Abstract
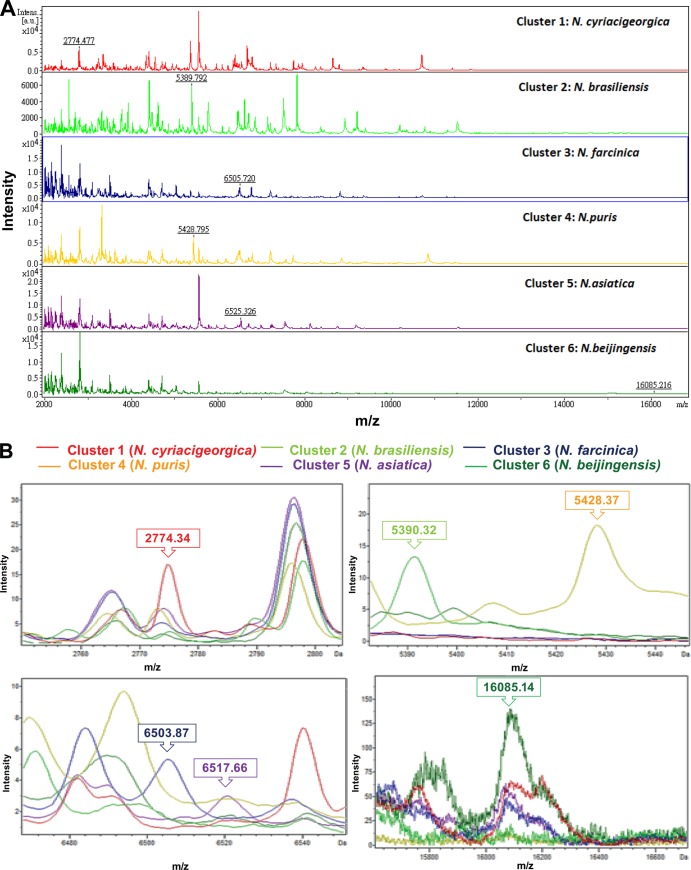
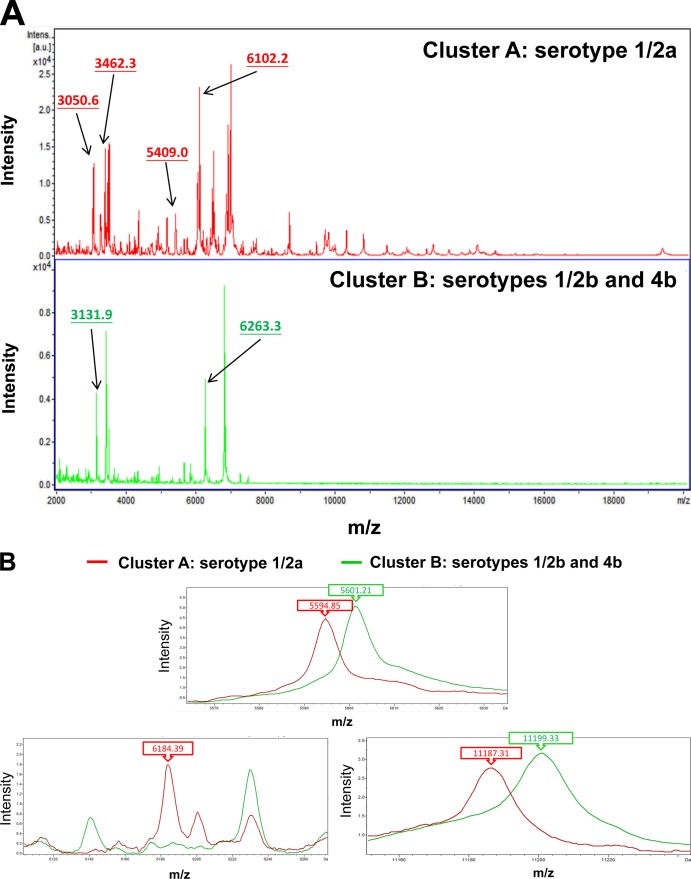
We evaluated whether the Bruker Biotyper matrix-associated laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) system provides accurate species-level identifications of 147 isolates of aerobically growing Gram-positive rods (GPRs). The bacterial isolates included Nocardia (n = 74), Listeria (n = 39), Kocuria (n = 15), Rhodococcus (n = 10), Gordonia (n = 7), and Tsukamurella (n = 2) species, which had all been identified by conventional methods, molecular methods, or both. In total, 89.7% of Listeria monocytogenes, 80% of Rhodococcus species, 26.7% of Kocuria species, and 14.9% of Nocardia species (n = 11, all N. nova and N. otitidiscaviarum) were correctly identified to the species level (score values, ≥2.0). A clustering analysis of spectra generated by the Bruker Biotyper identified six clusters of Nocardia species, i.e., cluster 1 (N. cyriacigeorgica), cluster 2 (N. brasiliensis), cluster 3 (N. farcinica), cluster 4 (N. puris), cluster 5 (N. asiatica), and cluster 6 (N. beijingensis), based on the six peaks generated by ClinProTools with the genetic algorithm, i.e., m/z 2,774.477 (cluster 1), m/z 5,389.792 (cluster 2), m/z 6,505.720 (cluster 3), m/z 5,428.795 (cluster 4), m/z 6,525.326 (cluster 5), and m/z 16,085.216 (cluster 6). Two clusters of L. monocytogenes spectra were also found according to the five peaks, i.e., m/z 5,594.85, m/z 6,184.39, and m/z 11,187.31, for cluster 1 (serotype 1/2a) and m/z 5,601.21 and m/z 11,199.33 for cluster 2 (serotypes 1/2b and 4b). The Bruker Biotyper system was unable to accurately identify Nocardia (except for N. nova and N. otitidiscaviarum), Tsukamurella, or Gordonia species. Continuous expansion of the MALDI-TOF MS databases to include more GPRs is necessary.
INTRODUCTION
Aerobically growing Gram-positive rods (GPRs) constitute a very heterogeneous and extensive group of bacterial species (1, 2). Some of them, such as Listeria monocytogenes and Nocardia, Kocuria, Rhodococcus, Gordonia, and Tsukamurella species, are associated with severe community-acquired infections, including meningitis, bacteremia, pneumonia, brain abscesses, and skin and soft tissue infections (3–16). These organisms also cause various health care-associated infections, including catheter-related bacteremia (5, 9, 12–14). Some species, especially L. monocytogenes, can cause outbreaks (17, 18). Since these GPRs differ in the clinical spectrum of diseases they cause and in their susceptibilities to antimicrobial agents, it is important to precisely identify these isolates beyond the genus level for both therapeutic and infection control measures (9, 19–23).
