Abstract
Multidrug-resistant Acinetobacter baumannii isolates, particularly those that produce carbapenemases, are increasingly reported worldwide. The biochemically based Carba NP test, extensively validated for the detection of carbapenemase producers among Enterobacteriaceae and Pseudomonas spp., has been modified to detect carbapenemase production in Acinetobacter spp. A collection of 151 carbapenemase-producing and 69 non-carbapenemase-producing Acinetobacter spp. were tested using the Carba NP test and a modified Carba NP protocol (the CarbAcineto NP test) in this study. The CarbAcineto NP test requires modified lysis conditions and an increased bacterial inoculum compared to those of the original Carba NP test. The Carba NP test detects metallo-β-lactamase producers but failed to detect the production of other carbapenemase types among Acinetobacter spp. In contrast, the newly designed CarbAcineto NP test, which is rapid and reproducible, detects all types of carbapenemases with a sensitivity of 94.7% and a specificity of 100%. This cost-effective technique offers a reliable and affordable technique for identifying carbapenemase production in Acinetobacter spp., which is a marker of multidrug resistance in those species. Its use will facilitate the recognition of these carbapenemases and prevent their spread.
INTRODUCTION
Acinetobacter spp., and particularly the A. baumannii-A. calcoaceticus complex, are opportunistic pathogens frequently involved in nosocomial outbreaks that occur mostly in intensive care units (ICU). Those infections range from septicemia to pneumonia and urinary tract infections (1). Due to their ability to develop rapid resistance to new antibiotics, multidrug-resistant strains belonging to the A. baumannii-A. calcoaceticus complex have been increasingly reported during the last decade (2). Consequently, carbapenems are often considered to be the antibiotic of last resort for treating infections caused by those strains. Therefore, resistance to carbapenems in those species is considered to define the isolate as highly resistant.
In recent years, the spread of carbapenem-resistant Acinetobacter spp. has become a worldwide issue. In Acinetobacter spp., resistance to carbapenems may result from (i) decreased permeability of the outer membrane due to the loss or modification of porins, (ii) modification of penicillin-binding proteins (which is rare), and (iii) production of a carbapenemase (most of the cases) (2). Those carbapenemases identified in Acinetobacter spp. belong to Ambler class A, B, or D. Some class A carbapenemases have been identified as being of the KPC or GES types (3, 4). Whereas KPC-producing isolates have rarely been described (4), the dissemination of GES-11- and GES-14-producing isolates was recently reported in the Middle East (5). Metallo-β-lactamases (MBLs) (Ambler class B) of the VIM, IMP, SIM, and NDM types have been also reported in Acinetobacter spp. (2, 6). Apart from SIM, those MBLs have all been reported in Enterobacteriaceae and Pseudomonas spp. (7). In addition, carbapenem-hydrolyzing Ambler class D β-lactamases (CHDLs) constitute the first source of acquired carbapenem resistance in Acinetobacter spp. Those CHDLs are divided into five different subgroups, namely, OXA-23, OXA-40, OXA-51, OXA-58, and OXA-143 (2, 8, 9). As opposed to KPC and MBLs, these CHDLs have been identified among Acinetobacter spp. isolates only. OXA-51-like enzymes are intrinsic and chromosomally encoded in the A. baumannii-calcoaceticus complex; they possess a weak carbapenemase activity and share a weak amino acid identity with the other known class D β-lactamases (10–12). Although the blaOXA-51-like genes are usually not expressed in a wild-type isolate, the insertion of ISAba1 at the 5′ end of blaOXA-51-like genes may lead to overexpression of the corresponding β-lactamase gene (13). On the other hand, genes encoding OXA-23, OXA-40, OXA-58, and OXA-143 CHDLs have been mainly identified on transferable genetic structures, such as plasmids or transposons, which contribute to their dissemination among the Acinetobacter genus (2).
