Abstract
Centrosomes ensure accurate chromosome segregation by directing spindle bipolarity. Loss of centrosome regulation results in centrosome amplification, multipolar mitosis and aneuploidy. Since centrosome amplification is common in premalignant lesions and breast tumors, it is proposed to play a central role in breast tumorigenesis, a hypothesis that remains to be tested. The coordination between the cell and centrosome cycles is of paramount importance to maintain normal centrosome numbers, and the E2Fs may be responsible for regulating these cycles. However, the role of E2F activators in centrosome amplification is unclear. Because E2Fs are deregulated in Her2+ cells displaying centrosome amplification, we addressed whether they signal this abnormal process. Knockdown of E2F1 or E2F3 in Her2+ cells decreased centrosome amplification without significantly affecting cell cycle progression, whereas the overexpression of E2F1, E2F2, or E2F3 increased centrosome amplification in MCF10A mammary epithelial cells. Our results revealed that E2Fs affect the expression of proteins, including Nek2 and Plk4, known to influence the cell/centrosome cycles and mitosis. Downregulation of E2F3 resulted in cell death and delays/blocks in cytokinesis, which was reversed by Nek2 overexpression. Nek2 overexpression enhanced centrosome amplification in Her2+ breast cancer cells silenced for E2F3, revealing a role for the E2F activators in maintaining centrosome amplification in part through Nek2.
INTRODUCTION
The E2F transcription factors regulate various biological functions, such as cell cycle progression, DNA repair, apoptosis, centrosome duplication, and differentiation (1–8). Eight E2F proteins have been identified and are categorized as activators E2F1 through E2F3a and repressors E2F3b through E2F8 (9, 10). Rb hyperphosphorylation by G1/S-phase cyclin/Cdk complexes releases the E2F activators, which bind promoters through consensus (T/C)TT(C/G)(G/C)CG(C/G) or noncanonical binding sites (11, 12) to activate a plethora of genes that regulate the aforementioned cellular activities (4, 13, 14). The E2Fs are deregulated and altered in most human cancers through various molecular mechanisms, including overstimulation of the G1/S-phase cyclin/Cdks that hyperphosphorylate and inactivate the Rb family (15). Another mode of deregulation is by overexpression, such as that of E2F1 in breast, lung, and prostate cancers (16–26) and E2F3 in various cancers, including breast cancers (18, 26–31).
Deregulated expression of the E2Fs in breast cancers influences outcome of survival, since patients overexpressing E2F1 and cyclin A displayed shorter disease-free survival (16). In addition, breast cancer cells with molecular alterations affecting the Rb pathway or E2F overexpression display altered chemotherapeutic responses (32–36), including resistance to the Cdk4/Cdk6 inhibitor PD-0332991 (37, 38). Mouse models demonstrated the requirement for E2Fs in mammary carcinogenesis, since ablation of E2F1 and E2F3 suppressed Her2/Neu and Myc-induced mammary tumorigenesis (26, 39, 40). Thus, studying E2F functions may provide clues not only to understanding how mammary tumors initiate and progress but also to how breast cancer cells fail to respond to common therapies.
The E2Fs may influence breast carcinogenesis by signaling various abnormal phenotypes, including centrosome amplification, defined as the acquisition of three or more centrosomes within a cell (6, 7). Centrosome amplification may initiate and sustain breast cancers by actively generating aneuploidy and chromosome instability (41), a hypothesis that remains to be tested. The centrosome must duplicate once in each cell cycle to maintain normal centrosome numbers, achieved by cell cycle and centrosome-specific regulators (42, 43). Faithful centrosome licensing (regulated in part by the phosphorylation of nucleophosmin [NPM] by Cdk2 and Cdk4), duplication (regulated by various kinases, including Plk4), and maturation and separation (regulated in part by Nek2) are essential to establish spindle bipolarity at mitosis and faithful segregation of chromosomes following cytokinesis (42–44). Deregulated centrosome duplication or cytokinesis defects are two major mechanisms leading to centrosome amplification, which results in aberrant pseudobipolar and multipolar mitotic spindles, chromosome losses/gains, and aneuploidy (7, 45–47).
Although various cancer types display elevated centrosome amplification (48, 49), the relationship between centrosome amplification and tumorigenesis is best understood in breast cancers, since a significant fraction of premalignant lesions and many breast tumors exhibit centrosome defects, including defects in numbers (centrosome amplification) or structure (size changes) (50–54). A major gap in knowledge is identifying pathways directly signaling centrosome amplification. Identifying the roles/functions and sources of centrosomal/mitotic kinases in signaling centrosome amplification is important to breast cancer control, since the overexpression of 16 centrosomal/mitotic kinases in breast cancer, including Nek2 and Plk4, represents a molecular signature that strongly associates with poorly prognostic breast cancers (55). In fact, Nek2 and Plk4 are overexpressed in low-prognosis breast cancer molecular subtypes, individually associating with accelerated time-to-metastasis and time-to-relapse of breast cancer patients (56). Major unanswered questions regarding the role of the E2Fs in centrosome amplification are addressed in the present study, and we provide direct evidence that the E2F activators induce and maintain centrosome amplification in breast cancer cells and that Nek2 drives centrosome amplification downstream of the E2F3 activator.
MATERIALS AND METHODS
Cell culture.
All cell lines were obtained from the ATCC (Manassas, VA) or from collaborators. The culture conditions for MCF10A, HCC1954, SKBR3, and JIMT1 cells have been described (57, 58). For serum starvation, cells were grown in media containing 0.2% fetal bovine serum (FBS) for 72 h. To develop stably silenced E2F cell populations, 2 μg of puromycin/ml was added to the media and 50 μg of hygromycin/ml was added in the media to develop MCF10A cells overexpressing E2Fs. Both puromycin and hygromycin were added to develop HCC1954 and JIMT1 cells stably knocked down for E2F3 (shE2F3) and overexpressing green fluorescent protein (GFP)-tagged Nek2 (shE2F3; GFP-Nek2).
Real-time PCR analysis.
Total RNA was isolated using TRIzol according to the manufacturer's protocol (Invitrogen, Grand Island, NY), and 2 μg of RNA was used to synthesize cDNA according to the manufacturer's protocol (Promega, Madison, WI). Then, 2 μl of 1:10-diluted cDNA was used for real-time PCR with iQ SYBR green Supermix (170-8880; Bio-Rad, Hercules, CA). Actin was used as an internal control, and the primer sequences are presented in Table 1.
TABLE 1.
Primer sequences
| Method and primer | Orientationa | Sequence (5′–3′) |
|---|---|---|
| Real-time PCR | ||
| Cyclin D1 | F | GGC TGG GTC TGT GCA TTT CT |
| R | AAC ATG CCG GTT ACT TGT TGG T | |
| Cyclin E1 | F | TGGATCTCTGTGTCCTGGATGTT |
| R | CAAGGCCGAAGCAGCAA | |
| E2F1 | F | GTT TGG GCC GGG TTT TG |
| R | GCA TTT CCC CAG CAA CCT T | |
| E2F2 | F | AGG GTG TCC CTT TTC CAC AGT A |
| R | CTT GAC CAC CTC CCT CTT CCT | |
| E2F3 | F | GCA TGA CAA CTC GTG TGT ATG AGA |
| R | CAA TTG CCA CCC GAC TTA CTC | |
| Nek2 | F | CGT GAG AGA CTA GCA GAG GAC AAA |
| R | TCC GTT CCT TTA GCA AGC TGT AG | |
| Plk4 | F | TGC ATA GTG CTG CTT CTC CAA |
| R | GAC CAA GTC CTT CAT TTG TAA CCA | |
| Actin | F | CGA GGC CCA GAG CAA GAG |
| R | CGT CCC AGT TGG TAA CAA TGC | |
| ChIP assay | ||
| Nek2 | F | TTG GCG ATC TCT ATC AGA GGG |
| R | AAA GTG TCA CTA GGC AAC CGC | |
| Plk4 | F | AGT GTC CCG AGG CAC TGC GGC TT |
| R | AGA TAA CCG CCA TCC CCT TGG A | |
| siRNA analysis | ||
| E2F1_2 | AGCAAAUCAAAGUGCAGAUUGGAGGGU | |
| E1F1_4 | CUCUGGAAACCCUGGUCCCUCCAAGCC | |
| E2F3a | UGAGGAUCUGGAUGUACGCUU | |
| E2F3a_4 | GUUCGUGGUGAGGAUCUGGAUGUACGC | |
| Cyclin D1 | CAAGAAUUACAUAGCCAAGAUGUGCAA | |
| Site-directed mutagenesis | ||
| Nek2 m1 | F | CCT CTC TCC ATC CCT CCG TTT GGC TTA GC |
| R | CGG AGG GAT GGA GAG AGG AAG CGG CAG | |
| Plk4 m1 | F | CAG CAA TCC ATC CCG AGC TAC CGC GTT AGA GC |
| R | GGT AGC TCG GGA TGG ATT GCT GAA AGA ACG | |
| Plk4 m2 | F | GCT ACC GAT TTA GAG CAG GGC AGG GCA GG |
| R | CTG CTC TAA ATC GGT AGC TCG GGC GGG | |
| Plk4 m3 | F | CGT TAG AAT AGG GCA GGG CTA CCT CC |
| R | CCT GCC CTA TTC TAA CGC GGT AGC TCG G | |
| Plk4 m4 | F | GGG CAG GAT TAC CTC CCA CTT CTC CAA GG |
| R | GGG AGG TAA TCC TGC CCT GCT CTA ACG C |
F, forward; R, reverse.
Transfection of siRNAs and BrdU incorporation assay.
Lipofectamine 2000 (11668-019; Invitrogen), along with 200 pmol of each E2F or cyclin D1 small interfering RNA (siRNA) constructs (Integrated DNA Technologies, Coralville, IA) or 5 μl of silencer negative-control siRNA 1 (50 μM, AM4611; Ambion, Grand Island, NY), was used. The primer sequences used for these experiments are presented in Table 1. Bromodeoxyuridine (BrdU) incorporation assay was performed according to our published protocols (59). The percentages of BrdU-positive cells in a population of at least 500 cells were calculated.
Generation of shE2F and shE2F3; GFP-Nek2 cell clones.
To generate stably silenced E2F cells, we used short hairpin RNA (shRNA)-mediated knockdown based on the lentiviral vector pLKO.1-puro (Addgene, Cambridge, MA). At 24 h after the second infection, cells were subjected to selection with 2 μg of puromycin/ml, and cell populations were obtained. Nek2 was subcloned into the pMONO-hygro-GFP plasmid (Invivogen, San Diego, CA) by the Emory DNA Custom Cloning Core Facility. pMONO-hygro-GFP-Nek2 was transfected using Lipofectamine 2000 into HCC1954 and JIMT1 cells stably downregulated for E2Fs.
Cell cycle analysis.
To analyze the cell cycle, we used fluorescein isothiocyanate BrdU/7-AAD flow cytometry kits (catalog no. 57891; BD Pharmingen, San Jose, CA). Briefly, 2 × 106 to 3 × 106 cells were plated on a 100-mm culture dish and cultured in serum starvation media (0.2% serum) for 72 h, released to 10% FBS-containing media, and harvested at 0, 12, 18, and 24 h. Before harvesting, the cells were pulse-labeled with 10 μM BrdU for 30 min at 37°C. The cells were processed and immunostained according to the manufacturer's protocol, acquired in a BD LSRII apparatus using flow cytometry, and analyzed with FlowJo software (Tree Star, Ashland, OR).
Immunostaining.
Centrosome amplification assays were done by plating cells on a four-well chamber slide and fixation in 4% paraformaldehyde for 10 min. Cells were permeabilized in 0.1% NP-40 for 10 min after being washed three times with phosphate-buffered saline. Cells were blocked in 10% normal goat serum (Invitrogen) for 1 h, following overnight primary antibody incubation against pericentrin (ab4448; Abcam, Cambridge, MA). Two hundred cells were counted, and cells with ≥3 pericentrin-positive cells are presented as percentages. For binucleation assays, the cytoskeleton was localized with α-tubulin antibody (sc-32293; Santa Cruz Biotechnology). Alexa Fluor-conjugated antibodies (catalog nos. A11008, A11001, or A21069; Invitrogen) were used as secondary antibodies. For counterstaining, DAPI (4′,6′-diamidino-2-phenylindole) at 1 mg/ml was applied. Two hundred cells were counted, or images were obtained at ×40 magnification using a Zeiss Axioplan-2 fluorescence microscope.
Chromatin immunoprecipitation (ChIP) assay.
Cells were plated on a 150-mm culture dish, and when they were 80 to 90% confluent the cells were cross-linked with 1% formaldehyde for 10 min on a shaker and quenched by adding 0.156 M glycine. After two washing steps, the cells were scraped off the plate for harvesting, and the rest of steps were followed as described previously (60). The following antibodies were used: E2F1 (3742; Cell Signaling, Danvers, MA), E2F2 (sc-633; Santa Cruz Biotechnology), and E2F3 (sc-878; Santa Cruz Biotechnology). Normal rabbit IgG antibody (2729; Cell Signaling) was used as a negative control. The sequences used in this assay are shown in Table 1.
Luciferase assay.
Approximately 1 and 1.2 kb of human Nek2 and Plk4 proximal-promoter regions were cloned into pGL3-Basic plasmid (E1751; Promega) and sequenced. E2F binding site mutants on Nek2 and Plk4 promoter regions were generated by site-directed mutagenesis using Phusion DNA polymerase (M0530; New England Biolabs) with the mutant primers listed in Table 1. Her2+ cell lines were cotransfected with either pGL3-Nek2, pGL3-Plk4 or mutant constructs along with pRL-CMV (E2261; Promega) as an internal control using TransIT-2020 transfection reagent (MIR5400; Mirus, Madison, WI) for 48 h, and cells were assayed for promoter activity by using a dual luciferase kit (E1910; Promega).
Western blotting.