In clinical microbiology laboratories, bacteria are traditionally identified using manual or automated phenotypic and biochemical methods. These methods are generally reliable for species-level identification but are often cumbersome and time-consuming to perform (1, 2). However, for some clinically important Gram-positive bacilli, performing these manual or automated methods is not always sufficient for identifying necessary organisms. Studies have clearly demonstrated that molecular methods, such as 16S rRNA gene sequencing analysis, are more accurate than conventional phenotypic methods for identifying unusual or rarely encountered pathogens, such as Nocardia, Rhodococcus, Kocuria, Gordonia, and Tsukamurella species (5, 9, 11–14).
Several commercially available matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) systems are now widely used in clinical microbiology laboratories for the rapid identification of commonly encountered bacteria and fungi (24–26). Numerous studies have compared the performances of MALDI-TOF MS systems with those of commonly used phenotypic testing and DNA sequence analysis techniques for identifying infrequently encountered bacterial pathogens; however, few studies have investigated the performance of MALDI-TOF MS for GPRs (1, 2, 27).
In this study, we assessed the performance of the Bruker Biotyper MALDI-TOF MS system for identifying a large collection of aerobically growing GPRs, including Nocardia, Listeria, Kocuria (which are coccoid but are always described as being rod-like), Rhodococcus, Gordonia, and Tsukamurella species, that were isolated from various clinical sources.
MATERIALS AND METHODS
Bacterial isolates.
A total of 147 nonduplicate isolates of GPRs that were recovered from various sources from patients treated at the National Taiwan University Hospital (NTUH) were evaluated. These included 74 isolates of Nocardia species, 39 isolates of L. monocytogenes, 15 isolates of Kocuria species, 10 isolates of Rhodococcus species, seven isolates of Gordonia species, and two isolates of Tsukamurella species. All of these isolates were associated various clinical infections from patients who were treated at NTUH, and some of these infections were previously reported (5–9, 11–16, 20–22).
