Abstract
Mycoplasma pneumoniae causes pneumonia, tracheobronchitis, pharyngitis, and asthma in humans. The pathogenesis of M. pneumoniae infection is attributed to excessive immune responses. We previously demonstrated that M. pneumoniae lipoproteins induced inflammatory responses through Toll-like receptor 2 (TLR2). In the present study, we demonstrated that M. pneumoniae induced strong inflammatory responses in macrophages derived from TLR2 knockout (KO) mice. Cytokine production in TLR2 KO macrophages was increased compared with that in the macrophages of wild-type (WT) mice. Heat-killed, antibiotic-treated, and overgrown M. pneumoniae failed to induce inflammatory responses in TLR2 KO macrophages. 3-Methyladenine and chloroquine, inhibitors of autophagy, decreased the induction of inflammatory responses in TLR2 KO macrophages. These inflammatory responses were also inhibited in macrophages treated with the TLR4 inhibitor VIPER and those obtained from TLR2 and TLR4 (TLR2/4) double-KO mice. Two mutants that lacked the ability to induce inflammatory responses in TLR2 KO macrophages were obtained by transposon mutagenesis. The transposons were inserted in atpC encoding an ATP synthase F0F1 ε subunit and F10_orf750 encoding hypothetical protein MPN333. These mutants showed deficiencies in cytadherence. These results suggest that cytadherence of M. pneumoniae induces inflammatory responses through TLR4 and autophagy.
INTRODUCTION
Mycoplasmas are wall-less parasitic bacteria and the smallest organisms capable of self-replication (1). Mycoplasma pneumoniae causes pneumonia, tracheobronchitis, pharyngitis, and asthma in humans (2–4). From 2010 to 2012, epidemics of M. pneumoniae infections were reported worldwide (e.g., in France, Israel, and Japan) (5). However, pathogenic agents such as endotoxins and exotoxins that cause such diseases have not been identified in M. pneumoniae. Cytadherence of invading mycoplasmas to the respiratory epithelium, localized host cell injury, and overaggressive inappropriate immune responses appear to contribute to the pathogenesis of M. pneumoniae infection (2, 6).
We previously identified 3 lipoproteins responsible for nuclear factor-kappa B (NF-κB) activation (7). One was MPN602, a b subunit of the F0F1-type ATPase (8). The activation of NF-κB by subunit b of the F0F1-type ATPase was dependent on the presence of TLR1, TLR2, and TLR6, indicating that subunit b of the F0F1-type ATPase is a diacylated lipoprotein. The others were predicted to be lipoproteins MPN611 and MPN162 and designated NF-κB-activating lipoprotein 1 (N-ALP1) and N-ALP2, respectively. N-ALP1 and N-ALP2 activated TLR signaling through TLR1 and TLR2, indicating that both are triacylated lipoproteins (9). Because mycoplasmas lack cell walls, they do not contain known pathogen-associated molecular patterns (PAMPS) such as those corresponding to lipopolysaccharide (LPS), peptidoglycan (PGN), or lipoteichoic acid. These findings suggested that lipoproteins are key factors of M. pneumoniae-induced inflammatory responses and facilitate the development of pneumonia in humans. However, the existence of lipoproteins in nonpathogenic mycoplasmas suggests the presence of an alternative mechanism by which M. pneumoniae induces inflammatory responses.
TLRs are a type of pattern-recognition receptor and play critical roles in early innate recognition and the inflammatory responses of the host to invading microbes (10, 11). Among the 10 reported TLR family members, TLR2, TLR4, TLR5, and TLR9 have been implicated in the recognition of different bacterial components. For example, PGN, lipoarabinomannan, zymosan, and lipoproteins from various microorganisms are recognized by TLR2 (12–18), whereas LPS, bacterial flagellin, and bacterial DNA are recognized by TLR4, TLR5, and TLR9, respectively (19–22). These TLR family members have been shown to activate NF-κB via interleukin-1R (IL-1R)-associated signal molecules, including myeloid differentiation protein (MyD88), interleukin-1 receptor-activated kinase (IRAK), tumor necrosis factor (TNF) receptor-associated factor 6 (TRAF6), and NF-κB-inducing kinase (NIK) (23).
Autophagy is a cellular response that involves sequestration of regions within the cytosol with double-membrane compartments. Autophagy has shown to play important roles in the response to starvation, cell death, removal of damaged organelles, and neurodegenerative diseases (24). Recently, it has been recognized that autophagy is involved in both innate and adaptive immunity to various microorganisms (25). However, the relationship between autophagy and mycoplasma species remains to be elucidated.
Cytadherence of M. pneumoniae in the respiratory tract is the initial event in infection and is mediated by P1 adhesin and other proteins such as P30 and high-molecular-weight (HMW) proteins (26–29). Although the cytadherence of M. pneumoniae is believed to be responsible for its pathogenesis (30, 31), the precise mechanisms by which cytadherence is involved in inflammatory responses remain unknown.
In this study, we demonstrated that live M. pneumoniae induced proinflammatory cytokines through a TLR2-independent pathway and investigated the mechanisms of the pathway. The activity of the TLR2-independent pathway was inhibited by the autophagy inhibitors and was also decreased in macrophages derived from TLR4 or MyD88 knockout (KO) mice. Moreover, mutant strains that failed to induce proinflammatory cytokines in TLR2 KO macrophages were isolated by transposon mutagenesis. These mutants showed deficiencies in cytadherence. Collectively, these data suggest that the cytadherence property of M. pneumoniae induces inflammatory responses through TLR4 and autophagy.
MATERIALS AND METHODS
M. pneumoniae strains.
M. pneumoniae wild-type (WT) strain M129 was cultured in PPLO broth (Difco, Franklin Lakes, NJ) containing 10% horse serum, 0.25% glucose, 0.25% yeast extract, and 0.002% phenol red at pH 7.6 until the beginning of a stationary phase (the medium color turned slightly orange). The bacterial concentration was adjusted according to the optical density at 595 nm (OD595) in phosphate-buffered saline (PBS). Heat-killed M. pneumoniae was obtained by heating at 60°C for 30 min. Sonication of M. pneumoniae was carried out for 30 s at output 5 using a Sonifier 200 cell disruptor (Branson, Danbury, CT). To obtain overgrown M. pneumoniae, bacteria in the stationary phase (orange color) were cultured for an additional 48 h. Antibiotic-killed M. pneumoniae was prepared by treatment of bacterial cultures with 50 μg/ml of gentamicin for 24 h. Under these conditions, heat-killed, sonicated, antibiotic-killed, and overgrown M. pneumoniae failed to grow on PPLO agar (Difco) containing 10% horse serum, 0.25% glucose, and 0.25% yeast extract, and colonies were not observed. To ensure the same amounts of M. pneumoniae at the same optical densities, the DNA amounts of M. pneumoniae were checked as previously described (32). Briefly, M. pneumoniae DNA was purified using a FastPure DNA kit (TaKaRa, Tokyo, Japan) and quantified using a spectrophotometer at OD280.
Mice and TLR KO mice.
C57BL mice were purchased from Kyudo (Saga, Japan). TLR2 and TLR4 KO mice originally established by S. Akira (Osaka University) (10) were purchased from Oriental Bio Service (Kyoto, Japan). The TLR2 and TLR4 (TLR2/4) double-KO mouse strain was generated by cross-breeding TLR2 KO mice with TLR4 KO mice. Genotyping was performed with the following primers: 5′-GTTTAGTGCCTGTATCCAGTCAGTGCG-3′, specific for the targeted TLR2 gene; 5′-TTGGATAAGTCTGATAGCCTTGCCTCC-3′, specific for the TLR2 gene downstream of the targeting construct; 5′-ATCGCCTTCTATCGCCTTCTTGACGAG-3′, specific for the neo resistance gene inserted in TLR2; 5′-CGTGTAAACCAGCCAGGTTTTGAAGGC-3′, specific for the targeted TLR4 gene; 5′-TGTTGCCCTTCAGTCACAGAGACTCTG-3′, specific for the TLR4 gene upstream of the targeting construct; and 5′-TGTTGGGTCGTTTGTTCGGATCCGTCG-3′, specific for the neo resistance gene inserted in TLR4. All experiments were conducted in compliance with the institutional guidelines of and were approved by Yamaguchi University and Kurume University.
Isolation of mouse peritoneal macrophages.
