics 174:575–584. 10.1534/genetics.106.060889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.den Hengst CD, Buttner MJ. 2008. Redox control in actinobacteria. Biochim. Biophys. Acta 1780:1201–1216. 10.1016/j.bbagen.2008.01.008 [DOI] [PubMed] [Google Scholar]
- 47.Graham JE, Clark-Curtiss JE. 1999. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS). Proc. Natl. Acad. Sci. U. S. A. 96:11554–11559. 10.1073/pnas.96.20.11554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Camacho LR, Constant P, Raynaud C, Laneelle MA, Triccas JA, Gicquel B, Daffe M, Guilhot C. 2001. Analysis of the phthiocerol dimycocerosate locus of Mycobacterium tuberculosis. Evidence that this lipid is involved in the cell wall permeability barrier. J. Biol. Chem. 276:19845–19854. 10.1074/jbc.M100662200 [DOI] [PubMed] [Google Scholar]
- 49.Goulding CW, Apostol MI, Gleiter S, Parseghian A, Bardwell J, Gennaro M, Eisenberg D. 2004. Gram-positive DsbE proteins function differently from Gram-negative DsbE homologs. A structure to function analysis of DsbE from Mycobacterium tuberculosis. J. Biol. Chem. 279:3516–3524. 10.1074/jbc.M311833200 [DOI] [PubMed] [Google Scholar]
- 50.Coynault C, Robbe-Saule V, Norel F. 1996. Virulence and vaccine potential of Salmonella typhimurium mutants deficient in the expression of the RpoS (sigma S) regulon. Mol. Microbiol. 22:149–160. 10.1111/j.1365-2958.1996.tb02664.x [DOI] [PubMed] [Google Scholar]
- 51.Hernandez Pando R, Aguilar LD, Smith I, Manganelli R. 2010. Immunogenicity and protection induced by a Mycobacterium tuberculosis sigE mutant in a BALB/c mouse model of progressive pulmonary tuberculosis. Infect. Immun. 78:3168–3176. 10.1128/IAI.00023-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O'Garra A, Redford PS, McNab FW, Bloom CI, Wilkinson RJ, Berry MP. 2013. The immune response in tuberculosis. Annu. Rev. Immunol. 31:475–527. 10.1146/annurev-immunol-032712-095939 [DOI] [PubMed] [Google Scholar]
- 53.Scandurra GM, de Lisle GW, Cavaignac SM, Young M, Kawakami RP, Collins DM. 2010. Assessment of live candidate vaccines for paratuberculosis in animal models and macrophages. Infect. Immun. 78:1383–1389. 10.1128/IAI.01020-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen JW, Faisal SM, Chandra S, McDonough SP, Moreira MA, Scaria J, Chang CF, Bannantine JP, Akey B, Chang YF. 2012. Immunogenicity and protective efficacy of the Mycobacterium avium subsp. paratuberculosis attenuated mutants against challenge in a mouse model. Vaccine 30:3015–3025. 10.1016/j.vaccine.2011.11.029 [DOI] [PubMed] [Google Scholar]
- 55.Jeyanathan M, Alexander DC, Turenne CY, Girard C, Behr MA. 2006. Evaluation of in situ methods used to detect Mycobacterium avium subsp. paratuberculosis in samples from patients with Crohn's disease. J. Clin. Microbiol. 44:2942–2950. 10.1128/JCM.00585-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seiler P, Ulrichs T, Bandermann S, Pradl L, Jorg S, Krenn V, Morawietz L, Kaufmann SH, Aichele P. 2003. Cell-wall alterations as an attribute of Mycobacterium tuberculosis in latent infection. J. Infect. Dis. 188:1326–1331. 10.1086/378563 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.