Abstract
The actin cytoskeleton is essential for cell adhesion and migration, functions important for tumor invasion. In addition to binding N-WASP/WASP, WIP binds and stabilizes F-actin. WIP−/− fibroblasts were used to test the role of WIP in F-actin function. WIP−/− cells had defective focal adhesion (FA), stress fiber assembly, and adherence to substrates, functions that were restored by transduction of wild-type WIP. Protein and mRNA levels of several FA constituents regulated by the myocardin-related transcription factor (MRTF)–serum response factor (SRF) transcription factor complex were reduced in WIP−/− fibroblasts. The level of G-actin, which sequesters MRTF in the cytoplasm, was increased, and nuclear localization of MRTF-A and SRF was reduced, in WIP−/− fibroblasts. Transfection of an MRTF-A mutant that constitutively translocates to the nucleus or transfection of constitutively active SRF restored FA and stress fiber assembly. Fibroblasts from knock-in mice expressing a WIP mutant that fails to bind actin phenocopied WIP−/− fibroblasts. Thus, WIP is a novel regulator of FA assembly and cell adhesion.
INTRODUCTION
A dynamic actin cytoskeleton is essential for many cellular functions, including adhesion, motility, endo/exocytosis, and cytokinesis. Actin cytoskeleton remodeling requires de novo actin assembly. The most important initiator of actin assembly in cells is the Arp2–Arp3 (Arp2/3) complex, a ubiquitously expressed heteropolymer of seven proteins. The Wiskott-Aldrich syndrome protein (WASP) and its homologue N-WASP are important activators of the Arp2/3 complex. In resting cells, WASP and N-WASP are kept in an autoinhibited inactive state. A number of proteins, including Cdc42 (1, 2), Src homology 3 (SH3) domain-containing proteins such as Nck (3), Grb2 (4), cortactin (5), and Toca-1 (6), have been shown to activate WASP/N-WASP. To date, only one protein, WASP-interacting protein (WIP), encoded by the Wipf1 gene, has been shown to be a physiological inhibitor of WASP/N-WASP-mediated actin assembly. In addition to regulating WASP/N-WASP activity, WIP is critical for the stability of WASP, but not for that of N-WASP (7).
The WASP-interacting protein WIP stabilizes WASP in T cells, as evidenced by the observation that WASP levels, but not WASP mRNA levels, are severely reduced in T cells from Wipf1−/− (WIP−/−) mice (7). Similarly, WASP levels, but not WASP mRNA levels, are severely reduced in T cells from Wiskott-Aldrich syndrome (WAS) patients with missense mutations in WASP that disrupt WIP binding (8). WIP also binds to actin (9, 10). This interaction stabilizes filamentous actin (F-actin), as demonstrated by the observation that the addition of recombinant WIP to F-actin inhibits its depolymerization in vitro (10). A purified glutathione S–transferase (GST)-WIP1–127 fusion protein interacts with purified actin in vitro (9), demonstrating a direct interaction. This interaction is mediated by a 12-amino acid (aa) sequence (aa 43 to 54) in the WASP homology 2 (WH2) domain of the verproline homology (VH) region of WIP, since deletion of this sequence abolishes the ability of GST-WIP1–127 to pull down actin. The WIP sequence includes the KLKK motif (aa 45 to 48), previously shown to mediate the binding of thymosin β4 to actin (11). Addition of recombinant WIP inhibits the depolymerization of purified F-actin in vitro (10). Deletion of WIP results in diminished F-actin levels in T cells from WIP−/− mice (12), while overexpression of WIP in the B cell line BJAB increases the cellular F-actin content (13).
Serum response factor (SRF) is a master regulator of the actin-based cytoskeleton and the cellular contractile apparatus (14). The SRF cofactors myocardin-related transcription factors A (MRTF-A; also called Mal) and B (MRTF-B) are critical for the induction of a coordinated set of genes whose protein products are important for cell adhesion and actin cytoskeleton assembly. In resting cells, globular actin (G-actin) sequesters MRTFs in the cytoplasm (15, 16). A shift in the G-actin↔F-actin equilibrium toward F-actin results in the dissociation of MRTF proteins from G-actin, allowing them to migrate into the nucleus, where they bind and activate SRF. The MRTF-SRF complex induces the transcription of genes that encode proteins critical for the assembly of focal adhesions (FAs) and the integrity of the actin cytoskeleton (14, 17).
We used fibroblasts from WIP−/− mice, as well as from knock-in mice that express a WIP mutant with impaired ability to bind actin, to test the hypothesis that the binding of WIP to actin regulates the activity of the MRTF-SRF complex, thereby regulating FA and stress fiber assembly.
MATERIALS AND METHODS
Reagents and antibodies.
Laminin and fibronectin were purchased from MP Biochemicals. Toluidine blue O was purchased from Fluka. Protease inhibitor cocktail and phosphatase inhibitor cocktail were obtained from G-Biosciences. Primary antibodies against various proteins were purchased as follows. Rabbit monoclonal antibodies against focal adhesion kinase (FAK), phospho-FAK, and vinculin were obtained from Cell Signaling Technology. Antibodies against actin, vinculin (MAb hvin1), and zyxin were purchased from Sigma. The following reagents were supplied as indicated: fluorescein isothiocyanate (FITC)-conjugated β1 integrin and tetramethyl rhodamine isothiocyanate (TRITC)-conjugated α integrin by eBiosciences, TRITC-conjugated α6 integrin and Alexa Fluor 488-conjugated anti-VP16 by Santa Cruz Biotechnology, monoclonal antibodies against α- and β-tubulin by MP Biochemicals, and anti-MRTF-A (anti MKL1) by Aviva. Alexa Fluor 488 (green)- or Alexa Fluor 580 (red)-conjugated secondary antibodies against mouse or rabbit antibodies were obtained from Molecular Probes.
Plasmids.
MRTF-A (MAL) and MRTF-AXXX (MALxxx) in the pEF vector were obtained from Richard Treisman. SRF-VP16 in pUC12 was also obtained from Richard Treisman. The insert was excised and was recloned into pCMV-Tag2B. Most of the DNA binding MADS (MCM1, Agamous, Deficiens, SRF) domain of SRF is encoded between nucleotides 421 and 732 (18). SRF with this region deleted (referred to below as SRFΔM-VP16) was constructed by PCR amplifying the sequence from nucleotide 1 to nucleotide 421 of the SRF gene with a BamHI site added to the 5′ end and a BglII site added to the 3′ end. The amplicon was digested with BamHI and BglII and was cloned into BamHI-BglII-digested SRF-VP16 in pUC12 to produce SRFΔM-VP16. The entire insert was excised and was recloned into pCMV-Tag2B.
Tissue culture conditions. (i) General growth conditions.
Cells were cultured in Dulbecco's modified Eagle medium (DMEM) containing 15% fetal calf serum (FCS), since a large amount of serum was required for long-term viability of WIP−/− fibroblasts. The day before all experiments, the cells were transferred to DMEM containing 10% FCS.
(ii) Establishment of WIP−/− mouse lung fibroblasts.
Lungs from neonatal mice (wild type [WT], WIP−/−, or WIP-ΔABD) were minced and were seeded in 6-well tissue culture clusters in DMEM containing 15% FCS. After a week, when fibroblasts were observed to crawl out of the seeded tissue, the lung tissue was removed, and the fibroblasts were allowed to grow until 90% confluent. The cells from one well were trypsinized, replated onto three wells of a 6-well cluster, and designated passage zero. When cells became 80% confluent, they were split and expanded. Mouse cells have been reported to undergo “spontaneous transformation” (19). Such “transformants” were observed around passage 10 to 12, and cell lines were generated from these cells by limiting dilution to obtain a homogenous cell population. Cells used in experiments had >25 passages. Experimental protocols using mice were approved by the Children's Hospital Animal Care and Use Committee.
Bright-field time-lapse microscopy.
WT and WIP−/− cells were plated in 6-well clusters at a density of 25,000 cells/well. Cells were allowed to attain 40 to 50% confluence, and time-lapse photomicrographs were taken every 10 s for 15 min using a Nikon Ti inverted microscope equipped with a warm stage.
Lentiviral preparations and cell transduction.
WIP-expressing lentiviral particles cloned under the control of the cytomegalovirus (CMV) promoter in the bicistronic lentiviral expression vector pLOC were purchased from Open Biosystems. The vector had the marker that localized Turbo-GFPnuc predominantly in the nucleus and was expressed independently of WIP using an internal ribosome entry site (IRES). The vector also has blasticidin selection to isolate cells that stably express the transgene. Similarly cloned lentiviral particles expressing glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were used as a control.
WIP−/− cells were treated with 0.5 μg/ml Polybrene for 30 min and were infected with lentivirus at a multiplicity of infection (MOI) of 10. WIP- or GAPDH-expressing cells were selected by applying blasticidin selection (10 μg/ml) for 1 week, with a medium change every day. The fraction of blasticidin-selected cells that was positive for green fluorescent protein (GFP) was quantified by flow cytometry (see Fig. S2 in the supplemental material).
For immunofluorescence, 75% confluent cells were harvested by trypsinization, and 40,000 cells were plated on 13-mm coverslips in 24-well plates and were cultured for an additional 14 h for optimal FA assembly. Cells were fixed and stained as described below (see “Immunofluorescence”).
Western blotting.