Thioglycolate broth (2 ml) (Sigma-Aldrich, St. Louis, MO) was injected into the peritoneal cavities of C57BL, TLR2 KO, TLR4 KO, and TLR2/4 double-KO mice. After 72 h later, peritoneal exudate macrophages were harvested by centrifugation. The cell pellets were suspended in RPMI 1640 medium (Sigma-Aldrich) containing 10% fetal calf serum (FCS) (Biowest, Nuaillé, France). The cells were allowed to adhere to 48-well culture plates for 2 h at 37°C in an atmosphere of 5% CO2. Nonadherent cells were removed by washing with PBS, and the remaining adherent cells were infected with M. pneumoniae.
Cell treatment and infection.
Peritoneal macrophages (5 × 105 cells/250 μl) were cultured for 2 h before infection in a 48-well plate and then treated with 100 μM chloroquine (Sigma-Aldrich), 2 μM cytochalasin D, 5 mM 3-methyladenine (3MA; Sigma-Aldrich), 50 μM VIPER (Imgenex, San Diego, CA), 10 μM OD2088 (Invivogen, San Diego, CA), 10 μM SB203580 (Wako, Osaka, Japan), 10 μM U0126 (Wako), or 50 μM SP600123 (Wako) for 30 min. Next, the cells were infected with 25 μl of M. pneumoniae (OD595 = 0.1) for 1, 3, or 6 h and the culture supernatants were collected, and concentrations of proinflammatory cytokines in the supernatants were measured using an enzyme-linked immunosorbent assay (ELISA). To ensure the same amounts of M. pneumoniae at the same optical densities, the DNA amounts of M. pneumoniae were checked as previously described (32).
Mouse infection model.
C57BL WT, TLR2 KO, and TLR2/4 double-KO mice were intranasally infected with 25 μl of M. pneumoniae (OD595 = 0.1). After 24 h, the mice were again infected with 25 μl of M. pneumoniae (OD595 = 0.1), and 24 h later, bronchoalveolar lavage fluid (BALF) was obtained by instilling 1 ml of PBS into the lungs and then aspirating the fluid from the trachea of the mice using a tracheal cannula. The cells that infiltrated in BALF were collected by centrifugation (3,000 rpm for 10 min), and the supernatants were stored at −80°C until determination of cytokine concentrations.
ELISA.
Concentrations of tumor necrosis factor alpha (TNF-α) and IL-6 in the supernatants of peritoneal macrophage were measured using standard ELISA development kits (Pepro Tech, Rocky Hill, NJ) according to the manufacturer's instructions. TNF-α concentrations in BALF of WT, TLR2 KO, and TLR2/4 double-KO mice were measured using an ELISA MAX standard kit (Biolegend, San Diego, CA) according to the manufacturer's instructions.
Real-time PCR.
Peritoneal macrophages (5 × 105 cells/250 μl) were cultured for 2 h in a 48-well plate and then infected with 25 μl of M. pneumoniae (OD595 = 0.1) for 1, 2, or 6 h. Total RNA was isolated from whole-lung tissue by using a NucleoSpin kit (Clonetech, Mountain View, CA), and 1 μg of total RNA was used to synthesize cDNA using a ReverTra Ace quantitative PCR (qPCR) reverse transcriptase (RT) kit (Toyobo, Osaka, Japan). PCR was performed using Thunderbird SYBR qPCR Mix (Toyobo). The following primer sets were purchased from TaKaRa: TNF-α, forward 5′-AAGCCTGTAGCCCACGTCGTA-3′ and reverse 5′-GGCACCACTAGTTGGTTGTCTTTG-3′; IL-6, forward 5′-CCACTTCACAAGTCGGAGGCTTA-3′ and reverse 5′-GCAAGTGCATCATCGTTGTTCATAC-3′; IL-10, forward 5′-GACCAGCTGGACAACATACTGCTAA-3′ and reverse 5′-GATAAGGCTTGGCAACCCAAGTAA-3′; IL-17, forward 5′-CTGATCAGGACGCGCAAAC-3′ and reverse 5′-TCGCTGCTGCCTTCACTGTA-3′; gamma interferon (IFN-γ), forward 5′-CGGCACAGTCATTGAAATCCTA-3′ and reverse 5′-GTTGCTGATGGCCTGATTGTC-3′; and β-actin, forward 5′-TGACAGGATGCAGAAGGAGA-3′ and reverse 5′-GCTGGAAGGTGGACAGTGAG-3′. All data were normalized to β-actin.
Immunofluorescence microscopy.
Peritoneal macrophages (2 × 106 cells/ml) were cultured for 2 h before infection on coverslips in a 12-well plate. Next, the cells were infected with 100 μl of M. pneumoniae M129 or green fluorescent protein (GFP)-expressing M. pneumoniae TK165 (33) (OD595 = 0.1) for 6 or 18 h. The samples were washed twice with PBS and fixed with 4% paraformaldehyde–PBS for 30 min at room temperature, washed three times with PBS, and incubated successively three times for 5 min each time in blocking buffer (3% bovine serum albumin–PBS) at room temperature. The samples were permeabilized in 0.1% Triton X-100 and washed three times with PBS, followed by treatment with 2 μg/ml antimicrotubule-associated protein 1A/1B-light chain 3 (LC3) polyclonal goat antibody (Santa Cruz, Dallas, TX) diluted in blocking buffer. After incubation for 1 h at room temperature, the samples were washed three times for 5 min with blocking buffer, stained with fluorescein isothiocyanate (FITC)- or rhodamine-labeled donkey anti-goat IgG (Santa Cruz) (4 μg/ml) in blocking buffer, and incubated for 1 h at room temperature. DNAs of macrophages and M. pneumoniae were stained with 4′,6-diamidino-2-phenylindole (DAPI). Fluorescent images were obtained using a LSM 710 confocal laser scanning microscope (Zeiss, Oberkochen, Germany).
Transposon mutagenesis.
Approximately 107 CFU/ml of M. pneumoniae were transfected with 5 μg of pISM2062 using a Gene Pulser system (Bio-Rad, Hercules, CA) at 100 Ω resistance, 25 F capacitance, and 2.5 kV. Transformed M. pneumoniae colonies were selected on PPLO agar containing 50 μg/ml of gentamicin. TLR2 KO peritoneal macrophages (5 × 105 cells/250 μl) were infected with 25 μl of transformed M. pneumoniae (OD595 = 0.02) for 3 h, and then the culture supernatants were collected. The strains that showed decreased TNF-α induction were screened by ELISA.
Verification of the transposon-inserted regions.
DNA of transposon-inserted M. pneumoniae was purified using a FastPure DNA kit (TaKaRa, Shiga, Japan). The first PCR amplification was performed with the following primers: specific for IS256, 5′-AAGTCCTCCTGGGTATGT-3′; and a random primer, 5′-GGCCACGCGTCGACTAGTACNNNNNNNNNNACGCC-3′. A second PCR amplification was performed with the following primers: specific for IS256, 5′-CGACTCTAGAGGATCCATTGTACCGTAAAAGGACTG-3′; and specific for a random primer, 5′-CGGTACCCGGGGATCGGCCACGCGTCGACTAGTAC-3′. The amplified PCR products were cloned into the pUC19 vector using an In-Fusion HD cloning kit (Clontech) and sequenced using an ABI3130 sequencer (Life Technologies, Carlsbad, CA).
Hemadsorption assay.
The hemadsorption assay was performed directly on an agar plate. In brief, the colonies were overlaid with 15 ml of fresh sheep blood, washed, and resuspended in PBS to reach a final concentration of 0.5% (vol/vol). After incubation at 37°C for 30 min, the suspension was carefully discarded and unbound erythrocytes were gently removed by washing 3 times with PBS.
Statistical analysis.
All data were compared using one-way analysis of variance, and the results are expressed as means and standard deviations. Differences between groups were compared by multiple comparisons using the Bonferroni t test. Differences were considered significant at a P value of <0.01 or 0.05.
RESULTS
TLR2-independent induction of proinflammatory cytokines by M. pneumoniae.
