Abstract
Prions are infectious proteins composed of the abnormal disease-causing isoform PrPSc, which induces conformational conversion of the host-encoded normal cellular prion protein PrPC to additional PrPSc. The mechanism underlying prion strain mutation in the absence of nucleic acids remains unresolved. Additionally, the frequency of strains causing chronic wasting disease (CWD), a burgeoning prion epidemic of cervids, is unknown. Using susceptible transgenic mice, we identified two prevalent CWD strains with divergent biological properties but composed of PrPSc with indistinguishable biochemical characteristics. Although CWD transmissions indicated stable, independent strain propagation by elk PrPC, strain coexistence in the brains of deer and transgenic mice demonstrated unstable strain propagation by deer PrPC. The primary structures of deer and elk prion proteins differ at residue 226, which, in concert with PrPSc conformational compatibility, determines prion strain mutation in these cervids.
Prions are protein-based transmissible agents causing lethal, incurable neurodegenerative diseases of mammals, including sheep scrapie, bovine spongiform encephalopathy, human Creutzfeldt-Jakob disease, and chronic wasting disease (CWD), a contagious prion disorder of cervids (a family of hoofed mammals, including deer and elk). During propagation, PrPSc, the β-sheet–rich disease-associated conformer of the prion protein (PrP), coerces the physiological form, PrPC, to adopt the PrPSc conformation. Prions share with nucleic acid–based pathogens the ability to propagate strain information. Although distinct conformers of PrPSc appear to encipher the characteristics of certain strains (1–5), it is unclear how prions mutate and adapt in the absence of nucleic acids. Strain mutation has been reported after interspecies prion transmission— for example, scrapie transmission to rodents (6). Prionmutationmay result in increased host range (7), a factor that complicates risk assessments for new hosts.
Although strain diversity has been recorded for sheep, human, and bovine prions, the existence of strains in CWD is unclear (8–10). Polymorphic residue 129 of human PrP and the corresponding elk PrP residue influence prion susceptibility at the level of strain selection (11, 12). PrP primary structures of CWD-susceptible species differ at residue 226, which is glutamic acid (E) in Rocky Mountain elk and glutamine (Q) in other susceptible cervids. To determine CWD strain prevalence and to assess the influence of these amino acids, we transmitted an extensive collection of CWD isolates (table S1) to transgenic mice expressing cervid PrP (CerPrP), referred to as Tg(CerPrP)1536+/−,which lack a species barrier to CWD (5, 8, 12–14).
We used standard criteria to differentiate strains, including distribution and severity of neuropathology and incubation time to disease onset (15–18).We identified two distinct neuropathological patterns in primary and secondary transmissions (Figs. 1 to 3). Different mean incubation times of mice with these distinct neuropathologies allowed us to classify mice as having been affected by one of two strains, referred to hereafter as CWD1 and CWD2.
Fig. 1.
Primary transmission of deer and elk CWD prions to Tg(CerPrP)1536+/− mice. (A) Blue and red circles, mice with CWD1 and CWD2 patterns of neuropathology, respectively, analyzed by IHC or histoblotting; open circles, mice not analyzed; gray circle, ambiguous strain pattern; squares, asymptomatic mice. Animal identification codes are shown on the x axis. Total CWD1 and CWD2, total number of mice with each strain pattern. Incubation time differences between groups were established using an unpaired, two-tailed t test (p < 0.0001). d, days. (B) Summary of strain profiles after primary passage of elk and deer. The mean (± SD) incubation times and numbers of all mice in each category are shown. The low titer CWD1 012-22012 elk isolate was excluded from the summary. (C) Neuronal vacuolation was quantified in nine brain regions: 1, medulla; 2, cerebellum; 3, midbrain; 4, hypothalamus; 5, thalamus; 6, hippocampus; 7, paraterminal body; 8, cerebral cortex at hippocampus; and 9, cerebral cortex at septum. Blue circles, eight mice with rapid incubation periods; red circles, seven mice with prolonged incubation periods; open circles, uninoculated, age-matched mice. Error bars represent the average number of vacuoles per field ± SEM. Differences were established using an unpaired, two-tailed t test; asterisks indicate the degree of significance.