Adherent cells were lysed at 4°C on culture plates using lysis buffer (50 mM Tris, 100 mM NaCl, 1% Triton X-100, 0.5% Sarkosyl plus protease inhibitor cocktail and phosphatase inhibitor cocktail [pH 7.4]). Lysates were clarified by spinning at 10,000 × g, and the supernatants were frozen in aliquots at −80°C. Lysates containing equal amounts of protein were loaded onto gels, transferred to supported nitrocellulose membranes, and probed with relevant antibodies using standard protocols. Membranes were developed using enhanced chemiluminescence (ECL) (PerkinElmer).
Quantitative PCR.
Total RNA was isolated from trypsinized cells using an RNeasy kit (Qiagen). Reverse transcription-PCR (RT-PCR) was performed by using an iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). 6-Carboxyfluorescein (FAM)-labeled specific TaqMan primers were purchased from Applied Biosystems. Quantitative PCRs were run on an ABI Prism 7300 sequence detection system platform (Applied Biosystems). The housekeeping gene β2-microglobulin was used as a control. The relative gene expression in the different samples was determined using the method described in reference 20. Quantities of all targets in test samples were normalized to the corresponding β2-microglobulin levels.
Transcription profile analysis.
RNA was extracted from fed and serum-starved (0.3% serum for 48 h) WT and WIP−/− fibroblasts by using the RNeasy kit (Qiagen). The mouse genome 430 AB Affymetrix GeneChip, containing more than 45,000 transcripts, was used for transcriptional profiling. Microarray analysis was conducted as described previously, using in vitro-transcribed biotin-labeled cRNA, by hybridization of cRNA with the mouse genome 430 AB Affymetrix GeneChip and scanning of image output files (21). The quality of scanned array images was determined on the basis of background values, percentage of present calls, scaling factors, and the 3′:5′ ratios of β-actin and GAPDH by using the arrayQualityMetrics package for R (22). The raw probe level data were normalized using a smoothing-spline invariant set method. The signal value for each transcript was summarized by using a perfect match (PM)-only-based signal modeling algorithm to reduce false-positive results and by filtering out transcripts that were absent in all samples (23). In comparisons of two groups of samples to identify differentially expressed genes, if the 90% lower confidence bound (LCB) of the fold change (FC) between the two groups was above 1.5, the gene was considered differentially expressed. The LCB is a stringent estimate of FC and has been shown to be the better-ranking statistic. It has been suggested that the criterion of an LCB above 1.5 most likely selects genes with an “actual” fold change of at least 2 in expression (23, 24).
Adhesion assay.
The adhesion assay was performed essentially as described previously (25) with minor modifications. One hundred microliters of freshly trypsinized fibroblasts (WT, WIP−/−, or WIPΔABD) in warm DMEM plus 10% FCS at a density of 106 cells/ml was plated in sextuplicate on 96-well flat-bottom culture plates (Nunc) and was allowed to adhere for 30 min at 37°C. Nonadherent cells were washed out using warm DMEM containing no FCS. Cells were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) at room temperature for 30 min and were stained with toluidine blue O (1% in PBS) for 60 min at room temperature. The plate was washed extensively with water (5 to 6 times) to remove excess stain and was air dried; the cell-bound dye was solubilized using 2% SDS; and the optical density was read at 620 nm.
FAK activation.
WT or WIP−/− cells were trypsinized, washed, replated on 24-well clusters in warm DMEM plus 10% FCS, and incubated at 37°C for 30, 60, 90, and 120 min. At the end of the incubation, plates were rinsed with DMEM to remove nonadherent cells, and lysates were as prepared as described above.
Transfection.
MRTF-A, MRTF-AXXX, SRF-VP16, and SRFΔM-VP16 were introduced into WIP−/− fibroblasts by transfection using Fugene HD (Promega). Briefly, 3 μl of Fugene HD transfection reagent was diluted into 100 μl of DMEM with no FCS and was incubated for 5 min at room temperature, and 500 μg of plasmid DNA was added, mixed, and incubated at room temperature for an additional 15 min. The entire transfection mixture was added dropwise to WIP−/− fibroblasts and was incubated at 37°C for 48 h for gene expression. The transfection efficiency was about 0.5%.
Nucleofection.
One microgram of WIP cloned into the pcDNA6 vector was nucleofected into 0.75 × 106 fibroblasts with the Amaxa Nucleofector kit (Lonza) using the A-23 program, and fibroblasts were plated on 6-well clusters. After overnight incubation, dead cells and debris were washed out by rinsing the adherent cells with serum-free DMEM; then the cells were trypsinized, plated onto 13-mm coverslips, and allowed to adhere for 15 h, and MRTF-A was visualized by incubation with an anti MRTF-A antibody (anti-MKL; Aviva) followed by an Alexa Fluor 588-conjugated anti-rabbit antibody. Coverslips were mounted as described in the next section.
Immunofluorescence.
Cells (104) were plated on washed glass coverslips (13 mm) and were allowed to adhere and spread for 14 h in DMEM plus 10% FCS. The coverslips were washed with PBS to remove dead cells and debris, fixed for 30 min in 4% paraformaldehyde in PBS (EM Sciences), washed again, and blocked with 10% FCS in PBS for 60 min. Coverslips were incubated with the appropriate primary antibody for 45 min at 37°C, washed, and incubated with the relevant secondary antibody for 45 min at 37°C. To visualize actin filaments, phalloidin conjugated to Alexa Fluor 488 or Alexa Fluor 588 was used (Molecular Probes). Coverslips were washed, mounted on slides by using ProLong Gold antifade agent with 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen), air dried, and sealed with nail polish. Immunofluorescence observation, image capture, and analysis were performed using a Nikon Eclipse E800 or Nikon Ti 90 microscope and NIS Elements software. A minimum of 10 cells from 10 different fields were observed for each condition in each experiment.
Flow cytometry.
To analyze integrin expression, cells were dislodged using an enzyme-free cell dissociation buffer (GIBCO). Single-cell suspensions were stained using antibodies against α5, α6, and β1 integrin (Biolegend) and were analyzed by flow cytometry using an LSRFortessa flow cytometer (Becton, Dickinson). The data were analyzed using FlowJo software (TreeStar Inc.).
Intracellular F-actin and G-actin contents were measured in trypsinized single-cell suspensions. Cells were washed, fixed, permeabilized in Cytofix/Cytoperm buffer (BD Biosciences), and stained either with TRITC-conjugated phalloidin (for F-actin) or with FITC-conjugated DNase I (Sigma). Data were analyzed using FlowJo as described above.
SRF activity assay.
A lentivirus-based SRE luciferase reporter assay kit was purchased from Qiagen (Cignal SRE [luc] assay kit). The kit contains the firefly luciferase gene under the control of SRE (serum response element) promoter elements and an internal standard of Renilla luciferase under the control of the CMV promoter. Cell lines stably transfected with the SRE-luciferase reporter were generated by infecting WIP−/− fibroblasts with SRE-firefly luciferase and Renilla luciferase in a 40:1 ratio, and stable transfectants were selected with puromycin (3 μg/ml). Antibiotic-selected fibroblasts were plated at 5,000 per well on 96-well plates and were allowed to rest overnight. The medium was replaced with fresh medium the next day, and after a 2-h incubation, the medium was aspirated; the cells were washed three times with PBS; the cells were lysed; and SRF activity was assayed using the Dual-Luciferase assay kit (Promega) as described by the manufacturer. The light signal was read using a Paradigm luminescence reader (Beckman). The result was expressed as the ratio of firefly luciferase to Renilla luciferase.
RESULTS
WIP−/− fibroblasts have impaired FAs and actin stress fibers.
We demonstrated previously that WIP−/− T cells have a low F-actin content and a disrupted actin cytoskeleton (12). However, because WIP stabilizes WASP in T cells (7), WASP levels are very much diminished in WIP−/− T cells, making it impossible to ascribe the cytoskeletal abnormalities in these cells to WIP deficiency alone. WASP is not expressed in fibroblasts, and WIP is not necessary for the stability of N-WASP in fibroblasts, since the levels of N-WASP in lung fibroblasts derived from WIP−/− mice and their WT littermates are comparable (Fig. 1A). We therefore used fibroblasts to examine the role of WIP in the structure and function of the actin cytoskeleton. In all studies, we compared two fibroblast lines each from two WIP−/− mice and two WT littermates. Similar results were obtained for each pair. We show representative results for single lines and present the pooled data obtained from two lines.
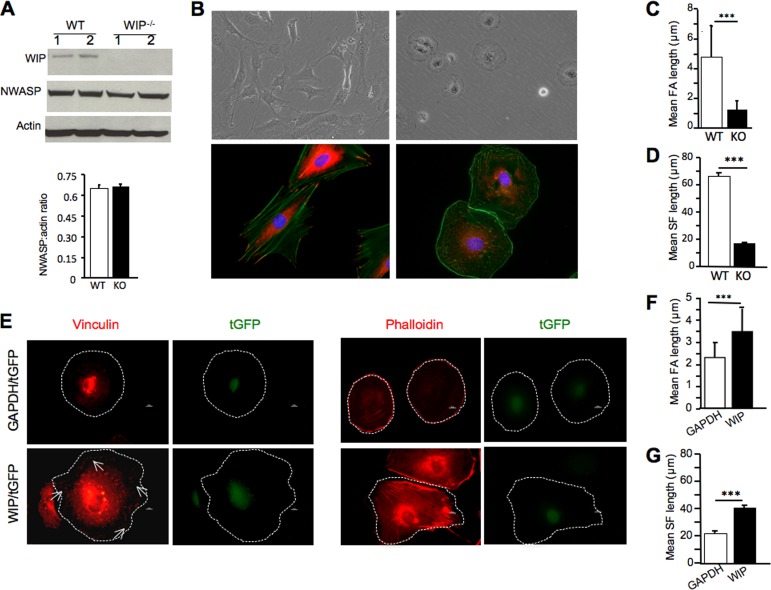
FIG 1.