Since most of the carbapenemases identified in Acinetobacter spp. are located on mobile genetic elements that may be transferred to other clinical relevant species (i.e., other Acinetobacter species, Enterobacteriaceae, and P. aeruginosa), it is critical to identify those carbapenemase-producing Acinetobacter organisms and consequently differentiate them from isolates that are carbapenem resistant due to nontransferable mechanisms (i.e., permeability defects and overexpression of efflux pumps). The rapid identification of the NDM-producing A. baumannii-A. calcoaceticus complex may help to limit the dissemination of carbapenemase genes not only in the Acinetobacter genus but also in Enterobacteriaceae through the rapid identification of their potential reservoirs (7).
Due to its intrinsic low permeability, the detection of carbapenemase production in the Acinetobacter genus is considered to be more difficult than in Enterobacteriaceae and Pseudomonas spp. Several phenotypic techniques have been proposed to detect carbapenemase-producing Acinetobacter spp. (14). The modified Hodge test has largely been used for this purpose. It is based on the in vitro detection of carbapenemase activity, therefore inactivating the antibiotic effect. Although this test is efficient for detecting IMP and VIM producers, NDM and CHDL producers may remain undetected, leading to false-negative results (15). Several techniques using the inhibition properties of EDTA were also proposed for detecting MBL-producing Acinetobacter spp. Those techniques include the combined disk test and the Etest MBL strip (15, 16). However, those techniques are not highly sensitive or specific, and they require an additional period of growth of 24 h.
Biochemical detection of carbapenemase production using ultraviolet (UV) spectrophotometry has also been proposed as a suitable method (17). This technique efficiently detects VIM, IMP, and SIM producers, but NDM and CHDL producers remain difficult to detect (15). Recently, the detection of carbapenemase production using matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry has been proposed for Acinetobacter spp. (18–20). It is based on the detection of the spectra of imipenem and of its hydrolyzed product. It shows good sensitivity and specificity but requires trained microbiologists and expensive equipment. Finally, molecular-based techniques using specific primers are useful for identifying carbapenemase genes. Simplex or multiplex PCR methods are available (21), as well as real-time PCR approaches that have the advantage of providing a result in 3 h (22). More recently, a DNA microarray has been developed to detect 91 target sequences associated with antibiotic resistance within 4 h from bacterial culture to results (23). Although those molecular-based tests are highly sensitive and specific, they fail to detect unknown carbapenemase genes or those that are not included in the panel of the test used.
One of the most promising techniques for the rapid and accurate detection of any carbapenemase producer is the Carba NP test. This test is based on the biochemical detection of the hydrolysis of the β-lactam ring of imipenem (24, 25). Although the Carba NP test has been extensively validated worldwide for the detection of carbapenemase producers among Enterobacteriaceae and Pseudomonas spp. (24, 26), it has not been validated for the detection of carbapenemase-producing Acinetobacter spp. The imipenem hydrolysis rates actually obtained with those CHDLs are too low to be detected by the Carba NP test in its original version. The aim of this study was to further evaluate the Carba NP test for detecting carbapenemase producers in Acinetobacter spp. and to settle a modified version of the Carba NP test, the CarbAcineto NP test, for optimal detection of this activity in Acinetobacter spp.
MATERIALS AND METHODS
Strain collection.
A total of 220 strains were used to evaluate the performance of the CarbAcineto NP test. They were from various clinical origins (blood culture, urine, and sputum samples) and of worldwide origins. Those strains had previously been characterized at the molecular level for their carbapenemase content (Tables 1 and 2). This strain collection included the most frequently acquired carbapenemases identified in human A. baumannii clinical isolates (Table 1). Those carbapenemases were OXA-23 (n = 68), OXA-24/OXA-40-like (n = 19), OXA-58-like (n = 26), OXA-143-like (n = 3), GES (n = 8), IMP (n = 2), VIM (n = 1), SIM (n = 1), and NDM (n = 14). Nine isolates coproduced two carbapenemases: GES-11 plus OXA-23 (n = 6) and NDM-1 plus OXA-23 (n = 3). The negative controls (n = 69) included (i) wild-type strains (n = 51), (ii) narrow-spectrum β-lactamase producers (n = 3), and (iii) extended-spectrum β-lactamase producers (n = 12) (Table 2). Three strains overexpressing their chromosome-encoded blaOXA-51-like carbapenemase genes were also tested (Table 2).
