Abstract
While glucocorticoids (GCs) are used clinically to treat many conditions, their neonatal and prenatal usage is increasingly controversial due to reports of delayed adverse outcomes, especially their effects on brain development. Such alterations may reflect the impact of GCs on neural progenitor/stem cell (NPSC) function. We previously demonstrated that the lipid raft protein caveolin-1 (Cav-1) was required for rapid GC signaling in embryonic mouse NPSCs operating through plasma membrane-bound glucocorticoid receptors (GRs). We show here that genomic GR signaling in NPSCs requires Cav-1. Loss of Cav-1 impacts the transcriptional response of many GR target genes (e.g., the serum- and glucocorticoid-regulated kinase 1 gene) that are likely to mediate the antiproliferative effects of GCs. Microarray analysis of wild-type C57 or Cav-1-deficient NPSCs identified approximately 100 genes that are differentially regulated by GC treatment. These changes in hormone responsiveness in Cav-1 knockout NPSCs are associated with the loss of GC-regulated phosphorylation of GR at serine 211 but not at serine 226. Chromatin recruitment of total GR to regulatory regions of target genes such as Fkbp-5, RhoJ, and Sgk-1, as well as p211-GR recruitment to Sgk-1, are compromised in Cav-1 knockout NPSCs. Cav-1 is therefore a multifunctional regulator of GR in NPSCs influencing both rapid and genomic action of the receptor to impact cell proliferation.
INTRODUCTION
Glucocorticoid (GC) hormones are essential for multiple postnatal processes, including immune responses, glucose metabolism, blood pressure regulation, and central nervous system function (1, 2). Given their potent anti-inflammatory actions, GCs are widely used therapeutically in children and adults to treat a variety conditions and diseases such as asthma, allergies, rheumatoid arthritis, chronic inflammation, and induction treatment for acute leukemias (2).
Dexamethasone (Dex) is one of the synthetic glucocorticoids recommended by the American College of Obstetricians and Gynecologists for antenatal therapy (betamethasone being the other) to limit adverse respiratory and vascular events in premature babies (3). Antenatal GC therapy is also used to reduce virilization of the external genitalia for female infants with congenital adrenal hyperplasia. However, this therapy remains highly controversial given the early gestational age required for clinical benefit (4).
Despite their efficacy in the treatment of many diseases, the diverse action of GCs in various tissues and cell types results in many side effects (2). Although the use of synthetic prenatal GCs to promote lung maturation in preterm infants is undisputed, clinical and preclinical studies have revealed the potential for prolonged or delayed side effects linked to antenatal or neonatal GC usage (5–7). In several species, including humans, these studies indicate that exogenous treatment with GCs during fetal or neonatal development or fetal exposure to high levels of endogenous GCs produced as a result of maternal stress can be associated with neurological, cognitive, and affective disorders that emerge later in childhood (7, 8). Although the mechanism(s) responsible for these affects are not well established, they may involve alterations in neural progenitor/stem cell (NPSC) proliferation and/or differentiation, thereby disrupting the development of neuronal circuits essential for higher-order cognitive or behavioral function.
Studies examining GC effects on NPSC function in a variety of experimental systems uncovered a number of affected pathways and signaling molecules. For example, in rat embryonic neural stem cells, GCs decreased proliferation by enhanced degradation of the cell cycle regulator cyclin D1 (9). In the mouse cerebellum, neonatal treatment with Dex increased apoptosis of the neural progenitor cells within the extra granule cell layer and decreased overall numbers of internal granule layer neurons (10). GC effects on the differentiation of NPSCs are varied and conflicting; reduced differentiation of glial cells by GCs was reported in mesencephalic NPSCs and adult rat hippocampal progenitor cells, while GCs increased the differentiation of glia in human NPSC cultures (11–13).
GC responses are mediated primarily by the glucocorticoid receptor (GR), a member of the nuclear receptor superfamily of transcription factors (1, 14), through the actions of two separate but interrelated mechanisms. In classical or genomic signaling, binding of ligand results in GR translocation to the nucleus, where it can either activate or repress the transcription of target genes (15). In contrast, rapid or nonclassical signaling involves a plasma membrane-associated GR that is capable of activating kinases, such as mitogen-activated protein kinase (MAPK). These kinases can then influence GR transcriptional response through direct modification of the receptor or its associated transcriptional coregulators (16). Although both rapid and genomic GR signaling operates in NPSCs (17), the degree of interaction between the two pathways and how they intersect to influence GC effects on NPSC biology are unknown.
Caveolin-1 (Cav-1) is a major protein subunit of specialized regions of the plasma membrane known as caveolae. Caveolae can act as signaling organizers, and previous work from our laboratory established a role for Cav-1 in a rapid GC signaling pathway that triggers MAPK activation in embryonic mouse NPSC cultures (17). One of the consequences of Cav-1-dependent activation of MAPK by GCs is an inhibition of gap junction intercellular communication (GJIC) between coupled NPSCs (17). Studies with other steroid hormone receptors, such as the androgen and estrogen receptor, have revealed mechanisms of cross talk between genomic and rapid actions of the receptors (reviewed in reference 18).
Given the role of Cav-1 in mediating the rapid response of GR (16, 17), we set out to examine whether Cav-1 could also participate in classical genomic GR signaling and impact the antiproliferative response of NPSCs to Dex. Here we show that loss of the Dex-mediated antiproliferative response in NPSCs derived from Cav-1 knockout (KO) mice is associated with the alterations in Dex-induced expression of an established negative regulator of the cell cycle in NPSCs, the serum- and GC-induced kinase 1 gene, Sgk-1 (19). We also revealed more global effects of Cav-1 deletion on the GR transcriptome and uncovered a mechanism for Cav-1-mediated cross talk between rapid and genomic GR signaling that impacts site-specific GR phosphorylation and chromatin recruitment of GR.
MATERIALS AND METHODS
Mouse NPSC cultures.
NPSCs were derived from embryonic (E14.5) cerebral cortex of wild-type C57BL/6 (C57) or Cav-1 knockout (KO) mice and grown as three-dimensional neurosphere cultures (17). Cells were passaged every 7 days, and experiments were performed at passage 3 unless indicated otherwise.
NPSC BrdU assays.
Passage 1 neurospheres were cultured for 3 days after passaging before replenishment with fresh epidermal growth factor and fibroblast growth factor 1. Approximately 6 h later, cells were treated with 100 nM Dex (Sigma Chemicals, St. Louis, MO) or vehicle (ethanol [EtOH]). Then, 10 μM bromodeoxyuridine (BrdU; Sigma Chemicals, catalog no. B-9285) was added for 1 h after 23 h of Dex treatment. Neurospheres were then dissociated into single cells and attached to poly-d-lysine-treated coverslips prior to fixation with 4% paraformaldehyde. Fixed cells were processed for immunocytochemistry according to standard methods. A rat anti-BrdU antibody (Abcam; catalog no. ab6326) was used at 1:500 and anti-rat antibody conjugated to Alexa Flour 488 (Invitrogen; catalog no. A21208) at 1:1,000. Images were captured using a Nikon Eclipse E400 microscope and a Photometrics Cool Snap E52 camera. For SGK-1 inhibitor experiments, GSK650394 (Tocris Bioscience) was used at a concentration of 50 nM and added coincident with Dex.
Microarrays.
Total RNA was extracted by using a Qiagen RNeasy kit (catalog no. 74104) from cell pellets comprising biologically distinct neurosphere cultures from C57 or Cav-1 KO embryos treated with 100 nM Dex or vehicle for 4 h (n = 5/treatment condition). Total RNA of the highest quality and integrity was subjected to further processing after purification, as defined by a 260/280 absorption ratio of ≥1.8 using spectrophotometry on the NanoDrop 1000 (NanoDrop, Wilmington, DE) and an RNA integrity number of ≥8.0 determined via electrophoretic analysis on a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). Each of the samples met these standards, and in vitro transcription was performed using the MessageAmp Premier Enhanced assay protocol (Ambion, Inc., Austin, TX) starting with 500 ng of purified total RNA. Confirmation of cRNA diversity was obtained using the Bioanalyzer 2100 to generate an electrophoretogram for each in vitro transcription reaction regarding sample yield, integrity, and size diversity against a Universal Human Reference RNA (Stratagene, La Jolla, CA). A 15-μg portion of purified, amplified, biotin-labeled cRNA was fragmented and hybridized onto Affymetrix Mouse Genome 430A 2.0 arrays (13,687 genes, 22,690 probes; Affymetrix Corp., Santa Clara, CA) for 18 h. Washing, staining, and scanning of arrays was performed on an Affymetrix Fluidics Station 450 and Scanner 3000 immediately after completion of hybridization.
Bioinformatic analysis.
The data were normalized and summarized using the robust multichip average (RMA) method (20). For the genes represented by multiple probe sets, the probe set with the highest interquartile ratio (a descriptive statistic used to summarize the extent of the spread of the data) was selected to represent the gene. An additional filtering step was performed to remove the genes that were flagged as absent across all 20 samples by the MAS5 normalization method. This reduces the data set to 9,916 genes that were used for further statistical analysis.
Statistical tests were performed using BRB-ArrayTools developed by Richard Simon and the BRB-ArrayTools Development Team (21). A Student t test was used to identify genes significantly different in the following comparisons: Cav-1 KO Dex versus vehicle, C57 Dex versus vehicle, and Cav-1 KO versus C57 vehicle. To find genes differentially regulated by Dex treatment of Cav-1 KO versus C57 NPSCs but not by the KO effect alone, a one-way analysis of variance (ANOVA) was performed. Genes that were found significantly different (false discovery rate, 10%) in the pairwise comparisons were further analyzed for canonical pathways, networks and biological functions using the Ingenuity pathway analysis (IPA) software (Ingenuity Systems, Redwood City, CA). Hierarchical clustering figures and Venn diagrams were produced in R-Bioconductor.