Decreased FAs and actin stress fibers in WIP−/− fibroblasts. (A) Immunoblot analysis (top) and quantitation (bottom) of N-WASP in WIP−/− (knockout [KO])and WT fibroblasts. (B) Representative bright-field differential interference contrast images (top) and immunofluorescence images (bottom) of WIP−/− (right) and WT (left) fibroblasts. Fibroblasts were stained with vinculin (red) and actin (green), as well as with DAPI (blue). (C and D) Mean lengths of FAs (C) and actin stress fibers (SF) (D) in WIP−/− and WT fibroblasts. (E) Representative immunofluorescence images of WIP−/− fibroblasts transduced with a lentivirus expressing either WIP and turbo GFP (tGFP) or GAPDH and tGFP and then stained for vinculin or phalloidin (red) and with DAPI (blue). The outlines of the cells are indicated by white dotted lines. Arrows point to focal adhesions. (F and G) Mean lengths of FAs (F) and actin stress fibers (G) in transduced WIP−/− fibroblasts. The data represent three independent experiments, each performed on two fibroblast lines from WIP−/− mice and two from WT controls. In panels A, C, D, F, and G, bars represent means; error bars, standard deviations. ***, P < 0.001.
WT fibroblasts cultured in 10% FCS and plated on plastic assumed a polygonal shape and extended robust ruffles (Fig. 1B; see also Movie S1 in the supplemental material). In contrast, WIP−/− fibroblasts assumed a rounded shape, and the ruffles they extended were attenuated (see Movie S2 in the supplemental material). This finding is consistent with our previous observation of decreased ruffle formation in murine fibroblasts microinjected with an anti-WIP antibody (10). The shape of adherent fibroblasts is determined by their spreading, which depends on FAs (26, 27). The morphology and distribution of FAs were examined by staining for the FA marker vinculin. WT fibroblasts exhibited elongated FAs that anchored long actin stress fibers that ran throughout the cell length (Fig. 1B). In contrast, FAs in WIP−/− fibroblasts were significantly reduced in length and anchored significantly shorter stress fibers that were irregularly aligned (Fig. 1B to D). Similar findings were obtained using paxillin staining to identify FAs (see Fig. S1 in the supplemental material). Lentiviral transduction of WIP, but not of GAPDH (used as a control), restored WIP levels in WIP−/− fibroblasts (see Fig. S2 in the supplemental material) and corrected their defective FA and stress fiber formation (Fig. 1E and F). These results suggest that WIP is essential for the formation of FAs and stress fibers in fibroblasts.
WIP−/− fibroblasts exhibit defective adherence to substrates.
FAs are important for the adherence of fibroblasts to substrates (28). The ability of WIP−/− fibroblasts to adhere to plastic and to laminin- or fibronectin-coated surfaces was significantly impaired compared to that of WT fibroblasts (Fig. 2A). Integrin α5β1 is implicated in the serum-dependent binding of fibroblasts to plastic, and integrins α5β1 and α6β1 are implicated in binding to fibronectin and laminin (29, 30). Fluorescence-activated cell sorter (FACS) analysis demonstrated that the surface expression of α5, α6, and β1 integrins was comparable in WIP−/− and WT fibroblasts (Fig. 2B). Thus, the failure of WIP−/− fibroblasts to adhere to substrates was not due to impaired expression of integrins on their surfaces.
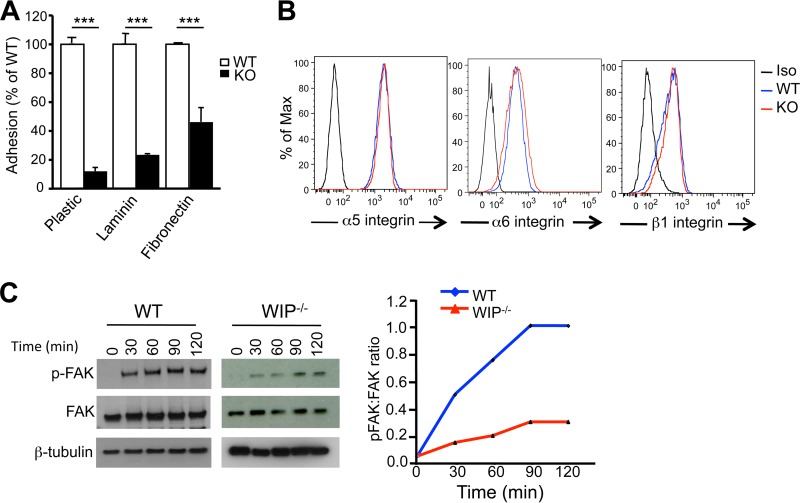
FIG 2.
Defective adherence and adhesion-driven phosphorylation of FAK in WIP−/− fibroblasts. (A) Adherence of WIP−/− and WT fibroblasts to substrates. Bars represent means; error bars, standard deviations. ***, P < 0.001. (B) FACS analysis of surface expression of α5, α6, and β1 integrins in WIP−/− and WT fibroblasts. (C) Representative immunoblot and quantitative analysis of FAK phosphorylation (p-FAK) following plating of WIP−/− and WT fibroblasts on plastic. The immunoblot for β-tubulin, used as a loading control, was developed using a different film, which explains the different background. The quantitative results in panel C are expressed as the ratio of the intensity of phosphoprotein to total protein. The data in panels A and B are representative of results of three independent experiments, and the data in panel C represent the results of two independent experiments, each performed on two fibroblast lines from WIP−/− mice and two from WT controls.
The clustering of integrins following their binding to substrates results in the recruitment of the nonreceptor tyrosine kinase FA kinase (FAK) and its activation by autophosphorylation at residue tyrosine 397 (Y397) (31). Enzymatically active FAK phosphorylates several FA proteins, a step critical for FA assembly (32). Plating of WT fibroblasts on plastic resulted in time-dependent phosphorylation of FAK in WT fibroblasts (Fig. 2C and D). The level of FAK phosphorylation was markedly lower in WIP−/− fibroblasts than in WT fibroblasts, in agreement with the attenuation of FAs in these cells.
Decreased expression of FA proteins in WIP−/− fibroblasts.
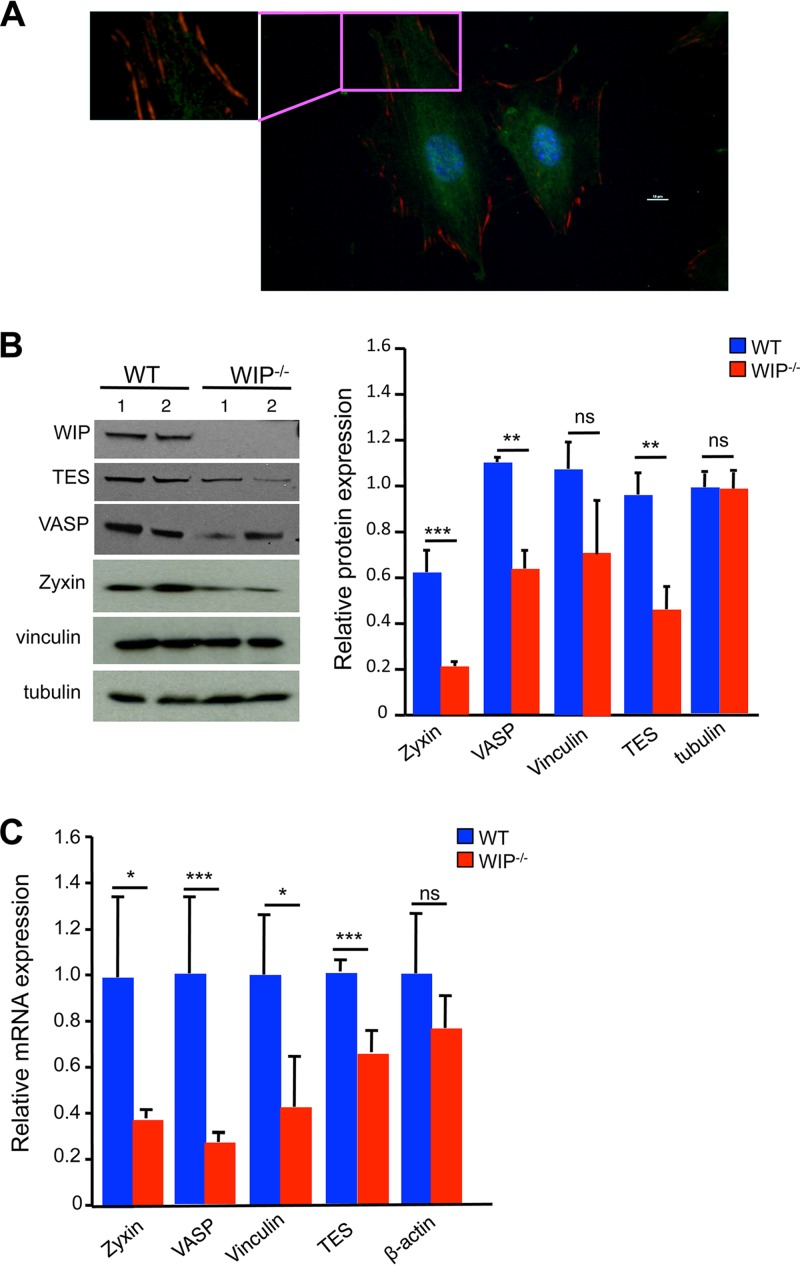
A potential explanation for the disruption of FAs in WIP−/− fibroblasts is that WIP is an essential component of FAs. Immunofluorescence examination of WT fibroblasts revealed that WIP does not localize to FAs (Fig. 3A). We tested the hypothesis that WIP deficiency results in decreased expression of FA proteins. Immunoblot analysis revealed that the levels of zyxin, vasodilator-associated phosphoprotein (VASP), and testin lim3 (TES) were significantly lower in WIP−/− fibroblasts than in WT controls (Fig. 3B). The level of vinculin was lower in WIP−/− fibroblasts than in WT fibroblasts, but the difference did not reach statistical significance. The levels of the non-FA protein tubulin were comparable in WIP−/− and WT fibroblasts (Fig. 3B). Quantitative PCR analysis revealed significantly lower zyxin, VASP, vinculin, and TES mRNA levels in WIP−/− fibroblasts than in WT controls (Fig. 3C). These results suggest that decreased expression of genes that encode key FA proteins contributes to impaired FA assembly in WIP−/− cells.