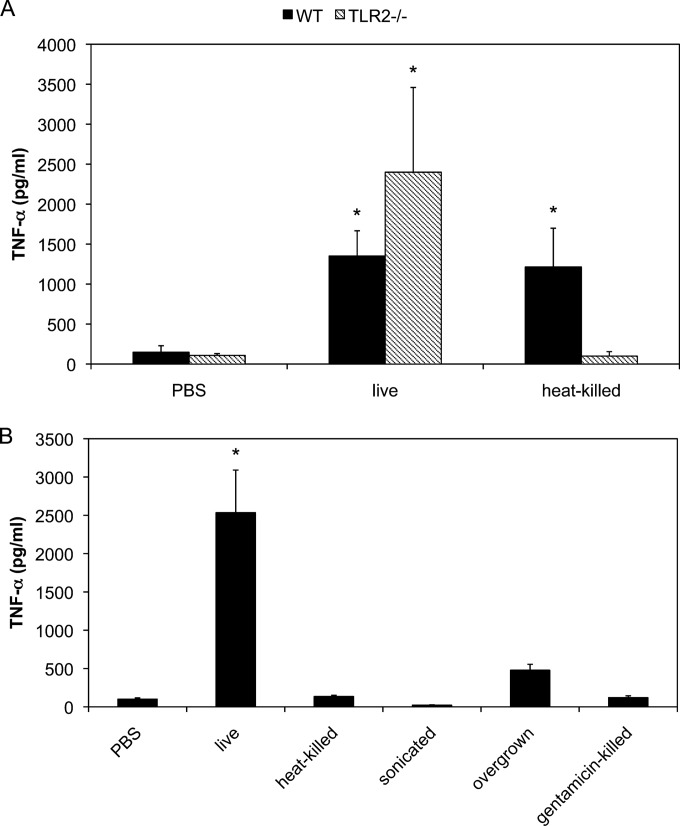
We previously demonstrated that purified or synthesized lipoproteins of M. pneumoniae induced inflammatory responses through TLR2. To examine whether TLR2 is an important receptor for the induction of inflammatory responses in live M. pneumoniae infection, peritoneal macrophages derived from TLR2 KO mice were infected with M. pneumoniae and induced TNF-α concentrations were measured using ELISA (Fig. 1A). In WT macrophages, infection by both live and heat-killed M. pneumoniae induced TNF-α expression. Contrary to expectations, live M. pneumoniae induced TNF-α expression in TLR2 KO macrophages which was slightly increased compared with that in WT macrophages infected with live M. pneumoniae. These results indicate that live M. pneumoniae induces inflammatory responses through a TLR2-independent pathway. Similar to lipoproteins of M. pneumoniae, heat-killed M. pneumoniae induced TNF-α expression in WT macrophages; however, they failed to do so in TLR2 KO macrophages, suggesting that lipoproteins and TLR2 are important to induce inflammatory responses in the infection by heat-killed M. pneumoniae. To rule out the possibility that heating of M. pneumoniae resulted in conformational changes in the surface components of M. pneumoniae leading to a reduction in TNF-α production, TLR2 KO macrophages were infected with sonication-killed, overgrown, and gentamicin-killed M. pneumoniae (Fig. 1B). All treatments decreased the induction of TNF-α expression in TLR2 KO macrophages. These results suggest that biological activity of M. pneumoniae plays an important role in the TLR2-independent pathway.
FIG 1.
TNF-α induction in TLR2 KO macrophages. (A) Peritoneal macrophages derived from WT and TLR2 KO mice were infected with live or heat-killed M. pneumoniae. After 6 h of incubation, TNF-α concentrations in the culture medium were measured using ELISA. (B) Peritoneal macrophages derived from TLR2 KO mice were infected with heat-, sonication-, or antibiotic-killed or overgrown M. pneumoniae. After 6 h of incubation, TNF-α concentrations in the culture medium were measured using ELISA. All values are presented as the means and standard deviations (SD) of the results of 3 assays. *, P < 0.01 compared with PBS results by multiple comparison.
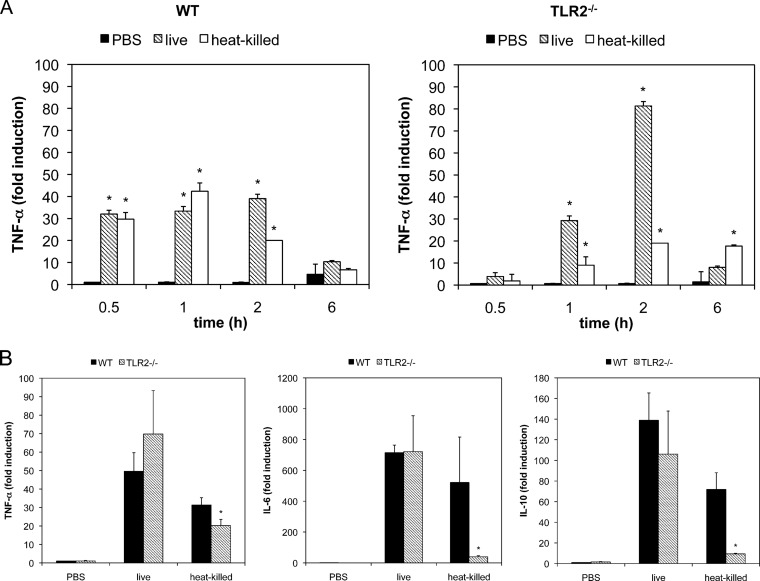
To investigate the temporal kinetics of the M. pneumoniae-induced inflammatory response, peritoneal macrophages from WT and TLR KO mice were infected with live and heat-killed M. pneumoniae and TNF-α mRNA expression levels at 0.5, 1, 2, or 6 h postinfection were measured by real-time PCR (Fig. 2A). TNF-α was expressed in WT macrophages infected with both live and heat-killed M. pneumoniae. TNF-α expression was observed at 0.5 h postinfection, and the same level was sustained until 2 h. However, TNF-α expression was not observed at 6 h postinfection. In TLR2 KO macrophages, TNF-α expression was induced by live M. pneumoniae, whereas the expression was reduced in the case of heat-killed M. pneumoniae infection. TNF-α expression by live M. pneumoniae was observed at 1 h postinfection and reached a maximum level at 2 h. The expression level at 2 h postinfection in TLR2 KO macrophages was approximately 2.5-fold higher than that in WT macrophages. To confirm whether the TLR2-independent pathway was important for the induction of other pro- and anti-inflammatory cytokines, the expression levels of TFN-α, IL-6, IL-17, IFN-γ, and IL-10 mRNA were measured (Fig. 2B). Although live and heat-killed M. pneumoniae induced TNF-α, IL-6, and IL-10 expression in WT macrophages, heat-killed M. pneumoniae failed to induce expression of these cytokines in TLR2 KO macrophages. M. pneumoniae-induced expression of IFN-γ and IL-17 was not detected in either WT or TLR2 KO macrophages (data not shown).
FIG 2.
mRNA expression of proinflammatory cytokines by M. pneumoniae infection. (A) Peritoneal macrophages derived from WT and TLR2 KO mice were infected with live or heat-killed M. pneumoniae. After 1, 2, 3, and 6 h of incubation, total RNA was isolated and TNF-α mRNA expression levels were measured using real-time PCR. All values are represented as the means and SD of the results of 3 assays. *, P < 0.01 compared with PBS results by multiple comparison. (B) Peritoneal macrophages derived from WT and TLR2 KO mice were infected with live and heat-killed M. pneumoniae. After 6 h of incubation, total RNA was isolated and the expression levels of TNF-α, IL-6, and IL-10 mRNA were measured using real-time PCR. All values are presented as the means and SD of the results of 3 assays. *, P < 0.01 compared with WT macrophages by multiple comparison.
Autophagy- and endocytosis-dependent induction of proinflammatory cytokines.