Meta-analysis was performed in NextBio Research (Cupertino, CA), a web-based data search and analysis engine, to compare the GC signature in our studies to 12 other individual publicly available data sets selected using relevant keywords. Differentially expressed genes from each of the data sets were combined to create the master list of GC-regulated genes. This list was then further filtered to identify genes in a subset of studies, for example, genes differentially expressed only in neuronal cells. The data sets used are shown in Table 1.
TABLE 1.
Data sets utilized for NextBio analysis
| Set ID no. | Cell type | Hormone | Concn (μM) | Duration of treatment (h) | Source or reference |
|---|---|---|---|---|---|
| 1 | E14.5 cerebral cortex from Cav-1 KO mice | Dex | 0.1 | 4 | This study |
| 2 | E14.5 cerebral cortex from C57 (wild type) mice | Dex | 0.1 | 4 | This study |
| 3 | AtT-20 mouse pituitary | Dex | 0.1 | 4 | 22 |
| 4 | Multipotent human fetal hippocampal progenitor cell line (HPC03A/07) | Cortisol | 0.1 | 12 | 23 |
| 5 | 3134 mouse mammary adenocarcinoma | Dex | 0.1 | 4 | 22 |
| 6 | C2C12 mouse myotubes | Dex | 1 | 24 | 24 |
| 7 | C2C12 mouse myotubes | Dex | 1 | 6 | 24 |
| 8 | Multipotent human fetal hippocampal progenitor cell line (HPC03A/07) | Cortisol | 100 | 12 | 23 |
| 9 | E14.5 cerebral cortex from rat | Corticosterone | 1 | 28 | 25 |
| 10 | Mouse oligodendrocyte precursor cell line (Oli-Neu) | Dex | 1 | 10 | 26 |
| 11 | Mouse oligodendrocyte precursor cell line (Oli-Neu) | Dex | 1 | 24 | 26 |
| 12 | Mouse 3T3-L1 preadipocytes | Dex | 1 | 2 | 27 |
| 13 | Mouse 3T3-L1 preadipocytes | Dex | 1 | 36 | 27 |
| 14 | Mouse C3H10T1/2 pluripotent stem cell line | Dex | 1 | 1.5 | 28 |
qRT-PCR.
For microarray validations, RNAs treated with Dex or ethanol for 4 h were isolated from biologically independent samples of each genotype using Macherey-Nagel Nucleospin RNA II kit. cDNA synthesis was performed using iScript Select cDNA synthesis kit (Bio-Rad; catalog no. 170-8897). Quantitative real-time PCRs (qRT-PCRs) were performed on a Stratagene Mx3000P and used iTaq Universal SYBR green Supermix (Bio-Rad; catalog no. 172-5121) and primers, with efficiencies calculated to be >80%.
Phospho-GR Westerns.
Neurospheres from C57 or Cav-1 KO mice were treated with 100 nM Dex for 1 h. After treatment, cells were lysed in radioimmunoprecipitation assay buffer (10 mM Tris-Cl [pH 8.0], 1 mM EDTA, 0.5 mM EGTA, 140 mM NaCl, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS]) supplemented with HALT protease and phosphatase inhibitors (Thermo, catalog no. 1861281) at 1:1,000. Lysate was subjected to immunoprecipitation using the BuGR2 mouse monoclonal antibody or incubated with nonimmune IgG using standard laboratory conditions. Immunoprecipitated material was run on SDS–7.5% PAGE gel and transferred to a polyvinylidene difluoride membrane (Immobilon-P, catalog no. IPVH00010; Millipore). Western blot analysis was performed using the p211-GR antibody (29) and an anti-rabbit horseradish peroxidase-conjugated secondary antibody (Bio-Rad, catalog no. 170-6515) using a chemiluminescence detection system (Advansta Western Bright ECL; catalog no. K-12045-D50).
Nuclear fractionation.
Subcellular fractionation was performed according to guidelines provided in the NE-PER kit (Thermo; catalog no. 78833) with lysates from C57 or Cav-1 KO neurospheres treated with Dex or ethanol for 4 h. Fractions were then separated on SDS–10% PAGE gels, and Western blotting was performed to detect total GR using the M20 rabbit polyclonal antibody (Santa Cruz; catalog no. sc-1004) and Cav-1 (BD Biosciences; catalog no. 610059). The purity of fractions was assessed using GAPDH (glyceraldehyde-3-phosphate dehydrogenase) as a marker for cytoplasmic (Santa Cruz; catalog no. sc-32233) and Lamin-B1 for nuclear proteins (Abcam; catalog no. ab16048).
ChIP.
NPSCs isolated from C57 or Cav-1 KO mice were grown as described above and treated with Dex or vehicle for 1.5 h. Cells were then fixed in 1% formaldehyde for 10 min at room temperature. After fixation, 0.125 M glycine was added for 5 min to quench the cross-linking reaction. Cells were washed with phosphate-buffered saline twice and frozen at −80 until immunoprecipitation. Prior to sonication, cell pellets were thawed on ice and lysed in chromatin immunoprecipitation (ChIP) lysis buffer (50 mM HEPES, 1 mM EDTA, 140 mM NaCl, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100) with freshly added HALT protease and phosphatase inhibitor cocktail (Thermo-Fisher). Cell lysate was centrifuged at 5,000 rpm for 5 min at 4°C, and crude nuclear pellets were collected and washed once with ChIP wash buffer (10 mM Tris-Cl, 1 mM EDTA, 200 mM NaCl) with freshly added HALT protease and phosphatase inhibitor cocktail. The pellet was then resuspended in ChIP sonication buffer (10 mM Tris-Cl, 1 mM EDTA, 0.5 mM EGTA, 0.5% N-lauroylsarcosine) with freshly added HALT protease and phosphatase inhibitor cocktail and chromatin sonicated using a Bioruptor into fragments of approximately 500 to 1,000 bp. Immunoprecipitation of GR was performed with 8 μg of a GR antibody cocktail (2 μg each of M20 [Santa Cruz], P20 [Santa Cruz], H300 [Santa Cruz] and BuGR2), 8 μg of phosphoserine-211 (ab55189), or normal rabbit IgG (Santa Cruz; catalog no. sc-2027) overnight at 4°C. A 1:1 mix of anti-rabbit and anti-mouse antibodies cross-linked to Dynabeads (Life Technologies) were used according to the manufacturer's instructions. After incubation, the beads were washed with low-salt immune complex buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-Cl [pH 8.1], 150 mM NaCl), high-salt immune complex buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-Cl [pH 8.1], 500 mM NaCl), LiCl immune complex buffer (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-Cl [pH 8.1]), and finally 1× Tris-EDTA. Cross-links were then removed via an overnight proteinase K digestion (Ambion; catalog no. AM2546), and DNA was purified using a standard phenol-chloroform extraction protocol. Purified DNA was then analyzed using qRT-PCR. Primers were designed based on GR ChIP-Seq data published previously (30–33), and the coordinates used were as follows: Fkbp-5, chromosome 17, 28556870 to 28557847; RhoJ, chromosome 12, 76404969 to 76405723; and Sgk-1, chromosome 10, 21683041 to 21683190.
Microarray data accession number.
The CEL files have been submitted to GEO under accession number GSE49804.
RESULTS
The antiproliferative effects of Dex in NPSCs require Cav-1.
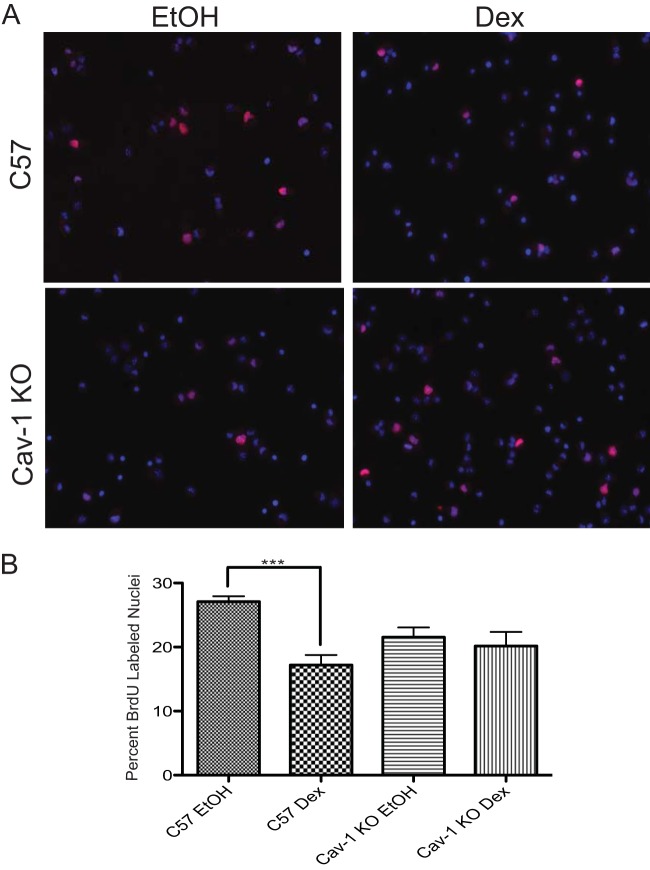
The contribution of rapid GR signaling to the antiproliferative effects of GCs exerted on a variety of tissue and cell types remains unresolved (9, 11, 16). Since rapid signaling effects of GCs on MAPK activation and GJIC are lost upon Cav-1 deletion in embryonic NPSCs (17), we tested whether the antiproliferative effects of GCs were altered in Cav-1 KO NPSCs. As shown in Fig. 1, Dex did not significantly impact proliferation of NPSCs in Cav-1 KO neurospheres, which contrasts with C57 NPSCs, where proliferation was reduced by 10%. Furthermore, basal proliferation of embryonic NPSCs was not affected by Cav-1 deletion (Fig. 1). Therefore, in embryonic NPSC cultures, Cav-1 was an essential component of a GR signaling pathway that operated to limit proliferation.