FIG 3.
Decreased expression of FA proteins in WIP−/− fibroblasts. (A) Representative immunofluorescence image of WT fibroblasts stained for vinculin (red) and WIP (green) at ×400 magnification. The inset demonstrates the absence of WIP in vinculin-rich FAs. (B) Representative immunoblot and quantitative analysis of the FA proteins zyxin, VASP, TES, and vinculin in WIP−/− and WT fibroblasts. β-Tubulin was used as loading control. Quantitative data represent the ratio of each protein to tubulin, normalized to the ratio in WT fibroblasts. (C) Quantitative PCR analysis of mRNA expression of zyxin, VASP, TES, and vinculin in WIP−/− and WT fibroblasts. Results are expressed relative to the expression of the housekeeping gene β2-microglobulin and are normalized to expression in WT fibroblasts. Results in panel A are representative of three independent experiments on each of two WT fibroblast lines. Results in panels B and C are representative of three independent experiments, each performed on two WIP−/− lines and two WT lines. Bars represent means; error bars, standard deviations. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
Decreased F-actin and increased G-actin levels in WIP−/− fibroblasts.
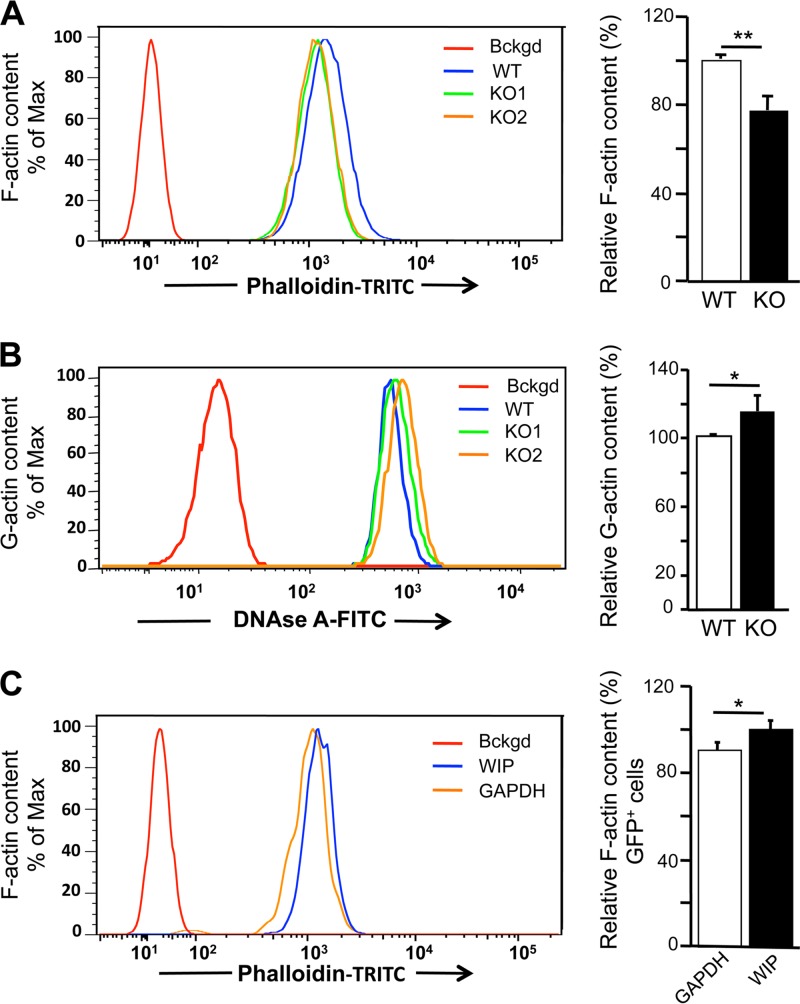
FACS analysis of trypsinized fibroblasts intracellularly stained with TRITC-conjugated phalloidin revealed that WIP−/− cells had a significantly lower F-actin content than WT controls (Fig. 4A). Trypsinization can drive actin polymerization by activating RhoA through protease-associated receptors (33, 34). Rho activities were comparable in WIP−/− and WT fibroblasts (data not shown), indicating that the difference in F-actin content between them is not an in vitro artifact.
FIG 4.
Altered F-actin and G-actin contents in WIP−/− fibroblasts. (A and B) Representative FACS and quantitative analyses of F-actin (A) and G-actin (B) in WIP−/− and WT fibroblasts. Unstained cells were used as a background (Bckgd) control. Quantitative results are expressed relative to the means for WT fibroblasts. (C) Representative FACS and quantitative analyses of the effect of transduction with bicistronic vectors encoding WIP and tGFP, or GAPDH and tGFP as a control, on F-actin contents in WIP−/− fibroblasts. Analysis was performed on cells gated for tGFP (green) expression. Quantitative results are expressed relative to the means for fibroblasts transduced with the vector encoding GAPDH and tGFP. Results in panels A and B represent three independent experiments performed on two WIP−/− lines and two WT lines. Results in panel C represent two independent experiments, each performed on two WIP−/− lines. Bars represent means; error bars, standard deviations. *, P < 0.05; **, P < 0.01.
Intracellular Alexa Fluor 488-conjugated DNase I staining revealed that the G-actin content was significantly higher in WIP−/− fibroblasts than in WT controls (Fig. 4B). The G-actin/F-actin ratio was 1.83-fold ± 0.44-fold higher in WIP−/− fibroblasts than in WT controls (n, 3; P, <0.001). Transduction of WIP in WIP−/− fibroblasts caused a significant increase in the F-actin content compared to transduction of GAPDH, used as a control (Fig. 4C). These results indicate that WIP is important for maintaining the G↔F actin equilibrium in fibroblasts.
Decreased SRF level and activity in WIP−/− fibroblasts.
Serum response factor (SRF) drives the expression of genes that encode FA proteins (14). Srf−/− mouse embryonic fibroblasts have impaired formation of FAs and stress fibers, reduced levels of several FA proteins, and diminished F-actin content (18), a phenotype similar to that of WIP−/− fibroblasts. Gene array analysis revealed decreased expression in WIP−/− fibroblasts of 11 genes known to be upregulated by SRF and increased expression of 2 genes known to be downregulated by SRF (35–37) (Table 1). These findings suggested that SRF activity might be impaired in WIP−/− fibroblasts.
TABLE 1.
Relative mRNA expression of SRF-regulated genes in WIP−/− fibroblasts compared to WT fibroblastsa
| Gene | Fold change in expression from WTb |
|---|---|
| Acta2 (alpha actin 2) | −2.2 |
| Cnn3 (calponin) | −2.2 |
| Cyr61 (cysteine-rich protein 61) | −2.0 |
| Fhl2 (four and a half lim domains 2) | −2.5 |
| Myh9 (nonmuscle myosin heavy chain) | −2.5 |
| Myl9 (nonmuscle myosin light chain) | −2.3 |
| Sdpr (serum deprivation response) | −2.6 |
| Tpm2 (tropomyosin 2) | −2.0 |
| Vasp (vasodilator-stimulated phosphoprotein) | −1.7 |
| Vcl (vinculin) | −1.5 |
| Zyx (zyxin) | −1.7 |
| Ctgf (connective tissue growth factor) | +1.8 |
| Nppb (natriuretic peptide type B) | +4.1 |
Only experimentally verified targets are listed (35–37). All genes listed are positively regulated by SRF, except for Ctgf and Npp, which are negatively regulated by SRF. The complete transcriptome profile is available on request.
Values are means of results from two independent experiments using fibroblasts from two WIP−/− mice and their WT littermates. The threshold cutoff was set at 1.5-fold.