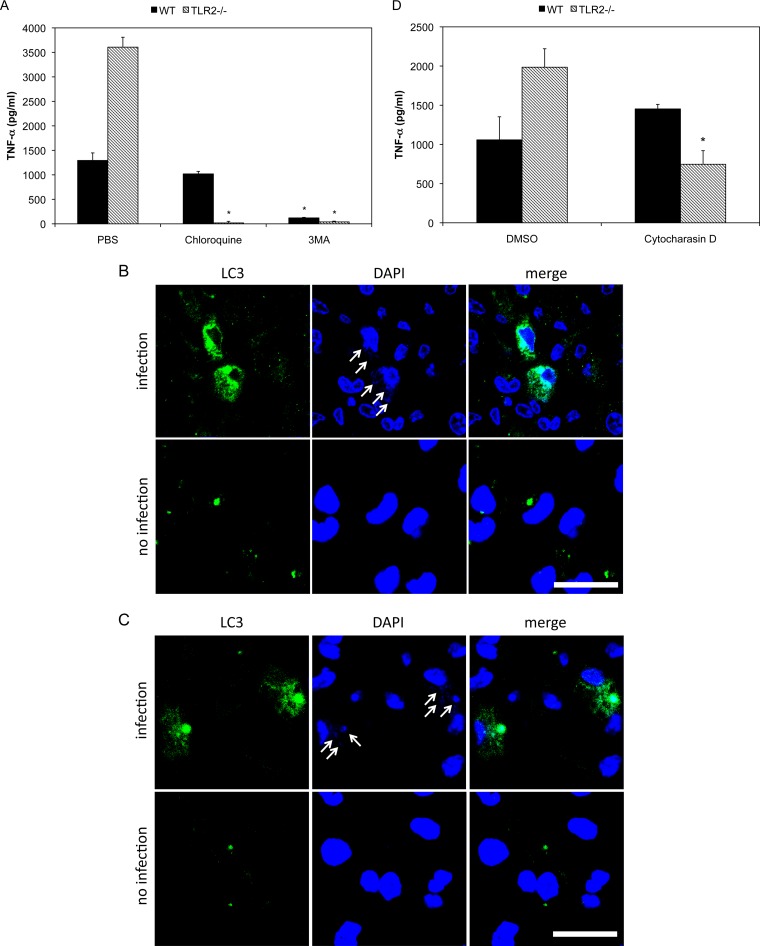
To elucidate the mechanism by which live M. pneumoniae induces TLR2-independent proinflammatory cytokine production, we examined the involvement of autophagy and endocytosis. Peritoneal macrophages were treated with autophagy and endocytosis inhibitors and then infected with live M. pneumoniae (Fig. 3A). 3-MA inhibits autophagy by blocking autophagosome formation via the inhibition of type III phosphatidylinositol 3-kinases (PI-3K) (34). When macrophages derived from WT and TLR2 KO mice were treated with 3-MA and infected with live M. pneumoniae, TNF-α induction was inhibited in comparison with that of control PBS-treated cells. Chloroquine is a lysosomotropic agent that prevents both fusion of autophagosome with lysosome and lysosomal protein degradation (35). In the presence of chloroquine, TNF-α induction in TLR2 KO macrophages was completely inhibited; however, the induction in WT macrophages was not decreased (Fig. 3A). To further examine whether the autophagy participates in induction of proinflammatory cytokines, localization of autophagy in M. pneumoniae-infected macrophages was examined. WT and TLR2 KO macrophages were infected with live M. pneumoniae, and the localizations of DNA of M. pneumoniae and autophagy marker protein LC3 were observed with confocal microscopy. After 6 h of infection, M. pneumoniae was observed as small particles of DNA in macrophages (Fig. 3B and C, arrows in upper panels), whereas the small particles of DNA were not observed in macrophages without infection (Fig. 3B and C, lower panels). The small DNA particles were colocalized with LC3 in both WT and TLR2 KO mouse (Fig. 3A and B; see also Video S1 and S2 in the supplemental material), whereas the small particles of DNA was completely removed and colocalization with LC3 was not observed at 18 h postinfection (see Fig. S1A and B). To confirm that the small particles of DNA represented M. pneumoniae, WT and TLR2 KO macrophages were infected with GFP-expressing M. pneumoniae TK165, and the colocalization of M. pneumoniae and LC3 was examined. The colocalization of GFP-expressing M. pneumoniae and LC3 was also observed at 6 h postinfection (see Fig. S2A and B). These results suggest that autophagy is necessary for TLR2-independent induction of inflammatory responses.
FIG 3.
Autophagy-dependent induction of TNF-α. (A) Peritoneal macrophages derived from TLR2 KO mice were treated with 100 μM chloroquine or 5 mM 3MA for 30 min. The treated cells were infected with live M. pneumoniae. After 6 h of incubation, TNF-α concentrations in the culture medium were measured using ELISA. (B and C) Peritoneal macrophages were infected with live M. pneumoniae for 6 h. LC3 was stained with anti-LC3 antibody and FITC-labeled secondary antibody (green). DNAs of macrophages and M. pneumoniae were stained with DAPI (blue). Small DNA particles derived from M. pneumoniae are indicated with arrows. Bar, 20 μm. (D) Peritoneal macrophages derived from TLR2 KO mice were treated with 2 μM cytochalasin D for 30 min and then infected with M. pneumoniae. After 6 h of incubation, TNF-α concentrations in the culture medium were measured using ELISA. All values are presented as the means and SD of the results of 3 assays. *, P < 0.01 compared with PBS or DMSO treatment by multiple comparison.
Cytochalasin D is an inhibitor of phagocytosis and macropinocytosis that disrupts actin filaments (36, 37). When TLR2 KO macrophages were treated with cytochalasin D and infected with live M. pneumoniae, TNF-α induction was inhibited in comparison with that seen with control dimethyl sulfoxide (DMSO)-treated cells, whereas the induction was not decreased in WT macrophages (Fig. 3D). These results suggest that phagocytosis is also important for TLR2-independent inflammatory responses.
TLR4-dependent induction of proinflammatory cytokines.
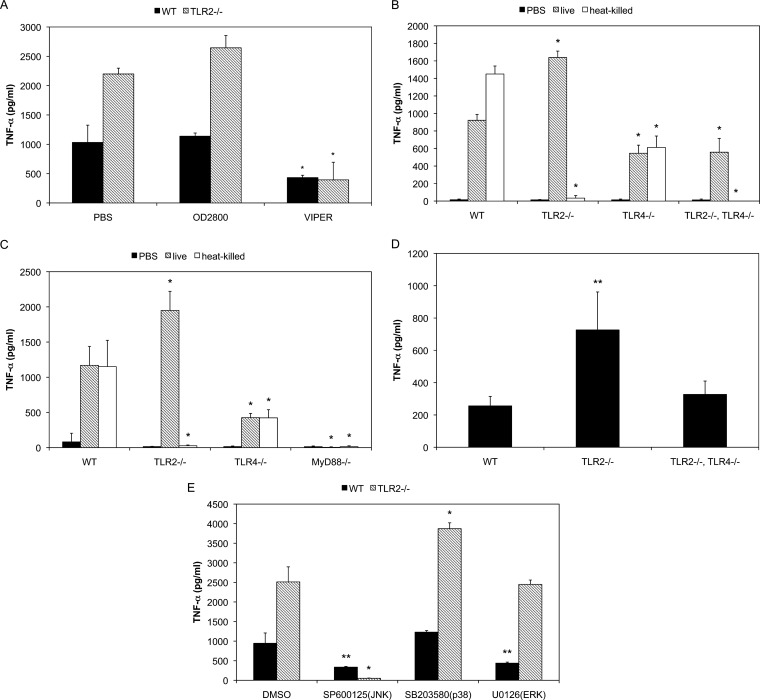
Some of TLRs such as TLR4 and TLR7 serve as sensors for autophagy (38–40). In addition, TLR4, TLR7, TLR8, and TLR9 are reported to be activated in mature endosomes and the acidification of endosomes is necessary to induce the downstream signaling (41–43). Therefore, the involvement of TLR4 and TLR9 in proinflammatory cytokine induction was examined. Macrophages were treated with the TLR4 and TLR9 antagonists VIPER and OD2088, respectively, and infected with M. pneumoniae (Fig. 4A). Although OD2088 administration did not affect TNF-α induction, VIPER decreased the induction markedly, suggesting the involvement of TLR4 in the TLR2-independent pathway.
FIG 4.
TLR4- and MyD88-dependent TNF-α induction. (A) Peritoneal macrophages derived from TLR2 KO mice were treated with 10 μM OD2088 or 50 μM VIPER for 30 min and then infected with M. pneumoniae. After 6 h of incubation, TNF-α concentrations in the culture medium were measured using ELISA. All values are presented as the means and SD of the results of 3 assays. *, P < 0.01 compared with PBS treatment by multiple comparison. (B) Peritoneal macrophages derived from TLR2 KO, TLR4 KO, and TLR2/4 double-KO mice were infected with M. pneumoniae. After 6 h of incubation, TNF-α concentrations in the culture medium were measured using ELISA. All values are presented as the means and SD of the results of 3 assays. *, P < 0.01 compared with WT macrophages by multiple comparison. (C) Peritoneal macrophages derived from TLR2 KO, TLR4 KO, and MyD88 KO mice were infected with M. pneumoniae (OD595 = 0.1). After 6 h of incubation, TNF-α concentrations in the culture medium were measured using ELISA. All values are presented as the means and SD of the results of 3 assays. *, P < 0.01 compared with WT macrophages by multiple comparison. (D) WT, TLR2 KO, and TLR2/4 double-KO mice were intranasally infected with M. pneumoniae. After 24 h, the mice were infected with same amounts of M. pneumoniae again for an additional 24 h. TNF-α concentrations in the BALF were measured using ELISA. All values are presented as the means and SD of the results of 3 assays. **, P < 0.05 compared with WT mice by multiple comparison. (E) Peritoneal macrophages derived from TLR2 KO mice were treated with 10 μM SB203580, 10 μM U0126, or 50 μM SP600123 for 30 min. The treated cells were infected with M. pneumoniae. After 6 h of incubation, TNF-α concentration in the culture medium were measured using ELISA. All values are presented as the means and SD of the results of 3 assays. *, P < 0.01 compared with DMSO treatment by multiple comparison. **, P < 0.05 compared with DMSO treatment by multiple comparison.