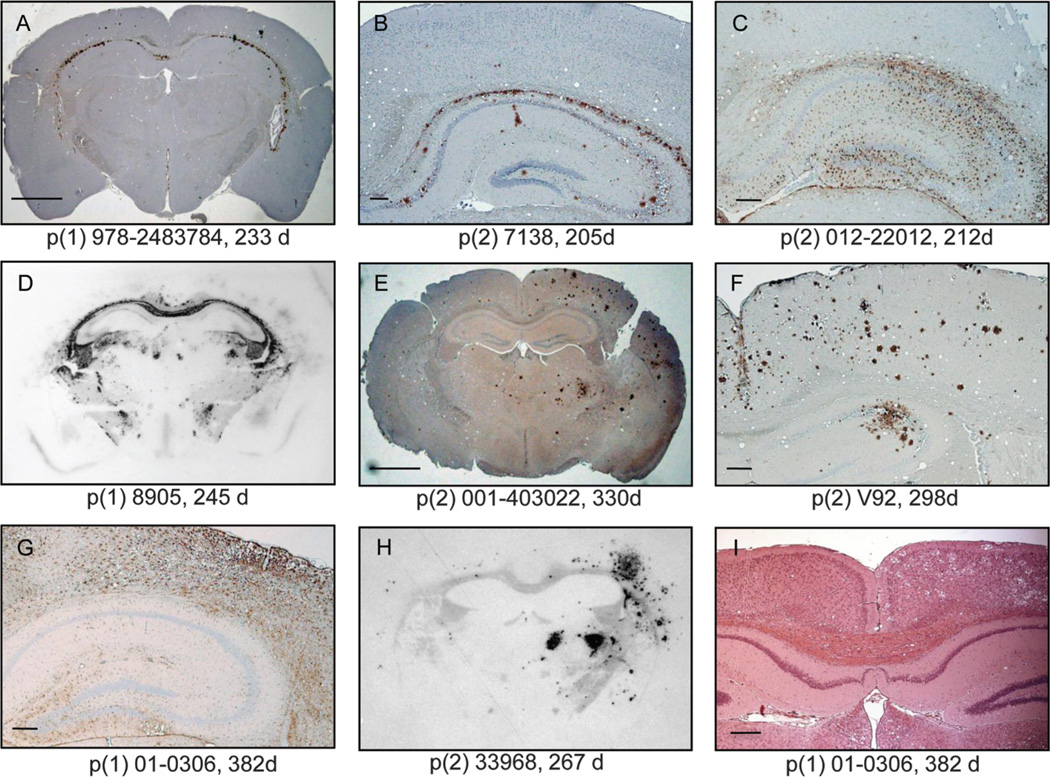
Fig. 3.
Representative distributions of CerPrPSc in the brains of diseased Tg(CerPrP)1536+/− mice. Sections encompassing the hippocampus and cortex were analyzed by IHC using anti-PrP mAb 6H4 (A, B, E, and F) or by using antibodies against glial fibrillary acidic protein (C and G). (D and H) CerPrPSc distribution was evaluated by histoblotting of coronal sections of similar regions. (I) Asymmetrical distribution of cortical florid plaques and associated neuronal vacuolation in CWD2-infected mice. Scale bars in (A) and (E), 1 mm; in (B), 50 µm; in (C), (F), (G), and (I), 100 mm. Isolate passage number (p) and incubation times of mice are shown (d, days).
Differences in the profiles of vacuolar degeneration (19) in mice affected by the short–incubation time CWD1 strain and the long–incubation time CWD2 strain were pronounced in the hippocampus (Fig. 1C). Immunohistochemically (IHC) stained sections of mice infected by CWD1 were characterized by continuous, symmetrical CerPrPSc deposits throughout the hippocampal alveus and, often, the CA3 region of the pyramidal cell layer with coincident astrocytic gliosis (Fig. 3, A to C). This distribution of CerPrPSc was recapitulated in histoblots (Fig. 3D) (20). The bilaterally symmetrical lesions in CWD1-infected mice are consistent with the expected neuropathology of prion infection by most prion strains. Though unusual, asymmetrical distribution of CerPrPSc and pathology in the inoculated hemisphere of mice infected by the long–incubation time CWD2 strain (Fig. 3, E to I) is also a characteristic of certain scrapie strains, such as 87A (21).