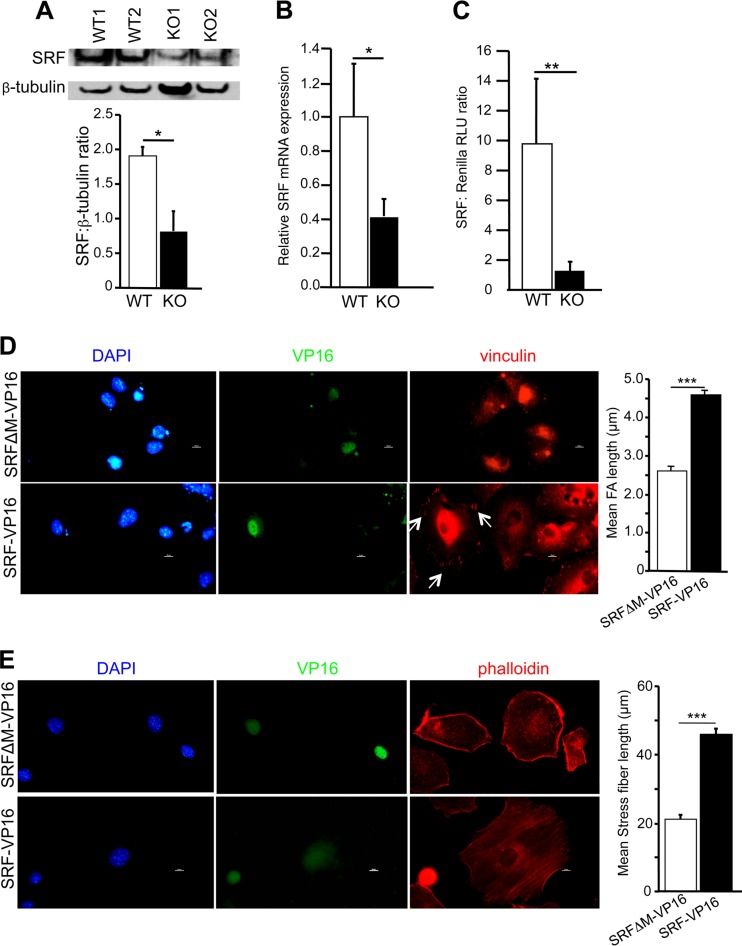
SRF autoregulates its expression (14, 37). SRF protein and mRNA expression was significantly lower in WIP−/− fibroblasts than in WT controls (Fig. 5A and B). Furthermore, a luciferase gene reporter assay revealed that SRF activity was markedly lower in WIP−/− fibroblasts than in WT fibroblasts (Fig. 5C). A fusion protein consisting of SRF and the transactivation domain of herpes simplex virus (HSV) protein VP16, comprising aa 412 to 490 (SRF-VP16), has been demonstrated to be constitutively active (18, 38). Expression of SRF-VP16 induced the formation of robust FAs and stress fibers in WIP−/− fibroblasts (Fig. 5D and E). In contrast, expression of a mutant SRF lacking most of the DNA binding MADS (MCM1, Agamous, Deficiens, SRF) domain fused to VP-16 (SRFΔM-VP16) failed to correct FA assembly and stress fiber formation in WIP−/− fibroblasts (Fig. 5D and E). These results strongly suggest that reduced SRF activity in WIP−/− fibroblasts contributes to their impaired FA assembly and stress fiber formation.
FIG 5.
Decreased SRF expression and activity in WIP−/− fibroblasts. (A and B) Representative immunoblot analysis and quantitation of SRF (A), and quantitative PCR analysis of Srf mRNA expression (B), in WIP−/− and WT fibroblasts. (C) Reporter gene assay of SRF activity in WIP−/− and WT fibroblasts cultured in 10% FCS. RLU, relative light units. (D) (Left) Representative immunofluorescence images of WIP−/− fibroblasts transfected with SRF-VP16 or with SRFΔM-VP16 as a control and stained for vinculin (red) and VP16 (green) and with DAPI (blue). (Right) Quantitative analysis of FA length in VP16-expressing cells. (E) (Left) Representative immunofluorescence images of WIP−/− fibroblasts transfected with SRF-VP16 or with SRFΔM-VP16 as a control and stained for phalloidin (red) and VP16 (green) and with DAPI (blue). (Right) Quantitative analysis of stress fiber length in VP16-expressing cells. Results in panels A to C represent three independent experiments, each performed on two WIP−/− and two WT lines. Results in panels D and E represent two independent experiments, each performed on two WIP−/− lines. Bars represent means; error bars, standard deviations. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Impaired nuclear translocation of MRTF-A in WIP−/− fibroblasts.
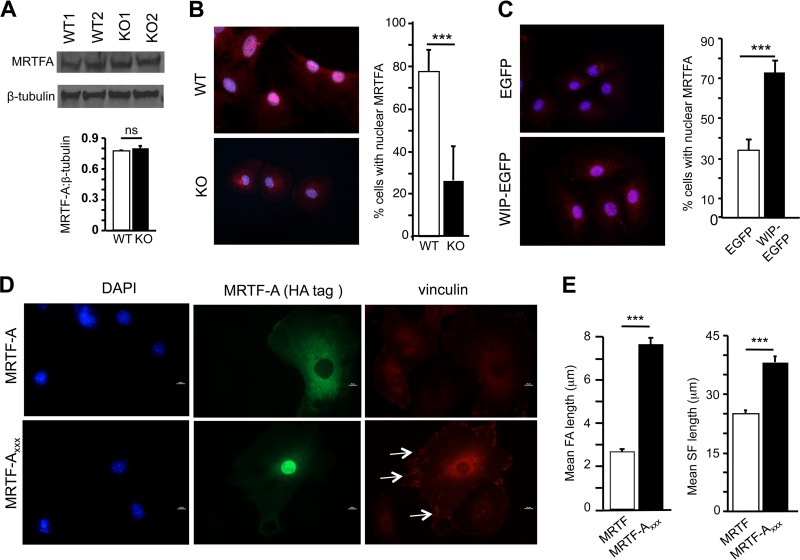
Myocardin-related transcription factors (MRTFs) A and B are encoded by separate genes, bind to SRF, and are essential coactivators of SRF (14, 39). MRTFs normally shuttle between the cytoplasm and the nucleus in cells cultured in serum (15, 16). G-actin binds to MRTFs and sequesters them in the cytoplasm (15, 16). Consequently, the G-actin/F-actin ratio controls the nuclear translocation of MRTFs and SRF activity. Since WIP−/− fibroblasts have elevated G-actin levels, we tested the hypothesis that the nuclear translocation of MRTFs is impaired in WIP−/− fibroblasts. Given the high degree of homology between MRTF-A and MRTF-B, we focused our studies on MRTF-A. Immunoblot analysis revealed that levels of MRTF-A were comparable in WIP−/− fibroblasts and WT controls (Fig. 6A). Intracellular staining revealed that MRTF-A was distributed in both the nucleus and the cytoplasm in WT fibroblasts (Fig. 6B). In contrast, MRTF-A was mostly confined to the cytoplasm in WIP−/− fibroblasts. Nucleofection of WIP fused to enhanced GFP (WIP-EGFP), but not EGFP alone, caused MRTF-A to localize to the nucleus in WIP−/− fibroblasts (Fig. 6C), demonstrating that WIP plays a critical role in the nuclear localization of MRTF-A. The binding of MRTF-A to G-actin is mediated by three REPL motifs (15). An MRTF-A triple point mutant, MRTF-AXXX, in which the arginine residue in the three REPL motifs is replaced by alanine, fails to bind G-actin but retains the ability to bind SRF (40). Consequently, MRTF-AXXX resides in the nucleus and causes constitutive activation of SRF. We investigated whether SRF activation by MRTF-AXXX restores the defective FA assembly in WIP−/− cells. WIP−/− fibroblasts were transfected with hemagglutinin (HA)-tagged MRTF-AXXX or WT MRTF-A. HA-tagged WT MRTF-A localized almost exclusively in the cytoplasm. In contrast, HA-tagged MRTF-AXXX localized almost exclusively in the nucleus (Fig. 6D). WIP−/− fibroblasts that expressed MRTF-AXXX had significantly longer FAs than those that expressed WT MRTF-A (Fig. 6D and E). In addition, they also had significantly longer stress fibers (Fig. 6E; see also Fig. S3 in the supplemental material). These results indicated that forced nuclear localization of MRTF-A corrects the defective FA assembly in WIP−/− fibroblasts.
FIG 6.
Impaired nuclear localization of MRTF-A in WIP−/− fibroblasts. (A) Representative immunoblot analysis (top) and quantification (bottom) of MRTF-A protein expression in WIP−/− and WT fibroblasts. Results are shown relative to β-tubulin expression. (B) Representative immunofluorescence images (left) and quantitative analysis (right) of the subcellular distribution of MRTF-A in WIP−/− and WT fibroblasts stained for MRTF-A (red) and with DAPI (blue). Purple nuclei after the merge indicate the presence of MRTF-A in the nucleus. (C) Representative immunofluorescence images (left) and quantitative analysis (right) of nuclear localization of MRTF-A in WIP−/− fibroblasts nucleofected with WIP-EGFP, or with EGPF as a control, and stained for MRTF-A (red) and with DAPI (blue). (D) Representative immunofluorescence images of WIP−/− fibroblasts transfected with HA-tagged MRTF-A or HA-tagged MRTF-AXXX and stained for vinculin (red) and HA-tagged MRTF-A (green) and with DAPI (blue). Arrows indicate FA. (E) Mean lengths of FAs and actin stress fibers in transfected WIP−/− fibroblasts expressing HA-tagged MRTF-A or HA-tagged MRTF-AXXX. Results in panels A to C represent three independent experiments, each performed on two WIP−/− and two WT lines. Results in panels D and E represent two independent experiments, each performed on two WIP−/− lines. Bars represent means; error bars, standard deviations. ***, P < 0.001; ns, not significant.