To confirm the involvement of TLR4, peritoneal macrophages derived from TLR4 KO mice and TLR2/4 double-KO mice were infected with M. pneumoniae (Fig. 4B). In TLR4 KO macrophages infected with live M. pneumoniae, TNF-α induction was decreased to approximately 60% of that in WT macrophages. On the other hand, TNF-α induction in TLR2/4 double-KO macrophages was decreased to 40% of that in TLR2 KO macrophages. In contrast, TNF-α induction by heat-killed M. pneumoniae was completely dependent on TLR2. These results indicate the involvement of TLR4 in TLR2-independent induction of inflammatory responses.
To further examine the association of TLRs, the involvement of MyD88, a critical adapter protein of TLRs, was examined. Peritoneal macrophages from MyD88 KO mice were infected with live or heat-killed M. pneumoniae (Fig. 4C). The induction of TFN-α by both live and heat-killed M. pneumoniae was completely impaired, suggesting that MyD88 is a critical factor for the induction of TLR2-independent inflammatory responses.
To determine whether the TLR2-independent pathway is involved in the development of pneumonia, inflammatory responses in the lungs of mice were investigated. WT, TLR2 KO, and TLR2/4 double-KO mouse were infected with M. pneumoniae intranasally and TNF-α induction in the bronchoalveolar lavage fluid (BALF) was measured using ELISA (Fig. 4D). In TLR2 KO mouse, TNF-α induction was increased compared with that in WT mice. TNF-α induction in TLR2/4 double-KO mice was decreased compared with that in TLR2 KO mice. These results suggest that the TLR2-independent pathway is involved in lung inflammation and that TLR4 is an important receptor.
Since mitogen-activated protein kinase (MAPK) is thought to be involved in downstream signaling of TLRs to induce autophagy, the involvement of MAPK was investigated (Fig. 4E). When WT macrophages were infected with live M. pneumoniae, TNF-α induction was decreased by the c-Jun N-terminal kinase (JNK) inhibitor SP600125 and the extracellular signal-regulated kinase (ERK) inhibitor U0126. In contrast, TNF-α induction in TLR2 KO macrophages was inhibited by SP600125 but not by U0126. SB203580, an inhibitor of p38, failed to reduce the TNF-α induction in both WT and TLR2 KO macrophages. These results indicate that TLR2-independent induction of inflammatory responses is MAPK dependent and that JNK is a key factor of this signaling pathway.
Cytadherence-dependent induction of inflammatory responses.
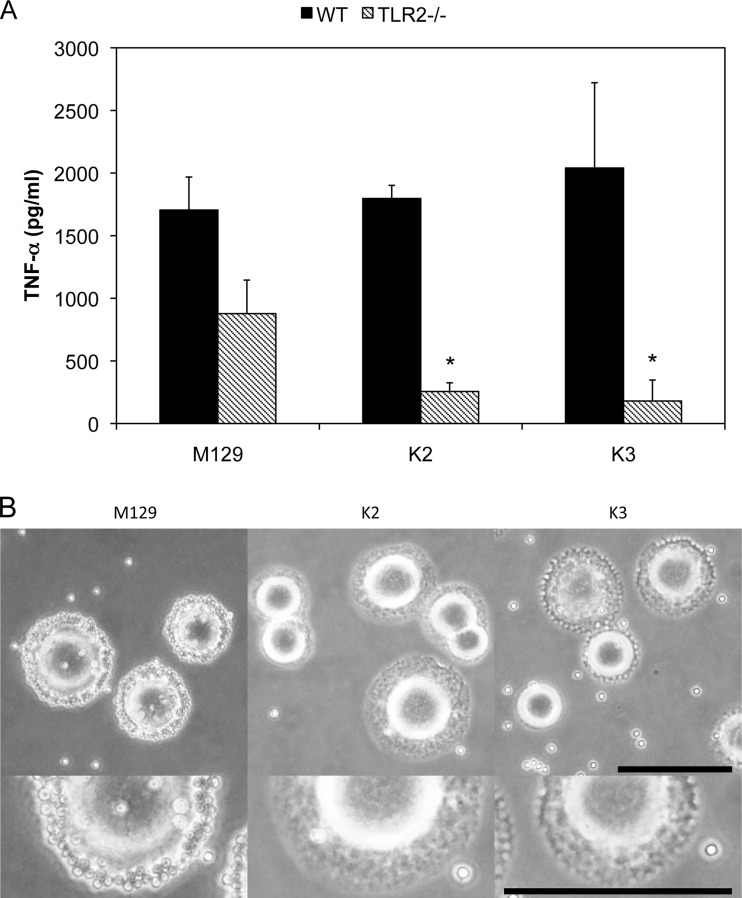
Next, transposon mutagenesis was conducted to identify bacterial factors that related to the TLR2-independent pathway. M. pneumoniae M129 was transformed with pISM2062 plasmids containing the IS256 transposon. TLR2 KO macrophages were infected with M. pneumoniae mutants, and those with a reduced ability to induce TNF-α expression were selected. Of 2,880 mutants, 2 strains, K2 and K3, were isolated as TNF-α-noninducible mutants in TLR2 KO macrophages (Fig. 5A). To identify the genes responsible for the TLR2-independent induction of inflammatory responses, transposon-inserted regions in the DNA of the K2 and K3 strains were amplified by PCR, cloned into pUC19 plasmids, and sequenced (Table 1). In the K2 strain, the transposon was inserted in atpC encoding an ATP synthase F0F1 ε subunit. In the K3 strain, the transposon was inserted within F10_orf750 encoding the hypothetical protein MPN333. The N-terminal sequence of MPN333 had similarity to that of the ATP-binding cassette-2 (ABC-2) family transporter protein. RNA expression of genes downstream of atpC and F10_orf750 was not impaired. Although WT M. pneumoniae normally binds to the culture flask through sialylated proteins contained in serum (44), these 2 mutants floated in the medium. Therefore, the cytadherence properties of these mutants were examined using the hemadsorption assay (Fig. 5B). WT M. pneumoniae was able to bind to sheep erythrocytes, but K2 and K3 did not exhibit binding activity, indicating that these mutants lacked cytadherence properties.
FIG 5.
Cytadherence-dependent TNF-α induction. (A) Transposon mutagenesis of M. pneumoniae was performed as described in Materials and Methods. Peritoneal macrophages derived from TLR2 KO mice were infected with transformed M. pneumoniae for 3 h. TNF-α concentrations in the culture medium were measured usig ELISA. All values are presented as the means and SD of the results of 3 assays. *, P < 0.01 compared with M129 by multiple comparison. (B) PPLO agar plates were overlaid with 15 ml of fresh sheep blood, washed, and resuspended in PBS to reach a final concentration of 0.5% (vol/vol). After incubation at 37°C for 30 min, the plates were washed with PBS. Bar, 1 mm.
TABLE 1.
Transposon-inserted genes in M. pneumoniae mutants
| Strain | Locus tag | Gene name | Inserted position | Function |
|---|---|---|---|---|
| K2 | MPN597 | atpC | 31 | ATP synthase F0F1 subunit ε |
| K3 | MPN333 | F10_orf750 | 768 | ABC-2 family transporter proteina |
The N-terminal sequence of the MPN333 protein is similar to that of the ABC-2 family transporter protein.
DISCUSSION
Mycoplasma species lack cell walls, and the cells are surrounded by cell membranes (1). Moreover, mycoplasma cells do not contain TLR ligands such as LPS, PGN, and lipoteichoic acid but contain an abundance of acylated proteins as cell-surface antigens, and many putative lipoprotein-encoding genes have been identified in sequenced mycoplasma genomes (45, 46). These findings suggest that lipoproteins are main components of M. pneumoniae that induce inflammatory responses and cause pneumonia in humans. We previously reported that the purified or synthesized lipoproteins of mycoplasma species induce inflammatory responses through TLR2 (7–9). Moreover, lipoproteins derived from various mycoplasmas have been reported to act as PAMPS (47–50). However, the existence of lipoproteins in nonpathogenic mycoplasmas suggests the presence of another mechanism by which M. pneumoniae induces inflammatory responses. In this study, we demonstrated that live M. pneumoniae was able to induce inflammatory responses even in the lung and macrophage cells of TLR2 KO mice (Fig. 1, 2, and 4). Notably, M. pneumoniae inactivated by heat, sonication, antibiotics, and overgrowth failed to induce inflammatory responses in TLR2 KO macrophages (Fig. 1B), suggesting that some biological activities of M. pneumoniae are necessary to induce TLR2-independent inflammatory responses.