FIG 1.
Antiproliferative effects of Dex in NPSCs require Cav-1. (A) C57 or Cav-1 KO NPSCs were treated with Dex or vehicle (ethanol [EtOH]) for 24 h and pulsed with BrdU during the last hour of treatment. Immunocytochemistry was performed to detect BrdU-positive nuclei (pink). Nuclei were visualized by DAPI staining (blue). (B) Three independent coverslips per biological replicate were counted to ascertain the percentage of cells that passed through S-phase (i.e., BrdU-positive nuclei). Error bars represent the standard error of the mean (SEM; n = three biological replicates). P < 0.001 (one-way ANOVA with Tukey's posttest).
Cav-1 deletion abolishes GR induction of Sgk-1, a gene required for the antiproliferative effect of Dex in NPSCs.
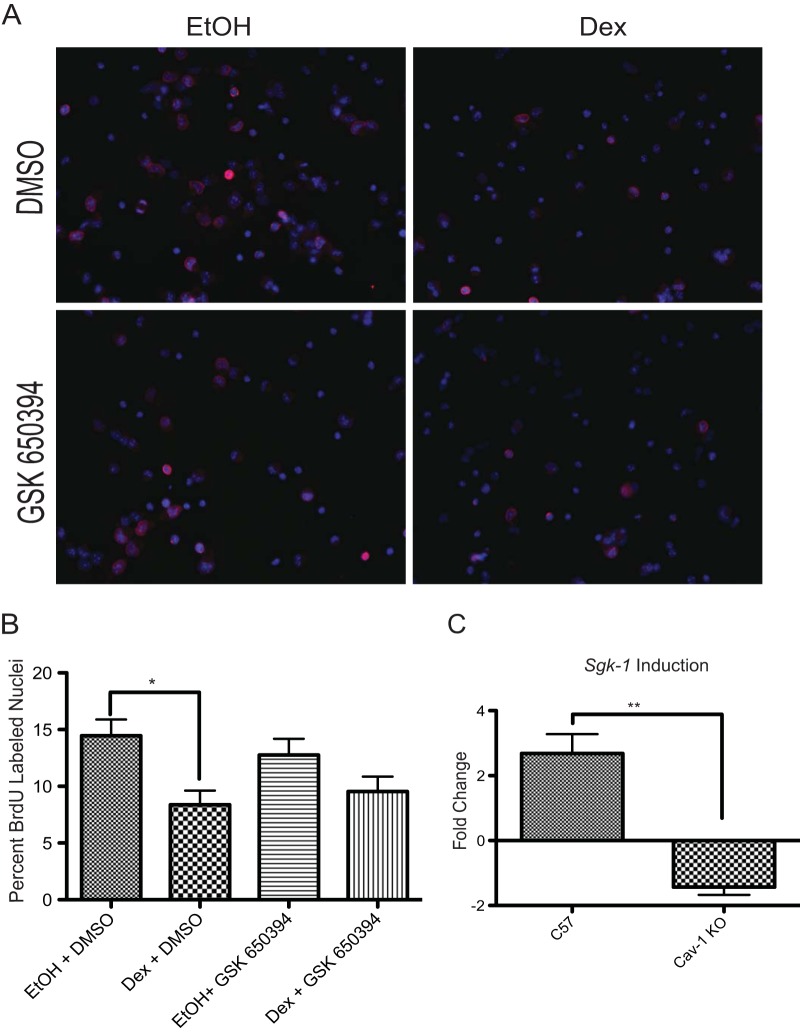
We previously established that inhibition of GJIC by the rapid action of GR was associated with an inhibition of S-phase progression in cultured NPSCs (17). Our observation that transient pharmacologic inhibition of GJIC is also sufficient to reduce NPSC proliferation (17) was recently confirmed in cultured embryonic stem cell-derived neural progenitors and in embryonic neural progenitors isolated from the cerebral cortex (34). Recent studies in human hippocampal neural progenitor cells indicate that the antiproliferative properties of GCs are dependent upon at least one genomic GR target gene, Sgk-1 (19). Consistent with these findings, Sgk-1 is also required for the antiproliferative effect of GCs in our NPSC cultures, since treatment with an SGK-1 inhibitor (GSK650394) blunts the growth-inhibitory affect of Dex (Fig. 2A and B). Sgk-1 was a GR target gene in C57 but not in Cav-1 KO NPSCs, as revealed by qRT-PCR analysis following a 4-h Dex treatment (Fig. 2C). Thus, the role of Cav-1 in GC-mediated effects on the cell cycle may extend beyond its impact on rapid GR signaling and influence genomic action of the receptor.
FIG 2.
Antiproliferative effects of Dex in NPSCs require SGK-1. (A) C57 or Cav-1 KO cells were treated with Dex and/or the SGK-1 inhibitor, GSK650394, and the appropriate vehicle (ethanol [EtOH] or dimethyl sulfoxide [DMSO]) for 24 h and pulsed with BrdU during the last hour of treatment. Immunocytochemistry was performed to detect BrdU-positive nuclei (pink). Nuclei were visualized by DAPI staining (blue). (B) Three independent coverslips per biological replicate were counted to ascertain the percentage of cells that passed through S phase (i.e., BrdU-positive nuclei). Error bars represent the SEM (n = 3 biological replicates). P < 0.05 (one-way ANOVA with Tukey's posttest). (C) qRT-PCR analysis of Sgk-1 mRNA indicates that significant induction of Sgk-1 occurs after a 4-h Dex treatment in C57 but not Cav-1 KO NPSCs (Student's t test, P < 0.01).
Analysis of Cav-1 effects on the GR transcriptome in NPSCs.
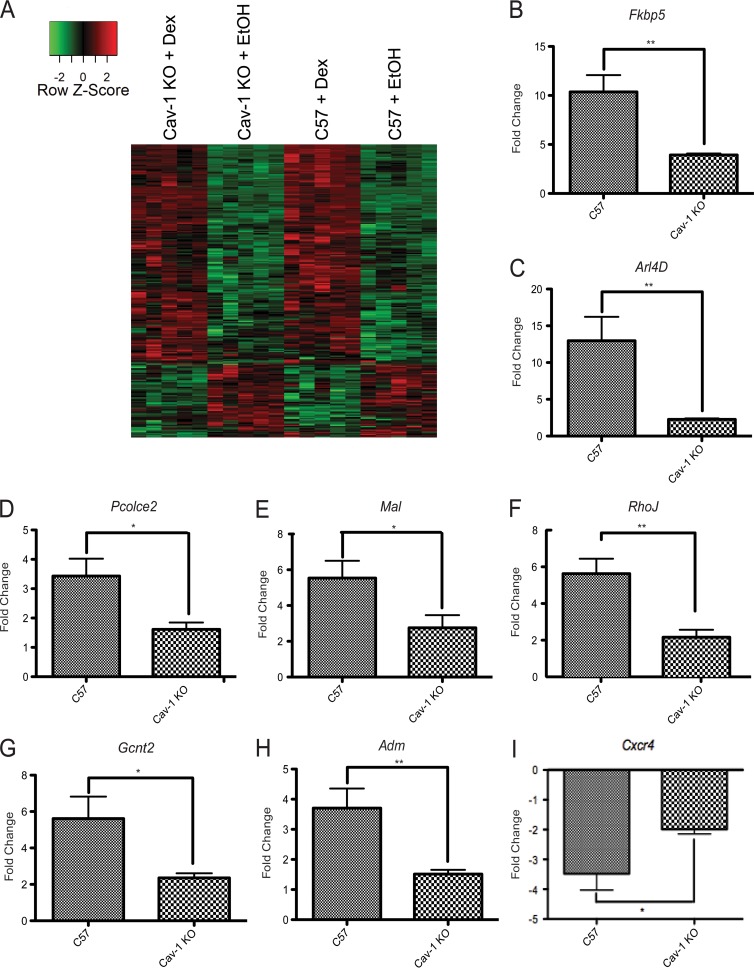
To examine Cav-1 regulation of genome-wide GR transcriptional responses, C57 and Cav-1 KO NPSCs were subjected to microarray expression analysis. Compared to C57 NPSCs, Cav-1 KO cells demonstrate differential basal expression of 186 genes (false discovery rate < 0.1) with 44 genes exhibiting higher expression in Cav-1 KO cells and 20 genes lower expression by at least 1.5-fold (Table S1). The transcripts with lowest expression in the Cav-1 KO versus C57 cells were Cav-1, as expected, and the ribosomal protein S9 (Rps9) gene, whose expression had previously been shown to be highly dependent upon Cav-1 (35). When Dex treated Cav-1 KO and C57 NPSCs were compared for gene expression, 568 genes (excluding Cav-1 and Rps9) were differentially Dex responsive (false discovery rate < 0.1) (see Table S1 in the supplemental material). In general, Dex-responsive genes were regulated in the same direction in both C57 and Cav-1 KO NPSCs as shown in a heat map (Fig. 3A). However, individual genes do exhibit differences in Dex responsiveness between the two genotypes.