Western blotting was performed according to our published protocols (59, 61, 62). The following primary antibodies were used in this experiment: E2F1 (3742; Cell Signaling), E2F2 (sc-633; Santa Cruz Biotechnology), E2F3 (sc-878; Santa Cruz Biotechnology), cyclin D1 (2922; Cell Signaling), cyclin E (sc-481; Santa Cruz Biotechnology), Nek2 (610593; BD Biosciences, San Jose, CA), phospho-NPMT199 (3541; Cell Signaling), and Plk4 (ab56752; Abcam). β-Actin antibody (4970; Cell Signaling) was used as a loading control. For secondary antibodies, either goat anti-rabbit antibody (sc-2004) or goat anti-mouse antibody (sc-2005) conjugated to horseradish peroxidase (Santa Cruz Biotechnology) was used. Signals were detected by using a Lumigen TMA-6 reagent (Lumigen, Inc., Southfield, MI). ImageJ software (National Institutes of Health, Bethesda, MD) was used to quantify protein levels.
Live-cell image analysis.
HCC1954 cells transduced with pLKO.1, shE2F3, or shE2F3; GFP-Nek2 (1 × 104 to 2 × 104 cells/well) were plated on an eight-chambered #1.5 German coverglass system (155409; Thermo Scientific). Cells were placed in Perkin-Elmer Ultra-View microscope (Perkin-Elmer, Waltham, MA) set at 37°C and 5% CO2, with a differential interference contrast filter, and live-cell images were captured every 5 min for 45 to 48 h under a 10× objective lens and compiled into movies for analysis. All image capture and analysis was done using Volocity 3D image analysis software (Perkin-Elmer).
Bioinformatic analysis.
The Cancer Genome Atlas (TCGA) RNASeq data from 922 breast cancer adenocarcinoma (BRCA) patients (Illumina HiSeq RNASeqv2 level 3 RSEM normalized level 3 gene data) were downloaded from the Broad Institute Firehose Standard Data set portal (63). Clinical metadata on the same data set were also obtained from the same source. Clinical subtypes were taken from previously published analyses of TCGA BRCA samples (64). Pearson correlation coefficients and associated P values were computed for E2F1, E2F2, and E2F3 relative to NEK2 and PLK4 for the entire data set and for clinically relevant subtypes. Scatterplots were generated by two-gene RNAseq analysis of the provisional TCGA breast data set on the cBio Cancer Genomics Portal website (65).
Statistical analysis.
The Student t test was applied to compare significance between groups, and P value of <0.05 are indicated by an asterisk. For promoter analysis, we applied the Mann-Whitney U test (nonparametric test). For live cell imaging analysis, either the chi-square test or the Fisher exact test was applied to compare the proportion of each type between each pair of cell lines, and the SAS statistical package (v9.3; SAS Institute, Inc., Cary, NC) was used for analyses with a significance level of 0.05.
RESULTS
The E2F activators and proteins regulating the cell and centrosome cycles are deregulated in Her2+ breast cancer cells harboring centrosome amplification.
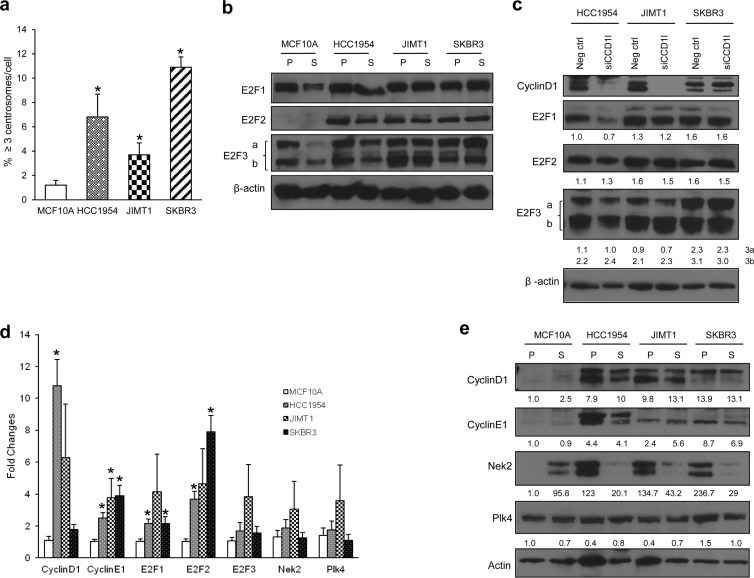
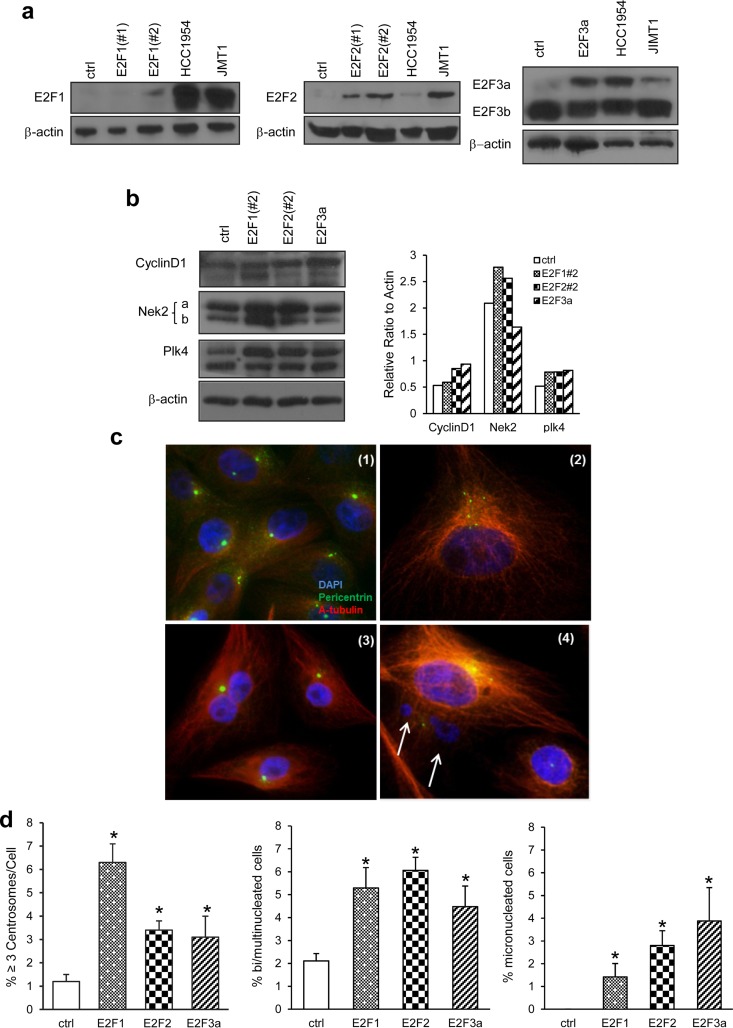
We selected Her2+ breast cancer cells that display centrosome amplification to establish whether E2F downregulation diminishes this abnormal phenotype. Using data presented in Fig. 1a and in two previous publications (56, 62), we screened cell lines of different molecular subtypes, including Her2+, triple-negative (ER− PR− Her2−) and luminal (ER+ Her2+ or Her2−) subtypes, and found centrosome amplification in roughly 50% of Her2+ breast cancer cells. Although there is significant elevation of centrosome amplification in Her2+ cells (SKBR3, HCC1954, and JIMT-1), other Her2+ cells (BT4T4, HCC1419, and HCC1569), and luminal ER+ PR− Her2+ (MDA-MB-361), luminal ER+ PR+ Her2− (MCF7 and T47D), and triple-negative (MDA-MB-231 and MDA-MB-468) cell lines do not display centrosome amplification (56, 62). MCF10A cells are used as a control, since they are nontransformed, immortalized human mammary epithelial cells that lack the p16INK4A and p14INK4B tumor suppressors and display normal p53 activity (66, 67). The genetic characteristics and origins of these cell lines have been described (58, 66). To investigate whether the E2F activators (E2F1, E2F2, and E2F3a) are deregulated in Her2+ breast cancer cells and drive centrosome amplification, their protein levels were analyzed in three ER− PR− Her2+ breast cancer cell lines (henceforth referred to as Her2+ cells): HCC1954, JIMT1, and SKBR3 (Fig. 1b). In proliferating MCF10A cells, E2F1 and E2F3 were robustly expressed. Unlike the decreases in E2F1 and E2F3a expression achieved under serum starvation of MCF10A cells, the E2F protein levels in Her2+ cells were unaffected by serum starvation, showing deregulation of E2Fs. Since E2Fs are under the control of cyclin D1, the upregulation of which has been reported in many types of breast cancer cells (68), we investigated whether the deregulation of E2F proteins seen in Her2+ cells is caused by deregulated cyclin D1. Thus, we transiently knocked down cyclin D1 by siRNA and measured the E2F levels. Cyclin D1 knockdown was efficient in HCC1954 and JIMT1 cells (Fig. 1c). The levels of E2F1, E2F2, or E2F3 were not significantly changed in cyclin D1 knockdown cells except for slight decreases of E2F1 in HCC1954 cells and of E2F3a in HCC1954 and JIMT1 cells. Nevertheless, the protein levels of E2Fs remained robust in the absence of cyclin D1. Her2+ cell lines displaying deregulated E2F activators also displayed centrosome amplification (Fig. 1a). We chose various canonical or potential E2F targets known to play roles in regulating the cell and centrosome duplication cycles based on our previously published reports in which we screened premalignant mouse mammary epithelial lesions expressing K-RasG12D or K-RasG12D and c-Myc for the expression of most known molecules regulating the cell and centrosome cycles (69). Those studies showed conservation in signaling between premalignant lesions expressing such oncogenes and Her2+ breast cancer cells (62). We found significantly elevated mRNA levels of cyclin D1, cyclin E1, E2F1, and E2F2 in Her2+ cells compared to MCF10A cells, whereas the levels of E2F3, Nek2, and Plk4 were slightly, but not significantly, increased in HCC1954 and JIMT1 cells (Fig. 1d). Western blots confirmed real-time PCR data showing high levels of cyclin D1 and cyclin E1, and there were no major changes in Plk4 with the exception of its overexpression in SKBR3 cells (Fig. 1e). In contrast to the real-time PCR data, Nek2 protein was consistently overexpressed in all three Her2+ breast cancer cell lines relative to MCF10A. Our experiments show that the specific breast cancer cell lines used in the present study display centrosome amplification, deregulated E2F activators, and canonical and potential E2F transcriptional targets.
FIG 1.
Centrosome amplification is elevated in Her2+ cells displaying deregulated E2F activators. (a) Centrosome amplification was assayed by pericentrin antibody staining. Two hundred cells/replicate were counted, and the percentages of cells with more than three centrosomes are presented. The assay was repeated five times and the graph represents means ± the standard errors of the mean (SEM; *, P ≤ 0.05). (b) Protein lysates were collected under proliferating (P) or serum starvation (S) conditions, and E2F1, E2F2, and E2F3 (a and b) protein levels were compared between MCF10A control cells and three Her2+ breast cancer cell lines by Western blotting. β-Actin was used as a loading control. (c) Cyclin D1 was transiently knocked down by siRNA, and E2F1, E2F2, and E2F3 protein levels were compared to parental cells by Western blotting. Numbers represent the fold induction relative to the respective controls, normalized for β-actin. (d) Real-time PCR was performed to measure and compare gene expression levels of potential E2F targets (cyclin D1, cyclin E1, E2Fs, Nek2, and Plk4) between MCF10A control and three Her2+ cells. Total RNA was isolated from proliferating cells three independent times, and the results were normalized relative to β-actin. The level of transcripts in MCF10A cells was normalized to 1, and the results for each gene represent fold induction over MCF10A; the graph represents means ± the SEM (*, P ≤ 0.05). (e) Cyclin D1, E1, Nek2, and Plk4 protein levels were compared between MCF10A control cells and three Her2+ breast cancer cell lines by Western blotting. Numbers represent the fold induction normalized to β-actin relative to MCF10A controls.
Nek2 and Plk4 are strongly correlated with E2F factors in breast cancer patients.
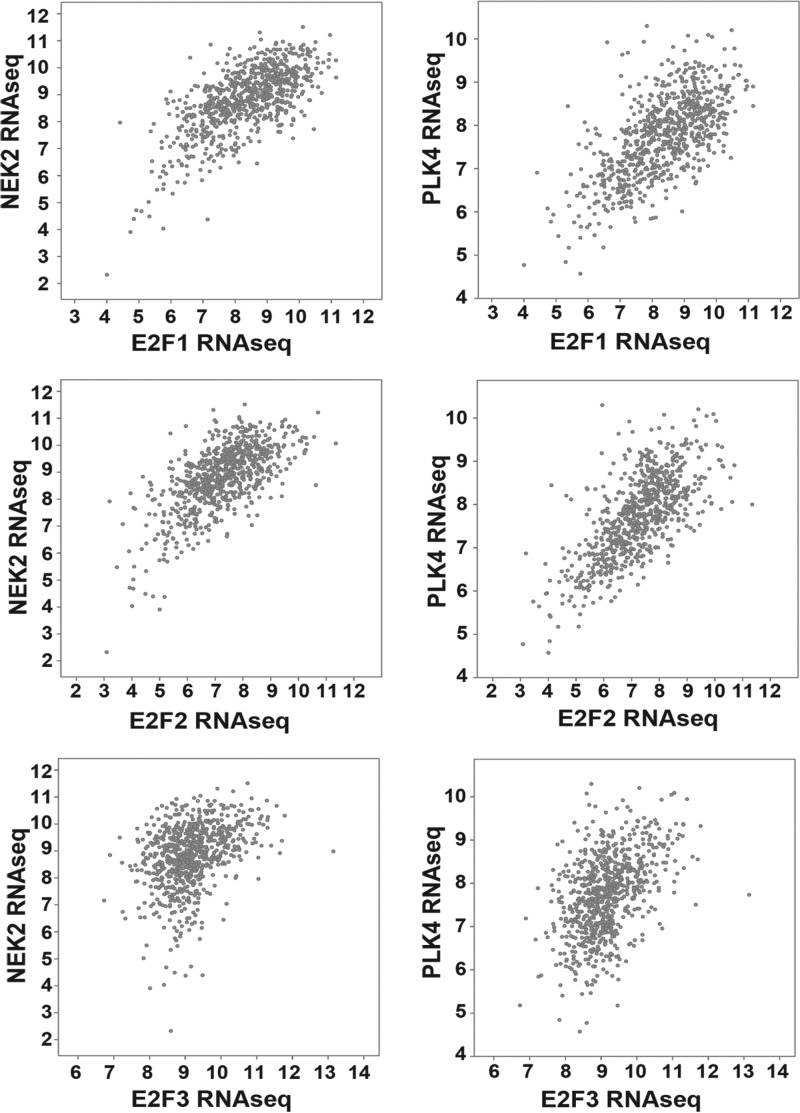
To determine whether the relationship between E2F family transcription factors and Nek2/Plk4 is observed in breast cancer patients, we mined publicly available RNAseq data from the TCGA project in BRCA data sets. We performed scatterplot analysis of Nek2 and Plk4 against E2F1, E2F2, and E2F3 using the cBio Cancer Genomics Portal (www.cbioportal.org) and observed a strong correlation of each E2F factor with their target genes (Fig. 2). To quantify the correlation, we downloaded normalized gene RNASeq data from the TCGA Data Coordination Center (63) and computed the Pearson correlation coefficients and corresponding P values (Table 2) for all 922 TCGA patients, as well as for basal, Her2+, luminal A, and luminal B subsets based on published classifications (64). Correlations were highly statistically significant, except for the Her2+ subtype, suggesting that E2F family transcription factors regulate Nek2 and Plk4 in breast cancer patients.