Bacterial identification.
The 108 isolates of Nocardia, Kocuria, Rhodococcus, Gordonia, and Tsukamurella species were obtained from a clinical microbiology laboratory and were initially identified to the genus and/or species level by conventional methods. Due to the previously reported high misidentification rates by routine identification systems for these GPRs (5–9, 11–16, 20–22), these isolates were further confirmed to the species level using several molecular methods. For isolates of Nocardia, Kocuria, Rhodococcus, and Tsukamurella species, partial sequencing analysis of the 16S rRNA gene was performed using the primers Noc1 (5′-GCTTAACACATGCAAGTCG-3′) (positions 46 to 64, Escherichia coli numbering system) and Noc2 (5′-GAATTCCAGTCTCCCCTG-3′). The sequences were compared with published sequences in the 16S rRNA database. For Nocardia species, a second molecular method by sequencing the subunit A of SecA preprotein translocase gene (secA1) was performed using a pair of primers, NsecA1 (5′-GTAAAACGACGGCCAGGACAGYGAGTGGATGGGYCGSGTGCACCG-3′) and NsecA1 R (5′-CAGGAAACAGCTATGACGCGGACGATGTAGTCCTTGTC-3′) (28).
Two molecular methods were used to identify Gordonia and Tsukamurella organisms to the species level (11, 12). The first molecular method was a previously described PCR restriction fragment length polymorphism (RFLP) identification scheme that used an amplified 440-bp segment of the 65-kDa heat shock protein gene (hsp65) performed using two primers, TB11 (5′-ACCAACGATGGTGTGTCCAT-3′) and TB12 (5′-CTTGTCGAACCGCATACCCT-3′) (12–14), and digestion by MspI and HinfI. The second molecular method was 16S rRNA gene sequencing using a pair of universal primers, 8FLP (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492RPL (5′-GGTTACCTTGTTACGACTT-3′) (12–14). The sequences (1,080 bp) obtained were compared with published sequences in the GenBank database using the BLASTn algorithm (http://www.ncbi.nlm.nih.gov/blast). The accession number and closest match observed for each sequence were obtained. The criteria used to determine the closest match for identification using 16S rRNA sequencing were >99.5% sequence similarity to the unknown strain and the difference between the closest and next closest match having >0.5% divergence (29). The identification of Gordonia and Tsukamurella isolates to species level was based on >99.5% similarity by 16S rRNA gene sequencing and compatible RFLP results.
L. monocytogenes (n = 39) was identified using conventional identification methods, including hemolysis on sheep blood agar plates and the Christie-Atkins-Munch-Petersen (CAMP) reaction, as well as the API Coryne system (bioMérieux, Marcy l'Etoile, France). The serotypes of the isolates were determined by PCR, as previously described (15, 30).
Performance of the MALDI Bruker Biotyper.
For analysis of the 147 GPRs by the Bruker MALDI-TOF Biotyper system, the samples were prepared as previously described (27). All isolates were incubated on Trypticase soy agar with 5% sheep blood (BAP) (Becton, Dickinson Microbiology Systems, Sparks, MD, USA) and incubated for 48 h at 37°C. Two to three colonies were transferred to a 1.5-ml screw-cap Eppendorf tube containing 300 μl of distilled water and then mixed with 900 μl of ethanol by pipetting. The suspension was pelleted by centrifugation at 13,000 rpm for 2 min, evaporated to dryness, and then reconstituted in 50 μl of 70% formic acid. After incubation for 30 s, 50 μl of acetonitrile (Sigma-Aldrich) was added. The suspension was then centrifuged at 13,000 rpm for 2 min. Next, 1.0 μl of the supernatant was applied to a 96-spot polished steel target plate (Bruker Daltonik GmbH, Bremen, Germany) and dried. A saturated solution of 1.0 μl of MALDI matrix (α-cyano-4-hydroxycinnamic acid [HCCA]; Bruker Daltonik GmbH) was applied to each sample and dried. Measurements were performed with the Bruker microflex LT MALDI-TOF MS (Bruker Daltonik GmbH) using FlexControl software with Compass Flex Series version 1.3 software and a 60-Hz nitrogen laser (337 nm wavelength). The spectra were collected in the linear positive mode in a mass range covering m/z 1,960 to 20,132. Spectra ranging from the mass-to-charge ratio (m/z) 2,000 to 20,000 were analyzed using Bruker Biotyper automation control and the Bruker Biotyper 3.1 software and library (database [DB] 5627 with 5,627 entries). Identification scores of ≥2.000 indicated species-level identification, scores of 1.700 to 1.999 indicated genus-level identification, and scores of <1.700 indicated no reliable identification. All isolates with discrepant identification results between the molecular and Bruker Biotyper methods were retested twice.