TABLE 1.
Results of the Carba NP and CarbAcineto NP tests when performed on Acinetobacter spp. producing an acquired carbapenemase
| Carbapenemase | Acinetobacter species | No. of isolates | MIC (μg/ml) ofa: |
Carba NP test result | CarbAcineto NP test result | |
|---|---|---|---|---|---|---|
| IMP | MER | |||||
| Ambler class A | ||||||
| GES-type | ||||||
| GES-11 | A. baumanniib | 3 | 2 | 4 | − | − |
| GES-14 | A. baumannii | 5 | 24–32 | 16–32 | − | − |
| Ambler class B | ||||||
| NDM-type | ||||||
| NDM-1 | A. baumannii | 13 | >32 | >32 | + | + |
| NDM-2 | A. baumannii | 1 | >32 | >32 | + | + |
| IMP-type | ||||||
| IMP-1 | A. baumannii | 1 | 4 | 6 | + | + |
| IMP-4 | A. baumannii | 1 | 24 | 16 | + | + |
| VIM-type | ||||||
| VIM-4 | Acinetobacter sp. genomospecies 16 | 1 | >32 | >32 | + | + |
| SIM-type | ||||||
| SIM-1 | A. baumannii | 1 | >32 | >32 | + | + |
| Ambler class D | ||||||
| OXA-23 group | ||||||
| OXA-23 | A. baumannii | 68 | 24 to >32 | 8 to >32 | − | + |
| OXA-40 group | ||||||
| OXA-24/OXA-40 | A. baumannii | 8 | >32 | >32 | − | + |
| OXA-25 | A. baumannii | 1 | >32 | >32 | − | + |
| OXA-26 | A. baumannii | 1 | >32 | >32 | − | + |
| OXA-72 | A. baumannii | 9 | >32 | >32 | − | + |
| OXA-58 group | ||||||
| OXA-58 | A. baumannii | 24 | 16 to >32 | 8 to >32 | − | + |
| OXA-58 | A. haemolyticus | 1 | >32 | 8 | − | + |
| OXA-97 | A. baumannii | 1 | >32 | >32 | − | + |
| OXA-143 group | ||||||
| OXA-143 | A. baumannii | 2 | >32 | >32 | − | + |
| OXA-253 | A. baumannii | 1 | >32 | >32 | − | + |
| Multiple | ||||||
| GES-11 + OXA-23 | A. baumannii | 6 | 32 | >32 | − | + |
| NDM-1 + OXA-23 | A. baumannii | 3 | >32 | >32 | + | + |
IMP, imipenem; MER, meropenem.
A. baumannii indicates A. baumannii-A. calcoaceticus complex.
TABLE 2.
Results of the Carba NP and CarbAcineto NP tests when testing Acinetobacter spp. strains that do not possess acquired carbapenemases
| Acquired β-lactamases | Acinetobacter species | No. of isolates | MIC (μg/ml) ofa: |
Carba NP test result | CarbAcineto NP test result | |
|---|---|---|---|---|---|---|
| IMP | MER | |||||
| None | ||||||
| A. baumanniib | 36 | 0.19–2 | 0.12–3 | − | − | |
| A. junii | 3 | 0.05–0.09 | 0.05–0.09 | − | − | |
| A. lwoffii | 4 | 0.09–0.19 | 0.09–0.19 | − | − | |
| A. ursingii | 2 | 0.06–0.09 | 0.06–0.09 | − | − | |
| A. johnsonii | 4 | 0.06–0.09 | 0.06–0.09 | − | − | |
| A. baumannii | 1 | 0.5 | 0.75 | NIc | − | |
| A. johnsonii | 1 | 0.09 | 0.09 | NI | − | |
| Narrow-spectrum β-lactamases | ||||||
| SCO-1 | A. baumannii | 1 | 0.38 | 0.5 | − | − |
| RTG-4 | A. baumannii | 1 | 0.38 | 0.38 | − | − |
| OXA-21 | A. baumannii | 1 | 0.75 | 0.5 | − | − |
| Extended-spectrum β-lactamases | ||||||
| SHV-5 | A. baumannii | 1 | 6 | 4 | − | − |
| PER-1 | A. baumannii | 1 | 1.5 | 1.5 | − | − |
| VEB-1 | A. baumannii | 9 | 0.38–1 | 0.5–1.5 | − | − |
| GES-12 | A. baumannii | 1 | 32 | 32 | − | − |
| Overexpressed chromosome-encoded OXA-51-like β-lactamases | ||||||
| ISAba1 + OXA-51 | A. baumannii | 2 | 2–3 | 3 | − | − |
| ISAba1 + OXA-66 | A. baumannii | 1 | 4 | 6 | − | − |
IMP, imipenem; MER, meropenem.