FIG 2.
Scatterplots of RNASeq data for NEK2 and PLK4 relative to E2F factors. Scatterplots were generated using the cBioPortal Cancer Genomics website (www.cbioportal.org) analyzing the provisional complete set of TCGA BRCA samples (last accessed 14 January 14). Each axis shows the log2 value of the RNASeq-V2-RSEM mRNA data from TCGA.
TABLE 2.
Correlation of NEK2 and PLK4 with E2F factors in TCGA BRCA samplesa
| TCGA sample group (n) and E2F factor | NEK2 |
PLK4 |
||
|---|---|---|---|---|
| Correlation | P | Correlation | P | |
| All BRCA (922) | PLK4 | |||
| E2F1 | 0.60 | 3.22E–92 | 0.54 | 6.13E–71 |
| E2F2 | 0.51 | 1.45E–62 | 0.50 | 2.21E–58 |
| E2F3 | 0.37 | 1.65E–31 | 0.43 | 8.38E–44 |
| Basal (96) | ||||
| E2F1 | 0.43 | 1.04E–05 | 0.19 | 6.28E–02 |
| E2F2 | 0.35 | 5.27E–04 | 0.19 | 5.82E–02 |
| E2F3 | 0.17 | 9.97E–02 | 0.30 | 3.24E–03 |
| Her2 (55) | ||||
| E2F1 | 0.07 | 6.07E–01 | 0.00 | 9.94E–01 |
| E2F2 | –0.08 | 5.45E–01 | –0.05 | 7.11E–01 |
| E2F3 | –0.03 | 8.02E–01 | –0.23 | 9.80E–02 |
| Luminal A (230) | ||||
| E2F1 | 0.45 | 8.18E–13 | 0.57 | 4.02E–21 |
| E2F2 | 0.46 | 1.86E–13 | 0.51 | 1.28E–16 |
| E2F3 | 0.21 | 1.23E–03 | 0.32 | 1.04E–06 |
| Luminal B (126) | ||||
| E2F1 | 0.31 | 4.97E–04 | 0.48 | 1.24E–08 |
| E2F2 | 0.27 | 2.69E–03 | 0.47 | 3.42E–08 |
| E2F3 | 0.13 | 1.46E–01 | 0.20 | 2.38E–02 |
Values indicated in boldface are significantly different.
Plk4 is a direct transcriptional target of the E2F activators.
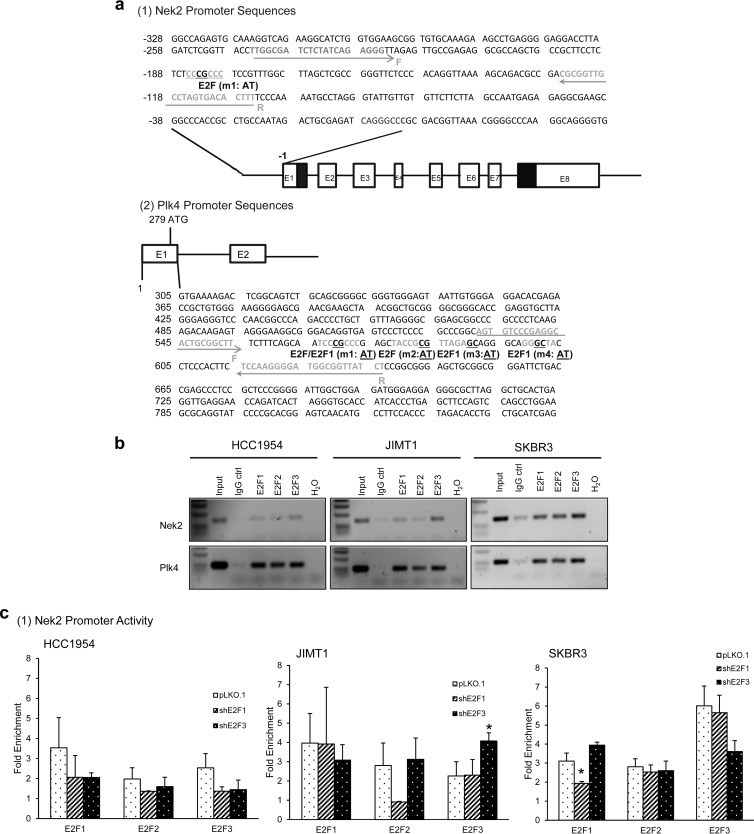
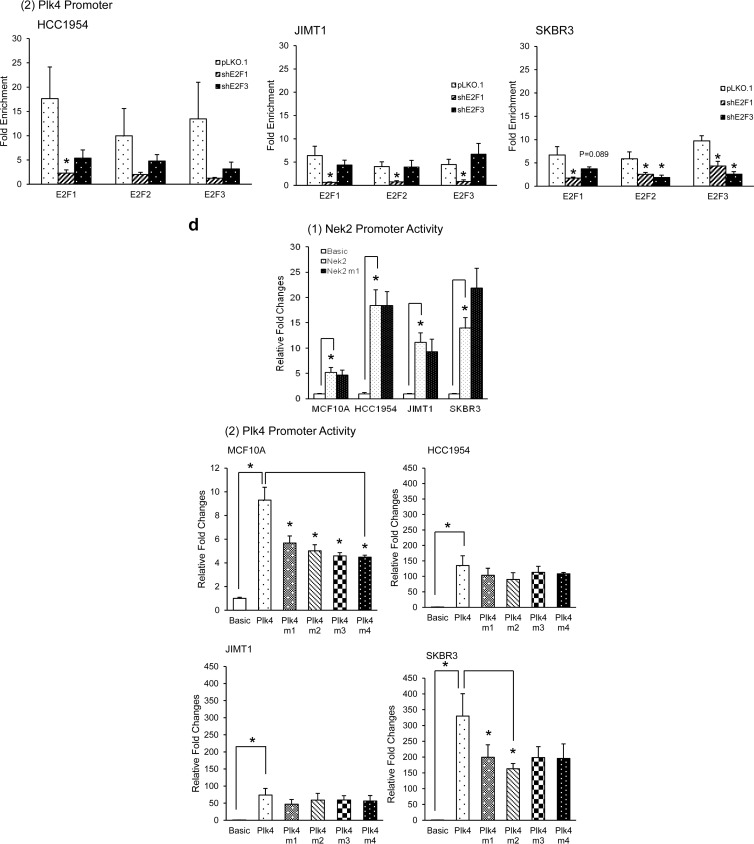
Cyclin D1, cyclin E1, and the E2F activators are known targets of the E2Fs (4, 13, 70). Although the data presented in Table 2 and Fig. 2 show strong correlations between overexpressed E2Fs and elevated Nek2/Plk4 transcripts in breast cancer, it is unknown whether the E2Fs are direct regulators of Nek2 and Plk4. Conserved transcription factor binding finder (CONFAC) analysis (71, 72) predicted one putative E2F site on the Nek2 promoter (70) and four putative E2F binding sites between exons 1 and 2 of Plk4 (Fig. 3a). To elucidate whether E2F1, E2F2, and E2F3 associate with regions on the Nek2 and Plk4 promoters, we used a ChIP assay. The data showed that all three E2F activators bound the predicted E2F binding sites in the Nek2 and Plk4 promoters (Fig. 3b). ChIP analysis in shE2F1 or shE2F3 cells showed no decreases in the occupancy of the E2F site by E2F1, E2F2, and E2F3 on the Nek2 promoter (Fig. 3c). On the other hand, downregulation of E2F1 significantly decreased binding of E2F1, E2F2, and E2F3 to the Plk4 promoter regions. E2F3 downregulation led to slight but not significant decreases in E2F occupancy in HCC1954 cells and significant decreases in SKBR3 cells. In addition, a luciferase assay showed higher Nek2 promoter activity in Her2+ cells (2- to 4-fold) compared to that of MCF10A control (Fig. 3d1). In regard to the Plk4 promoter, the activity was 15- to 30-fold higher in Her2+ cells relative to MCF10A cells (Fig. 3d2). To analyze this further, E2F binding site mutants of the Nek2 (Nek2 m1) and Plk4 promoter (Plk4 m1, m2, m3, and m4) were created by site-directed mutagenesis, by changing the core bases CG to AT. We did not detect decreased luciferase activity of the E2F binding site mutant on the Nek2 promoter (Fig. 3d1). We cloned more than 3 kb of the promoter region into the luciferase construct to detect nonconsensus E2F sites but could not detect additional sites or the high luciferase expression detected with the minimal promoter (data not shown). In contrast, all four mutations of E2F binding sites on the Plk4 promoter significantly decreased its promoter activities (ca. 40 to 50% that of parental construct) in MCF10A cells. Overall, Plk4 promoter activities were decreased ca. 15 to 40% in mutant constructs compared to the wild type in three Her2+ cells, but most of them were not statistically significant except Plk4 mutant 1 and 2 constructs in SKBR3 (Fig. 3d2). These data imply that mutation of single E2F binding sites on the Plk4 promoter was not enough to decrease E2F-dependent regulation of Plk4 transcription in all breast cancer cells. Together, these data show that Plk4 is under direct E2F transcriptional regulation, whereas Nek2 is not.
FIG 3.
The E2F activators bind the Nek2 and Plk4 promoter regions and increase their promoter activities. (a) Location of canonical E2F binding sites adjacent to the Nek2 and Plk4 promoters, as predicted by the CONFAC program. The primers used for amplification and mutant E2F promoter sequences are also indicated. (b) E2F1, E2F2, or E2F3 antibodies were used to immunoprecipitate chromatin displaying potential E2F binding sites located in proximity to the Nek2 and Plk4 promoters in HCC1954, JIMT1, and SKBR3 cells. A fraction of chromatin was used as a nonimmunoprecipitated input control. Normal rabbit IgG was used as a negative control for the ChIP assay, and H2O was used as control for PCR. (c) ChIP assay on shE2F1 and shE2F3 cells. The x axis indicates the antibody used for immunoprecipitation, while the y axis indicates the fold enrichment relative to IgG controls. (d) Nek2 and Plk4 promoter activities were measured using dual-luciferase assay in three Her2+ cell lines, as well as MCF10A cells. The graph represents means ± the SEM (*, P ≤ 0.05).
E2F overexpression induces centrosome amplification and chromosome instability in immortalized mammary epithelial cells.
We have reported that MCF10A cells have a low percentage of centrosome amplification (56, 62, 69). To address whether the overexpression of E2F1, E2F2, or E2F3 is sufficient to induce centrosome amplification in MCF10A cells, we developed stable populations of MCF10A cells overexpressing E2F1, E2F2, or E2F3a. Overexpression of the E2Fs was confirmed by Western blotting (Fig. 4a). This analysis revealed that E2F1, E2F2, and E2F3a are overexpressed relative to MCF10A parental controls at similar (E2F2 and E2F3a) or lower (E2F1) levels relative to Her2+ cells. The data presented in Fig. 1d and e revealed the overexpression of various cell and centrosome regulatory proteins, including Nek2 and cyclin D1 in all Her2+ cells, and significant overexpression of Plk4 in SKBR3 cells. Thus, we addressed whether single overexpression of E2Fs in MCF10A cells increased protein level of above targets. Populations expressing the highest E2F levels were chosen for this analyses and showed that MCF10A cells expressing single E2F1, E2F2, or E2F3a displayed upregulation of cyclin D1, Nek2, and Plk4 (Fig. 4b). Based on these observations, we further analyzed E2F-overexpressing cells to measure centrosome amplification by pericentrin (Fig. 4c2) and to quantify binucleated (Fig. 4c3) and micronucleated cells (Fig. 4c4) by α-tubulin/DAPI. Binucleation, an intermediate to tetraploidy (46) (Fig. 4c3), and micronucleation, a measure of whole chromosome or chromosome fragment losses (73) (Fig. 4c4), are measures of chromosome instability (73–75). Our results demonstrate that ectopic E2F expression in MCF10A cells significantly elevated frequencies of centrosome amplification, binucleation, and micronucleation relative to control cells (Fig. 4d). The results confirmed that Nek2 and Plk4 levels are under direct or indirect E2F activator control and demonstrate that single E2F activators are sufficient to trigger generators of aneuploidy and chromosome instability.
FIG 4.