To identify the bacterial factor that induces the TLR2-independent inflammation pathway, transposon mutagenesis was conducted. As a result, 2 mutants with decreased abilities to induce TNF-α expression in TLR2 KO macrophages were isolated (Fig. 5A and Table 1). The transposons were inserted into atpC and F10_orf750. atpC encodes an ATP synthase F0F1 ε subunit. ATP synthase F0F1 subunit ε is a regulatory protein of the F0F1 type ATPase and can inhibit the ATP hydrolysis in the absence of proton motive forces (51). F10_orf750 encodes a hypothetical protein, MPN333, with an N-terminal sequence similar to that of the ABC-2 family transporter protein. The ABC-2 family transporter protein is a member of a subfamily of ABC transporters and is related to capsular polysaccharide export (52). Notably, these mutants were deficient in cytadherence (Fig. 5B). The cytadherence of M. pneumoniae is mediated by attachment organelles, including P1 adhesin and other additional proteins such as P30 or HMW proteins (26–29). These proteins are unique to mycoplasma species, and their homologs have not been identified in any other bacterial species (53). The cytadherence of M. pneumoniae is closely linked to the unique movement specific to mycoplasma species called gliding motility (28). The gliding motility of some mycoplasmas such as M. mobile is ATP dependent (29). In addition, the ABC-2 family transporter protein is reportedly involved in the motility of Myxococcus xanthus (54). Considering that complementation of mutated genes is impossible in M. pneumoniae, we could not rule out the possibility that inactivation of downstream genes of transposon-inserted genes resulted in the deficiency in cytadherence. However, the mRNA expression of genes downstream of atpC and F10_orf750 (MPN596 and bcrA, respectively) was not impaired (data not shown). Taken together, these results indicate that AtpC and MPN333 may be the new virulence factors that are responsible for cytadherence and inflammation in M. pneumoniae infections. However, in this study, we failed to screen well-known cytadherence factors such as P1, P30, and HMW. This may suggest that the functions of AtpC or MPN333 itself are important for the induction of inflammatory responses. Although further screening of mutants is necessary to clarify the relationship between cytadherence and the induction of inflammatory responses, the relationship between cytadherence and inflammatory responses in host cells is consistent with our previous report that cytadherence of M. pneumoniae activates cytokine production in human monocyte cells (32) and with an earlier report that a protease treatment decreases the induction of proinflammatory cytokines by M. pneumoniae (31). Furthermore, M. pneumoniae cultured under nonadherent conditions fails to induce IL-4 expression in rodent mast cells (30). Moreover, an elongated infection time or a high concentration of AtpC and MPN333 mutants still induced proinflammatory cytokines (data not shown) in TLR2 KO macrophages. These results suggest that cytadherence is more likely to be involved in the TLR2-independent induction of inflammatory responses than the functions of AtpC and MPN333. However, the mechanism by which M. pneumoniae cytadherence activates the induction of proinflammatory responses has not been determined. In this study, we observed that the TLR2-independent induction of proinflammatory cytokines was dependent on endocytosis, because inhibition of endocytosis with cytochalasin D decreased the TNF-α induction in TLR2 KO macrophages (Fig. 3D). These result indicate that uptake of M. pneumoniae by macrophages is necessary for the activity of the TLR2-independent pathway and suggest that cytadherence of M. pneumoniae may enhance the uptake by macrophages.
The inductions of proinflammatory cytokines were inhibited by the inhibitors of autophagy, and M. pneumoniae was colocalized with autophagy marker protein LC3 in WT and TLR2 KO macrophages (Fig. 3). These results indicate that autophagy plays an important role in the induction of proinflammatory cytokines by M. pneumoniae. An autophagy inhibitor, chloroquine, inhibits the fusion of lysosomes and autophagosomes. In this study, chloroquine treatment completely eliminated the TLR2-independent induction of proinflammatory cytokines, suggesting that degradation of M. pneumoniae in autophagosomes is required for the induction. The TLR2-independent induction of proinflammatory cytokine was also dependent on TLR4 and MyD88 (Fig. 4). It was reported that some TLRs such as TLR4 and TLR7 serve as a sensor for autophagy in a MyD88-dependent manner and that MAPK is an important downstream signal of this pathway (38–40, 55). These results suggest that recognition of M. pneumoniae involving TLR4 induces autophagy, followed by induction of proinflammatory cytokines. The TLR2-independent pathway was also dependent on JNK MAPK in this study (Fig. 4E). This result was consistent with the report that JNK MAPK was shown to be involved in induction of autophagy and cell death (56).
Generally, bacteria that are retained in or escape from phagosomes can be targeted by autophagy. Recently, LC3-associated phagocytosis (LAP) was shown to take up and degrade the bacteria without the ability to retain or escape from the phagosome (57). Similar to autophagy, LAP is consistent with the presence of autophagy-related protein, including LC3, autophagy-related gene 5 (ATG5), and ATG7. However, unlike the results seen with autophagy, double-membrane structures are not formed around the LPA (58). Since it is still controversial whether M. pneumoniae can escape from phagosome and grow intracellularly (59), we could not rule out the possibility that the TLR2-independent induction of proinflammatory cytokines was dependent on LPA but not on autophagy.
In this study, TLR4 seemed to be the key receptor to induce autophagy following inflammatory responses (Fig. 4). TLR4 is essential for the recognition of LPS, which is composed of lipid A, a core oligosaccharide, and an O-antigen. TLR4 recognizes lipid A of LPS. Because mycoplasma species lack cell walls and do not contain LPS, the ligands for TLR4 in mycoplasmas remain unclear. Other than LPS, TLR4 also recognizes fungal mannan and glucuronoxylomannan (60), protozoan glycoinositolphospholipids (61, 62), and viral proteins (63, 64). Mycoplasma species also express unique glycolipids, phosphoglycolipids (65), and polysaccharides (66, 67). Although further studies are needed to determine the exact ligands of M. pneumoniae for TLR4, these molecules may be potential TLR4 ligands. TNF-α induction was decreased only partially in VIPER-treated TLR2 KO macrophages and TLR2/4 double-KO macrophages (Fig. 3 and 4A). In contrast, TNF-α induction was completely inhibited in MyD88 KO macrophages (Fig. 4B). Because MyD88 is an essential adaptor protein in TLR signaling, these results suggest that TLRs are necessary to induce inflammatory responses in TLR2 KO macrophages and that other TLRs are associated with this induction in concert with TLR2 and TLR4.
In conclusion, our results suggest that M. pneumoniae induces inflammatory responses in a TLR4- and autophagy-dependent manner and that the cytadherence property of M. pneumoniae is a key factor. Hence, the proteins involved in cytadherence, including AtpC and MPN333, or TLR4 ligands present potential targets for the development of alternative strategies to prevent and treat M. pneumoniae infection.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a Grant-in-Aid for Scientific Research (23790488) and a Grant-in-Aid for Scientific Research on Innovative Areas (25117530) from the Ministry of Education, Culture Sports, Science and Technology of Japan.
Footnotes
Published ahead of print 5 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01961-14.