A. baumannii indicates A. baumannii-A. calcoaceticus complex.
NI, not interpretable.
Susceptibility testing.
Susceptibility testing was performed by determining MIC values using the Etest (bioMérieux, La Balme-les-Grottes, France) on Mueller-Hinton agar plates at 37°C, and the results were recorded according to U.S. guidelines (CLSI), as updated in 2013. The breakpoints for imipenem and meropenem were ≤4 μg/ml for susceptibility (S) and ≥16 μg/ml for resistance (R).
Carba NP test.
The Carba NP test is based on the colorimetric and pH-based detection of hydrolysis of the β-lactam ring of imipenem. The updated protocol of the Carba NP test was performed on bacterial isolates grown on Trypticase soy (TS) agar (bioMérieux), as previously described (25).
CarbAcineto NP test.
The CarbAcineto NP test was adapted from the updated version of the Carba NP test used for the detection of carbapenemase-producing Enterobacteriaceae and Pseudomonas spp. (25), in order to use the test for Acinetobacter spp. In this updated version, the lysis buffer was replaced by a 5 M NaCl solution, avoiding any buffer effect, and the bacterial inoculum was doubled from one-third to one-half of a calibrated loop (10 μl) to a full calibrated loop in order to increase the enzyme quantity. Briefly, a full calibrated loop (10 μl) of the tested strain was recovered from TS agar plates and resuspended in two 1.5-ml Eppendorf tubes (A and B) containing 100 μl of 5 M NaCl. In tube A (internal control), 100 μl of the revealing solution containing a pH indicator (phenol red) was added. In tube B (test tube), 100 μl of an extemporaneously prepared revelation solution supplemented with 6 mg/ml imipenem was added. Tubes A and B were incubated at 37°C for a maximum of 2 h. Optical reading of the color change of each tube was performed. In tube B, the carbapenemase activity was detected by a color change of phenol red solution (red to yellow/orange) resulting from the hydrolysis of imipenem into a carboxylic derivative, leading to a decrease of the pH value. The results of the CarbAcineto NP test were interpreted as follows: (i) tube A red and tube B remaining red indicated a non-carbapenemase-producing isolate, (ii) tube A red and tube B turning yellow/orange indicated a carbapenemase-producing strain, and (iii) tube A and tube B both turning yellow/orange indicated a noninterpretable result. The phenol red revealing solution was prepared as previously described (24). The 5 M NaCl solution was prepared from a dilution of NaCl powder (Sigma-Aldrich, Saint-Quentin-Fallavier, France) in distilled water.
RESULTS
Using the updated protocol of the Carba NP test set up for the detection of carbapenemase activity in Enterobacteriaceae and Pseudomonas spp., positive results were obtained for MBL-producing Acinetobacter spp. only (Table 1). The Carba NP test failed to detect Acinetobacter spp. isolates producing carbapenemases of the GES and/or OXA types (Table 1). This test did not detect any carbapenemase activity among strains found to be resistant to carbapenems due to the overexpression of their intrinsic OXA-51-like enzyme (Table 2). Accordingly, the specificity and sensitivity of the Carba NP test for the detection of Acinetobacter spp. isolates producing an acquired carbapenemase were found to be 100% and only 11.9%, respectively.