Ectopic expression of E2F1, E2F2, or E2F3a results in overexpression of centrosome regulators, centrosome amplification and chromosome instability. (a) Cells overexpressing human E2F1, E2F2, or E2F3a (pBH-E2F1, pBH-E2F2, or pBH-E2F3a, respectively) were generated in MCF10A cells, and the E2F levels were measured by Western blotting. (b) Canonical and potential targets of E2Fs—cyclin D1, Nek2, and Plk4—were analyzed by Western blotting, and their levels were quantified. Nek2a and Nek2b were quantified in the same graph. (c) Microscope images of pericentrin/α-tubulin/DAPI staining (1, normal number of centrosomes in MCF10A cells; 2, centrosome amplification; 3, binucleation; 4, micronucleation) in E2F overexpressing MCF10A cells. Pictures were taken with a Zeiss Axioplan-2 microscope under ×40 magnification. (d) Centrosome amplification was assayed by pericentrin staining, binucleation by α-tubulin, and micronucleation by DAPI. The percentage of cells with three or more centrosomes was calculated in 200 cells per replicate per group. The assay was repeated three times and the graph was represented by mean percentages ± the SEM (*, P ≤ 0.05).
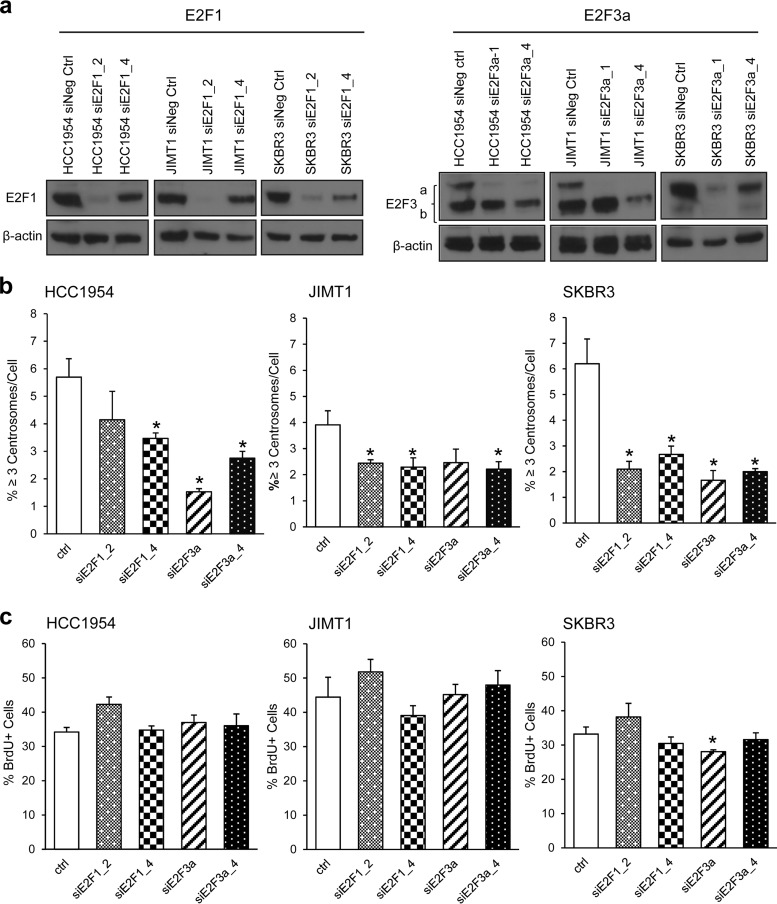
Transient silencing of the E2F activators in Her2+ breast cancer cells decreases centrosome amplification without significantly affecting DNA replication.
Having demonstrated that E2F overexpression triggers centrosome amplification in nontransformed mammary epithelial cells, we addressed whether the E2F activators maintain centrosome amplification in Her2+ breast cancer cells by transiently transfecting siRNAs to silence E2F1, E2F2, and E2F3a. We screened several siRNA constructs against E2F2 and did not achieve downregulation of E2F2 protein (data not shown). Two E2F1 siRNA constructs, siE2F1_2 and siE2F1_4, knocked down E2F1 efficiently in all three Her2+ cell lines (Fig. 5a, left panel). For E2F3, we designed siRNA constructs specifically targeting the E2F3a activator, and both constructs efficiently knocked down E2F3a in all three Her2+ cell lines (Fig. 5a, right panel). Transient downregulation of E2F1 and E2F3a was accompanied by decreased centrosome amplification in the three Her2+ cell lines (Fig. 5b). Since E2Fs play a significant role in controlling DNA replication and to establish whether the cause of the decrease in centrosome amplification was a result of a general block in DNA replication, we measured the percentage of cells in S phase with a BrdU incorporation assay. Neither knockdown of E2F1 nor knockdown of E2F3a affected DNA replication in Her2+ cells relative to cells transfected with control siRNA (Fig. 5c). The data indicate that transient knockdown of E2F1 and E2F3a diminishes centrosome amplification and that a block in S-phase progression is not a likely mechanism leading to reduced centrosome amplification. These data demonstrate a role for E2F1 and E2F3a in maintaining centrosome amplification in Her2+ breast cancer cells.
FIG 5.
Transient knockdown of E2F1 or E2F3a suppresses centrosome amplification without greatly affecting DNA replication. (a) siRNA-mediated gene silencing was used to transiently knockdown E2F1 and E2F3a in three Her2+ cells. Two independent siRNAs sequences against E2F1 (siRNAE2F1_2 and siRNAE2F1_4) or E2F3a (siRNAE2F3a and siRNAE2F3a_4) were used to transfect target cells. Western blotting was performed to detect protein levels; β-actin served as a loading control. (b) Cells were transfected with the indicated control or siRNA against E2F1 or E2F3a. Graphs represent the percentages of cells with more than three centrosomes (localized by pericentrin antibody staining). Each replicate was done in a population of at least 200 cells. The assay was repeated three times, and the graph shows the means ± the SEM (*, P ≤ 0.05). (c) A BrdU incorporation assay was performed to measure DNA synthesis in cells silenced for E2F1 or E2F3a. Cells were pulsed with BrdU and processed for BrdU staining. Graphs represent BrdU+ cells in a population of cells. A population of 500 cells was counted per group per replicate. Three independent experiments were performed, and the graph shows the means ± the SEM (*, P ≤ 0.05).
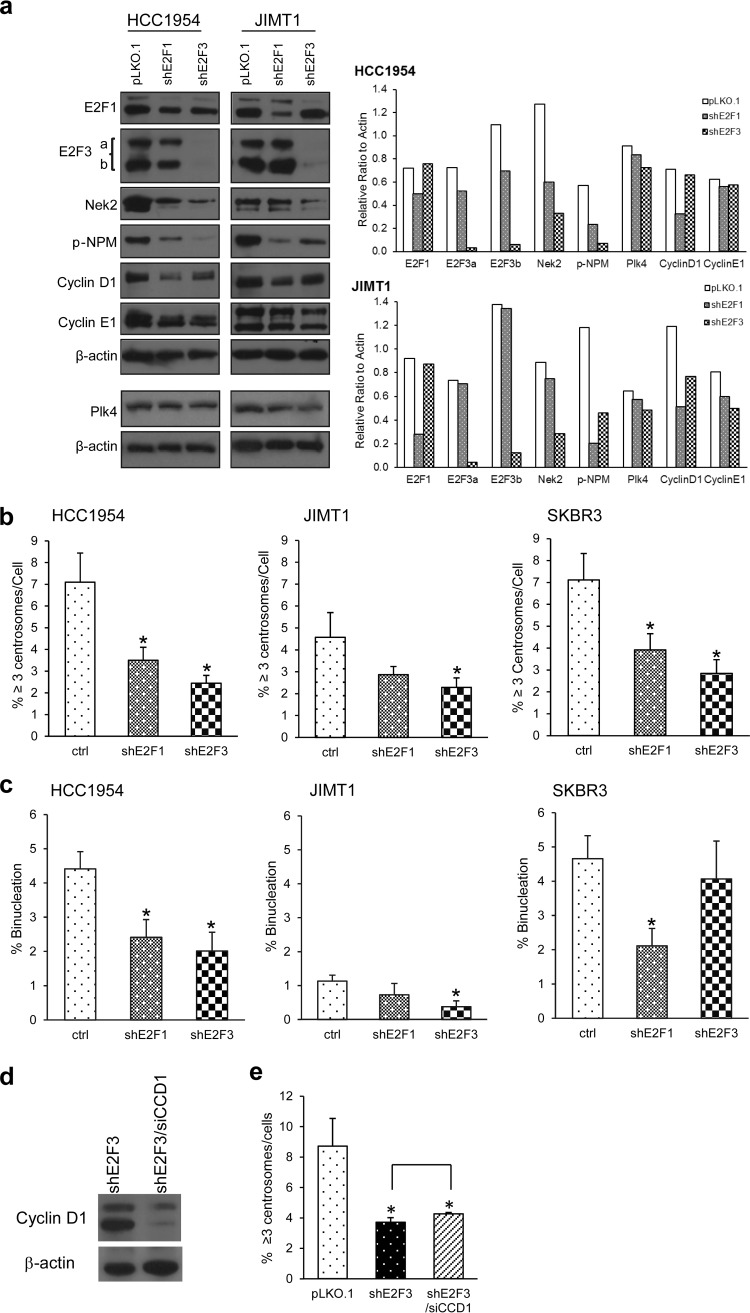
Stable E2F knockdowns in Her2+ breast cancer cells decrease centrosome amplification and binucleation but do not significantly affect cell cycle progression.
To explore the roles of E2Fs in centrosome amplification more closely, stable E2F1 and E2F3 knockdowns were generated in JIMT1, SKBR3, and HCC1954 cells using shRNA-mediated silencing techniques. E2F1 knockdown was partial, whereas E2F3 knockdown was almost complete (Fig. 6a). Next, we investigated whether knockdown of E2F1 and E2F3 in HCC1954 and JIMT1 cell lines impacts proteins involved in the centrosome duplication cycle. Silencing E2F1 or E2F3 led to decreased cyclin D1 levels in both cell lines (Fig. 6a), whereas changes in cyclin E1 or Plk4 were small. Silencing E2F1 or E2F3 also led to lower levels of Nek2 and phospho-NPMT199, a known phosphorylation target of Cdk2 and Cdk4 (59, 76, 77). Overall, these results confirm that the E2F activators modulate expression of central regulators of the cell and centrosome cycles, which may signal centrosome amplification in Her2+ breast cancer cells.
FIG 6.
Her2+ cells stably knocked down for E2F1 or E2F3 display lower levels of cyclin D1, Nek2, Plk4, and p-NPM199 and suppress centrosome amplification and binucleation. (a) E2F1 or E2F3 were stably downregulated using shRNA-mediated silencing technique in HCC1954 and JIMT1 cells. Potential E2F target protein levels (cyclin D1, Nek2, Plk4, and p-NPM199) were compared in these cell lines by Western blotting and quantified (right panel). (b) Centrosome amplification was assayed by immunostaining centrosomes with pericentrin antibodies. Two hundred cells per group per replicate were counted. The assay was repeated five times, and the graph shows the mean percent cells with three or more centrosomes ± the SEM (*, P ≤ 0.05). (c) A binucleation assay was performed by immunostaining the cytoskeleton with α-tubulin and visualizing nuclei with DAPI. Five independent replicates were performed, and the total binucleated cells in a population of 200 cells per replicate per group was calculated. The graph shows the mean percentages of binucleated cells ± the SEM (*, P ≤ 0.05). (d) HCC1954 cells expressing shE2F3 were transfected with either control siRNA or siRNA against cyclin D1, and the protein levels of cyclin D1 were assessed by Western blotting. (e) Frequencies of centrosome amplification in the indicated molecular groups were addressed as in panel b.
Mirroring transient knockdowns, stable knockdown of E2F1 or E2F3 suppressed centrosome amplification in all cell lines (Fig. 6b). To establish whether silencing of E2Fs diminished binucleation, we calculated its percentages in cells silenced for E2F1 and E2F3 (Fig. 6c). Noticeably, JIMT1 cells had a lower extent of binucleation. Silencing E2F1 significantly decreased the percentage of binucleated cells in HCC1954 and SKBR3 cells. On the other hand, silencing E2F3 significantly decreased the percentages of binucleated cells in HCC1954 and JIMT1 cells. Consistent with Fig. 1c, where transient knockdown of cyclin D1 did not affect the protein expression levels of E2F3, transient knockdown of cyclin D1 in shE2F3 cells did not decrease CA further compared to that of shE2F3 alone (Fig. 6d and e). These data imply that E2Fs and cyclin D1 do not cooperate in maintaining CA in Her2+ cells. To establish whether the decreases in binucleation and centrosome amplification were due to blocks in the cell cycle, we performed a flow cytometry-based DNA replication assay that measures BrdU incorporation and calculated the percentages of cells in various phases of the cell cycle (Table 3). Under proliferating conditions, E2F knockdown in HCC1954 cells showed a lower percentage of cells in S phase and in G2/M phase relative to pLKO.1. No such changes were observed in JIMT1 and SKBR3 cells. Nonetheless, HCC1954 and JIMT1 cells lines reached S phase ∼18 h after serum addition, whereas SKBR3 reached S phase 24 h after release. These results show that single E2F activator silencing in Her2+ cells does not greatly affect cell cycle progression.
TABLE 3.