FIG 3.
A subset of genes are differentially regulated by GR in C57 versus Cav-1 KO NPSCs. (A) Hierarchical gene clustering of 20 NPSC cultures using Dex-responsive genes in C57 and Cav-1 KO NPSCs (Student t test; false discovery rate < 0.1) using Pearson dissimilarity as the distance measure and the average linkage method for linkage analysis. The data are represented using a z-score normalized before plotting the heat map. For each gene (row), the z-score was calculated by subtracting the expression value by mean expression across all samples (centering) and dividing by the standard deviation (scaling). (B-I) C57 or Cav-1 KO NPSCs from tissues independent of those used for microarray analysis were treated for 4 h with 100 nM Dex and mRNA expression of indicated genes analyzed using qRT-PCR. Although all genes shown are significantly induced in response to Dex, activation is attenuated in the Cav-1 KO cells. Error bars represent the SEM (n = 6). *, P < 0.05; **, P < 0.01 (Student's t test).
A number of genes that were induced by Dex at least 2.5-fold and exhibited robust differences in Dex-regulated expression between C57 and Cav-1 KO NPSCs (see Table S1 in the supplemental material) were validated by qRT-PCR using independent NPSC cultures (Fig. 3B to I). For this analysis, we examined both well-established GR target genes (e.g., Fkbp5) and genes with limited reports of GC responsiveness (e.g., Gcnt2). Specifically, Dex induction of Fkbp5, Gcnt2, RhoJ, Pcolce2, Mal, Adm, and Arl4d mRNA expression was significantly reduced in Cav-1 KO versus C57 NPSCs (Fig. 3). We also observed that Cxcr4 did not attain appropriate level of repression in response to Dex treatment in Cav-1 KO NPSCs (Fig. 3). Therefore, Cav-1 effects on GR action are not limited to rapid signaling (17) but also extend globally to genomic responses.
IPA was used to examine components of molecular networks and pathways defined by Dex-regulated genes in C57 and Cav-1 KO NPSCs. As shown in Fig. S1C and D in the supplemental material, the most significant functional networks defined by Dex-responsive genes in C57 NPSCs were distinct from networks formed in Cav-1 KO NPSCs. For example, in C57 NPSCs, cell cycle genes comprise the top two highly rated networks; 9 of the next 10 highly rated gene networks included molecules involved in development, particularly in the nervous system (see Fig. S1C in the supplemental material). Figure S1A in the supplemental material shows the most highly rated cell cycle network in Dex-treated C57 NPSCs. Included in this network are genes such as Pcolce2 (Fig. 3D), which was validated as a GR target by qRT-PCR of independent samples. In addition, the top canonical pathways within this cell cycle network include a number of nuclear receptors: retinoid acid receptor, GR, peroxisome proliferation-activating receptor alpha/retinoid X receptor alpha (RXRα), estrogen receptor, and thyroid hormone receptor/RXRα.
In contrast to C57 Dex-regulated networks, Cav-1 KO NPSCs demonstrated only one highly rated network involved in nervous system development (see Fig. S1D in the supplemental material). Although GR signaling pathways were part of nervous system development networks in both Dex-treated C57 and Cav-1 KO NPSCs, the genes in the two networks were different (see Fig. S2A and B in the supplemental material). In addition, the only cell cycle network (sixth highest) in Dex-regulated Cav-1 KO NPSCs was unrelated to the highest-rated cell cycle network in Dex-treated C57 NPSCs (see Fig. S1A and B).
The analysis described above used Dex-regulated genes from C57 and Cav-1 KO NPSCs, including those that were similarly responsive to hormone in the two genotypes. To examine differences in Dex responsiveness between the two genotypes, IPA was performed using genes determined to be significantly different by ANOVA with regard to the Dex response. This analysis showed that approximately half (i.e., 13) of the 25 highly rated networks formed by differentially Dex-responsive genes were involved in some aspect of organ or tissue development (see Fig. S3C in the supplemental material). In fact, the most highly rated networks function in nervous system development (see Fig. S3A and B in the supplemental material), protein ubiquitylation (see Fig. S3C), and axon guidance or cellular/extracellular matrix interactions (see Fig. S3C). GC effects on neuronal migration and ubiquitin-proteasome mediated degradation have previously been observed in neural stem cells (9, 36) and may be influenced by Cav-1. To summarize, in addition to identifying many novel primary GC responsive genes in NPSCs, our microarray data implicated Cav-1 as a modulator of both rapid signaling and genomic action of GR.
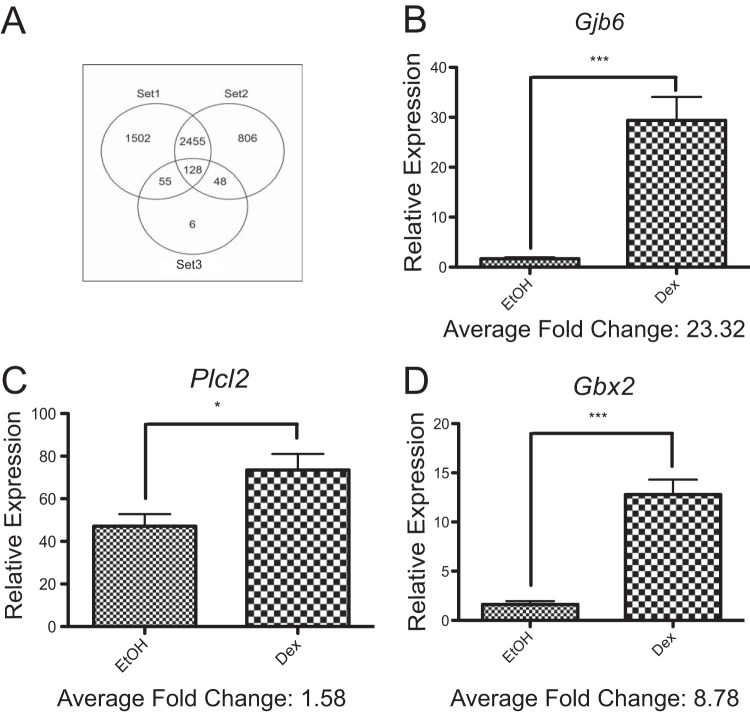
To identify Dex-regulated genes that are unique to neural stem cells, the NextBio data mining tools (see Materials and Methods) were used to compare the GR-regulated transcriptome in NPSCs to others from cell and tissue sources as diverse as the 3134 mouse mammary adenocarcinoma cell line (22) and a mouse oligodendrocyte progenitor cell line (26) (Table 1). Specifically, this analysis included 14 separate studies: a union of differentially expressed genes from each of these studies generated 5,000 GC-regulated genes (see Table S2 in the supplemental material). A comparison of the GC-regulated gene list in our NPSC cultures (Fig. 4A, set 3) with a subset of these studies that used closely related cell types (i.e., rat neural [25] and oligodendrocyte [26] progenitors; Fig. 4A, set 2) revealed 176 common genes (Fig. 4A). However, of the 5,000 GC-regulated genes from 14 data sets, only six were uniquely GC regulated in our mouse NPSC cultures: Gjb6, Gbx2, Card10, Plcl2, Smc5, and Rnf157. Three of these genes (Gjb6, Gbx2, and Plcl2) were validated as Dex regulated in qRT-PCR analysis of independent samples (Fig. 4B to D). Therefore, the GR transcriptome in three-dimensional cultures of mouse NPSCs only included a limited number of unique target genes.
FIG 4.
NextBio analysis reveals a subset of genes regulated by Dex in embryonic mouse NPSC cultures. (A) Venn diagram comparing the GC-regulated gene lists in the 14 studies used to perform meta-analysis in NextBio. The genes contained in set 1 are from study ID numbers 3 to 8 and 12 to 14 (Table 1) and derived from cell types most distant from our mouse embryonic NPSCs (set 3). The genes contained in set 2 are from study ID numbers 9 to 11 (Table 1) and derived from cell types closely related to our mouse embryonic NPSCs (i.e., rat neural progenitors or mouse oligodendrocyte progenitor cells). (B to D) C57 KO NPSCs from tissues independent of those used in the microarray were treated for 4 h with 100 nM Dex and induction of Gjb6 (B), Plcl2 (C), and Gbx2 (D) mRNA analyzed using qRT-PCR. Error bars represent the SEM (n = 6). *, P < 0.05; ***, P < 0.001 (Student's t test).
Cav-1 influences site-specific GR phosphorylation.