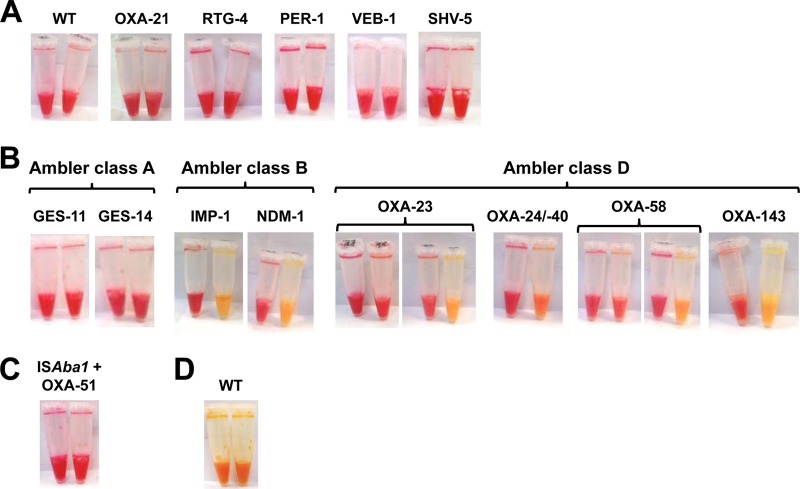
Conversely, the CarbAcineto NP test detected all isolates that produced an acquired carbapenemase, with the exception of some GES-type producers (Table 1 and Fig. 1B). Isolates that were carbapenem resistant due to overexpression of their chromosome-encoded OXA-51-like β-lactamase (Table 2, Fig. 1C) or to non-carbapenemase-mediated mechanisms, such as combined mechanisms of resistance (outer membrane permeability defect and/or associated with the overproduction of cephalosporinase and/or extended-spectrum β-lactamases [ESBLs]) also remained negative with this test (Table 2 and Fig. 1A). In most cases, positive results gave a frank color change from red to yellow (Fig. 1B). For MBL producers, a positive result was always obtained in <15 min. For ca. 13% (9/68) of the OXA-23 producers and ca. 23% (6/22) of the OXA-58-like producers, the color in the test tube (tube B) turned from red to orange but only after 2 h of incubation (Fig. 1B). However, this color change was easy to detect when looking at the upper part of the tubes after vortexing (Fig. 1B). Among the 211 tested strains of Acinetobacter spp., only two gave noninterpretable results (Table 2 and Fig. 1D). The specificity and sensitivity of the CarbAcineto NP test were therefore estimated to be 100% and 94.7%, respectively.
FIG 1.
Representative results obtained using the CarbAcineto NP test on Acinetobacter spp. isolates. (A) Representative results obtained using the CarbAcineto NP test on wild-type (WT), narrow-spectrum β-lactamase-producing (OXA-21 and RTG-4), and extended-spectrum β-lactamase-producing Acinetobacter spp. (PER-1, VEB-1, and SHV-5). (B) Representative results obtained using the CarbAcineto NP test on acquired carbapenemase-producing Acinetobacter spp. isolates. Ambler class A (GES-types), Ambler class B (IMP-type and NDM-type), and Ambler class D (OXA-23, OXA-24/OXA-40, OXA-58, and OXA-143) carbapenemases are represented. (C) Representative result obtained using the CarbAcineto test on a carbapenem-resistant Acinetobacter sp. overexpressing its chromosome-encoded OXA-51 β-lactamase. (D) Representative result of a noninterpretable result using the CarbAcineto NP on a wild-type Acinetobacter isolate.
DISCUSSION
The Carba NP test, initially set up for the detection of carbapenemase-producing Enterobacteriaceae and Pseudomonas spp., efficiently detects MBL-producing Acinetobacter spp. However, it fails to detect OXA-type carbapenemases, which are the most frequently identified carbapenemases among Acinetobacter spp. Since those CHDLs found in Acinetobacter spp. (OXA-23, OXA-24/OXA-40-like, OXA-58-like, and OXA-143-like) possess weaker carbapenemase activity than that usually measured in Enterobacteriaceae (mostly KPC, MBLs, and OXA-48) and Pseudomonas spp. (mostly MBLs), we made the hypothesis that the buffer used in the Carba NP test may buffer at a too high level and counteract the detection of any weak carbapenemase activity.