Primary transmissions of elk inocula indicated that roughly half were infected with either CWD1 or CWD2 (Fig. 1, A and B, and table S1). Of 22 analyzed mice from five elk CWD transmissions, 21 had short incubation times andCWD1 neuropathology, whereas only 1 had CWD2 pathology and delayed disease onset (Fig. 1, A and B). Titer-related delayed disease onset was a feature of one isolate with CWD1 properties (Fig. 1A and SOM). Of the 33 analyzed mice from five other elk inocula, 32 had long incubation times and CWD2 neuropathology, whereas only 1 had CWD1 pathology and rapid disease onset (Fig. 1, Aand B). Transmission of elk prions generated by protein misfolding cyclic amplification (PMCA) (22) also produced long incubation times and CWD2 pathology (Fig. 1A). In contrast to elk, transmission of 12 out of 15 deer inocula produced mixed intra-study incubation times and CWD1 and CWD2 neuropathologies (Fig. 1, A and B). Transmission of PMCA-generated deer prions (5) produced mixed CWD1 and CWD2 responses (Fig. 1A).
Though elk prions manifested predominantly as either CWD1 or CWD2 strains on primary passage (Fig. 1B), serial passage favored strain mixtures and indistinguishable aggregated mean incubation times for each elk group (Fig. 2B); serial transmission of deer prions also produced mixed strains (Fig. 2, A and B). Neuropathological profiles were available for 17 mice used as inocula in the 25 secondary transmissions. Profiles of six representative transmissions illustrate the stochastic stabilities of CWD1 and CWD2 strains (Fig. 2C). Also in this regard, CWD2 is reminiscent of the 87A scrapie strain and its more stable counterpart, ME7 (6), but with neither CWD strain being a more stable derivative of the other.
Fig. 2.
Serial transmission of deer and elk CWD prions to Tg(CerPrP)1536+/− mice. (A) Symbols are the same as in Fig. 1A. Animal identification codes are shown on the x axis. Asterisks indicate the degree of significance. (B) Summary of strain profiles on secondary passage. The mean (± SD) incubation times and numbers of mice in each category are shown. (C) Six representative transmissions demonstrating instability of CWD1 and CWD2. For each transmission, the single circles represent diseased mice after the first passage of CWD isolates (p1); boxed symbols represent mice resulting from serial passage of those brains (p2).
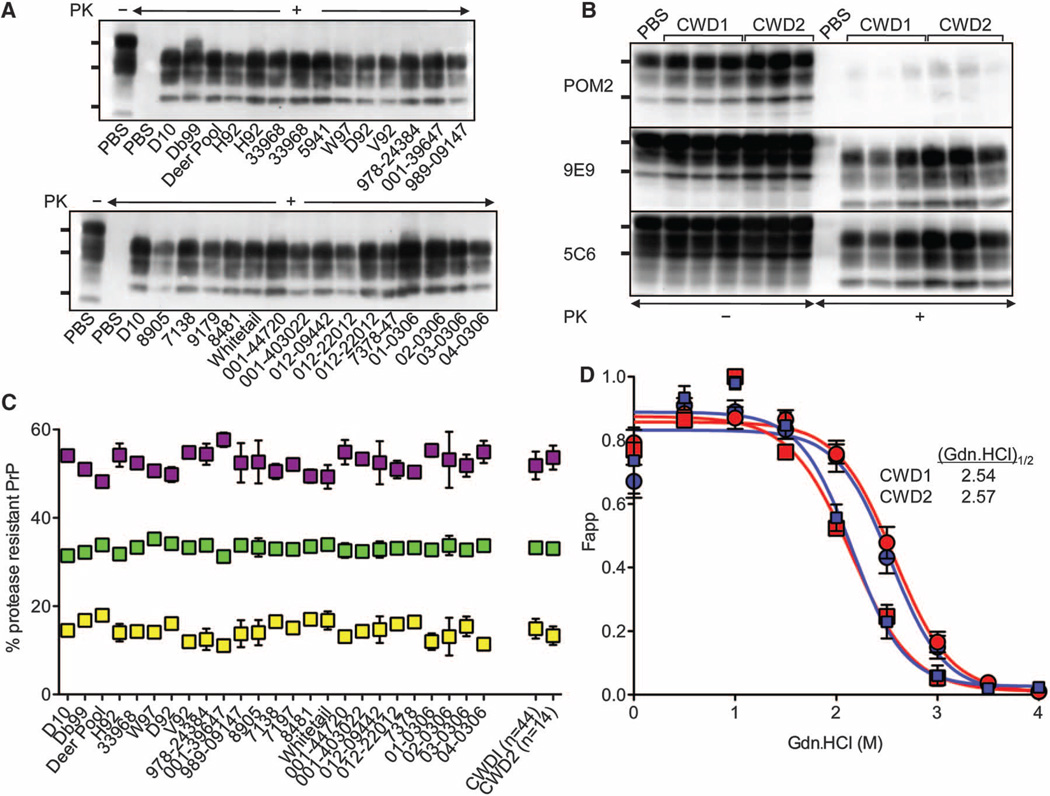