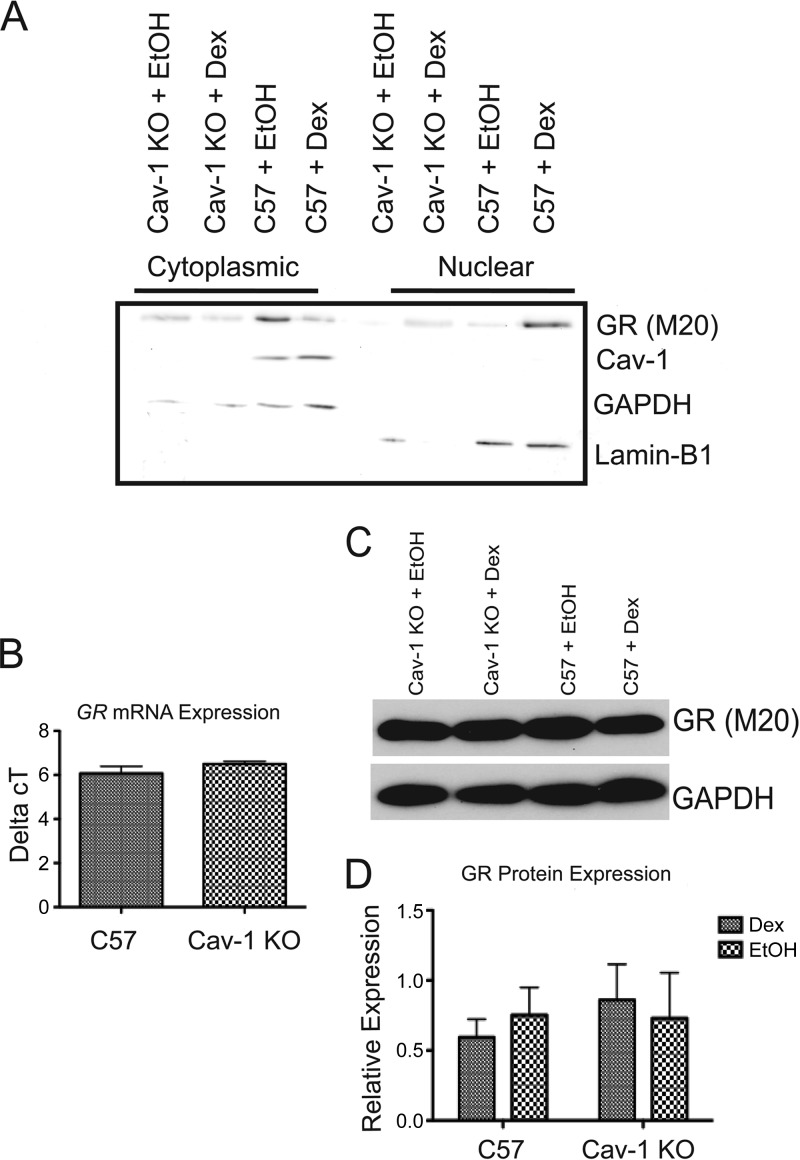
To determine a mechanism for Cav-1 effects on GR transcriptional activity, we first sought to determine whether Cav-1 was found in the nucleus of NPSCs. In ovarian carcinoma cell lines, Cav-1 regulates cell cycle regulatory gene expression through direct DNA binding (37). Cav-1 also affects transcription in lung epithelial Beas-2B cells by directly binding the transcription factor nuclear erythroid 2 p45-related factor 2 (Nrf2) (38). Our previous work (17) revealed an interaction between Cav-1 and GR in NPSCs. However, Cav-1 was undetectable in nuclear fractions prepared from untreated or Dex-treated NPSCs (Fig. 5A). Dex-dependent nuclear translocation of GR occurred in both genotypes (Fig. 5A). Furthermore, GR protein and mRNA levels were unaltered in Cav-1 KO neurospheres (Fig. 5D). Therefore, Cav-1 may indirectly affect GR transcriptional response via regulation of a cytoplasmic signaling pathway that ultimately impacts GR bound at specific gene targets.
FIG 5.
Cav-1 protein is not detectable in the nucleus, nor does the loss of Cav-1 affect GR protein or mRNA expression. (A) Cytoplasmic and nuclear fractions prepared from C57 and Cav-1 KO NPSCs treated for 1 h with 100 nM Dex or EtOH vehicle were subjected to Western blot analysis to detect GR, Cav-1, and markers for cytoplasmic (GAPDH) or nuclear (lamin B1) proteins. Cav-1 was not detected in nuclear fractions from C57 NPSCs lysates and in either cytoplasmic or nuclear fractions from Cav-1 KO cells. The blot shown is representative of three independent experiments. (B) qRT-PCR analysis indicated no difference in GR mRNA expression between C57 and Cav-1 KO cells (n = 5). (C and D) Western blot analysis also indicated no difference in GR protein (relative to GAPDH) expression between C57 and Cav-1 KO cells treated with 100 nM Dex or EtOH vehicle for 4 h (n = 4). Error bars represent the SEM.
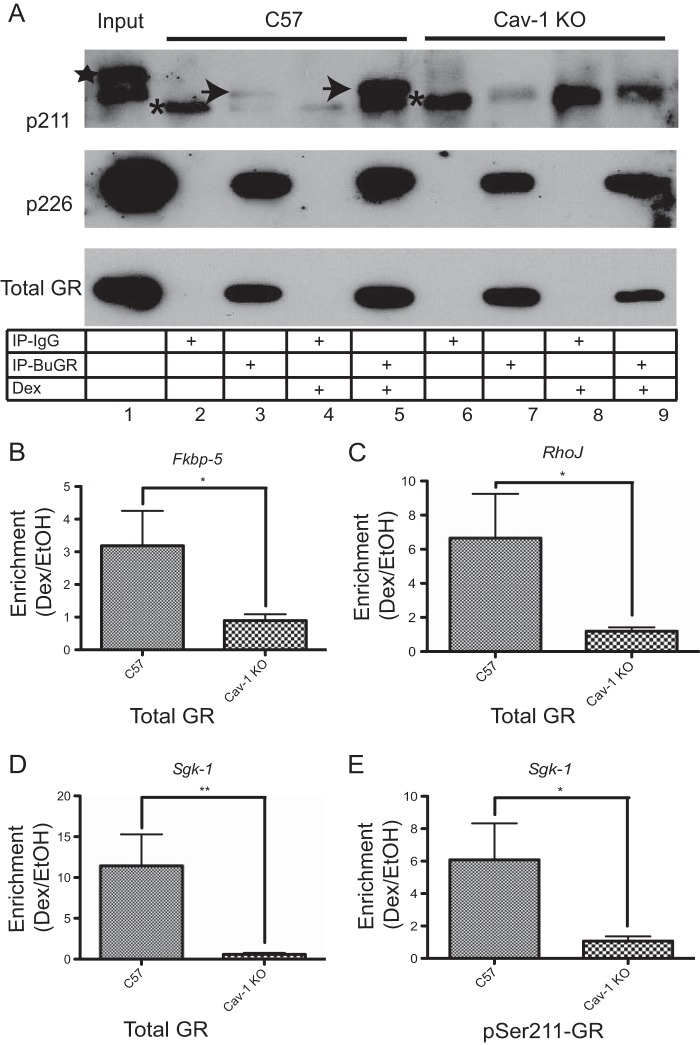
Site-specific GR phosphorylation influences its transcriptional regulatory activity. In fact, different phosphorylation patterns within the amino-terminal activation function-1 domain can dictate which target genes will be bound by GR (29). Both MAPKs, targets of rapid GR signaling and cyclin-dependent kinase 2 (CDK2), phosphorylate GR and influence its transcriptional activity (39). Therefore, we tested the impact of Cav-1 deletion on MAPK- and CDK2-dependent phosphorylation of GR at serine-211 (S211) and serine-226 (S226), respectively. As shown in Fig. 6, phosphorylation at S211, a Dex-responsive site, was undetectable in Cav-1 KO neurospheres. Since hormone-dependent phosphorylation at S211 is associated with transcriptionally active GR, the loss of phosphorylation at S211 may explain diminished Dex responsiveness of select genes in Cav-1 KO NPSCs (e.g., Fig. 3). GR phosphorylation at S226 is unaltered in Cav-1 KO NPSCs. Therefore, Cav-1 does not alter global GR phosphorylation.
FIG 6.
GR phosphorylation at S211 but not S226 is altered in Cav-1 KO NPSCs. (A) Whole-cell lysates from C57 and Cav-1 KO NPSCs treated for 1 h with 100 nM Dex or EtOH vehicle were subjected to immunoprecipitation with the BuGR-2 mouse monoclonal antibody against GR or nonimmune mouse IgG and then subjected to Western blot analysis to detect total GR or phospho-S211 or phospho-S226 GR isoforms. Asterisks show the nonspecific band detected in all lanes following pulldown with nonimmune IgG. The identity of the higher-molecular-weight band in the input lane (star) is unknown, but it was not detected in anti-GR antibody immunoprecipitates. The Dex-inducible phospho-S211 isoform, detectable in C57 but not Cav-1 KO NPSC lysates, is indicated by the arrows. GR phosphorylation at S226 is similar in C57 and Cav-1 KO lysates. The blot is representative of three biologically independent experiments. ChIP experiments using total GR (B to D) or phospho-S211 (E) antibodies indicates attenuated recruitment of GR to target genes in response to a 1.5-h treatment with 100 nM Dex. DNA was analyzed by using qRT-PCR, and final values are shown relative to total and IgG-negative controls before comparing enrichment in Dex versus EtOH (n = 3 biological replicates for C57 and n = 5 for Cav-1 KO cells). Error bars represent the SEM. *, P < 0.05; **, P < 0.01 (Student's t test).
Cav-1 influences recruitment of GR to chromatin of target genes.