Deletion of actin binding domain of WIP (WIPΔABD) phenocopies WIP deficiency in fibroblasts.
WIP contains in its N-terminal domain a conserved actin binding sequence (aa 43 to 54). Deletion of this sequence abolishes the ability of a GST-WIP1–127 fusion protein to pull down actin from fibroblast lysates (9). We tested the hypothesis that the binding of WIP to actin is essential for FA assembly and stress fiber formation. We used lung-derived fibroblasts from knock-in mice homozygous for a Wipf1 allele that encodes a WIP mutant lacking this sequence (WIPΔABD mice). The WIPΔABD mice will be described in detail elsewhere. We compared two fibroblast lines from two WIPΔABD mice and two WT littermates. Similar results were obtained for each pair. We show representative results on single lines and present the pooled data from two lines.
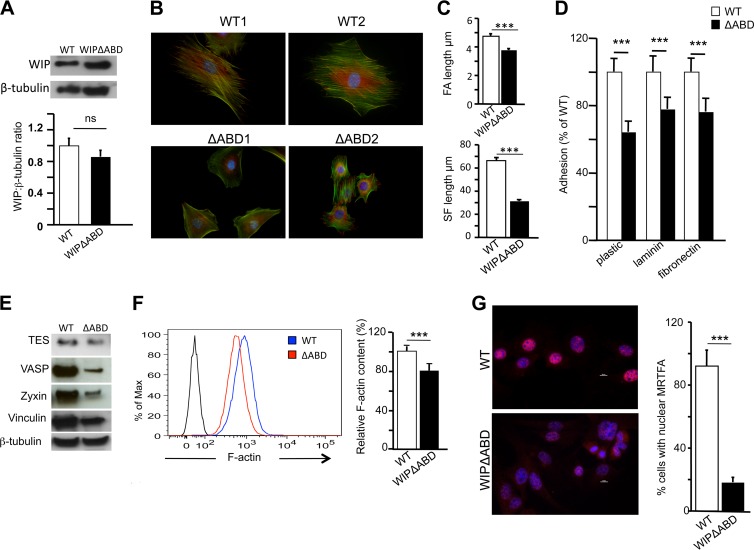
WIPΔABD fibroblasts expressed WIP with a molecular weight slightly lower than that of WT WIP (Fig. 7A). The level of expression of the mutant in WIPΔABD fibroblasts was slightly lower than that of WT WIP in control fibroblasts, but the difference was not significant (Fig. 7A). The phenotype of WIPΔABD fibroblasts replicated, albeit to a lesser degree, the phenotype of WIP−/− fibroblasts. WIPΔABD fibroblasts were less elongated and more rounded than WT controls and displayed multiple poles (Fig. 7B). They had significantly shorter FAs and actin stress fibers than WT controls (Fig. 7C). WIPΔABD fibroblasts adhered less to substrates (Fig. 7D) and expressed significantly less zyxin, VASP, and TES than WT fibroblasts (Fig. 7E). The F-actin content was significantly lower in WIPΔABD fibroblasts than in WT controls (Fig. 7F). Finally, less MRTF-A was localized to the nucleus in WIPΔABD fibroblasts than in WT controls (Fig. 7G). These results indicate that WIP binding to actin is important for maintaining a normal F-actin content, nuclear translocation of MRTF-A, expression of SRF-regulated FA genes, FA assembly, and actin stress fiber formation in fibroblasts.
FIG 7.
Deletion of the WIP acting domain (WIPΔABD) phenocopies WIP deficiency in fibroblasts. (A) Immunoblot analysis and quantification of WIP in WIPΔABD and WT fibroblasts. (B) Representative immunofluorescence images of WIPΔABD and WT fibroblasts stained for vinculin (red) and actin (green) and with DAPI (blue). (C) Mean lengths of FAs and actin stress fibers in WIPΔABD and WT fibroblasts. (D) Adherence of WIPΔABD and WT fibroblasts to substrates. (E) Representative immunoblots of TES, VASP, zyxin, and vinculin in WIPΔABD and WT fibroblasts. β-Tubulin was used as a loading control. (F) Representative FACS analysis and quantification of F-actin in WIPΔABD and WT fibroblasts. Quantitative results are expressed relative to the mean for WT fibroblasts. (G) Representative immunofluorescence images and quantitative analysis of the subcellular distribution of MRTF-A in WIPΔABD and WT fibroblasts stained for MRTF-A (red) and with DAPI (blue). Purple nuclei after the merge indicate the presence of MRTF-A in the nucleus. Bars represent means; error bars, standard deviations. ***, P < 0.001; ns, not significant.
DISCUSSION
We demonstrate that by virtue of its binding to F-actin, WIP plays a critical role in FA assembly, stress fiber formation, and adherence to substrates by fibroblasts. We show that WIP shifts the F↔G actin equilibrium away from G-actin, promoting the nuclear translocation of MRTFs and the activation of SRF, which drives the expression of genes that encode FA proteins.
Independently derived lines of WIP−/− fibroblasts assumed a round rather than a polygonal shape and demonstrated deficient FA assembly and stress fiber formation. The decreased FA length was unlikely to be an artifact of staining for vinculin, the level of which was lower in WIP−/− fibroblasts than in WT controls, because similar results were obtained by staining for paxillin, which was expressed at comparable levels in WIP−/− fibroblasts and WT controls. The morphological abnormalities of WIP−/− fibroblasts were reversed by the expression of WT WIP, indicating that these abnormalities are a consequence of WIP deficiency.
FAs play an important role in stress fiber assembly (41, 42). Conversely, stress fibers that are inserted into FAs exert tensile forces that cause the elongation and maturation of FAs (43, 44). Both FAs and stress fibers are important determinants of cell shape, and abnormalities in both likely contributed to the abnormal shape of WIP−/− fibroblasts. The fibroblasts we used had been maintained in culture for more than 12 passages, by which time they would have undergone spontaneous immortalization and attained uniformity (19). It was noted previously that the shape of WIP−/− fibroblasts at earlier passages was comparable to that of WT fibroblasts (45, 46), but FA length and stress fibers were not evaluated in those studies. Independent microarray analysis of transcripts from those low-passage-number WIP−/− fibroblasts revealed decreases in the expression of several genes encoding FA proteins comparable to those observed in the present study in high-passage-number WIP−/− fibroblasts (I. M. Anton, unpublished data), suggesting that multiple passages accentuate the difference in shape between WIP−/− and WT fibroblasts.
The importance of FAs for integrin-mediated adhesion to substrates and for integrin signaling in adherent cells has been extensively studied (47–49). The defective adhesion of WIP−/− fibroblasts to substrates despite normal surface expression of integrins and their defective adhesion-driven phosphorylation of FAK upon plating are consistent with the impaired assembly of FAs in these cells. We were unable to detect WIP in FAs, suggesting that WIP plays an indirect role in FA assembly. The levels of a number of FA proteins, including VASP, zyxin, TES, and vinculin, were decreased in WIP−/− fibroblasts. This was associated with a concomitant decrease in the mRNA expression of the genes that encode these FA proteins. Both vinculin and VASP have been demonstrated to be important for FA assembly and cell adhesion (25, 50), suggesting that deficiencies in these FA proteins contribute to defective FA assembly in WIP−/− fibroblasts. In contrast, zyxin-deficient fibroblasts show enhanced adhesion (19), suggesting that zyxin has important effects on cell adhesion other than those that contribute to FA assembly.
We provide evidence that WIP regulates the expression of genes that encode FA proteins by driving the G↔F actin equilibrium toward F-actin, thereby promoting the nuclear translocation of MRTF-A and consequently the activity of SRF, which regulates the expression of these genes. This mechanism was supported by the following observations. First, WIP−/− fibroblasts had significantly decreased levels of F-actin and, conversely, increased levels of G-actin, which is known to sequester MRTF-A in the cytosol. Reconstitution of WIP−/− fibroblasts with WIP reversed the decrease in F-actin content. The decreased F-actin content in WIP−/− fibroblasts is most likely a direct consequence of the loss of the stabilizing effect of WIP on F-actin. This is supported by our previous observation that WIP binds to F-actin and inhibits its depolymerization (10). Second, nuclear localization of MRTF-A was impaired in WIP−/− fibroblasts and was restored upon the introduction of WIP. Reconstitution with a MRTF-A mutant that does not bind G-actin and that constitutively translocates to the nucleus restored FA assembly and stress fiber formation in WIP−/− fibroblasts. Pharmacologic agents that interfere with MRTF-A nuclear translocation by shifting the G↔F actin equilibrium toward G-actin inhibit the expression of a number of genes that are important for FA assembly and exhibit decreased levels in WIP−/− fibroblasts. These include Acta2, Cyr61, Fhl2, Myh9, Myl9, Sdpr, Srf, Tagln, Tpm, Vcl, and Zyx (35). Third, the level and activity of SRF were decreased in WIP−/− fibroblasts. Furthermore, the phenotypes of SRF−/− and WIP−/− fibroblasts are strikingly similar, with defective FA assembly and stress fiber formation, decreased F-actin content and increased G-actin content, decreased expression of a number of genes that encode FA proteins, and reduced adhesion and activation of FAK (18). More importantly, the introduction of constitutively active SRF in the form of an SRF-VP16 fusion protein rescued FA assembly and stress fiber formation in WIP−/− fibroblasts.