Cluster analysis by the Bruker Biotyper for six Nocardia species and L. monocytogenes isolates.
A clustering analysis of the isolates was performed using ClinProTools 3.0 (Bruker Daltonics GmbH) (31). Fifty-five isolates belonging to six Nocardia species (N. cyriacigeorgica [n = 18], N. brasiliensis [n = 20], N. farcinica [n = 8], N. puris [n = 5], N. asiatica [n = 4], and N. beijingensis [n = 3] were identified by molecular methods, and 39 isolates of L. monocytogenes, including 10 isolates of serotype 1/2a, 17 of serotype 1/2b, and 12 of serotype 4b, were analyzed for specific signals by clustering analysis (31, 32). A model based on a genetic algorithm was created with the parameter of 15 as the maximum number of best peaks, 15 as the maximum number of generation, a mutation rate of 0.2%, and a crossover rate of 0.5%.
RESULTS
Bacterial identification.
Table 1 shows the identification results using molecular methods for the 108 isolates of Nocardia, Kocuria, Rhodococcus, Gordonia, and Tsukamurella species. A total of 14 different Nocardia species, two Kocuria species, two Rhodococcus species, three Gordonia species, and one Tsukamurella species were identified by various molecular methods. All isolates were identified to the species level, with >99.5% sequence similarity at the level of the 16S rRNA sequence. All isolates of N. cyriacigeorgica, N. farcinica, N. beijingensis, N. otitidiscaviarum, N. puris, Kocuria marina, Gordonia bronchialis, Gordonia amicalis, Gordonia sputi, Rhodococcus equi, and Rhodococcus kroppenstedtii exhibited 100% sequence similarity by 16S rRNA sequence analysis. All but three Nocardia species identified by 16S rRNA sequence analysis possessed compatible identification results by secA1 gene sequencing (>95.0% [range, 97.0% to 100%] similarity). The three Nocardia species with discordant results by 16S rRNA and secA1 gene sequencing included N. transvalensis (99.6% similarity by 16S rRNA) identified as N. wallacei (99.5% similarity by secA1; GenBank accession no. JN562389.1), N. beijingensis (100% similarity by 16S rRNA) identified as N. arthritidis (97.6% similarity by secA1; GenBank accession no. DQ360262.1), and N. abscessus (99.5% similarity by 16S rRNA) identified as N. asiatica (100% similarity by secA1; GenBank accession no. JQ773453.1). As for the 39 L. monocytogenes isolates, 17 (43.6%) belonged to serotype 1/2b, 12 (30.8%) were serotype 4b, and 10 (25.6%) were serotype 1/2a (Table 2).
TABLE 1.