We next sought to determine whether any of the genes differentially regulated in C57 and Cav-1 KO NPSCs (Fig. 3) were direct targets of GR and whether recruitment of GR to gene regulatory regions was altered in Cav-1 KO cells. Directed ChIP assays were therefore used to reveal GR recruitment to select target genes affected by Cav-1 KO, using previously reported GR binding sites in other cell lines (30–33). As shown in Fig. 6, three genes validated as differentially expressed in C57 versus Cav-1 KO NPSCs have significantly decreased chromatin recruitment of GR in response to Dex treatment in Cav-1 KO NPSCs (Fig. 6B to D). Since GR phosphorylation at Ser211 is undetectable in Cav-1 KO cells (Fig. 6A), we did not expect to detect any recruitment of pSer211-GR to GC-regulated genes. Nonetheless, this we confirmed by ChIP assays with the pSer211-GR antibody, as shown in Fig. 6E, which also demonstrated diminished recruitment of pSer211-GR to the Sgk-1 promoter in Cav-1 KO NPSCs. Therefore, reduced transcriptional responses to Dex in Cav-1 KO NPSCs can be mediated by diminished recruitment of select GR phosphoisoforms to target gene regulatory sites.
DISCUSSION
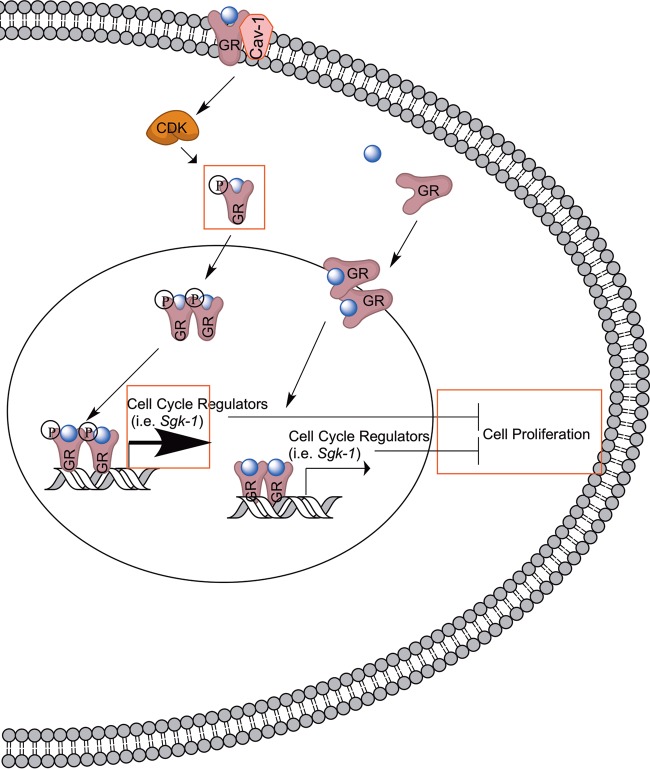
The nonclassical rapid activation of nuclear receptors residing at the plasma membrane mobilizes various cytoplasmic signaling pathways that can either directly alter cellular physiology (16, 17) or indirectly modulate transcriptional responses (40). Such cross talk between rapid nuclear receptor signaling and classical genomic action can utilize nuclear receptors at target genes or independent transcription factors. In this report, we demonstrated that cross talk between GR rapid and genomic pathways in embryonic NPSCs utilized Cav-1, a lipid raft protein. Cav-1 did not act as a classical coregulator of GR on genomic targets but rather altered the phosphorylation of GR on at least one site, S211. Phosphorylation of S211 in U2OS cells influences the GC transcriptional program (29) and may likewise impact target genes regulated by GR in NPSCs. In fact, alterations of the GR transcriptome in NPSCs upon Cav-1 deletion define unique networks that potentially alter GC regulation of the cell cycle (see model in Fig. 7).
FIG 7.
Model for Cav-1-mediated effects on the genomic actions of GR. As shown on the right, previous work demonstrated that GR binds to the promoter of Sgk-1 (52), a GR target implicated in the antiproliferative effects of GC in neural progenitor cells (19). New findings from the present study are surrounded by rectangles.
The most prominent pathways defined by the GR transcriptome in C57 NPSCs regulated cell cycle progression and contain a number of genes that could contribute to the antiproliferative effects of GCs (9, 16, 17). Notably, one validated GR target gene, Sgk-1, is required for the antiproliferative effects of GC in embryonic mouse NPSCs (the present study) and a human hippocampal progenitor cell line (19) but did not reach significance as GC regulated in our microarray analysis or that reported in hippocampal progenitors (see Table S2 in the supplemental material) (23). The high stringency applied to microarray data sets, while reducing the number of false positives, may miss bona fide GR targets that play an important role in the biological effects of GCs. The inability of GR to induce Sgk-1 expression in Cav-1 KO NPSCs may underlie in part the loss of an antiproliferative response to Dex. Importantly, a number of cell cycle-regulated genes were also found to be differentially responsive to Dex in C57 versus Cav-1 KO NPSCs, so there are likely to be multiple GR targets that participate in the complex regulation of proliferation in NPSCs, which are influenced by Cav-1. Since the GR transcriptome in C57 and Cav-1 KO embryonic NPSC cultures also defines a number of pathways implicated in neuronal development, future studies may reveal a role for Cav-1 operating through either rapid signaling (17) or cross talk with genomic GR pathways to influence antenatal effects of GCs on neurodevelopment.
Our study supports a role for Cav-1 in mediating the genomic action of GR in addition to its role in rapid GR signaling. NPSCs null for Cav-1 contain a subset of genes that are differentially regulated by GR in response to Dex. Of the subset that we validated, Fkbp5 is a known target of GR involved in negative regulation of GR and the stress response. Improper activity of Fkbp5 can contribute to neuropsychiatric diseases such as posttraumatic stress disorder (41). Alterations in the regulation of Fkbp5 may explain why adult Cav-1 KO animals tend to exhibit more anxious behaviors than their wild-type counterparts (42).
Gjb6, Gbx2, and Plcl2 were novel validated GR target genes in NPSC cultures but their role in regulating neural stem or progenitor cell function has yet to be determined. Gjb6 encodes connexin 30, a gap junction protein expressed primarily in astrocytes but not until postnatal day 10 in mice (43). Therefore, Dex induction of Gjb6 in our neurosphere cultures may be reflective of GR action in astrocyte progenitor cells. Gbx2 is a homeobox transcriptional factor that has been shown to regulate various aspects of neural stem cell differentiation (44) and may contribute to the reported effects of GCs on differentiation of distinct neuroprogenitors (11–13). No role for phospholipase C-related protein (Plcl2) in neurodevelopment or GC action has been reported, although Plcl2 was identified in exome sequencing analysis as one of 40 genes with protein coding sequence variations in schizophrenia patients (45).
Of the 25 highly rated networks identified by our analysis of genes differentially responsive to Dex in C57 versus Cav-1 KO NPSCs, approximately half were related to organ or tissue development—the top two from our data set involving nervous system development. Notably, one of these pathways involves protein ubiquitinylation. Multiple studies have revealed an impact of GCs on the protein degradation of NPSCs. For example, GC treatment in rat embryonic neural stem cells results in decreased proliferation by enhancing ubiquitin-mediated degradation of the cell cycle regulator cyclin D1 (9). In embryonic rat cells, Dex treatment increases the expression of a deubiquitinating enzyme, Usp8/Ubpy that can indirectly cause increased degradation of the BRUCE/Apollo inhibitor of apoptosis protein. As a result of increased BRUCE degradation, there is a consequent decrease in cell proliferation (36). Interestingly, Cav-1 interacts with and regulates the polyubiquitinylation of active Rac1 (46). Rac1 is a member of the Rho-like GTPases and also involved in actin polymerization and protrusion and cell migration.
The second most highly regulated pathway that we uncovered during our network analysis involved axon guidance. Interestingly, RhoJ and Arl4D were validated as genes differentially regulated between C57 and Cav-1 KO and are both implicated in actin-mediated cell migration (47, 48). Furthermore, Cxcr4 was also validated as a differentially regulated gene and is implicated in dopaminergic cell migration (49). GC effects on neural stem cell migration are well documented (25). Therefore, GR signaling mediated by Cav-1 could have a critical impact on the biology and development of NPSCs by regulating the degradation of cell cycle regulators or migration of differentiated cells derived from NPSCs to their final position in the cortex.
Although Cav-1 was not found in the nucleus of NPSCs, it influenced site-specific GR phosphorylation, which in turn could impact the GR transcriptome. Specifically, phosphorylation at S211 but not S226 was affected in Cav-1 KO NPSCs. Therefore, the GC response of genes that require pSer211-GR to reach peak activation may be altered in Cav-1 KO NPSCs. For example, pSer211-GR is recruited to promoters of genes such as Gilz and Tat to activate their transcription (29). Since GR phosphorylated at S211 is associated with activated gene transcription, alterations in pSer211-GR are consistent with the attenuated activation of select GR target genes in Cav-1 KO NPSCs. The results from ChIP assays on select GR target genes provide additional mechanistic insights regarding the role of Cav-1 in GC-regulated transcription and show reduced recruitment of GR (and pSer211-GR) to select regulatory regions of genes differentially responsive to Dex in C57 versus Cav-1 KO NPSCs. Since GR is recruited to multiple binding sites of its target genes, a more comprehensive analysis of the GR cistrome in C57 versus Cav-1 KO NPSCs using ChIP-seq will be required to uncover other mechanistic features of site-specific Cav-1-dependent GR recruitment. Since the majority of Dex-regulated genes in NPSCs were unaffected by Cav-1 KO, GR chromatin recruitment is not globally regulated by Cav-1. Finally, other future experiments directed toward identifying genes occupied by distinct GR phosphoisoforms (e.g., pSer211-GR) in NPSCs could identify specific GC-regulated gene networks that are influenced by cross talk with cytoplasmic signaling pathways. In fact, alterations in GR phosphorylation in specific adult rat brain regions are generated in response to specific stresses and may impact select genes that modulate behavioral responses to stressful states (50).