Percentage of cells in each cell cycle phase
| Cell category | Mean % cells ± SEMa |
||
|---|---|---|---|
| G0/G1 | S | G2+M | |
| HCC1954 cells | |||
| Proliferating | |||
| HCC1954 pLKO. 1 | 18.4 ± 0.530 | 60.6 ± 0.417 | 21 ± 0.727 |
| HCC1954 shE2F1 | 29.5 ± 1.192* | 56.6 ± 1.096* | 13.9 ± 1.070* |
| HCC1954 shE2F3 | 30 ± 0.59* | 51 ± 0.603* | 19 ± 1.071* |
| 0 h after release | |||
| HCC1954 pLKO.1 | 64 ± 4.67 | 12.4 ± 1.995 | 23.5 ± 4.947 |
| HCC1954 shE2F1 | 63.2 ± 5.151 | 18.9 ± 1.178* | 18.1 ± 4.496 |
| HCC1954 shE2F3 | 80.7 ± 2.56* | 7.2 ± 0.978* | 12.1 ± 1.808* |
| 12 h after release | |||
| HCC1954 pLKO.1 | 72.1 ± 2.611 | 12 ± 0.46 | 15.9 ± 2.463 |
| HCC1954 shE2F1 | 72.9 ± 0.598 | 18.6 ± 1.341* | 8.6 ± 0.843* |
| HCC1954 shE2F3 | 84.9 ± 3.008* | 9 ± 2.077 | 6 ± 0.963* |
| 18 h after release | |||
| HCC1954 pLKO.1 | 35.7 ± 8.413 | 52.6 ± 7.584 | 11.7 ± 0.844 |
| HCC1954 shE2F1 | 34.3 ± 9.225 | 58.1 ± 6.92 | 7.6 ± 2.394 |
| HCC1954 shE2F3 | 34.9 ± 9.658 | 59.9 ± 8.555 | 5.1 ± 1.408* |
| 24 h after release | |||
| HCC1954 pLKO.1 | 15.6 ± 1.476 | 74.8 ± 1.694 | 9.6 ± 1.019. |
| HCC1954 shE2F1 | 21.3 ± 2.417* | 70 ± 2.675 | 8.7 ± 0.313 |
| HCC1954 shE2F3 | 13.9 ± 1.458 | 79.6 ± 1.003* | 6.5 ± 1.123* |
| JIMT1 cells | |||
| Proliferating | |||
| JIMT1 pLKO.1 | 33 ± 4.723 | 53.1 ± 2.944 | 13.9 ± 2.065 |
| JIMT1 shE2F1 | 28.6 ± 3.266 | 49.4 ± 3.076 | 22 ± 2.103* |
| JIMT1 shE2F3 | 38.8 ± 3.683 | 49.6 ± 2.538 | 11.6 ± 1.453 |
| 0 h after release | |||
| JIMT1 pLKO.1 | 60.7 ± 3.24 | 25 ± 1.887 | 14.3 ± 1.541 |
| JIMT1 shE2F1 | 50.3 ± 1.647* | 20.7 ± 1.118* | 29 ± 2.235* |
| JIMT1 shE2F3 | 59.9 ± 5.962 | 23 ± 4.995 | 17.1 ± 1.014 |
| 12 h after release | |||
| JIMT1 pLKO.1 | 71.2 ± 3.023 | 12.2 ± 1.302 | 16.7 ± 2.38 |
| JIMT1 shE2F1 | 60.6 ± 0.604* | 8.3 ± 0.727* | 31 ± 0.947* |
| JIMT1 shE2F3 | 71.8 ± 2.982 | 12.7 ± 1.439 | 15.6 ± 1.615 |
| 18 h after release | |||
| JIMT1 pLKO.1 | 53.8 ± 1.536 | 36.4 ± 2.635 | 9.8 ± 0.778 |
| JIMT1 shE2F1 | 41.4 ± 0.107* | 40.6 ± 3.062 | 18 ± 2.043* |
| JIMT1 shE2F3 | 50 ± 3.354 | 39.9 ± 1.55 | 10.1 ± 0.824 |
| 24 h after release | |||
| JIMT1 pLKO.1 | 39 ± 1.446 | 51 ± 2.568 | 10 ± 1.324. |
| JIMT1 shE2F1 | 28.2 ± 1.095* | 54.2 ± 2.925 | 17.6 ± 1.89* |
| JIMT1 shE2F3 | 40.1 ± 0.61 | 48.4 ± 1.716 | 11.4 ± 1.474 |
| SKBR3 cells | |||
| Proliferating | |||
| SKBR3 pLKO.1 | 37.8 ± 2.175 | 36.7 ± 1.839 | 25.5 ± 0.339 |
| SKBR3 shE2F1 | 49.3 ± 3.089* | 32.5 ± 3.636 | 18.2 ± 2.679* |
| SKBR3 shE2F3 | 38.9 ± 4.522 | 35.1 ± 2.984 | 26 ± 1.574 |
| 0 h after release | |||
| SKBR3 pLKO.1 | 52.2 ± 2.812 | 11.9 ± 0.271 | 35.9 ± 2.96 |
| SKBR3 shE2F1 | 52.4 ± 4.021 | 20.3 ± 0.942* | 27.3 ± 4.526 |
| SKBR3 shE2F3 | 45.9 ± 7.804 | 17.9 ± 3.644 | 36.2 ± 4.224 |
| 12 h after release | |||
| SKBR3 pLKO.1 | 58 ± 3.416 | 18.9 ± 0.948 | 23.1 ± 2.674 |
| SKBR3 shE2F1 | 62.2 ± 1.392 | 20 ± 0.889 | 17.8 ± 2.195 |
| SKBR3 shE2F3 | 61.6 ± 4.36 | 14.2 ± 2.323 | 24.2 ± 2.2 |
| 18 h after release | |||
| SKBR3 pLKO.1 | 53.8 ± 2.239 | 22.4 ± 1.012 | 23.8 ± 2.185 |
| SKBR3 shE2F1 | 60.3 ± 0.611* | 20.6 ± 2.565 | 19.2 ± 2.329 |
| SKBR3 shE2F3 | 58.4 ± 6.384 | 15.3 ± 2.118* | 26.3 ± 4.27 |
| 24 h after release | |||
| SKBR3 pLKO.1 | 45.8 ± 1.542 | 32.1 ± 1.669 | 22.1 ± 3.187 |
| SKBR3 shE2F1 | 59.1 ± 6.398 | 22.2 ± 2.887* | 18.6 ± 3.999 |
| SKBR3 shE2F3 | 54.3 ± 7.184 | 20.9 ± 2.479* | 24.8 ± 5.195 |
*, P < 0.05 (as determined by Student t test).
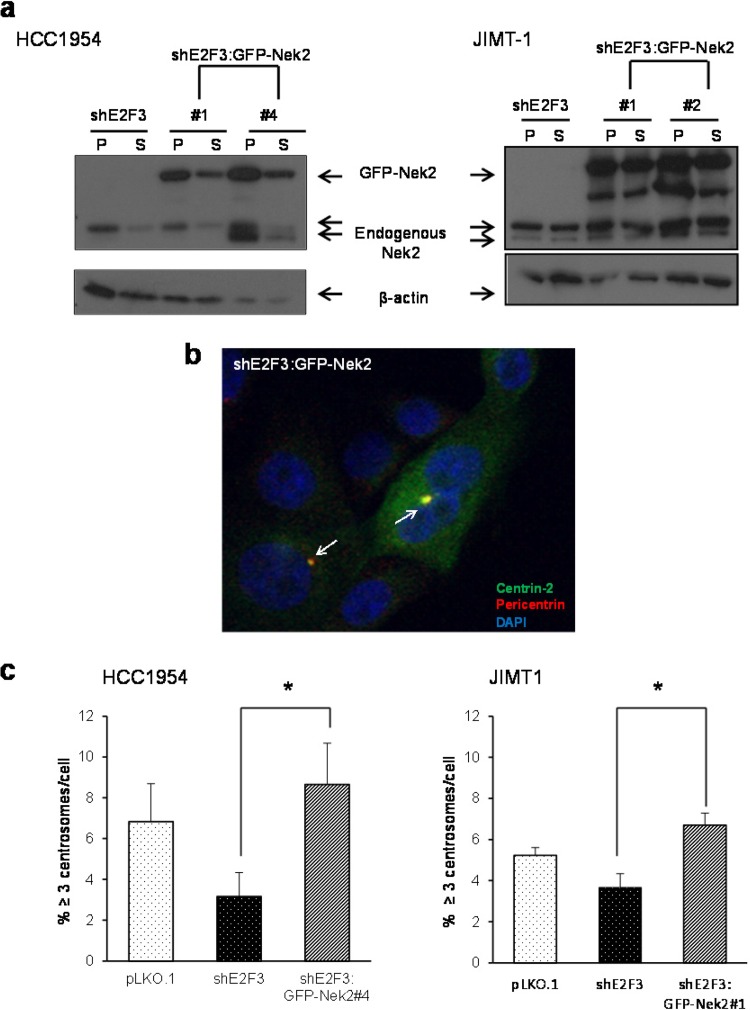
Nek2 overexpression enhances centrosome amplification in Her2+ breast cancer cells silenced for E2F3.
Western blots presented in Fig. 6a showed that decreases in centrosome amplification were associated with the suppression of Nek2 and cyclin D1. Since Nek2 specifically regulates centrosomal functions (78) relative to cyclin D1, which regulates both the cell and centrosome cycles (59, 69, 79), we selected to overexpress Nek2 to establish whether it was sufficient to rescue centrosome amplification in HCC1954 and JIMT1 cells silenced for E2F3. At least two clones of stable cell lines overexpressing GFP-tagged Nek2 were generated: clone 4 was chosen for HCC1954, and clone 1 was chosen for JIMT1, since they overexpressed GFP-Nek2 at similar levels (Fig. 7a). Immunofluorescence microscopy images confirmed GFP-Nek2 expression and localization to centrosomes in interphase (Fig. 7b). Expression of GFP-Nek2 significantly increased centrosome amplification in both HCC1954 and JIMT1 cells downregulated for E2F3 (Fig. 7c). The data show that Nek2 is indeed an important target mediating E2F-dependent centrosome amplification in breast cancer cells.
FIG 7.
Overexpression of Nek2 in Her2+ cells stably downregulated for E2F3 triggers centrosome amplification. (a) Western blots show shE2F3 HCC1954, and JIMT1 cell populations overexpressing GFP-tagged Nek2 and endogenous Nek2. Western blots were probed with antibodies against Nek2 and β-actin as a loading control. (b) shE2F3 cells overexpressing GFP-Nek2 were visualized by fluorescence microscopy. The centrosomes were detected with antipericentrin antibodies, and nuclei were detected by using DAPI. Arrows indicate colocalization of GFP-Nek2 and pericentrin. Images were obtained under ×40 magnification, cropped, and enlarged for visualization purposes. (c) Centrosome amplification was assayed by pericentrin antibody staining. The assay was repeated three times, and the graph shows the mean percentages ± the SEM (*, P ≤ 0.05 [comparison of shE2F3 versus shE2F3; GFP-Nek2]).
Downregulation of E2F3 triggers cell death and delays cytokinesis in Her2+ cells.
To analyze further the functions of E2Fs in Her2+ breast cancer cells, we investigated the timing of the initiation of mitosis from the previous cytokinesis (visually indicated by the rounding up of cells following interphase) and the timing of the completion of cytokinesis (time from initiation of mitosis to physical separation of cells following cytokinesis) in HCC1954 cells by live cell imaging analysis for 48 h, representing two cell cycles. First, we categorized events into three: divided (successful cell division), not divided (no cell division), and dead cells (Table 4). Our results indicate that fewer shE2F3 and shE2F3; GFP-Nek2 cells underwent cell division compared to the pLKO.1 control. We observed a modest, yet significant decrease in the percentage of shE2F3; GFP-Nek2 cells that did not divide relative to shE2F3 cells. In addition, a significantly higher percentage of shE2F3 and shE2F3; GFP-Nek2 cells were eliminated by cell death before the completion of the experiment compared to pLKO.1 control. Among divided cells, we expanded our analysis into three more subcategories, the first being delayed only in initiation of mitosis. Since most cells start mitosis at approximately 20 to 24 h after the previous cytokinesis, we define delayed initiation of mitosis when it takes more than 28 h to reach mitosis after a previous cytokinesis. Another category was delayed only in cytokinesis (i.e., it takes more than 1 to 2 h from initiation of mitosis to achieve cell division) or cells displaying both delays in initiating mitosis and in completing cytokinesis (Table 4). Overall, we did not detect differences in percentages of cells with delayed initiation of mitosis between controls and shE2F3 cells. In the “delayed only in cytokinesis” and the “delayed in both initiation of mitosis and cytokinesis” categories, the shE2F3 group showed significant increases relative to the controls. On the other hand, GFP-Nek2 overexpression in shE2F3 cells resulted in a timing of completion of cytokinesis similar to that of pLKO.1 cells.
TABLE 4.
Analysis of live cell imaging
| Comparison and cell categorya | Cell line | % |
Pb |
|
|---|---|---|---|---|
| Chi-square test | Fisher exact test | |||
| Comparison of the proportion of each event among cell lines | ||||
| Divided | pLKO.1 | 80.39 | R | |
| shE2F3 | 59.66 | <0.001 | R | |
| shE2F3; GFP-Nek2 | 63 | <0.001 | 0.267 | |
| Not divided | pLKO.1 | 9.63 | R | |
| shE2F3 | 25.64 | <0.001 | R | |
| shE2F3; GFP-Nek2 | 20.08 | <0.001 | 0.033 | |
| Dead | pLKO.1 | 5.08 | R | |
| shE2F3 | 10.6 | <0.001 | R | |
| shE2F3; GFP-Nek2 | 12.26 | <0.001 | 0.396 | |
| Comparison of the proportion of each subevent among divided cells | ||||
| Delayed only in initiation of mitosis | pLKO.1 | 32.68 | R | |
| shE2F3 | 37.54 | 0.151 | R | |
| shE2F3; GFP-Nek2 | 27.52 | 0.132 | 0.007 | |
| Delayed only in cytokinesis | pLKO.1 | 6.75 | R | |
| shE2F3 | 11.75 | 0.014 | R | |
| shE2F3; GFP-Nek2 | 6.38 | 0.838 | 0.019 | |
| Delayed both in initiation of mitosis and in cytokinesis | pLKO.1 | 3.92 | R | |
| shE2F3 | 1.43 | 0.035 | R | |
| shE2F3; GFP-Nek2 | 3.69 | 0.872 | 0.065 | |
| Comparison of the proportion of subevent of the binucleated cells | ||||
| Divided | pLKO.1 | 65.52 | R | |
| shE2F3 | 50 | 0.254 | R | |
| shE2F3; GFP-Nek2 | 42.42 | 0.069 | 0.571 | |
| Binucleated cells divided and died | pLKO.1 | 31.58 | R | |
| shE2F3 | 58.33 | 0.141 | R | |
| shE2F3; GFP-Nek2 | 21.43 | 0.698 | 0.105 | |
| Not divided | pLKO.1 | 31.03 | R | |
| shE2F3 | 25 | 0.627 | R | |
| shE2F3; GFP-Nek2 | 30.3 | 0.950 | 0.660 | |
| Dead | pLKO.1 | 22.22 | R | |
| shE2F3 | 50 | 0.329 | R | |
| shE2F3; GFP-Nek2 | 30 | 1.000 | 0.607 | |
The percentages of cells whose fate was not determined (mostly because of the time limit) were not incorporated into the table.