Identification results and scores of 108 clinical isolates of Nocardia, Kocuria, Rhodococcus, Gordonia, and Tsukamurella species with the MALDI Bruker Biotyper database (DB 5627)a
| Isolate no. by genus (n) | Identification results by 16S rRNA sequencing |
Organism (best match) | Score | Organism (second best match) | Score | |
|---|---|---|---|---|---|---|
| Bacterial species | GenBank accession no. | |||||
| Nocardia species (74) | ||||||
| 1 | N. brasiliensis | FJ172109.1 | Nocardia sp. | 1.957 | N. brasiliensis | 1.856 |
| 2 | N. brasiliensis | FJ172109.1 | Nocardia sp. | 1.985 | N. brasiliensis | 1.748 |
| 3 | N. brasiliensis | FJ172109.1 | Nocardia sp. | 1.904 | Lactobacillus paracasei (NRI) | 1.570 |
| 4 | N. brasiliensis | FJ172109.1 | Nocardia sp. | 2.041 | N. brasiliensis | 1.852 |
| 5 | N. brasiliensis | FJ172109.1 | Nocardia sp. | 1.967 | N. brasiliensis | 1.773 |
| 6 | N. brasiliensis | FJ172109.1 | Nocardia sp. | 2.093 | N. brasiliensis | 1.938 |
| 7 | N. brasiliensis | FJ172109.1 | Nocardia sp. | 1.97 | N. brasiliensis | 1.715 |
| 8 | N. brasiliensis | FJ172109.1 | Nocardia sp. | 1.912 | N. brasiliensis | 1.754 |
| 9 | N. brasiliensis | FJ172109.1 | Nocardia sp. | 2.048 | N. brasiliensis | 1.897 |
| 10 | N. brasiliensis | FJ172109.1 | Nocardia sp. | 2.019 | N. brasiliensis (NRI) | 1.68 |
| 11 | N. brasiliensis | FJ172109.1 | Nocardia sp. | 2.009 | N. brasiliensis | 1.807 |
| 12 | N. brasiliensis | FJ172109.1 | Clostridium bifermentans (NRI)b | 1.455 | Lactobacillus paracasei (NRI) | 1.406 |
| 13 | N. brasiliensis | FJ172109.1 | Nocardia sp. | 1.867 | N. brasiliensis | 1.691 |
| 14 | N. brasiliensis | FJ172109.1 | N. brasiliensis | 1.872 | N. brasiliensis | 1.845 |
| 15 | N. brasiliensis | FJ172109.1 | Nocardia sp. | 2.041 | N. brasiliensis | 1.845 |
| 16 | N. brasiliensis | FJ172109.1 | Nocardia sp. | 1.995 | N. brasiliensis | 1.973 |
| 17 | N. brasiliensis | FJ172109.1 | Nocardia sp. | 2.074 | N. brasiliensis | 1.923 |
| 18 | N. brasiliensis | FJ172109.1 | Nocardia sp. | 2.068 | N. brasiliensis | 1.857 |
| 19 | N. brasiliensis | FJ172109.1 | Nocardia sp. | 1.971 | N. brasiliensis | 1.867 |
| 20 | N. brasiliensis | FJ172109.1 | Nocardia sp. | 2.033 | N. brasiliensis | 1.858 |
| 21 | N. cyriacigeorgica | GQ376180.1 | N. cyriacigeorgica | 1.918 | N. cyriacigeorgica | 1.908 |
| 22 | N. cyriacigeorgica | GQ376180.1 | N. cyriacigeorgica | 1.842 | N. cyriacigeorgica | 1.841 |
| 23 | N. cyriacigeorgica | GQ376180.1 | N. cyriacigeorgica | 1.747 | N. cyriacigeorgica | 1.722 |
| 24 | N. cyriacigeorgica | GQ376180.1 | N. cyriacigeorgica | 1.745 | N. cyriacigeorgica | 1.719 |
| 25 | N. cyriacigeorgica | GQ376180.1 | N. cyriacigeorgica | 1.844 | N. cyriacigeorgica | 1.839 |
| 26 | N. cyriacigeorgica | GQ376180.1 | N. cyriacigeorgica (NRI) | 1.601 | N. cyriacigeorgica | 1.551 |
| 27 | N. cyriacigeorgica | GQ376180.1 | N. cyriacigeorgica | 1.816 | N. cyriacigeorgica | 1.73 |
| 28 | N. cyriacigeorgica | GQ376180.1 | Pseudomonas jinjuensis (NRI) | 1.305 | Aromatoleum tolulyticus (NRI) | 1.256 |
| 29 | N. cyriacigeorgica | GQ376180.1 | N. cyriacigeorgica | 1.712 | NRI | 1.611 |