Although certain strains are associated with distinct conformers of PrPSc (1–5), not all strains that can be biologically distinguished are composed of PrPSc with recognizably different biochemical properties (4, 23, 24). The electrophoretic migration patterns of cervid PrPSc (CerPrPSc) from the brains of mice infected by either strain were indiscernible (Fig. 4A), even when immunoblots were probed with monoclonal antibodies (mAbs) recognizing epitopes within or immediately downstream of the octarepeat region or in the structured carboxyl terminus of PrP (Fig. 4B), arguing against the coexistence of distinct CerPrPSc populations (25). CerPrPSc associated with CWD1 and CWD2 was composed of equivalent proportions of di-, mono-, and a-glycosyl forms (Fig. 4C). Finally, CerPrPSc fromCWD1- and CWD2-infected mice had similar unfolding characteristics after treatment with guanidinium hydrochloride (Gdn.HCl). When CWD1 or CWD2 elk isolates were transmitted to transgenic mice expressing elk PrP (14), stabilities of the resulting CerPrPSc encoding E at residue 226 (CerPrPSc-E226) remained indistinguishable, but were distinct from those of CerPrPSc encoding Q (CerPrPSc-Q226) in Tg(CerPrP)1536+/− mice expressing deer PrP (Fig. 4D). The indistinguishable CerPrPSc conformations and unstable strain transitions ofCWD1 andCWD2 in deer and Tg(CerPrP)1536+/− mice are consistent with their separation by relatively low energy barriers. Our inability to resolve subtle biochemical properties of CerPrPSc is reminiscent of the properties of postulated swainsonine sensitive and resistant mutant substrains, or quasi-species, that emerged in response to selective pressure on 22L prions in cell culture (23).
Fig. 4.
Biochemical properties of CerPrPSc associated with CWD1 and CWD2. (A) Immunoblots probed with mAb 6H4. (B) Immunoblots of samples from mice infected with CWD1 or CWD2 probed with mAbs as indicated. Samples were treated as indicated with proteinase K (PK). The bands on immunoblots show the various glycoforms of proteaseresistant CerPrPSc and, in samples from phosphate-buffered saline–inoculated mice not treated with PK, CerPrPC. Positions of protein molecular mass markers at 37, 25, and 20 kD (top to bottom) are shown. (C) Ratio of CerPrPSc glycoforms. Data points represent the mean (± SD) relative proportions from three or four animals of di-(purple squares), mono-(green squares), and unglycosylated (yellow squares) CerPrPSc. Aggregated values for all mice exhibiting CWD1 or CWD2 strain properties are also shown. n, number of analyzed mice of each strain type. (D) Percentage of CerPrPSc as a function of Gdn.HCl concentration in 17 mice infected with CWD1 (blue circles) and 20 mice affected with CWD2 (red circles). (Gdn.HCl)1/2 values are shown for Tg(CerPrP)1536+/− mice. Tg(CerPrP-E226)5037+/− mice were infected with elk isolates that produced CWD1 (12389, blue squares) or CWD2 (04-0306, red squares). Fapp, fraction of apparent PK-resistant PrPSc = (maximum signal – individual signal)/(maximum signal – minimum signal); error bars indicate SD of mean values from three animals analyzed in each study group.