We show that the binding of WIP to actin controls the actin dynamics→MRTF-A→SRF→FA assembly signaling cascade. Fibroblasts from knock-in mice that express a WIP mutant lacking the actin-binding domain phenocopied WIP−/− fibroblasts. They had decreased F-actin content, impaired nuclear translocation of MRTF-A, and decreased expression of FA proteins, as well as decreased FA assembly, shorter stress fibers, and decreased adhesion, underscoring the importance of the actin binding function of WIP in regulating the actin dynamics→FA assembly axis. WIP has two homologues, CR16 and WIRE/WICH. CR16 is expressed almost exclusively in the brain. WIRE/WICH contains the actin-binding motif KLKK and has been shown to stabilize F-actin (51). WIRE/WICH is expressed predominantly in the brain and colon and to a lesser extent in the lung and stomach (51); it is expressed minimally in fibroblasts compared to WIP, as determined by RT-PCR (unpublished observations). The observation that fibroblasts from WIP−/− and WIPΔABD mice exhibit a distinct morphological phenotype indicates that actin binding by WIP plays a dominant role in the genesis of this phenotype. However, the possibility that WIRE/WICH might partially compensate for WIP deficiency in fibroblasts cannot be ruled out.
We show that a number of genes that play important roles in cancer cell migration, invasion, and metastasis or are tumor suppressors are regulated by WIP, as well as by the MRTF-SRF complex. These include Vasp, vinculin, zyxin, Myh9, Myl9, Tes, and Cyr61 (36, 52–58). In addition, FAK, the activation of which is regulated by WIP, plays an important role in tumor invasion and metastatic cell adhesion and has recently been a target for cancer therapy (59, 60). It would be of interest to explore the potential role of WIP in cancer cell invasion and metastasis.
Supplementary Material
ACKNOWLEDGMENTS
We thank Richard Treisman for generously providing MRTF, MRTFxxx, and SRF-VP16 plasmids. We thank Pallavi Banerjee for her help.
This work was supported by USPHS grant HL-059561.
Footnotes
Published ahead of print 5 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00017-14.
REFERENCES
- 1.Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, Kirschner M. 1999. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell 97:221–231. 10.1016/S0092-8674(00)80732-1 [DOI] [PubMed] [Google Scholar]
- 2.Symons M, Derry JMJ, Kariak B, Jiang S, Lemahieu V, McCormick F, Francke U, Abo A. 1996. Wiskott-Aldrich syndrome protein, a novel effector for the GTPase Cdc42Hs, is implicated in actin polymerization. Cell 84:723–734. 10.1016/S0092-8674(00)81050-8 [DOI] [PubMed] [Google Scholar]
- 3.Rohatgi R, Nollau P, Ho HY, Kirschner MW, Mayer BJ. 2001. Nck and phosphatidylinositol 4,5-bisphosphate synergistically activate actin polymerization through the N-WASP-Arp2/3 pathway. J. Biol. Chem. 276:26448–26452. 10.1074/jbc.M103856200 [DOI] [PubMed] [Google Scholar]
- 4.She HY, Rockow S, Tang J, Nishimura R, Skolnik EY, Chen M, Margolis B, Li W. 1997. Wiskott-Aldrich syndrome protein is associated with the adapter protein Grb2 and the epidermal growth factor receptor in living cells. Mol. Biol. Cell 8:1709–1721. 10.1091/mbc.8.9.1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weaver AM, Karginov AV, Kinley AW, Weed SA, Li Y, Parsons JT, Cooper JA. 2001. Cortactin promotes and stabilizes Arp2/3-induced actin filament network formation. Curr. Biol. 11:370–374. 10.1016/S0960-9822(01)00098-7 [DOI] [PubMed] [Google Scholar]
- 6.Ho HY, Rohatgi R, Lebensohn AM, Le M, Li J, Gygi SP, Kirschner MW. 2004. Toca-1 mediates Cdc42-dependent actin nucleation by activating the N-WASP-WIP complex. Cell 118:203–216. 10.1016/j.cell.2004.06.027 [DOI] [PubMed] [Google Scholar]
- 7.de la Fuente MA, Sasahara Y, Calamito M, Anton IM, Elkhal A, Gallego MD, Suresh K, Siminovitch K, Ochs HD, Anderson KC, Rosen FS, Geha RS, Ramesh N. 2007. WIP is a chaperone for Wiskott-Aldrich syndrome protein (WASP). Proc. Natl. Acad. Sci. U. S. A. 104:926–931. 10.1073/pnas.0610275104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Massaad MJ, Ramesh N, Le Bras S, Giliani S, Notarangelo LD, Al-Herz W, Notarangelo LD, Geha RS. 2011. A peptide derived from the Wiskott-Aldrich syndrome (WAS) protein-interacting protein (WIP) restores WAS protein level and actin cytoskeleton reorganization in lymphocytes from patients with WAS mutations that disrupt WIP binding. J. Allergy Clin. Immunol. 127:998–1005. 10.1016/j.jaci.2011.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Antón IM, Saville SP, Byrne MJ, Curcio C, Ramesh N, Hartwig JH, Geha RS. 2003. WIP participates in actin reorganization and rufle formation induced by PDGF. J. Cell Sci. 116:2443–2451. 10.1242/jcs.00433 [DOI] [PubMed] [Google Scholar]
- 10.Martinez-Quiles N, Rohatgi R, Anton IM, Medina M, Saville SP, Miki H, Yamaguchi H, Takenawa T, Hartwig JH, Geha RS, Ramesh N. 2001. WIP regulates N-WASP-mediated actin polymerization and filopodium formation. Nat. Cell Biol. 3:484–491. 10.1038/35074551 [DOI] [PubMed] [Google Scholar]
- 11.van Troys M, Dewitte D, Goethas M, Carlier MF, Vanderkerckhove J, Ampe C. 1996. The actin binding site of thymosin β4 mapped by mutational analysis. EMBO J. 15:201–210 [PMC free article] [PubMed] [Google Scholar]
- 12.Antón IM, de la Fuente MA, Sims TN, Freeman S, Ramesh N, Hartwig JH, Dustin ML, Geha RS. 2002. WIP deficiency reveals a differential role for WIP and the actin cytoskeleton in T and B cell activation. Immunity 16:193–204. 10.1016/S1074-7613(02)00268-6 [DOI] [PubMed] [Google Scholar]
- 13.Ramesh N, Anton IM, Hartwig JH, Geha RS. 1997. WIP, a protein associated with Wiskott-Aldrich syndrome protein, induces actin polymerization and redistribution in lymphoid cells. Proc. Natl. Acad. Sci. U. S. A. 94:14671–14676. 10.1073/pnas.94.26.14671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miano JM, Long X, Fujiwara K. 2007. Serum response factor: master regulator of the actin cytoskeleton and contractile apparatus. Am. J. Physiol. Cell Physiol. 292:C70–C81. 10.1152/ajpcell.00386.2006 [DOI] [PubMed] [Google Scholar]
- 15.Miralles F, Posern G, Zaromytidou AI, Treisman R. 2003. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell 113:329–342. 10.1016/S0092-8674(03)00278-2 [DOI] [PubMed] [Google Scholar]
- 16.Sotiropoulos A, Gineitis D, Copeland J, Treisman R. 1999. Signal-regulated activation of serum response factor is mediated by changes in actin dynamics. Cell 98:159–169. 10.1016/S0092-8674(00)81011-9 [DOI] [PubMed] [Google Scholar]
- 17.Sun Q, Chen G, Streb JW, Long X, Yang Y, Stoeckert CJ, Jr, Miano JM. 2006. Defining the mammalian CArGome. Genome Res. 16:197–207. 10.1101/gr.4108706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schratt G, Philippar U, Berger J, Schwarz H, Heidenreich O, Nordheim A. 2002. Serum response factor is crucial for actin cytoskeletal organization and focal adhesion assembly in embryonic stem cells. J. Cell Biol. 156:737–750. 10.1083/jcb.200106008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffman LM, Jensen CC, Kloeker S, Wang CL, Yoshigi M, Beckerle MC. 2006. Genetic ablation of zyxin causes Mena/VASP mislocalization, increased motility, and deficits in actin remodeling. J. Cell Biol. 172:771–782. 10.1083/jcb.200512115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhasin MK, Dusek JA, Chang BH, Joseph MG, Denninger JW, Fricchione GL, Benson H, Libermann TA. 2013. Relaxation response induces temporal transcriptome changes in energy metabolism, insulin secretion and inflammatory pathways. PLoS One 8:e62817. 10.1371/journal.pone.0062817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kauffmann A, Gentleman R, Huber W. 2009. arrayQualityMetrics—a bioconductor package for quality assessment of microarray data. Bioinformatics 25:415–416. 10.1093/bioinformatics/btn647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li C, Wong WH. 2001. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. U. S. A. 98:31–36. 10.1073/pnas.98.1.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuen T, Wurmbach E, Pfeffer RL, Ebersole BJ, Sealfon SC. 2002. Accuracy and calibration of commercial oligonucleotide and custom cDNA microarrays. Nucleic Acids Res. 30:e48. 10.1093/nar/30.10.e48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu W, Baribault H, Adamson ED. 1998. Vinculin knockout results in heart and brain defects during embryonic development. Development 125:327–337 [DOI] [PubMed] [Google Scholar]