REFERENCES
- 1.Razin S. 1992. Peculiar properties of mycoplasmas: the smallest self-replicating prokaryotes. FEMS Microbiol. Lett. 100:423–431 [DOI] [PubMed] [Google Scholar]
- 2.Waites KB, Talkington DF. 2004. Mycoplasma pneumoniae and its role as a human pathogen. Clin. Microbiol. Rev. 17:697–728. 10.1128/CMR.17.4.697-728.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gil JC, Cedillo RL, Mayagoitia BG, Paz MD. 1993. Isolation of Mycoplasma pneumoniae from asthmatic patients. Ann. Allergy 70:23–25 [PubMed] [Google Scholar]
- 4.Kraft M, Cassell GH, Henson JE, Watson H, Williamson J, Marmion BP, Gaydos CA, Martin RJ. 1998. Detection of Mycoplasma pneumoniae in the airways of adults with chronic asthma. Am. J. Respir. Crit. Care Med. 158:998–1001. 10.1164/ajrccm.158.3.9711092 [DOI] [PubMed] [Google Scholar]
- 5.Pereyre S, Charron A, Hidalgo-Grass C, Touati A, Moses AE, Nir-Paz R, Bebear C. 2012. The spread of Mycoplasma pneumoniae is polyclonal in both an endemic setting in France and in an epidemic setting in Israel. PLoS One 7:e38585. 10.1371/journal.pone.0038585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tryon VV, Baseman JB. 1992. Pathogenic determinant and mechanisms, p 457–489 In Maniloff J, McElhaney RN, Finch LR, Baseman JB. (ed), Mycoplasmas—molecular biology and pathogenesis. American Society for Microbiology, Washington, DC [Google Scholar]
- 7.Shimizu T, Kida Y, Kuwano K. 2008. Mycoplasma pneumoniae-derived lipopeptides induce acute inflammatory responses in the lungs of mice. Infect. Immun. 76:270–277. 10.1128/IAI.00955-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shimizu T, Kida Y, Kuwano K. 2005. A dipalmitoylated lipoprotein from Mycoplasma pneumoniae activates NF-kappa B through TLR1, TLR2, and TLR6. J. Immunol. 175:4641–4646. 10.4049/jimmunol.175.7.4641 [DOI] [PubMed] [Google Scholar]
- 9.Shimizu T, Kida Y, Kuwano K. 2007. Triacylated lipoproteins derived from Mycoplasma pneumoniae activate nuclear factor-kappaB through toll-like receptors 1 and 2. Immunology 121:473–483. 10.1111/j.1365-2567.2007.02594.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akira S, Takeda K. 2004. Toll-like receptor signaling. Nat. Rev. Immunol. 4:499–511. 10.1038/nri1391 [DOI] [PubMed] [Google Scholar]
- 11.Kopp EB, Medzhitov R. 1999. The Toll-receptor family and control of innate immunity. Curr. Opin. Immunol. 11:13–18. 10.1016/S0952-7915(99)80003-X [DOI] [PubMed] [Google Scholar]
- 12.Aliprantis AO, Yang RB, Mark MR, Suggett S, Devaux B, Radolf JD, Klimpel GR, Godowski P, Zychlinsky A. 1999. Cell activation and apoptosis by bacterial lipoproteins through toll-like receptor-2. Science 285:736–739. 10.1126/science.285.5428.736 [DOI] [PubMed] [Google Scholar]
- 13.Brightbill HD, Libraty DH, Krutzik SR, Yang RB, Belisle JT, Bleharski JR, Maitland M, Norgard MV, Plevy SE, Smale ST, Brennan PJ, Bloom BR, Godowski PJ, Modlin RL. 1999. Host defense mechanisms triggered by microbial lipoproteins through toll-like receptors. Science 285:732–736. 10.1126/science.285.5428.732 [DOI] [PubMed] [Google Scholar]
- 14.Lien E, Sellati TJ, Yoshimura A, Flo TH, Rawadi G, Finberg RW, Carroll JD, Espevik T, Ingalls RR, Radolf JD, Golenbock DT. 1999. Toll-like receptor 2 functions as a pattern recognition receptor for diverse bacterial products. J. Biol. Chem. 274:33419–33425. 10.1074/jbc.274.47.33419 [DOI] [PubMed] [Google Scholar]
- 15.Means TK, Lien E, Yoshimura A, Wang S, Golenbock DT, Fenton MJ. 1999. The CD14 ligands lipoarabinomannan and lipopolysaccharide differ in their requirement for Toll-like receptors. J. Immunol. 163:6748–6755 [PubMed] [Google Scholar]
- 16.Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, Takeda K, Akira S. 1999. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity 11:443–451. 10.1016/S1074-7613(00)80119-3 [DOI] [PubMed] [Google Scholar]
- 17.Takeuchi O, Kaufmann A, Grote K, Kawai T, Hoshino K, Morr M, Muhlradt PF, Akira S. 2000. Preferentially the R-stereoisomer of the mycoplasmal lipopeptide macrophage-activating lipopeptide-2 activates immune cells through a toll-like receptor 2- and MyD88-dependent signaling pathway. J. Immunol. 164:554–557. 10.4049/jimmunol.164.2.554 [DOI] [PubMed] [Google Scholar]
- 18.Underhill DM, Ozinsky A, Hajjar AM, Stevens A, Wilson CB, Bassetti M, Aderem A. 1999. The Toll-like receptor 2 is recruited to macrophage phagosomes and discriminates between pathogens. Nature 401:811–815. 10.1038/44605 [DOI] [PubMed] [Google Scholar]
- 19.Hayashi F, Smith KD, Ozinsky A, Hawn TR, Yi EC, Goodlett DR, Eng JK, Akira S, Underhill DM, Aderem A. 2001. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410:1099–1103. 10.1038/35074106 [DOI] [PubMed] [Google Scholar]
- 20.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S. 2000. A Toll-like receptor recognizes bacterial DNA. Nature 408:740–745. 10.1038/35047123 [DOI] [PubMed] [Google Scholar]
- 21.Hoshino K, Takeuchi O, Kawai T, Sanjo H, Ogawa T, Takeda Y, Takeda K, Akira S. 1999. Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J. Immunol. 162:3749–3752 [PubMed] [Google Scholar]
- 22.Poltorak A, He X, Smirnova I, Liu MY, Huffel CV, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. 1998. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282:2085–2088. 10.1126/science.282.5396.2085 [DOI] [PubMed] [Google Scholar]
- 23.Medzhitov R, Preston-Hurlburt P, Kopp E, Stadlen A, Chen C, Ghosh S, Janeway CA., Jr 1998. MyD88 is an adaptor protein in the hToll/IL-1 receptor family signaling pathways. Mol. Cell 2:253–258. 10.1016/S1097-2765(00)80136-7 [DOI] [PubMed] [Google Scholar]
- 24.Levine B. 2005. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell 120:159–162. 10.1016/j.cell.2005.01.005 [DOI] [PubMed] [Google Scholar]
- 25.Schmid D, Munz C. 2007. Innate and adaptive immunity through autophagy. Immunity 27:11–21. 10.1016/j.immuni.2007.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krause DC, Balish MF. 2001. Structure, function, and assembly of the terminal organelle of Mycoplasma pneumoniae. FEMS Microbiol. Lett. 198:1–7. 10.1111/j.1574-6968.2001.tb10610.x [DOI] [PubMed] [Google Scholar]
- 27.Balish MF, Krause DC. 2002. Cytadherence and the cytoskelton, p 491–518 In Razin S, Herrmann R. (ed), Molecular biology and pathogenicity of mycoplasmas. Kluwer Academic/Plenum Publishers, New York, NY [Google Scholar]
- 28.Miyata M. 2008. Centipede and inchworm models to explain Mycoplasma gliding. Trends Microbiol. 16:6–12. 10.1016/j.tim.2007.11.002 [DOI] [PubMed] [Google Scholar]
- 29.Miyata M. 2008. Molecular mechanism of mycoplasma gliding—a novel cell motility system, p 137–175 In Lenz P. (ed), Cell Motility. Springer Science, New York, NY [Google Scholar]
- 30.Hoek KL, Duffy LB, Cassell GH, Dai Y, Atkinson TP. 2005. A role for the Mycoplasma pneumoniae adhesin P1 in interleukin (IL)-4 synthesis and release from rodent mast cells. Microb. Pathog. 39:149–158. 10.1016/j.micpath.2005.07.004 [DOI] [PubMed] [Google Scholar]
- 31.Yang J, Hooper WC, Phillips DJ, Talkington DF. 2002. Regulation of proinflammatory cytokines in human lung epithelial cells infected with Mycoplasma pneumoniae. Infect. Immun. 70:3649–3655. 10.1128/IAI.70.7.3649-3655.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimizu T, Kida Y, Kuwano K. 2011. Cytoadherence-dependent induction of inflammatory responses by Mycoplasma pneumoniae. Immunology 133:51–61. 10.1111/j.1365-2567.2011.03408.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kenri T, Seto S, Horino A, Sasaki Y, Sasaki T, Miyata M. 2004. Use of fluorescent-protein tagging to determine the subcellular localization of Mycoplasma pneumoniae proteins encoded by the cytadherence regulatory locus. J. Bacteriol. 186:6944–6955. 10.1128/JB.186.20.6944-6955.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blommaart EF, Krause U, Schellens JP, Vreeling-Sindelarova H, Meijer AJ. 1997. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur. J. Biochem. 243:240–246. 10.1111/j.1432-1033.1997.0240a.x [DOI] [PubMed] [Google Scholar]