Here we have identified two strains in a large number of CWD-affected deer and elk. If additional strains exist, they would be likely to represent a minor component of the CWD. The generally mixed CWD strain responses from deer and the relatively uniform manifestation of CWD1 or CWD2 strains from elk appear to reflect strain constitutions in the natural host, rather than adaptation and divergence of progenitor strains in recipient mice, which are alternatives considered in the conformational selection model (26, 27) that could not be resolved by previous limited CWD transmissions to rodent models (28, 29).
Differences in deer and elk PrP primary structures provide a framework for understanding these strain profiles. Propagation of either strain by CerPrPC-Q226 in deer brain is unstable, and both strains are manifested as mixtures after primary transmissions of deer CWD to Tg(CerPrP) 1536+/− mice. Regardless of whether prions originated as stable strains in elk or as mixtures in deer, the predominantly mixed strain profiles on serial passage indicate that strain mixtures are also generated during unstable conversion of CerPrPC-Q226 in Tg(CerPrP)1536+/− mice. In contrast, the almost exclusive manifestation of CWD1 or CWD2 strains after primary transmissions of elk CWD prions reflects relatively stable strain propagation by CerPrPC-E226 in elk. However, whereas the ultimate phenotype of diseased mice reflects the originating elk CWD1 or CWD2 strain, its manifestation is counterbalanced by the tendency of transgene-expressed CerPrPC-Q226 to unstably generate mixed strains. Accordingly, strain switching was detected in 4% of recipient mice during primary transmissions of CWD1 and CWD2 from elk (Fig. 1A). Because of the substantial role played by residue 226 in determining CWDstrain properties, the description of a lysine polymorphism at this position in deer (30) is of considerable additional interest.
The identification and characterization of distinct CWD strains with similar conformations and the influence of PrP primary structure on their stabilities are of importance when considering the potential for transmission to species outside the family cervidae. Although CWD prions have reassuringly failed to induce disease in transgenic mice expressing human PrP (10, 31), because of the risk of prion exposure from contaminated venison (13) and other infected materials (14), systematically addressing the tissue distributions of CWD1 and CWD2 and their zoonotic potentials would appear to be high priorities.
Supplementary Material
Acknowledgments
We thank Prionics for mAb 6H4, V. Saylor for technical help, A. Aguzzi for mAb POM2, P. Nelson for neuropathological expertise, R. Kryscio and J. Marcum for biostatistical advice, and C. Ryou for reading the manuscript. C.S. is currently Founder and Chief Scientific Officer of Amprion Inc., a privately held company focusing on exploiting PMCA for sensitive detection of prions and early disease diagnosis. These studies were supported by grant 2RO1 NS040334-04 from the National Institute of Neurological Disorders and Stroke, grants N01-AI-25491 and 1P01AI077774-01 from the National Institute of Allergy and Infectious Diseases, and grant V180003 from the U.S. Department of Defense. R.C.A. and S.B. were supported by T32 DA022738 and T32 AI49795 Training Programs from the National Institute on Drug Abuse and the National Institute of Allergy and Infectious Diseases, respectively.