The P value was calculated using a chi-square test or the Fisher exact test where appropriate. Boldfacing indicates statistical significance. R, reference.
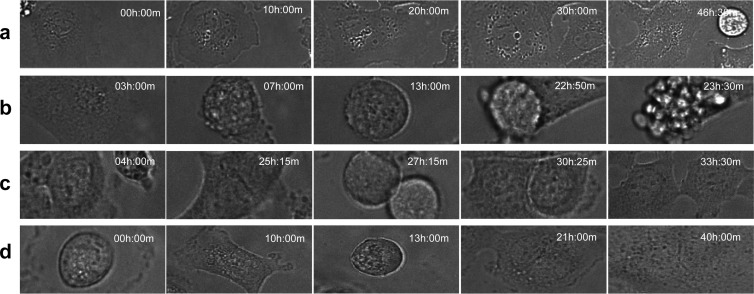
A major controversy in cancer biology is whether binucleated cells can stably undergo mitosis and progress through the cell cycle (46). Thus, we analyzed the fate of binucleated cells (Fig. 8 and Table 4). For this purpose, we included cells already binucleated at 0 h of recording, as well as cells binucleated after cytokinesis. Again, we categorized them into two events: divided and not divided. Among divided cells, we further analyzed the portion of binucleated cells that die after mitosis. Under the “not divided” category, we also quantified the portion of binucleated cells dying before reaching mitosis. Although more binucleated cancer cells divided in pLKO.1 control cells (65.52%) compared to shE2F3 (50%) or shE2F3; GFP-Nek2 (42.42%) cells, the results were not statistically significant. Likewise, no significant differences were found in the percentages of binucleated cells dying following mitosis or before reaching mitosis. Overall, these data indicate that E2F3 knockdown compromised survival in the overall population of cells, diminished the fraction of dividing cells, and delayed cytokinesis of HCC1954 cells. On the other hand, Nek2 overexpression in this setting resulted in timing of initiation of mitosis and completion of cytokinesis more closely resembling those of HCC1954 control cells. Also, it is striking to find that a significant fraction of binucleated cells divide, indicating that Her2+ breast cancer cells can tolerate and successfully divide a highly polyploid genome.
FIG 8.
Time-sequential images of the fate of binucleated cells. Several fates of binucleated cells captured at the designated time points are presented. (a) No mitosis; (b) death after mitosis; (c) normal mitosis; (d) generation of binucleated cell after mitosis.
DISCUSSION
The frequent deregulation of the E2Fs and the presence of centrosome amplification in the vast majority of breast cancers suggest that they play central roles in breast cancer initiation and/or progression (51–53). Although E2F1 and E2F3 are important mediators of Neu and Myc-initiated mammary tumorigenesis (26, 39), the E2F-dependent activities (including centrosome amplification) contributing to mammary tumors are unclear. Identifying these mechanisms would help to establish the role played by centrosome amplification in breast tumorigenesis. Most progress has been achieved in understanding how centrosomal proteins that include Plk4 and γ-tubulin are regulated by ubiquitination and degradation (80, 81). Another area where major progress has been achieved is that phosphorylation of centrosomal proteins by the G1-phase Cdks modifies their function, with NPM, CP110, and Mps1 being under such regulation (7, 59, 76, 82, 83). Our laboratory has made major progress in unraveling oncogenic signals and G1/S regulatory molecules responsible for centrosome amplification in mammary epithelial cells. We showed that centrosome amplification in Her2+ cells and MCF10A cells expressing H-RasG12V is signaled through cyclin D/Cdk4 and Nek2 (62, 69). While this suggests a role for the E2Fs, the major transcriptional effectors of cyclin D/Cdk4, in centrosome amplification, this remained to be tested. The data shown here, wherein cyclin D1 downregulation does not modify E2F levels or affect the percentages of CA in cells silenced for E2F3, suggest that E2F deregulation in the Her2+ breast cancer cells analyzed here has become independent of cyclin D regulation.
Results presented here indicate that overexpression of E2F1, E2F2, or E2F3a in MCF10A resulted in centrosome amplification and two intermediates to aneuploidy and chromosome instability: binucleation and micronucleus formation. Consistent with this finding, the downregulation of E2F1 or E2F3 significantly diminish centrosome amplification and binucleation in Her2+ cells. Together, these results demonstrate that the E2Fs are sufficient to trigger centrosome amplification and chromosome instability in nontransformed mammary epithelial cells and maintain centrosome amplification and chromosome instability in Her2+ cells. E2F-dependent cancer-driving activities, including hyperproliferation and centrosome amplification, are redundant or unique, dependent on the cellular or tissue context. For example, while the overexpression of E2F1, E2F2, and E2F3 shows redundancy in stimulating the S phase in cultured cells (84–87) and in the context of MMTV-Neu mammary tumors (26, 39), knockout technology indicated the specificity of the functions. For instance, E2F3a/b knockout mouse embryo fibroblasts (MEFs) show cell proliferation, cell cycle, and centrosome cycle defects compared to wild-type, E2F1−/−, or E2F2−/− MEFs (7, 88–90). In contrast to experimental mouse models, in our Her2+ breast cancer model, E2F3a or E2F3a/b knockdowns did not affect the S phase, suggesting that this activity is compensated for by the deregulation of E2F1 and E2F2. On the other hand, E2F1 or E2F3 knockdown in Her2+ cells alleviated centrosome amplification, which was not compensated for by the remaining E2F activators.
Our previous study showed that cyclin D1/Cdk4 and Nek2 are required for centrosome amplification in Her2+ cells (62) and in MCF10A cells expressing the H-RasG12V or H-RasG12V and c-Myc oncogenes (69), with a positive autoregulatory loop between Cdk4 and Nek2 (62). However, a major unanswered question was whether these targets are regulated via E2F-dependent transcriptional mechanisms. There is evidence that these genes are under E2F control, since E2F sites were reported in the cyclin D1 promoter, and this binding is required to induce cyclin D1 in Her2+ breast cancer cells (91). In addition, Plk4 promoter activity was increased by overexpressing E2Fs in A549 lung carcinoma cells (92), whereas Nek2 is under the repressive control of p107/p130/E2F4 (70). Exploration of the TCGA database showed that overexpression of E2F1, E2F2, and E2F3 in breast cancers strongly associates with the overexpression of the Plk4 and Nek2 transcripts, particularly in ER+ tumors. Expression data in the present manuscript in control cells and cells silenced for E2F1 or E2F3 shows that the E2Fs deregulate various molecules that control the cell/centrosome duplication cycles, including cyclin D1, Plk4, and Nek2, which correlate with centrosome amplification. These data strongly suggest a unique function of E2Fs in centrosome amplification in Her2+ cells by disturbing genes involved in both the cell and centrosome cycles (e.g., cyclin D1) and cells specific to the centrosome cycle (e.g., Nek2 and Plk4).
We demonstrated that Plk4 is under the direct control of the E2F activators in MCF10A and in the Her2+ breast cancer cell line SKBR3. A ChIP assay on putative E2F binding sites on Plk4 promoter in shE2F1 or shE2F3 cells indicated significantly decreased occupancy of E2F1, E2F2, or E2F3 protein binding on these sites compared to pLKO.1 control cells. On the other hand, mutating individual E2F sites in the Plk4 promoter significantly suppressed promoter activity in MCF10A and SKBR3, whereas no significant effects were observed in HCC1954 and JIMT1. However, although the overexpression of E2Fs resulted in elevated Plk4 protein levels, no major changes were observed in two Her2+ cells downregulated for E2F1 or E2F3; a potential explanation is that Plk4 protein levels are under tight ubiquitination and degradation control (93, 94). Another explanation is that given the deep deregulation of E2Fs in Her2+ breast cancer cells, the presence of the remaining E2Fs occupy the E2F sites in the promoter and increase basal transcription of the reporter promoter. Our data showed that whereas the Nek2 promoter activity is significantly elevated in Her2+ cells relative to MCF10A controls, deletion of the only canonical E2F site in the Nek2 proximal promoter did not affect promoter activity. One potential explanation for this lack of regulation by E2Fs is that another distal E2F site controls Nek2 transcription; however, there are no other E2F consensus sites in >3 kb of Nek2 promoter region. Other potential explanations are that noncanonical E2F sites are required for Nek2 transcription, that E2F regulates another transcription factor directly regulating Nek2 transcription, or that E2F controls other molecules responsible for the stability of the Nek2 transcript/protein levels. Further experimentation is needed to address the source of the E2F-dependent overexpression of Nek2.
Centrosome amplification is caused by various mechanisms such as premature centriole separation/duplication or binucleation (44, 95, 96). Here, we show that there is a close relationship between the E2F activators, centrosome amplification, and binucleation, since the overexpression of E2F1, E2F2, and E2F3a induced these phenotypes, whereas the silencing of E2F1 or E2F3 suppressed them. While we expected that silencing of E2F3 would suppress centrosome amplification by preventing defective cytokinesis in Her2+ cells, live imaging demonstrated that silenced E2F3 results in a significant increase in dead cells and delayed cytokinesis. On the other hand, the overexpression of Nek2 reversed the cytokinesis delays and blocks observed in shE2F3 cells. Our results suggest that silencing of E2F3 restricts unregulated mitosis and cytokinesis in Her2+ cells by restoring checkpoint controls.
We show that Nek2 is a mediator of centrosome amplification in Her2+ breast cancer cells downstream of E2F3. Even though our results indicate that centrosome amplification is a redundant function of the E2F activators, our findings are relevant to the understanding of breast carcinogenesis, since E2F1, E2F3, and Nek2 are frequently overexpressed in breast tumors (26, 97) and negatively impact the outcomes of survival (16, 56, 98). In fact, interference with Nek2 overexpression suppresses tumorigenesis of breast cancer cells (99). However, it is unknown whether the suppression of tumorigenesis relates to centrosome amplification or to the role of Nek2 in regulating various aspects of mitosis (100). Our work has future therapeutic implications, given the published data that deregulation of E2F represents the basis of resistance to various therapeutic agents. Whether the E2Fs signal the resistance via deregulating the cell cycle or through generating genetic diversity through signaling centrosome amplification-aneuploidy remains to be investigated. This research provides data showing that inhibiting the E2F activators and their centrosomal target Nek2 could prevent or reverse centrosome amplification and possibly the occurrence of aneuploidy in Her2+ breast tumors.
ACKNOWLEDGMENTS
We thank Ruth O'Regan and Rita Nahta for various Her2+ breast cancer cell lines. We thank Gustavo Leone and Andrew Fry for the E2F and Nek2 constructs, respectively. We thank Mihaela Marina and Jamie King for editing and helpful discussions, M. Marina for the generation and design of the GFP-Nek2, and Oskar Laur from the Emory DNA Custom Cloning Core construct for cloning GFP-Nek2. We thank the Emory University Integrated Cellular Imaging Microscopy Core and the Biostatistics core of the Winship Cancer Institute.
This research project was supported by R01 CA151521 from the National Institutes of Health.