Phosphorylation of GR at S211 and S226 is mediated by two different kinases—CDK2 at S211 and MAPK at S226. Others have suggested that Cav-1 is a tumor suppressor gene due to its role in mitogenic signaling (51). Ablation of Cav-1 in metastatic lung cancer cell lines results in proliferation arrest and decreased expression of cyclin D1 and CDK4 (51). Therefore, Cav-1 loss may independently or in conjunction with rapid GR signaling impact CDK2 and consequently genomic action of GR (Fig. 7).
In summary, Cav-1 is a multifunctional regulator of GR action in NPSCs. Cav-1 influences both GR-dependent rapid changes in intercellular communication through gap junctions (17), which is required for the establishment of cerebral cortical architecture (34) and the GR transcriptome, including genes responsible for regulating NPSC proliferation. The wide variety of GR gene networks affected by Cav-1 also suggests that this novel regulator of receptor action could impact the ultimate fate and laminar position of cells derived from embryonic cerebral cortical NPSCs exposed prematurely to GCs during fetal development.
Supplementary Material
ACKNOWLEDGMENTS
We thank Teresa Liu, Marcia Lewis, Christin Sculli, and Janie Zhang for technical assistance in these studies. In addition, we thank Bill LaFramboise and Selma Witchel for helpful critiques and suggestions.
This study was supported by Public Health Service grants DK078394 from the National Institute of Diabetes and Digestive and Kidney Diseases (D.B.D.), MH086651 from the National Institute of Mental Health (M.J.G.), AG030636 from the National Institute on Aging (F.G.), and a T32 training grant T32GM008424 from the National Institute of General Medical Sciences (M.E.P.). In addition, support was received from the American Heart Association (13GRNT16560012 to F.G. and 12SDG8800012 to D.V.). This project used the University of Pittsburgh Cancer Institute's Cancer Informatics and Cancer Biomarkers Facility Cores, which are supported in part by Public Health Service award P30CA047904.
Footnotes
Published ahead of print 28 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01121-13.
REFERENCES
- 1.George AA, Schiltz RL, Hager GL. 2009. Dynamic access of the glucocorticoid receptor to response elements in chromatin. Int. J. Biochem. Cell Biol. 41:214–224. 10.1016/j.biocel.2008.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rhen T, Cidlowski JA. 2005. Anti-inflammatory action of glucocorticoids: new mechanisms for old drugs. N. Engl. J. Med. 353:1711–1723. 10.1056/NEJMra050541 [DOI] [PubMed] [Google Scholar]
- 3.ACOG. 2011. ACOG Committee opinion no. 475: antenatal corticosteroid therapy for fetal maturation. Obstet. Gynecol. 117:422–424. 10.1097/AOG.0b013e31820eee00 [DOI] [PubMed] [Google Scholar]
- 4.Miller WL, Witchel SF. 2013. Prenatal treatment of congenital adrenal hyperplasia: risks outweigh benefits. Am. J. Obstet. Gynecol. 208:354–359. 10.1016/j.ajog.2012.10.885 [DOI] [PubMed] [Google Scholar]
- 5.Crowther CA, Doyle LW, Haslam RR, Hiller JE, Harding JE, Robinson JS. 2007. Outcomes at 2 years of age after repeat doses of antenatal corticosteroids. N. Engl. J. Med. 357:1179–1189. 10.1056/NEJMoa071152 [DOI] [PubMed] [Google Scholar]
- 6.Wapner RJ, Sorokin Y, Mele L, Johnson F, Dudley DJ, Spong CY, Peaceman AM, Leveno KJ, Malone F, Caritis SN, Mercer B, Harper M, Rouse DJ, Thorp JM, Ramin S, Carpenter MW, Gabbe SG. 2007. Long-term outcomes after repeat doses of antenatal corticosteroids. N. Engl. J. Med. 357:1190–1198. 10.1056/NEJMoa071453 [DOI] [PubMed] [Google Scholar]
- 7.Damsted SK, Born AP, Paulson OB, Uldall P. 2011. Exogenous glucocorticoids and adverse cerebral effects in children. Eur. J. Paediatr. Neurol. 15:465–477. 10.1016/j.ejpn.2011.05.002 [DOI] [PubMed] [Google Scholar]
- 8.Davis EP, Sandman CA, Buss C, Wing DA, Head K. 2013. Fetal glucocorticoid exposure is associated with preadolescent brain development. Biol. Psychiatr. 74:647–655. 10.1016/j.biopsych.2013.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sundberg M, Savola S, Hienola A, Korhonen L, Lindholm D. 2006. Glucocorticoid hormones decrease proliferation of embryonic neural stem cells through ubiquitin-mediated degradation of cyclin D1. J. Neurosci. 26:5402–5410. 10.1523/JNEUROSCI.4906-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noguchi KK, Walls KC, Wozniak DF, Olney JW, Roth KA, Farber NB. 2008. Acute neonatal glucocorticoid exposure produces selective and rapid cerebellar neural progenitor cell apoptotic death. Cell Death Differ. 15:1582–1592. 10.1038/cdd.2008.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moors M, Bose R, Johansson-Haque K, Edoff K, Okret S, Ceccatelli S. 2012. Dickkopf 1 mediates glucocorticoid-induced changes in human neural progenitor cell proliferation and differentiation. Toxicol. Sci. 125:488–495. 10.1093/toxsci/kfr304 [DOI] [PubMed] [Google Scholar]
- 12.Sabolek M, Herborg A, Schwarz J, Storch A. 2006. Dexamethasone blocks astroglial differentiation from neural precursor cells. Neuroreport 17:1719–1723. 10.1097/01.wnr.0000236862.08834.50 [DOI] [PubMed] [Google Scholar]
- 13.Wagner K, Couillard-Despres S, Lehner B, Brockhoff G, Rivera FJ, Blume A, Neumann I, Aigner L. 2009. Prolactin induces MAPK signaling in neural progenitors without alleviating glucocorticoid-induced inhibition of in vitro neurogenesis. Cell Physiol. Biochem. 24:397–406. 10.1159/000257432 [DOI] [PubMed] [Google Scholar]
- 14.Oitzl MS, Champagne DL, van der Veen R, de Kloet ER. 2010. Brain development under stress: hypotheses of glucocorticoid actions revisited. Neurosci. Biobehav. Rev. 34:853–866. 10.1016/j.neubiorev.2009.07.006 [DOI] [PubMed] [Google Scholar]
- 15.Heitzer MD, Wolf IM, Sanchez ER, Witchel SF, DeFranco DB. 2007. Glucocorticoid receptor physiology. Rev. Endocrinol. Metab. Disord. 8:321–330. 10.1007/s11154-007-9059-8 [DOI] [PubMed] [Google Scholar]
- 16.Matthews L, Berry A, Ohanian V, Ohanian J, Garside H, Ray D. 2008. Caveolin mediates rapid glucocorticoid effects and couples glucocorticoid action to the antiproliferative program. Mol. Endocrinol. 22:1320–1330. 10.1210/me.2007-0154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samarasinghe RA, Di Maio R, Volonte D, Galbiati F, Lewis M, Romero G, DeFranco DB. 2011. Nongenomic glucocorticoid receptor action regulates gap junction intercellular communication and neural progenitor cell proliferation. Proc. Natl. Acad. Sci. U. S. A. 108:16657–16662. 10.1073/pnas.1102821108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammes SR, Levin ER. 2011. Minireview: recent advances in extranuclear steroid receptor actions. Endocrinology 152:4489–4495. 10.1210/en.2011-1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anacker C, Cattaneo A, Musaelyan K, Zunszain PA, Horowitz M, Molteni R, Luoni A, Calabrese F, Tansey K, Gennarelli M, Thuret S, Price J, Uher R, Riva MA, Pariante CM. 2013. Role for the kinase SGK1 in stress, depression, and glucocorticoid effects on hippocampal neurogenesis. Proc. Natl. Acad. Sci. U. S. A. 110:8708–8713. 10.1073/pnas.1300886110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bolstad BM, Irizarry RA, Astrand M, Speed TP. 2003. A comparison of normalization methods for high density oligonucleotide array databased on variance and bias. Bioinformatics 19:185–193. 10.1093/bioinformatics/19.2.185 [DOI] [PubMed] [Google Scholar]
- 21.Simon R, Lam A, Li MC, Ngan M, Menenzes S, Zhao Y. 2007. Analysis of gene expression data using BRB-ArrayTools. Cancer Inform. 3:11–17 [PMC free article] [PubMed] [Google Scholar]
- 22.John S, Johnson TA, Sung MH, Biddie SC, Trump S, Koch-Paiz CA, Davis SR, Walker R, Meltzer PS, Hager GL. 2009. Kinetic complexity of the global response to glucocorticoid receptor action. Endocrinology 150:1766–1774. 10.1210/en.2008-0863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anacker C, Cattaneo A, Luoni A, Musaelyan K, Zunszain PA, Milanesi E, Rybka J, Berry A, Cirulli F, Thuret S, Price J, Riva MA, Gennarelli M, Pariante CM. 2013. Glucocorticoid-related molecular signaling pathways regulating hippocampal neurogenesis. Neuropsychopharmacology 38:872–883. 10.1038/npp.2012.253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuo T, Lew MJ, Mayba O, Harris CA, Speed TP, Wang JC. 2012. Genome-wide analysis of glucocorticoid receptor-binding sites in myotubes identifies gene networks modulating insulin signaling. Proc. Natl. Acad. Sci. U. S. A. 109:11160–11165. 10.1073/pnas.1111334109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fukumoto K, Morita T, Mayanagi T, Tanokashira D, Yoshida T, Sakai A, Sobue K. 2009. Detrimental effects of glucocorticoids on neuronal migration during brain development. Mol. Psychiatr. 14:1119–1131. 10.1038/mp.2009.60 [DOI] [PubMed] [Google Scholar]