References and Notes
- 1.Bessen RA, Marsh RF. J. Virol. 1994;68:7859. doi: 10.1128/jvi.68.12.7859-7868.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Telling GC, et al. Science. 1996;274:2079. doi: 10.1126/science.274.5295.2079. [DOI] [PubMed] [Google Scholar]
- 3.Collinge J, Sidle KCL, Meads J, Ironside J, Hill AF. Nature. 1996;383:685. doi: 10.1038/383685a0. [DOI] [PubMed] [Google Scholar]
- 4.Peretz D, et al. Protein Sci. 2001;10:854. doi: 10.1110/ps.39201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green KM, et al. PLoS Pathog. 2008;4:e1000139. doi: 10.1371/journal.ppat.1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruce ME, Dickinson AG. J. Gen. Virol. 1987;68:79. doi: 10.1099/0022-1317-68-1-79. [DOI] [PubMed] [Google Scholar]
- 7.Bartz JC, Marsh RF, McKenzie DI, Aiken JM. Virology. 1998;251:297. doi: 10.1006/viro.1998.9427. [DOI] [PubMed] [Google Scholar]
- 8.Browning SR, et al. J. Virol. 2004;78:13345. doi: 10.1128/JVI.78.23.13345-13350.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.LaFauci G, et al. J. Gen. Virol. 2006;87:3773. doi: 10.1099/vir.0.82137-0. [DOI] [PubMed] [Google Scholar]
- 10.Tamgüney G, et al. J. Virol. 2006;80:9104. doi: 10.1128/JVI.00098-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wadsworth JDF, et al. Science. 2004;306:1793. doi: 10.1126/science.1103932. published online 11 November 2004 (10.1126/science.1103932). [DOI] [PubMed] [Google Scholar]
- 12.Green KM, et al. J. Gen. Virol. 2008;89:598. doi: 10.1099/vir.0.83168-0. [DOI] [PubMed] [Google Scholar]
- 13.Angers RC, et al. Science. 2006;311:1117. doi: 10.1126/science.1122864. published online 26 January 2006 (10.1126/science.1122864). [DOI] [PubMed] [Google Scholar]
- 14.Angers RC, et al. Emerg. Infect. Dis. 2009;15:696. doi: 10.3201/eid1505.081458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dickinson AG, Meikle VMH. Mol. Gen. Genet. 1971;112:73. doi: 10.1007/BF00266934. [DOI] [PubMed] [Google Scholar]
- 16.Bruce ME, Dickinson AG. In: Slow Transmissible Diseases of the Nervous System. Prusiner SB, Hadlow WJ, editors. Vol. 2. New York: Academic Press; 1979. pp. 71–86. [Google Scholar]
- 17.Bruce ME, McBride PA, Farquhar CF. Neurosci. Lett. 1989;102:1. doi: 10.1016/0304-3940(89)90298-x. [DOI] [PubMed] [Google Scholar]
- 18.Materials and methods are available as supporting material on Science Online
- 19.Fraser H, Dickinson AG. J. Comp. Pathol. 1968;78:301. doi: 10.1016/0021-9975(68)90006-6. [DOI] [PubMed] [Google Scholar]
- 20.Taraboulos A, et al. Proc. Natl. Acad. Sci. U.S.A. 1992;89:7620. doi: 10.1073/pnas.89.16.7620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bruce ME, Fraser H. Acta Neuropathol. 1982;58:133. doi: 10.1007/BF00691654. [DOI] [PubMed] [Google Scholar]
- 22.Castilla J, Saá P, Hetz C, Soto C. Cell. 2005;121:195. doi: 10.1016/j.cell.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 23.Li J, Browning S, Mahal SP, Oelschlegel AM, Weissmann C. Science. 2010;327:869. doi: 10.1126/science.1183218. published online 31 December 2009 (10.1126/science.1183218). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Legname G, et al. Proc. Natl. Acad. Sci. U.S.A. 2006;103:19105. doi: 10.1073/pnas.0608970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Polymenidou M, et al. Lancet Neurol. 2005;4:805. doi: 10.1016/S1474-4422(05)70225-8. [DOI] [PubMed] [Google Scholar]
- 26.Collinge J. Lancet. 1999;354:317. doi: 10.1016/S0140-6736(99)05128-4. [DOI] [PubMed] [Google Scholar]
- 27.Collinge J, Clarke AR. Science. 2007;318:930. doi: 10.1126/science.1138718. [DOI] [PubMed] [Google Scholar]
- 28.Sigurdson CJ, et al. J. Virol. 2006;80:12303. doi: 10.1128/JVI.01120-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raymond GJ, et al. J. Virol. 2007;81:4305. doi: 10.1128/JVI.02474-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson C, et al. J. Gen. Virol. 2006;87:2109. doi: 10.1099/vir.0.81615-0. [DOI] [PubMed] [Google Scholar]
- 31.Kong Q, et al. J. Neurosci. 2005;25:7944. doi: 10.1523/JNEUROSCI.2467-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.