Footnotes
Published ahead of print 5 May 2014
REFERENCES
- 1.Asp P, Acosta-Alvear D, Tsikitis M, van Oevelen C, Dynlacht BD. 2009. E2f3b plays an essential role in myogenic differentiation through isoform-specific gene regulation. Genes Dev. 23:37–53. 10.1101/gad.1727309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chong JL, Tsai SY, Sharma N, Opavsky R, Price R, Wu L, Fernandez SA, Leone G. 2009. E2f3a and E2f3b contribute to the control of cell proliferation and mouse development. Mol. Cell. Biol. 29:414–424. 10.1128/MCB.01161-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu T, Ghazaryan S, Sy C, Wiedmeyer C, Chang V, Wu L. 2012. Concomitant inactivation of Rb and E2f8 in hematopoietic stem cells synergizes to induce severe anemia. Blood 119:4532–4542. 10.1182/blood-2011-10-388231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishida S, Huang E, Zuzan H, Spang R, Leone G, West M, Nevins JR. 2001. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol. Cell. Biol. 21:4684–4699. 10.1128/MCB.21.14.4684-4699.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee EY, Yuan TL, Danielian PS, West JC, Lees JA. 2009. E2F4 cooperates with pRB in the development of extra-embryonic tissues. Dev. Biol. 332:104–115. 10.1016/j.ydbio.2009.05.541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meraldi P, Lukas J, Fry AM, Bartek J, Nigg EA. 1999. Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-cyclin A. Nat. Cell Biol. 1:88–93. 10.1038/10054 [DOI] [PubMed] [Google Scholar]
- 7.Saavedra HI, Maiti B, Timmers C, Altura R, Tokuyama Y, Fukasawa K, Leone G. 2003. Inactivation of E2F3 results in centrosome amplification. Cancer Cell 3:333–346. 10.1016/S1535-6108(03)00083-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trikha P, Sharma N, Opavsky R, Reyes A, Pena C, Ostrowski MC, Roussel MF, Leone G. 2011. E2F1–3 are critical for myeloid development. J. Biol. Chem. 286:4783–4795. 10.1074/jbc.M110.182733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen HZ, Tsai SY, Leone G. 2009. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat. Rev. Cancer 9:785–797. 10.1038/nrc2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu L, Timmers C, Maiti B, Saavedra HI, Sang L, Chong GT, Nuckolls F, Giangrande P, Wright FA, Field SJ, Greenberg ME, Orkin S, Nevins JR, Robinson ML, Leone G. 2001. The E2F1, E2F2, and E2F3 transcription factors are essential for cellular proliferation. Nature 414:457–462. 10.1038/35106593 [DOI] [PubMed] [Google Scholar]
- 11.Rabinovich A, Jin VX, Rabinovich R, Xu X, Farnham PJ. 2008. E2F in vivo binding specificity: comparison of consensus versus nonconsensus binding sites. Genome Res. 18:1763–1777. 10.1101/gr.080622.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wells J, Boyd KE, Fry CJ, Bartley SM, Farnham PJ. 2000. Target gene specificity of E2F and pocket protein family members in living cells. Mol. Cell. Biol. 20:5797–5807. 10.1128/MCB.20.16.5797-5807.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muller H, Bracken AP, Vernell R, Moroni MC, Christians F, Grassilli E, Prosperini E, Vigo E, Oliner JD, Helin K. 2001. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 15:267–285. 10.1101/gad.864201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu W, Giangrande PH, Nevins JR. 2004. E2Fs link the control of G1/S and G2/M transcription. EMBO J. 23:4615–4626. 10.1038/sj.emboj.7600459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nevins JR. 2001. The Rb/E2F pathway and cancer. Hum. Mol. Genet. 10:699–703. 10.1093/hmg/10.7.699 [DOI] [PubMed] [Google Scholar]
- 16.Baldini E, Camerini A, Sgambato A, Prochilo T, Capodanno A, Pasqualetti F, Orlandini C, Resta L, Bevilacqua G, Collecchi P. 2006. Cyclin A and E2F1 overexpression correlate with reduced disease-free survival in node-negative breast cancer patients. Anticancer Res. 26:4415–4421 [PubMed] [Google Scholar]
- 17.Banerjee D, Gorlick R, Liefshitz A, Danenberg K, Danenberg PC, Danenberg PV, Klimstra D, Jhanwar S, Cordon-Cardo C, Fong Y, Kemeny N, Bertino JR. 2000. Levels of E2F-1 expression are higher in lung metastasis of colon cancer as compared with hepatic metastasis and correlate with levels of thymidylate synthase. Cancer Res. 60:2365–2367 [PubMed] [Google Scholar]
- 18.De Meyer T, Bijsmans IT, Van de Vijver KK, Bekaert S, Oosting J, Van Criekinge W, van Engeland M, Sieben NL. 2009. E2Fs mediate a fundamental cell-cycle deregulation in high-grade serous ovarian carcinomas. J. Pathol. 217:14–20. 10.1002/path.2452 [DOI] [PubMed] [Google Scholar]
- 19.Ebihara Y, Miyamoto M, Shichinohe T, Kawarada Y, Cho Y, Fukunaga A, Murakami S, Uehara H, Kaneko H, Hashimoto H, Murakami Y, Itoh T, Okushiba S, Kondo S, Katoh H. 2004. Overexpression of E2F-1 in esophageal squamous cell carcinoma correlates with tumor progression. Dis. Esophagus 17:150–154. 10.1111/j.1442-2050.2004.00393.x [DOI] [PubMed] [Google Scholar]
- 20.Eymin B, Gazzeri S, Brambilla C, Brambilla E. 2001. Distinct pattern of E2F1 expression in human lung tumours: E2F1 is upregulated in small cell lung carcinoma. Oncogene 20:1678–1687. 10.1038/sj.onc.1204242 [DOI] [PubMed] [Google Scholar]
- 21.Imai MA, Oda Y, Oda M, Nakanishi I, Kawahara E. 2004. Overexpression of E2F1 associated with LOH at RB locus and hyperphosphorylation of RB in non-small-cell lung carcinoma. J. Cancer Res. Clin. Oncol. 130:320–326. 10.1007/s00432-003-0538-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iwamoto M, Banerjee D, Menon LG, Jurkiewicz A, Rao PH, Kemeny NE, Fong Y, Jhanwar SC, Gorlick R, Bertino JR. 2004. Overexpression of E2F-1 in lung and liver metastases of human colon cancer is associated with gene amplification. Cancer Biol. Ther. 3:395–399 [PubMed] [Google Scholar]
- 23.Lai R, Medeiros LJ, Coupland R, McCourty A, Brynes RK. 1998. Immunohistochemical detection of E2F-1 in non-Hodgkin's lymphomas: a survey of 124 cases. Modern Pathol. 11:457–463 [PubMed] [Google Scholar]
- 24.Suh DS, Yoon MS, Choi KU, Kim JY. 2008. Significance of E2F-1 overexpression in epithelial ovarian cancer. Int. J. Gynecol. Cancer 18:492–498. 10.1111/j.1525-1438.2007.01044.x [DOI] [PubMed] [Google Scholar]
- 25.Volante M, Croce S, Pecchioni C, Papotti M. 2002. E2F-1 transcription factor is overexpressed in oxyphilic thyroid tumors. Modern Pathol. 15:1038–1043. 10.1097/01.MP.0000028645.36632.A8 [DOI] [PubMed] [Google Scholar]
- 26.Wu L, de Bruin A, Wang H, Simmons T, Cleghorn W, Goldenberg LE, Sites E, Sandy A, Trimboli A, Fernandez SA, Eng C, Shapiro C, Leone G. 2013. Selective roles of E2Fs for ErbB2- and Myc-mediated mammary tumorigenesis. Oncogene 10.1038/onc.2013.511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andre F, Job B, Dessen P, Tordai A, Michiels S, Liedtke C, Richon C, Yan K, Wang B, Vassal G, Delaloge S, Hortobagyi GN, Symmans WF, Lazar V, Pusztai L. 2009. Molecular characterization of breast cancer with high-resolution oligonucleotide comparative genomic hybridization array. Clin. Cancer Res. 15:441–451. 10.1158/1078-0432.CCR-08-1791 [DOI] [PubMed] [Google Scholar]
- 28.Cooper CS, Nicholson AG, Foster C, Dodson A, Edwards S, Fletcher A, Roe T, Clark J, Joshi A, Norman A, Feber A, Lin D, Gao Y, Shipley J, Cheng SJ. 2006. Nuclear overexpression of the E2F3 transcription factor in human lung cancer. Lung Cancer 54:155–162. 10.1016/j.lungcan.2006.07.005 [DOI] [PubMed] [Google Scholar]
- 29.Foster CS, Falconer A, Dodson AR, Norman AR, Dennis N, Fletcher A, Southgate C, Dowe A, Dearnaley D, Jhavar S, Eeles R, Feber A, Cooper CS. 2004. Transcription factor E2F3 overexpressed in prostate cancer independently predicts clinical outcome. Oncogene 23:5871–5879. 10.1038/sj.onc.1207800 [DOI] [PubMed] [Google Scholar]
- 30.Reimer D, Hubalek M, Kiefel H, Riedle S, Skvortsov S, Erdel M, Hofstetter G, Concin N, Fiegl H, Muller-Holzner E, Marth C, Altevogt P, Zeimet AG. 2011. Regulation of transcription factor E2F3a and its clinical relevance in ovarian cancer. Oncogene 30:4038–4049. 10.1038/onc.2011.119 [DOI] [PubMed] [Google Scholar]
- 31.Wu Q, Hoffmann MJ, Hartmann FH, Schulz WA. 2005. Amplification and overexpression of the ID4 gene at 6p22.3 in bladder cancer. Mol. Cancer 4:16. 10.1186/1476-4598-4-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee KY, Lee JW, Nam HJ, Shim JH, Song Y, Kang KW. 2011. PI3-kinase/p38 kinase-dependent E2F1 activation is critical for pin1 induction in tamoxifen-resistant breast cancer cells. Mol. Cells 32:107–111. 10.1007/s10059-011-0074-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Louie MC, McClellan A, Siewit C, Kawabata L. 2010. Estrogen receptor regulates E2F1 expression to mediate tamoxifen resistance. Mol. Cancer Res. 8:343–352. 10.1158/1541-7786.MCR-09-0395 [DOI] [PubMed] [Google Scholar]
- 34.Louie MC, Zou JX, Rabinovich A, Chen HW. 2004. ACTR/AIB1 functions as an E2F1 coactivator to promote breast cancer cell proliferation and antiestrogen resistance. Mol. Cell. Biol. 24:5157–5171. 10.1128/MCB.24.12.5157-5171.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silver DP, Richardson AL, Eklund AC, Wang ZC, Szallasi Z, Li Q, Juul N, Leong CO, Calogrias D, Buraimoh A, Fatima A, Gelman RS, Ryan PD, Tung NM, De Nicolo A, Ganesan S, Miron A, Colin C, Sgroi DC, Ellisen LW, Winer EP, Garber JE. 2010. Efficacy of neoadjuvant cisplatin in triple-negative breast cancer. J. Clin. Oncol. 28:1145–1153. 10.1200/JCO.2009.22.4725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tordai A, Wang J, Andre F, Liedtke C, Yan K, Sotiriou C, Hortobagyi GN, Symmans WF, Pusztai L. 2008. Evaluation of biological pathways involved in chemotherapy response in breast cancer. Breast Cancer Res. 10:R37. 10.1186/bcr2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dean JL, Thangavel C, McClendon AK, Reed CA, Knudsen ES. 2010. Therapeutic CDK4/6 inhibition in breast cancer: key mechanisms of response and failure. Oncogene 29:4018–4032. 10.1038/onc.2010.154 [DOI] [PubMed] [Google Scholar]
- 38.Finn RS, Dering J, Conklin D, Kalous O, Cohen DJ, Desai AJ, Ginther C, Atefi M, Chen I, Fowst C, Los G, Slamon DJ. 2009. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. 11:R77. 10.1186/bcr2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andrechek ER. 2013. HER2/Neu tumorigenesis and metastasis is regulated by E2F activator transcription factors. Oncogene 10.1038/onc.2013.540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fujiwara K, Yuwanita I, Hollern DP, Andrechek ER. 2011. Prediction and genetic demonstration of a role for activator E2Fs in Myc-induced tumors. Cancer Res. 71:1924–1932. 10.1158/0008-5472.CAN-10-2386 [DOI] [PubMed] [Google Scholar]
- 41.Salisbury JL, D'Assoro AB, Lingle WL. 2004. Centrosome amplification and the origin of chromosomal instability in breast cancer. J. Mammary Gland Biol. Neoplasia 9:275–283. 10.1023/B:JOMG.0000048774.27697.30 [DOI] [PubMed] [Google Scholar]
- 42.Fukasawa K. 2007. Oncogenes and tumour suppressors take on centrosomes. Nat. Rev. Cancer 7:911–924. 10.1038/nrc2249 [DOI] [PubMed] [Google Scholar]
- 43.Harrison MK, Adon AM, Saavedra HI. 2011. The G1 phase Cdks regulate the centrosome cycle and mediate oncogene-dependent centrosome amplification. Cell Division 6:2. 10.1186/1747-1028-6-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fukasawa K. 2005. Centrosome amplification, chromosome instability and cancer development. Cancer Lett. 230:6–19. 10.1016/j.canlet.2004.12.028 [DOI] [PubMed] [Google Scholar]
- 45.Godinho SA, Kwon M, Pellman D. 2009. Centrosomes and cancer: how cancer cells divide with too many centrosomes. Cancer Metastasis Rev. 28:85–98. 10.1007/s10555-008-9163-6 [DOI] [PubMed] [Google Scholar]
- 46.Krzywicka-Racka A, Sluder G. 2011. Repeated cleavage failure does not establish centrosome amplification in untransformed human cells. J. Cell Biol. 194:199–207. 10.1083/jcb.201101073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tarapore P, Horn HF, Tokuyama Y, Fukasawa K. 2001. Direct regulation of the centrosome duplication cycle by the p53-p21Waf1/Cip1 pathway. Oncogene 20:3173–3184. 10.1038/sj.onc.1204424 [DOI] [PubMed] [Google Scholar]
- 48.Pihan GA, Purohit A, Wallace J, Knecht H, Woda B, Quesenberry P, Doxsey SJ. 1998. Centrosome defects and genetic instability in malignant tumors. Cancer Res. 58:3974–3985 [PubMed] [Google Scholar]
- 49.Zyss D, Gergely F. 2009. Centrosome function in cancer: guilty or innocent? Trends Cell Biol. 19:334–346. 10.1016/j.tcb.2009.04.001 [DOI] [PubMed] [Google Scholar]
- 50.D'Assoro AB, Barrett SL, Folk C, Negron VC, Boeneman K, Busby R, Whitehead C, Stivala F, Lingle WL, Salisbury JL. 2002. Amplified centrosomes in breast cancer: a potential indicator of tumor aggressiveness. Breast Cancer Res. Treat 75:25–34. 10.1023/A:1016550619925 [DOI] [PubMed] [Google Scholar]