- 35.Shintani T, Klionsky DJ. 2004. Autophagy in health and disease: a double-edged sword. Science 306:990–995. 10.1126/science.1099993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mimura N, Asano A. 1976. Synergistic effect of colchicine and cytochalasin D on phagocytosis by peritoneal macrophages. Nature 261:319–321. 10.1038/261319a0 [DOI] [PubMed] [Google Scholar]
- 37.Dutta D, Donaldson JG. 2012. Search for inhibitors of endocytosis: intended specificity and unintended consequences. Cell Logist. 2:203–208. 10.4161/cl.23967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu Y, Jagannath C, Liu XD, Sharafkhaneh A, Kolodziejska KE, Eissa NT. 2007. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity 27:135–144. 10.1016/j.immuni.2007.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Delgado MA, Deretic V. 2009. Toll-like receptors in control of immunological autophagy. Cell Death Differ. 16:976–983. 10.1038/cdd.2009.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu Y, Liu XD, Gong X, Eissa NT. 2008. Signaling pathway of autophagy associated with innate immunity. Autophagy 4:110–112 [DOI] [PubMed] [Google Scholar]
- 41.Gay NJ, Gangloff M. 2007. Structure and function of Toll receptors and their ligands. Annu. Rev. Biochem. 76:141–165. 10.1146/annurev.biochem.76.060305.151318 [DOI] [PubMed] [Google Scholar]
- 42.Gangloff M. 2012. Different dimerisation mode for TLR4 upon endosomal acidification? Trends Biochem. Sci. 37:92–98. 10.1016/j.tibs.2011.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Latz E, Schoenemeyer A, Visintin A, Fitzgerald KA, Monks BG, Knetter CF, Lien E, Nilsen NJ, Espevik T, Golenbock DT. 2004. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat. Immunol. 5:190–198. 10.1038/ni1028 [DOI] [PubMed] [Google Scholar]
- 44.Kasai T, Nakane D, Ishida H, Ando H, Kiso M, Miyata M. 2013. Role of binding in Mycoplasma mobile and Mycoplasma pneumoniae gliding analyzed through inhibition by synthesized sialylated compounds. J. Bacteriol. 195:429–435. 10.1128/JB.01141-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chambaud I, Wroblewski H, Blanchard A. 1999. Interactions between mycoplasma lipoproteins and the host immune system. Trends Microbiol. 7:493–499. 10.1016/S0966-842X(99)01641-8 [DOI] [PubMed] [Google Scholar]
- 46.Wieslamder A, Boyer MJ, Wroblewski H. 1992. Membrane protein structure, p 93–112 In Maniloff J, McElhaney RN, Finch LR, Baseman JB. (ed), Mycoplasmas—molecular biology and pathogenesis. American Society for Microbiology, Washington, DC [Google Scholar]
- 47.Mühlradt PF, Kiess M, Meyer H, Süssmuth R, Jung G. 1997. Isolation, structure elucidation, and synthesis of a macrophage stimulatory lipopeptide from Mycoplasma fermentans acting at picomolar concentration. J. Exp. Med. 185:1951–1958. 10.1084/jem.185.11.1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shibata K, Hasebe A, Into T, Yamada M, Watanabe T. 2000. The N-terminal lipopeptide of a 44-kDa membrane-bound lipoprotein of Mycoplasma salivarium is responsible for the expression of intercellular adhesion molecule-1 on the cell surface of normal human gingival fibroblasts. J. Immunol. 165:6538–6544. 10.4049/jimmunol.165.11.6538 [DOI] [PubMed] [Google Scholar]
- 49.Mühlradt PF, Kiess M, Meyer H, Süssmuth R, Jung G. 1998. Structure and specific activity of macrophage-stimulating lipopeptides from Mycoplasma hyorhinis. Infect. Immun. 66:4804–4810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jan G, Brenner C, Wroblewski H. 1996. Purification of Mycoplasma gallisepticum membrane proteins p52, p67 (pMGA), and p77 by high-performance liquid chromatography. Protein Expr. Purif. 7:160–166. 10.1006/prep.1996.0023 [DOI] [PubMed] [Google Scholar]
- 51.Feniouk BA, Junge W. 2005. Regulation of the F0F1-ATP synthase: the conformation of subunit epsilon might be determined by directionality of subunit gamma rotation. FEBS Lett. 579:5114–5118. 10.1016/j.febslet.2005.08.030 [DOI] [PubMed] [Google Scholar]
- 52.Reizer J, Reizer A, Saier MH., Jr 1992. A new subfamily of bacterial ABC-type transport systems catalyzing export of drugs and carbohydrates. Protein Sci. 1:1326–1332. 10.1002/pro.5560011012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Himmelreich R, Hilbert H, Plagens H, Pirkl E, Li BC, Herrmann R. 1996. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 24:4420–4449. 10.1093/nar/24.22.4420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu SS, Wu J, Cheng YL, Kaiser D. 1998. The pilH gene encodes an ABC transporter homologue required for type IV pilus biogenesis and social gliding motility in Myxococcus xanthus. Mol. Microbiol. 29:1249–1261. 10.1046/j.1365-2958.1998.01013.x [DOI] [PubMed] [Google Scholar]
- 55.Shi CS, Kehrl JH. 2008. MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. J. Biol. Chem. 283:33175–33182. 10.1074/jbc.M804478200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu L, Alva A, Su H, Dutt P, Freundt E, Welsh S, Baehrecke EH, Lenardo MJ. 2004. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science 304:1500–1502. 10.1126/science.1096645 [DOI] [PubMed] [Google Scholar]
- 57.Romao S, Münz C. 7 January 2014. LC3-associated phagocytosis. Autophagy 10.4161/auto.27606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, Komatsu M, Tanaka K, Cleveland JL, Withoff S, Green DR. 2007. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 450:1253–1257. 10.1038/nature06421 [DOI] [PubMed] [Google Scholar]
- 59.Baseman JB, Lange M, Criscimagna NL, Giron JA, Thomas CA. 1995. Interplay between mycoplasmas and host target cells. Microb. Pathog. 19:105–116. 10.1006/mpat.1995.0050 [DOI] [PubMed] [Google Scholar]
- 60.Netea MG, Van der Graaf C, Van der Meer JW, Kullberg BJ. 2004. Recognition of fungal pathogens by Toll-like receptors. Eur. J. Clin. Microbiol. Infect. Dis. 23:672–676. 10.1007/s10096-004-1192-7 [DOI] [PubMed] [Google Scholar]
- 61.Netea MG, Van Der Graaf CA, Vonk AG, Verschueren I, Van Der Meer JW, Kullberg BJ. 2002. The role of toll-like receptor (TLR) 2 and TLR4 in the host defense against disseminated candidiasis. J. Infect. Dis. 185:1483–1489. 10.1086/340511 [DOI] [PubMed] [Google Scholar]
- 62.Oliveira AC, Peixoto JR, de Arruda LB, Campos MA, Gazzinelli RT, Golenbock DT, Akira S, Previato JO, Mendonca-Previato L, Nobrega A, Bellio M. 2004. Expression of functional TLR4 confers proinflammatory responsiveness to Trypanosoma cruzi glycoinositolphospholipids and higher resistance to infection with T. cruzi. J. Immunol. 173:5688–5696. 10.4049/jimmunol.173.9.5688 [DOI] [PubMed] [Google Scholar]
- 63.Haynes LM, Moore DD, Kurt-Jones EA, Finberg RW, Anderson LJ, Tripp RA. 2001. Involvement of toll-like receptor 4 in innate immunity to respiratory syncytial virus. J. Virol. 75:10730–10737. 10.1128/JVI.75.22.10730-10737.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rassa JC, Meyers JL, Zhang Y, Kudaravalli R, Ross SR. 2002. Murine retroviruses activate B cells via interaction with toll-like receptor 4. Proc. Natl. Acad. Sci. U. S. A. 99:2281–2286. 10.1073/pnas.042355399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kornspan JD, Rottem S. 2012. The phospholipid profile of mycoplasmas. J. Lipids 2012:640762. 10.1155/2012/640762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wilson MH, Collier AM. 1976. Ultrastructural study of Mycoplasma pneumoniae in organ culture. J. Bacteriol. 125:332–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Daubenspeck JM, Bolland JR, Luo W, Simmons WL, Dybvig K. 2009. Identification of exopolysaccharide-deficient mutants of Mycoplasma pulmonis. Mol. Microbiol. 72:1235–1245. 10.1111/j.1365-2958.2009.06720.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.