The specificity and sensitivity of the CarbAcineto NP test were found to be 100% and 94.7%, respectively. The CarbAcineto NP test efficiently detected OXA-type carbapenemase producers, leading to a significant improvement of the sensitivity (94.7% for the CarbAcineto NP test versus 11.9% for the Carba NP test). The better detection of CHDLs is due to two independent factors. First, the bacterial inoculum used in the CarbAcineto NP test was doubled compared to that in the Carba NP test, leading to an increased amount of enzyme released in the revealing solution. Second, the lysis buffer used for the Carba NP test (Bacterial Protein Extraction Reagent [B-PER II]; Thermo Scientific Pierce, Villebon-sur-Yvette, France) has been replaced by an hyperosmotic solution of 5 M NaCl. The use of the 5 M NaCl solution provides two main advantages. It does not buffer enough to interfere with slight pH changes, and its hyperosmotic properties lead to an efficient lysis of the bacteria. Water has also been tested as a substitute for the B-PER-II lysis buffer, but the color changes were less clear and the test failed to detect 19 carbapenemase producers (nine OXA-23 and six OXA-58-like), giving orange results (data not shown). This lack of detection might be explained by the weak lysis of the bacteria in water compared to the hyperosmotic solution of NaCl, resulting in lower enzyme release. Recently, a derivative version of the Carba NP test was developed. This test used water instead of the B-PER buffer and bromothymol blue instead of red phenol as the pH indicator (27). The authors claimed that the use of water instead of B-PER buffer led to the detection of all carbapenemase-producing Acinetobacter spp. However, in their study, only 14 OXA-23 and six OXA-58-like producers were tested, which is not sufficient to assess the reliability of that test, considering that only 13.2% of the OXA-23 producers and 23.1% of the OXA-58-like producers were detected when using water instead of NaCl. As previously observed using the Carba NP test with P. aeruginosa strains, the GES-type carbapenemase producers were not detected using the CarbAcineto NP test (26). The GES-type carbapenemases are point mutant analogues of the ESBL GES-1 and possess an extended but very weak carbapenemase activity that might explain this lack of detection. This lack of detection of GES-type carbapenemases in P. aeruginosa may be also due to a weaker release or production of GES-type carbapenemases in P. aeruginosa (26), since a GES-5-producing Enterobacter cloacae isolate was identified with the Carba NP test (24).
Although the number of tested strains was low (3 isolates), the CarbAcineto NP test gave negative results when testing carbapenem-resistant Acinetobacter spp. overexpressing their chromosome-encoded OXA-51-like β-lactamase (Table 2). In fact, the lack of detection of OXA-51-like overproducers may be interesting when considering the usefulness of the CarbAcineto NP test. Indeed, those chromosomally encoded resistance mechanisms are not supposed to be transferable to other organisms, in contrast to plasmid-encoded mechanisms, which therefore constitutes a much less important clinical issue. Thus, clearly distinguishing acquired from nonacquired carbapenemase producers provides an added value to the test.
In conclusion, the CarbAcineto NP test offers a rapid and cost-effective solution for detecting acquired carbapenemase producers in Acinetobacter species. It might contribute to preventing the spread of those multidrug-resistant strains and also of several carbapenemase genes, considering, for example, that blaNDM-1-like genes that initially spread among Acinetobacter spp. then targeted Enterobacteriaceae and Pseudomonas spp. (7). The use of the CarbAcineto NP test will be interesting particularly for ICU patients, for whom multidrug-resistant isolates belonging to the A. baumannii-A. calcoaceticus complex are a common source of severe infections. It particularly makes sense to test for carbapenemase production considering that the acquisition of a carbapenemase in A. baumannii is a marker of multidrug resistance.
Finally, by using both the Carba NP and CarbAcineto NP tests, any microbiology laboratory worldwide may have the opportunity to efficiently identify one of the most important clinical resistance traits of modern microbiology, i.e., carbapenem resistance-mediated mechanisms leading to multidrug or pandrug resistance in clinically significant Gram negatives.
ACKNOWLEDGMENTS
We are grateful to Y. Chong and K. Lee for providing us with the SIM-1-producing isolate and to D. M. Livermore for the gift of the OXA-25- and OXA-26-producing isolates.