- 51.Guo HQ, Gao M, Ma J, Xiao T, Zhao LL, Gao Y, Pan QJ. 2007. Analysis of the cellular centrosome in fine-needle aspirations of the breast. Breast Cancer Res. 9:R48. 10.1186/bcr1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lingle WL, Barrett SL, Negron VC, D'Assoro AB, Boeneman K, Liu W, Whitehead CM, Reynolds C, Salisbury JL. 2002. Centrosome amplification drives chromosomal instability in breast tumor development. Proc. Natl. Acad. Sci. U. S. A. 99:1978–1983. 10.1073/pnas.032479999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lingle WL, Lutz WH, Ingle JN, Maihle NJ, Salisbury JL. 1998. Centrosome hypertrophy in human breast tumors: implications for genomic stability and cell polarity. Proc. Natl. Acad. Sci. U. S. A. 95:2950–2955. 10.1073/pnas.95.6.2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schneeweiss A, Sinn HP, Ehemann V, Khbeis T, Neben K, Krause U, Ho AD, Bastert G, Kramer A. 2003. Centrosomal aberrations in primary invasive breast cancer are associated with nodal status and hormone receptor expression. Int. J. Cancer 107:346–352. 10.1002/ijc.11408 [DOI] [PubMed] [Google Scholar]
- 55.Finetti P, Cervera N, Charafe-Jauffret E, Chabannon C, Charpin C, Chaffanet M, Jacquemier J, Viens P, Birnbaum D, Bertucci F. 2008. Sixteen-kinase gene expression identifies luminal breast cancers with poor prognosis. Cancer Res. 68:767–776. 10.1158/0008-5472.CAN-07-5516 [DOI] [PubMed] [Google Scholar]
- 56.Marina M, Saavedra HI. 2014. Nek2 and Plk4: prognostic markers, drivers of breast tumorigenesis and drug resistance. Front. Biosci. 19:352–365. 10.2741/4212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eddy SF, Kane SE, Sonenshein GE. 2007. Trastuzumab-resistant HER2-driven breast cancer cells are sensitive to epigallocatechin-3 gallate. Cancer Res. 67:9018–9023. 10.1158/0008-5472.CAN-07-1691 [DOI] [PubMed] [Google Scholar]
- 58.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, Speed T, Spellman PT, DeVries S, Lapuk A, Wang NJ, Kuo WL, Stilwell JL, Pinkel D, Albertson DG, Waldman FM, McCormick F, Dickson RB, Johnson MD, Lippman M, Ethier S, Gazdar A, Gray JW. 2006. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10:515–527. 10.1016/j.ccr.2006.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Adon AM, Zeng X, Harrison MK, Sannem S, Kiyokawa H, Kaldis P, Saavedra HI. 2010. Cdk2 and Cdk4 regulate the centrosome cycle and are critical mediators of centrosome amplification in p53-null cells. Mol. Cell. Biol. 30:694–710. 10.1128/MCB.00253-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gillespie RF, Gudas LJ. 2007. Retinoid regulated association of transcriptional coregulators and the polycomb group protein SUZ12 with the retinoic acid response elements of Hoxa1, RARβ(2), and Cyp26A1 in F9 embryonal carcinoma cells. J. Mol. Biol. 372:298–316. 10.1016/j.jmb.2007.06.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hagen KR, Zeng X, Lee MY, Tucker Kahn S, Harrison Pitner MK, Zaky SS, Liu Y, O'Regan RM, Deng X, Saavedra HI. 2013. Silencing CDK4 radiosensitizes breast cancer cells by promoting apoptosis. Cell Division 8:10. 10.1186/1747-1028-8-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harrison Pitner MK, Saavedra HI. 2013. Cdk4 and nek2 signal binucleation and centrosome amplification in a her2+ breast cancer model. PLoS One 8:e65971. 10.1371/journal.pone.0065971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.TCGA-DCC. 2014. Firehose Dashboard-Stdata. The Cancer Genome Atlas-Data Coordinating Center, National Institutes of Health, Bethesda, MD: https://confluence.broadinstitute.org/display/GDAC/Dashboard-Stddata [Google Scholar]
- 64.Cancer Genome Atlas. 2012. Comprehensive molecular portraits of human breast tumours. Nature 490:61–70. 10.1038/nature11412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. 2012. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discovery 2:401–404. 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jonsson G, Staaf J, Olsson E, Heidenblad M, Vallon-Christersson J, Osoegawa K, de Jong P, Oredsson S, Ringner M, Hoglund M, Borg A. 2007. High-resolution genomic profiles of breast cancer cell lines assessed by tiling BAC array comparative genomic hybridization. Genes Chromosomes Cancer 46:543–558. 10.1002/gcc.20438 [DOI] [PubMed] [Google Scholar]
- 67.Worsham MJ, Pals G, Schouten JP, Miller F, Tiwari N, van Spaendonk R, Wolman SR. 2006. High-resolution mapping of molecular events associated with immortalization, transformation, and progression to breast cancer in the MCF10 model. Breast Cancer Res. Treat 96:177–186. 10.1007/s10549-005-9077-8 [DOI] [PubMed] [Google Scholar]
- 68.Gillett C, Fantl V, Smith R, Fisher C, Bartek J, Dickson C, Barnes D, Peters G. 1994. Amplification and overexpression of cyclin D1 in breast cancer detected by immunohistochemical staining. Cancer Res. 54:1812–1817 [PubMed] [Google Scholar]
- 69.Zeng X, Shaikh FY, Harrison MK, Adon AM, Trimboli AJ, Carroll KA, Sharma N, Timmers C, Chodosh LA, Leone G, Saavedra HI. 2010. The Ras oncogene signals centrosome amplification in mammary epithelial cells through cyclin D1/Cdk4 and Nek2. Oncogene 29:5103–5112. 10.1038/onc.2010.253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ren B, Takahashi CHY, Volkert T, Terragni J, Young RA, Dynlacht BD. 2002. E2F integrates cell cycle progression with DNA repair, replication, and G2/M checkpoints. Genes Dev. 16:245–256. 10.1101/gad.949802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Karanam S, Moreno CS. 2004. CONFAC: automated application of comparative genomic promoter analysis to DNA microarray datasets. Nucleic Acids Res. 32:W475–W484. 10.1093/nar/gkh353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rao JS, Karanam S, McCabe CD, Moreno CS. 2008. Genomic promoter analysis predicts functional transcription factor binding. Adv. Bioinformatics 2008:3698301–3698309. 10.1155/2008/369830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Saavedra HI, Fukasawa K, Conn CW, Stambrook PJ. 1999. MAPK mediates RAS-induced chromosome instability. J. Biol. Chem. 274:38083–38090. 10.1074/jbc.274.53.38083 [DOI] [PubMed] [Google Scholar]
- 74.Mukherjee M, Ge G, Zhang N, Edwards DG, Sumazin P, Sharan SK, Rao PH, Medina D, Pati D. 2013. MMTV-Espl1 transgenic mice develop aneuploid, estrogen receptor alpha (ERα)-positive mammary adenocarcinomas. Oncogene 10.1038/onc.2013.493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Saavedra HI, Knauf JA, Shirokawa JM, Wang J, Ouyang B, Elisei R, Stambrook PJ, Fagin JA. 2000. The RAS oncogene induces genomic instability in thyroid PCCL3 cells via the MAPK pathway. Oncogene 19:3948–3954. 10.1038/sj.onc.1203723 [DOI] [PubMed] [Google Scholar]
- 76.Okuda M, Horn HF, Tarapore P, Tokuyama Y, Smulian AG, Chan PK, Knudsen ES, Hofmann IA, Snyder JD, Bove KE, Fukasawa K. 2000. Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell 103:127–140. 10.1016/S0092-8674(00)00093-3 [DOI] [PubMed] [Google Scholar]
- 77.Tokuyama Y, Horn HF, Kawamura K, Tarapore P, Fukasawa K. 2001. Specific phosphorylation of nucleophosmin on Thr(199) by cyclin-dependent kinase 2-cyclin E and its role in centrosome duplication. J. Biol. Chem. 276:21529–21537. 10.1074/jbc.M100014200 [DOI] [PubMed] [Google Scholar]
- 78.Fry AM, Meraldi P, Nigg EA. 1998. A centrosomal function for the human Nek2 protein kinase, a member of the NIMA family of cell cycle regulators. EMBO J. 17:470–481. 10.1093/emboj/17.2.470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nelsen CJ, Kuriyama R, Hirsch B, Negron VC, Lingle WL, Goggin MM, Stanley MW, Albrecht JH. 2005. Short term cyclin D1 overexpression induces centrosome amplification, mitotic spindle abnormalities, and aneuploidy. J. Biol. Chem. 280:768–776 [DOI] [PubMed] [Google Scholar]
- 80.Rogers GC, Rusan NM, Roberts DM, Peifer M, Rogers SL. 2009. The SCF Slimb ubiquitin ligase regulates Plk4/Sak levels to block centriole reduplication. J. Cell Biol. 184:225–239. 10.1083/jcb.200808049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Starita LM, Machida Y, Sankaran S, Elias JE, Griffin K, Schlegel BP, Gygi SP, Parvin JD. 2004. BRCA1-dependent ubiquitination of gamma-tubulin regulates centrosome number. Mol. Cell. Biol. 24:8457–8466. 10.1128/MCB.24.19.8457-8466.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen Z, Indjeian VB, McManus M, Wang L, Dynlacht BD. 2002. CP110, a cell cycle-dependent CDK substrate, regulates centrosome duplication in human cells. Dev. Cell 3:339–350. 10.1016/S1534-5807(02)00258-7 [DOI] [PubMed] [Google Scholar]
- 83.Fisk HA, Winey M. 2001. The mouse Mps1p-like kinase regulates centrosome duplication. Cell 106:95–104. 10.1016/S0092-8674(01)00411-1 [DOI] [PubMed] [Google Scholar]
- 84.Johnson DG, Schwarz JK, Cress WD, Nevins JR. 1993. Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature 365:349–352. 10.1038/365349a0 [DOI] [PubMed] [Google Scholar]
- 85.Paulson QX, McArthur MJ, Johnson DG. 2006. E2F3a stimulates proliferation, p53-independent apoptosis and carcinogenesis in a transgenic mouse model. Cell Cycle 5:184–190. 10.4161/cc.5.2.2307 [DOI] [PubMed] [Google Scholar]
- 86.Pierce AM, Schneider-Broussard R, Gimenez-Conti IB, Russell JL, Conti CJ, Johnson DG. 1999. E2F1 has both oncogenic and tumor-suppressive properties in a transgenic model. Mol. Cell. Biol. 19:6408–6414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pierce AM, Schneider-Broussard R, Philhower JL, Johnson DG. 1998. Differential activities of E2F family members: unique functions in regulating transcription. Mol. Carcinog. 22:190–198. [DOI] [PubMed] [Google Scholar]
- 88.Humbert PO, Verona R, Trimarchi JM, Rogers C, Dandapani S, Lees JA. 2000. E2f3 is critical for normal cellular proliferation. Genes Dev. 14:690–703 [PMC free article] [PubMed] [Google Scholar]
- 89.Leone G, DeGregori J, Yan Z, Jakoi L, Ishida S, Williams RS, Nevins JR. 1998. E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes Dev. 12:2120–2130. 10.1101/gad.12.14.2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wu L, Timmers C, Maiti B, Saavedra HI, Sang L, Chong GT, Nuckolls F, Giangrande P, Wright FA, Field SJ, Greenberg ME, Orkin S, Nevins JR, Robinson ML, Leone G. 2001. The E2F1-3 transcription factors are essential for cellular proliferation. Nature 414:457–462. 10.1038/35106593 [DOI] [PubMed] [Google Scholar]
- 91.Lee RJ, Albanese C, Fu M, D'Amico M, Lin B, Watanabe G, Haines GK, III, Siegel PM, Hung MC, Yarden Y, Horowitz JM, Muller WJ, Pestell RG. 2000. Cyclin D1 is required for transformation by activated Neu and is induced through an E2F-dependent signaling pathway. Mol. Cell. Biol. 20:672–683. 10.1128/MCB.20.2.672-683.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tategu M, Nakagawa H, Sasaki K, Yamauchi R, Sekimachi S, Suita Y, Watanabe N, Yoshid K. 2008. Transcriptional regulation of human polo-like kinases and early mitotic inhibitor. J. Genet. Genomics 35:215–224. 10.1016/S1673-8527(08)60030-2 [DOI] [PubMed] [Google Scholar]
- 93.Holland AJ, Lan W, Niessen S, Hoover H, Cleveland DW. 2010. Polo-like kinase 4 kinase activity limits centrosome overduplication by autoregulating its own stability. J. Cell Biol. 188:191–198. 10.1083/jcb.200911102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Puklowski A, Homsi Y, Keller D, May M, Chauhan S, Kossatz U, Grunwald V, Kubicka S, Pich A, Manns MP, Hoffmann I, Gonczy P, Malek NP. 2011. The SCF-FBXW5 E3-ubiquitin ligase is regulated by PLK4 and targets HsSAS-6 to control centrosome duplication. Nat. Cell Biol. 13:1004–1009. 10.1038/ncb2282 [DOI] [PubMed] [Google Scholar]
- 95.Fukasawa K, Choi T, Kuriyama R, Rulong S, Vande Woude GF. 1996. Abnormal centrosome amplification in the absence of p53. Science 271:1744–1747. 10.1126/science.271.5256.1744 [DOI] [PubMed] [Google Scholar]
- 96.Tarapore P, Okuda M, Fukasawa K. 2002. A mammalian in vitro centriole duplication system: evidence for involvement of CDK2/cyclin E and nucleophosmin/B23 in centrosome duplication. Cell Cycle 1:75–81 [PubMed] [Google Scholar]
- 97.Hayward DG, Clarke RB, Faragher AJ, Pillai MR, Hagan IM, Fry AM. 2004. The centrosomal kinase Nek2 displays elevated levels of protein expression in human breast cancer. Cancer Res. 64:7370–7376. 10.1158/0008-5472.CAN-04-0960 [DOI] [PubMed] [Google Scholar]
- 98.Han S, Park K, Bae BN, Kim KH, Kim HJ, Kim YD, Kim HY. 2003. E2F1 expression is related with the poor survival of lymph node-positive breast cancer patients treated with fluorouracil, doxorubicin, and cyclophosphamide. Breast Cancer Res. Treat. 82:11–16. 10.1023/B:BREA.0000003843.53726.63 [DOI] [PubMed] [Google Scholar]
- 99.Tsunoda N, Kokuryo T, Oda K, Senga T, Yokoyama Y, Nagino M, Nimura Y, Hamaguchi M. 2009. Nek2 as a novel molecular target for the treatment of breast carcinoma. Cancer Sci. 100:111–116. 10.1111/j.1349-7006.2008.01007.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.O'Regan L, Blot J, Fry AM. 2007. Mitotic regulation by NIMA-related kinases. Cell Division 2:25. 10.1186/1747-1028-2-25 [DOI] [PMC free article] [PubMed] [Google Scholar]