- 26.Goldmann WH. 2012. Mechanotransduction and focal adhesions. Cell Biol. Int. 36:649–652. 10.1042/CBI20120184 [DOI] [PubMed] [Google Scholar]
- 27.Goldmann WH. 2012. Mechanotransduction in cells. Cell Biol. Int. 36:567–570. 10.1042/CBI20120071 [DOI] [PubMed] [Google Scholar]
- 28.Wolfenson H, Henis YI, Geiger B, Bershadsky AD. 2009. The heel and toe of the cell's foot: a multifaceted approach for understanding the structure and dynamics of focal adhesions. Cell Motil. Cytoskeleton 66:1017–1029. 10.1002/cm.20410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goodman SL. 1992. Alpha 6 beta 1 integrin and laminin E8: an increasingly complex simple story. Kidney Int. 41:650–656. 10.1038/ki.1992.100 [DOI] [PubMed] [Google Scholar]
- 30.Hynes RO. 2002. Integrins: bidirectional, allosteric signaling machines. Cell 110:673–687. 10.1016/S0092-8674(02)00971-6 [DOI] [PubMed] [Google Scholar]
- 31.Schaller MD, Borgman CA, Cobb BS, Vines RR, Reynolds AB, Parsons JT. 1992. pp125FAK, a structurally distinct protein-tyrosine kinase associated with focal adhesions. Proc. Natl. Acad. Sci. U. S. A. 89:5192–5196. 10.1073/pnas.89.11.5192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parsons JT, Martin KH, Slack JK, Taylor JM, Weed SA. 2000. Focal adhesion kinase: a regulator of focal adhesion dynamics and cell movement. Oncogene 19:5606–5613. 10.1038/sj.onc.1203877 [DOI] [PubMed] [Google Scholar]
- 33.Martin CB, Mahon GM, Klinger MB, Kay RJ, Symons M, Der CJ, Whitehead IP. 2001. The thrombin receptor, PAR-1, causes transformation by activation of Rho-mediated signaling pathways. Oncogene 20:1953–1963. 10.1038/sj.onc.1204281 [DOI] [PubMed] [Google Scholar]
- 34.Narumiya S, Tanji M, Ishizaki T. 2009. Rho signaling, ROCK and mDia1, in transformation, metastasis and invasion. Cancer Metastasis Rev. 28:65–76. 10.1007/s10555-008-9170-7 [DOI] [PubMed] [Google Scholar]
- 35.Descot A, Hoffmann R, Shaposhnikov D, Reschke M, Ullrich A, Posern G. 2009. Negative regulation of the EGFR-MAPK cascade by actin-MAL-mediated Mig6/Errfi-1 induction. Mol. Cell 35:291–304. 10.1016/j.molcel.2009.07.015 [DOI] [PubMed] [Google Scholar]
- 36.Medjkane S, Perez-Sanchez C, Gaggioli C, Sahai E, Treisman R. 2009. Myocardin-related transcription factors and SRF are required for cytoskeletal dynamics and experimental metastasis. Nat. Cell Biol. 11:257–268. 10.1038/ncb1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Selvaraj A, Prywes R. 2004. Expression profiling of serum inducible genes identifies a subset of SRF target genes that are MKL dependent. BMC Mol. Biol. 5:13. 10.1186/1471-2199-5-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dalton S, Treisman R. 1992. Characterization of SAP-1, a protein recruited by serum response factor to the c-fos serum response element. Cell 68:597–612. 10.1016/0092-8674(92)90194-H [DOI] [PubMed] [Google Scholar]
- 39.Posern G, Treisman R. 2006. Actin' together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 16:588–596. 10.1016/j.tcb.2006.09.008 [DOI] [PubMed] [Google Scholar]
- 40.Guettler S, Vartiainen MK, Miralles F, Larijani B, Treisman R. 2008. RPEL motifs link the serum response factor cofactor MAL but not myocardin to Rho signaling via actin binding. Mol. Cell. Biol. 28:732–742. 10.1128/MCB.01623-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burridge K, Fath K, Kelly T, Nuckolls G, Turner C. 1988. Focal adhesions: transmembrane junctions between the extracellular matrix and the cytoskeleton. Annu. Rev. Cell Biol. 4:487–525. 10.1146/annurev.cb.04.110188.002415 [DOI] [PubMed] [Google Scholar]
- 42.Schoenwaelder SM, Burridge K. 1999. Bidirectional signaling between the cytoskeleton and integrins. Curr. Opin. Cell Biol. 11:274–286. 10.1016/S0955-0674(99)80037-4 [DOI] [PubMed] [Google Scholar]
- 43.Hotulainen P, Lappalainen P. 2006. Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J. Cell Biol. 173:383–394. 10.1083/jcb.200511093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oakes PW, Beckham Y, Stricker J, Gardel ML. 2012. Tension is required but not sufficient for focal adhesion maturation without a stress fiber template. J. Cell Biol. 196:363–374. 10.1083/jcb.201107042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Banon-Rodriguez I, Saez de Guinoa J, Bernardini A, Ragazzini C, Fernandez E, Carrasco YR, Jones GE, Wandosell F, Anton IM. 2013. WIP regulates persistence of cell migration and ruffle formation in both mesenchymal and amoeboid modes of motility. PLoS One 8:e70364. 10.1371/journal.pone.0070364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lanzardo S, Curcio C, Forni G, Anton IM. 2007. A role for WASP interacting protein, WIP, in fibroblast adhesion, spreading and migration. Int. J. Biochem. Cell Biol. 39:262–274. 10.1016/j.biocel.2006.08.011 [DOI] [PubMed] [Google Scholar]
- 47.Campbell ID. 2008. Studies of focal adhesion assembly. Biochem. Soc. Trans. 36:263–266. 10.1042/BST0360263 [DOI] [PubMed] [Google Scholar]
- 48.Geiger B, Spatz JP, Bershadsky AD. 2009. Environmental sensing through focal adhesions. Nat. Rev. Mol. Cell Biol. 10:21–33. 10.1038/nrm2593 [DOI] [PubMed] [Google Scholar]
- 49.Wehrle-Haller B. 2012. Assembly and disassembly of cell matrix adhesions. Curr. Opin. Cell Biol. 24:569–581. 10.1016/j.ceb.2012.06.010 [DOI] [PubMed] [Google Scholar]
- 50.Shen K, Tolbert CE, Guilluy C, Swaminathan VS, Berginski ME, Burridge K, Superfine R, Campbell SL. 2011. The vinculin C-terminal hairpin mediates F-actin bundle formation, focal adhesion, and cell mechanical properties. J. Biol. Chem. 286:45103–45115. 10.1074/jbc.M111.244293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kato M, Miki H, Kurita S, Endo T, Nakagawa H, Miyamoto S, Takenawa T. 2002. WICH, a novel verproline homology domain-containing protein that functions cooperatively with N-WASP in actin-microspike formation. Biochem. Biophys. Res. Commun. 291:41–47. 10.1006/bbrc.2002.6406 [DOI] [PubMed] [Google Scholar]
- 52.Drusco A, Zanesi N, Roldo C, Trapasso F, Farber JL, Fong LY, Croce CM. 2005. Knockout mice reveal a tumor suppressor function for Testin. Proc. Natl. Acad. Sci. U. S. A. 102:10947–10951. 10.1073/pnas.0504934102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mierke CT, Kollmannsberger P, Zitterbart DP, Diez G, Koch TM, Marg S, Ziegler WH, Goldmann WH, Fabry B. 2010. Vinculin facilitates cell invasion into three-dimensional collagen matrices. J. Biol. Chem. 285:13121–13130. 10.1074/jbc.M109.087171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mise N, Savai R, Yu H, Schwarz J, Kaminski N, Eickelberg O. 2012. Zyxin is a transforming growth factor-beta (TGF-β)/Smad3 target gene that regulates lung cancer cell motility via integrin α5β1. J. Biol. Chem. 287:31393–31405. 10.1074/jbc.M112.357624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nürnberg A, Kitzing T, Grosse R. 2011. Nucleating actin for invasion. Nat. Rev. Cancer 11:177–187. 10.1038/nrc3003 [DOI] [PubMed] [Google Scholar]
- 56.Tong AH, Drees B, Nardelli G, Bader GD, Brannetti B, Castagnoli L, Evangelista M, Ferracuti S, Nelson B, Paoluzi S, Quondam M, Zucconi A, Hogue CW, Fields S, Boone C, Cesareni G. 2002. A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science 295:321–324. 10.1126/science.1064987 [DOI] [PubMed] [Google Scholar]
- 57.Tong X, O'Kelly J, Xie D, Mori A, Lemp N, McKenna R, Miller CW, Koeffler HP. 2004. Cyr61 suppresses the growth of non-small-cell lung cancer cells via the beta-catenin-c-myc-p53 pathway. Oncogene 23:4847–4855. 10.1038/sj.onc.1207628 [DOI] [PubMed] [Google Scholar]
- 58.Weeks RJ, Kees UR, Song S, Morison IM. 2010. Silencing of TESTIN by dense biallelic promoter methylation is the most common molecular event in childhood acute lymphoblastic leukaemia. Mol. Cancer 9:163. 10.1186/1476-4598-9-163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cance W, Kurenova E, Marlowe T, Golubovskaya V. 2013. Disrupting the scaffold to improve focal adhesion kinase-targeted cancer therapeutics. Sci. Signal. 6:pe10. 10.1126/scisignal.2004021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Duxbury MS, Ito H, Zinner MJ, Ashley SW, Whang EE. 2004. Focal adhesion kinase gene silencing promotes anoikis and suppresses metastasis of human pancreatic adenocarcinoma cells. Surgery 135:555–562. 10.1016/j.surg.2003.10.017 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.