- 26.Gobert RP, Joubert L, Curchod ML, Salvat C, Foucault I, Jorand-Lebrun C, Lamarine M, Peixoto H, Vignaud C, Fremaux C, Jomotte T, Francon B, Alliod C, Bernasconi L, Abderrahim H, Perrin D, Bombrun A, Zanoguera F, Rommel C, Hooft van Huijsduijnen R. 2009. Convergent functional genomics of oligodendrocyte differentiation identifies multiple autoinhibitory signaling circuits. Mol. Cell. Biol. 29:1538–1553. 10.1128/MCB.01375-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pantoja C, Huff JT, Yamamoto KR. 2008. Glucocorticoid signaling defines a novel commitment state during adipogenesis in vitro. Mol. Biol. Cell 19:4032–4041. 10.1091/mbc.E08-04-0420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.So AY, Cooper SB, Feldman BJ, Manuchehri M, Yamamoto KR. 2008. Conservation analysis predicts in vivo occupancy of glucocorticoid receptor-binding sequences at glucocorticoid-induced genes. Proc. Natl. Acad. Sci. U. S. A. 105:5745–5749. 10.1073/pnas.0801551105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blind RD, Garabedian MJ. 2008. Differential recruitment of glucocorticoid receptor phospho-isoforms to glucocorticoid-induced genes. J. Steroid Biochem. Mol. Biol. 109:150–157. 10.1016/j.jsbmb.2008.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu CY, Mayba O, Lee JV, Tran J, Harris C, Speed TP, Wang JC. 2010. Genome-wide analysis of glucocorticoid receptor binding regions in adipocytes reveal gene network involved in triglyceride homeostasis. PLoS One 5:e15188. 10.1371/journal.pone.0015188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steger DJ, Grant GR, Schupp M, Tomaru T, Lefterova MI, Schug J, Manduchi E, Stoeckert CJ, Jr, Lazar MA. 2010. Propagation of adipogenic signals through an epigenomic transition state. Genes Dev. 24:1035–1044. 10.1101/gad.1907110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA. 2011. Chromatin accessibility predetermines glucocorticoid receptor binding patterns. Nat. Genet. 43:264–268. 10.1038/ng.759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siersbaek R, Nielsen R, John S, Sung MH, Baek S, Loft A, Hager GL, Mandrup S. 2011. Extensive chromatin remodeling and establishment of transcription factor “hot spots” during early adipogenesis. EMBO J. 30:1459–1472. 10.1038/emboj.2011.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malmersjo S, Rebellato P, Smedler E, Planert H, Kanatani S, Liste I, Nanou E, Sunner H, Abdelhady S, Zhang S, Andang M, El Manira A, Silberberg G, Arenas E, Uhlen P. 2013. Neural progenitors organize in small-world networks to promote cell proliferation. Proc. Natl. Acad. Sci. U. S. A. 110:E1524–E1532. 10.1073/pnas.1220179110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mercier I, Casimiro MC, Zhou J, Wang C, Plymire C, Bryant KG, Daumer KM, Sotgia F, Bonuccelli G, Witkiewicz AK, Lin J, Tran TH, Milliman J, Frank PG, Jasmin JF, Rui H, Pestell RG, Lisanti MP. 2009. Genetic ablation of caveolin-1 drives estrogen-hypersensitivity and the development of DCIS-like mammary lesions. Am. J. Pathol. 174:1172–1190. 10.2353/ajpath.2009.080882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sippel M, Rajala R, Korhonen L, Bornhauser B, Sokka AL, Naito M, Lindholm D. 2009. Dexamethasone regulates expression of BRUCE/Apollon and the proliferation of neural progenitor cells. FEBS Lett. 583:2213–2217. 10.1016/j.febslet.2009.06.018 [DOI] [PubMed] [Google Scholar]
- 37.Sanna E, Miotti S, Mazzi M, De Santis G, Canevari S, Tomassetti A. 2007. Binding of nuclear caveolin-1 to promoter elements of growth-associated genes in ovarian carcinoma cells. Exp. Cell Res. 313:1307–1317. 10.1016/j.yexcr.2007.02.005 [DOI] [PubMed] [Google Scholar]
- 38.Li W, Liu H, Zhou JS, Cao JF, Zhou XB, Choi AM, Chen ZH, Shen HH. 2012. Caveolin-1 inhibits expression of antioxidant enzymes through direct interaction with nuclear erythroid 2 p45-related Factor-2 (Nrf2). J. Biol. Chem. 287:20922–20930. 10.1074/jbc.M112.352336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Galliher-Beckley AJ, Williams JG, Collins JB, Cidlowski JA. 2008. Glycogen synthase kinase 3β-mediated serine phosphorylation of the human glucocorticoid receptor redirects gene expression profiles. Mol. Cell. Biol. 28:7309–7322. 10.1128/MCB.00808-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wong WP, Tiano JP, Liu S, Hewitt SC, Le May C, Dalle S, Katzenellenbogen JA, Katzenellenbogen BS, Korach KS, Mauvais-Jarvis F. 2010. Extranuclear estrogen receptor-alpha stimulates NeuroD1 binding to the insulin promoter and favors insulin synthesis. Proc. Natl. Acad. Sci. U. S. A. 107:13057–13062. 10.1073/pnas.0914501107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hauger RL, Olivares-Reyes JA, Dautzenberg FM, Lohr JB, Braun S, Oakley RH. 2012. Molecular and cell signaling targets for PTSD pathophysiology and pharmacotherapy. Neuropharmacology 62:705–714. 10.1016/j.neuropharm.2011.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gioiosa L, Raggi C, Ricceri L, Jasmin JF, Frank PG, Capozza F, Lisanti MP, Alleva E, Sargiacomo M, Laviola G. 2008. Altered emotionality, spatial memory and cholinergic function in caveolin-1 knockout mice. Behav. Brain Res. 188:255–262 [DOI] [PubMed] [Google Scholar]
- 43.Ezan P, Andre P, Cisternino S, Saubamea B, Boulay AC, Doutremer S, Thomas MA, Quenech'du N, Giaume C, Cohen-Salmon M. 2012. Deletion of astroglial connexins weakens the blood-brain barrier. J. Cereb. Blood Flow Metab. 32:1457–1467. 10.1038/jcbfm.2012.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen L, Chatterjee M, Li JY. 2010. The mouse homeobox gene Gbx2 is required for the development of cholinergic interneurons in the striatum. J. Neurosci. 30:14824–14834. 10.1523/JNEUROSCI.3742-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu B, Roos JL, Dexheimer P, Boone B, Plummer B, Levy S, Gogos JA, Karayiorgou M. 2011. Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat. Genet. 43:864–868. 10.1038/ng.902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nethe M, Anthony EC, Fernandez-Borja M, Dee R, Geerts D, Hensbergen PJ, Deelder AM, Schmidt G, Hordijk PL. 2010. Focal-adhesion targeting links caveolin-1 to a Rac1-degradation pathway. J. Cell Sci. 123:1948–1958. 10.1242/jcs.062919 [DOI] [PubMed] [Google Scholar]
- 47.Li CC, Chiang TC, Wu TS, Pacheco-Rodriguez G, Moss J, Lee FJ. 2007. ARL4D recruits cytohesin-2/ARNO to modulate actin remodeling. Mol. Biol. Cell 18:4420–4437. 10.1091/mbc.E07-02-0149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ho H, Soto Hopkin A, Kapadia R, Vasudeva P, Schilling J, Ganesan AK. 2013. RhoJ modulates melanoma invasion by altering actin cytoskeletal dynamics. Pigment Cell Melanoma Res. 26:218–225. 10.1111/pcmr.12058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bodea GO, Spille JH, Abe P, Andersson AS, Acker-Palmer A, Stumm R, Kubitscheck U, Blaess S. 2014. Reelin and CXCL12 regulate distinct migratory behaviors during the development of the dopaminergic system. Development 141:661–673. 10.1242/dev.099937 [DOI] [PubMed] [Google Scholar]
- 50.Adzic M, Djordjevic J, Djordjevic A, Niciforovic A, Demonacos C, Radojcic M, Krstic-Demonacos M. 2009. Acute or chronic stress induce cell compartment-specific phosphorylation of glucocorticoid receptor and alter its transcriptional activity in Wistar rat brain. J. Endocrinol. 202:87–97. 10.1677/JOE-08-0509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pancotti F, Roncuzzi L, Maggiolini M, Gasperi-Campani A. 2012. Caveolin-1 silencing arrests the proliferation of metastatic lung cancer cells through the inhibition of STAT3 signaling. Cell Signal. 24:1390–1397. 10.1016/j.cellsig.2012.02.015 [DOI] [PubMed] [Google Scholar]
- 52.Itani OA, Liu KZ, Cornish KL, Campbell JR, Thomas CP. 2002. Glucocorticoids stimulate human sgk1 gene expression by activation of a GRE in its 5′-flanking region. Am. J. Physiol. Endocrinol. Metab. 283:E971–E979 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.