An international patent form for the Carba NP test (that included further developments, such as the CarbAcineto NP test) has been filed on behalf of INSERM Transfert (Paris, France).
This work was funded by a grant from the INSERM (UMR914).
Footnotes
Published ahead of print 23 April 2014
REFERENCES
- 1. Bergogne-Bérézin E, Towner KJ. 1996. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 9:148–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Poirel L, Nordmann P. 2006. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin. Microbiol. Infect. 12:826–836. 10.1111/j.1469-0691.2006.01456.x [DOI] [PubMed] [Google Scholar]
- 3. Bonnin RA, Rotimi VO, Al Hubail M, Gasiorowski E, Al Sweih N, Nordmann P, Poirel L. 2013. Wide dissemination of GES-type carbapenemases in Acinetobacter baumannii isolates in Kuwait. Antimicrob. Agents Chemother. 57:183–188. 10.1128/AAC.01384-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Robledo IE, Aquino EE, Santé MI, Santana JL, Otero DM, León CF, Vázquez GJ. 2010. Detection of KPC in Acinetobacter spp. in Puerto Rico. Antimicrob. Agents Chemother. 54:1354–1357. 10.1128/AAC.00899-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Moubareck C, Brémont S, Conroy MC, Courvalin P, Lambert T. 2009. GES-11, a novel integron-associated GES variant in Acinetobacter baumannii. Antimicrob. Agents Chemother. 53:3579–3581. 10.1128/AAC.00072-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bonnin RA, Poirel L, Naas T, Pirs M, Seme K, Schrenzel J, Nordmann P. 2012. Dissemination of New Delhi metallo-β-lactamase-1-producing Acinetobacter baumannii in Europe. Clin. Microbiol. Infect. 18:E362–E365. 10.1111/j.1469-0691.2012.03928.x [DOI] [PubMed] [Google Scholar]
- 7. Bonnin RA, Poirel L, Nordmann P. 2014. New Delhi metallo-β-lactamase-producing Acinetobacter baumannii: a novel paradigm for spreading antibiotic resistance genes. Future Microbiol. 9:33–41. 10.2217/fmb.13.69 [DOI] [PubMed] [Google Scholar]
- 8. Higgins PG, Poirel L, Lehmann M, Nordmann P, Seifert H. 2009. OXA-143, a novel carbapenem-hydrolyzing class D β-lactamase in Acinetobacter baumannii. Antimicrob. Agents Chemother. 53:5035–5038. 10.1128/AAC.00856-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mugnier PD, Poirel L, Naas T, Nordmann P. 2010. Worldwide dissemination of the blaOXA-23 carbapenemase gene of Acinetobacter baumannii. Emerg. Infect. Dis. 16:35–40. 10.3201/eid1601.090852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brown S, Young HK, Amyes SGB. 2005. Characterisation of OXA-51, a novel class D carbapenemase found in genetically unrelated clinical strains of Acinetobacter baumannii from Argentina. Clin. Microbiol. Infect. 11:15–23. 10.1111/j.1469-0691.2004.01016.x [DOI] [PubMed] [Google Scholar]
- 11. Evans BA, Brown S, Hamouda A, Findlay J, Amyes SGB. 2007. Eleven novel OXA-51-like enzymes from clinical isolates of Acinetobacter baumannii. Clin. Microbiol. Infect. 13:1137–1138. 10.1111/j.1469-0691.2007.01828.x [DOI] [PubMed] [Google Scholar]
- 12. Héritier C, Poirel L, Fournier PE, Claverie JM, Raoult D, Nordmann P. 2005. Characterization of the naturally occurring oxacillinase of Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:4174–4179. 10.1128/AAC.49.10.4174-4179.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Turton JF, Ward ME, Woodford N, Kaufmann ME, Pike R, Livermore DM, Pitt TL. 2006. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol. Lett. 258:72–77. 10.1111/j.1574-6968.2006.00195.x [DOI] [PubMed] [Google Scholar]
- 14. Bonnin RA, Nordmann P, Poirel L. 2013. Screening and deciphering antibiotic resistance in Acinetobacter baumannii: a state of the art. Expert Rev. Anti Infect. Ther. 11:571–583. 10.1586/eri.13.38 [DOI] [PubMed] [Google Scholar]
- 15. Bonnin RA, Naas T, Poirel L, Nordmann P. 2012. Phenotypic, biochemical, and molecular techniques for detection of metallo-β-lactamase NDM in Acinetobacter baumannii. J. Clin. Microbiol. 50:1419–1421. 10.1128/JCM.06276-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Franklin C, Liolios L, Peleg AY. 2006. Phenotypic detection of carbapenem-susceptible metallo-β-lactamase-producing gram-negative bacilli in the clinical laboratory. J. Clin. Microbiol. 44:3139–3144. 10.1128/JCM.00879-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bernabeu S, Poirel L, Nordmann P. 2012. Spectrophotometry-based detection of carbapenemase producers among Enterobacteriaceae. Diagn. Microbiol. Infect. Dis. 74:88–90. 10.1016/j.diagmicrobio.2012.05.021 [DOI] [PubMed] [Google Scholar]
- 18. Álvarez-Buylla A, Picazo JJ, Culebras E. 2013. Optimized method for Acinetobacter species carbapenemase detection and identification by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 51:1589–1592. 10.1128/JCM.00181-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kempf M, Bakour S, Flaudrops C, Berrazeg M, Brunel JM, Drissi M, Mesli E, Touati A, Rolain JM. 2012. Rapid detection of carbapenem resistance in Acinetobacter baumannii using matrix-assisted laser desorption ionization–time of flight mass spectrometry. PLoS One 7:e31676. 10.1371/journal.pone.0031676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee W, Chung HS, Lee Y, Yong D, Jeong SH, Lee K, Chong Y. 2013. Comparison of matrix-assisted laser desorption ionization-time-of-flight mass spectrometry assay with conventional methods for detection of IMP-6, VIM-2, NDM-1, SIM-1, KPC-1, OXA-23, and OXA-51 carbapenemase-producing Acinetobacter spp., Pseudomonas aeruginosa, and Klebsiella pneumoniae. Diagn. Microbiol. Infect. Dis. 77:227–230. 10.1016/j.diagmicrobio.2013.07.005 [DOI] [PubMed] [Google Scholar]
- 21. Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S, Amyes SG, Livermore DM. 2006. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 27:351–353. 10.1016/j.ijantimicag.2006.01.004 [DOI] [PubMed] [Google Scholar]
- 22. Huang XZ, Cash DM, Chahine MA, Nikolich MP, Craft DW. 2012. Development and validation of a multiplex TaqMan real-time PCR for rapid detection of genes encoding four types of class D carbapenemase in Acinetobacter baumannii. J. Med. Microbiol. 61:1532–1537. 10.1099/jmm.0.045823-0 [DOI] [PubMed] [Google Scholar]
- 23. Dally S, Lemuth K, Kaase M, Rupp S, Knabbe C, Weile J. 2013. DNA microarray for genotyping antibiotic resistance determinants in Acinetobacter baumannii clinical isolates. Antimicrob. Agents Chemother. 57:4761–4768. 10.1128/AAC.00863-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nordmann P, Poirel L, Dortet L. 2012. Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 18:1503–1507. 10.3201/eid1809.120355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dortet L, Bréchard L, Poirel L, Nordmann P. 2014. Impact of the isolation medium for detection of carbapenemase-producing Enterobacteriaceae using an updated version of the Carba NP test. J. Med. Microbiol., in press [DOI] [PubMed] [Google Scholar]
- 26. Dortet L, Poirel L, Nordmann P. 2012. Rapid detection of carbapenemase-producing Pseudomonas spp. J. Clin. Microbiol. 50:3773–3776. 10.1128/JCM.01597-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pires J, Novais A, Peixe L. 2013. Blue-Carba, an easy biochemical test for detection of diverse carbapenemase producers directly from bacterial cultures. J. Clin. Microbiol. 51:4281–4283. 10.1128/JCM.01634-13 [DOI] [PMC free article] [PubMed] [Google Scholar]