| 30 | N. cyriacigeorgica | GQ376180.1 | N. cyriacigeorgica | 1.924 | N. cyriacigeorgica | 1.862 |
| 31 | N. cyriacigeorgica | GQ376180.1 | N. cyriacigeorgica | 1.806 | NRI | 1.577 |
| 32 | N. cyriacigeorgica | GQ376180.1 | N. cyriacigeorgica | 1.787 | NRI | 1.607 |
| 33 | N. cyriacigeorgica | GQ376180.1 | Staphylococcus lutrae (NRI) | 1.414 | Lactobacillus fructivorans (NRI) | 1.412 |
| 34 | N. cyriacigeorgica | GQ376180.1 | N. cyriacigeorgica | 1.788 | NRI | 1.694 |
| 35 | N. cyriacigeorgica | GQ376180.1 | N. cyriacigeorgica (NRI) | 1.661 | N. cyriacigeorgica (NRI) | 1.59 |
| 36 | N. cyriacigeorgica | GQ376180.1 | N. cyriacigeorgica | 2.177 | N. cyriacigeorgica | 2.165 |
| 37 | N. cyriacigeorgica | GQ376180.1 | Staphylococcus epidermidis (NRI) | 1.404 | Staphylococcus aureus (NRI) | 1.387 |
| 38 | N. cyriacigeorgica | GQ376180.1 | N. cyriacigeorgica | 1.784 | NRI | 1.562 |
| 39 | N. farcinica | GQ853065.1 | N. farcinica | 2.036 | N. farcinica | 1.956 |
| 40 | N. farcinica | GQ853065.1 | Lactobacillus paracasei | 1.447 | Lactobacillus vini (NRI) | 1.391 |
| 41 | N. farcinica | GQ853065.1 | N. farcinica (NRI) | 1.632 | N. farcinica (NRI) | 1.608 |
| 42 | N. farcinica | GQ853065.1 | N. farcinica | 1.944 | NRI | 1.647 |
| 43 | N. farcinica | GQ853065.1 | N. farcinica | 1.943 | N. farcinica | 1.929 |
| 44 | N. farcinica | GQ853065.1 | N. farcinica | 1.773 | NRI | 1.615 |
| 45 | N. farcinica | GQ853065.1 | N. farcinica | 1.749 | NRI | 1.668 |
| 46 | N. farcinica | GQ853065.1 | N. farcinica (NRI) | 1.677 | N. farcinica (NRI) | 1.558 |
| 47 | N. asiatica | GQ217495.1 | N. cyriacigeorgica | 2.256 | N. cyriacigeorgica | 2.245 |
| 48 | N. asiatica | GQ217495.1 | N. cyriacigeorgica | 1.905 | N. cyriacigeorgica | 1.869 |
| 49 | N. asiatica | GQ217495.1 | N. asiatica | 1.843 | Aeromonas veronii (NRI) | 1.44 |
| 50 | N. asiatica | GQ217495.1 | N. asiatica | 1.859 | Lactobacillus paracasei (NRI) | 1.513 |
| 51 | N. nova | GQ376190.1 | N. nova | 2.221 | NRI | 1.571 |
| 52 | N. nova | AF430031.1 | N. nova | 2.245 | NRI | 1.513 |
| 53 | N. nova | GQ376190.1 | N. nova | 2.327 | NRI | 1.378 |
| 54 | N. nova | GQ376190.1 | N. nova | 2.342 | NRI | 1.479 |
| 55 | N. nova | GQ376190.1 | N. nova | 2.076 | NRI | 1.3 |
| 56 | N. nova | FJ172123.1 | N. nova | 2.437 | NRI | 1.638 |
| 57 | N. beijingensis | GQ217493.1 | N. asiatica (NRI) | 1.576 | Lactobacillus paracasei (NRI) | 1.552 |
| 58 | N. beijingensis | GQ217493.1 | N. asiatica (NRI) | 1.444 | Nocardia sp. (NRI) | 1.381 |
| 59 | N. beijingensis | GQ217493.1 | Nocardia sp. (NRI) | 1.328 | N. asiatica (NRI) | 1.305 |
| 60 | N. puris | GQ217500.1 | N. cyriacigeorgica (NRI) | 1.521 | NRI | 1.363 |
| 61 | N. puris | GQ217500.1 | Aromatoleum alkani (NRI) | 1.393 | Streptomyces lavendulae (NRI) | 1.362 |
| 62 | N. puris | GQ217500.1 | Lactobacillus aviarius (NRI) | 1.32 | Streptomyces hirsutus (NRI) | 1.306 |
| 63 | N. puris | GQ217500.1 | Rhodococcus equi (NRI) | 1.42 | Lactobacillus paracasei (NRI) | 1.404 |
| 64 | N. puris | AB097453.1 | Rhizobium radiobacter (NRI) | 1.418 | Xenorhabdus ehlersii (NRI) | 1.281 |
| 65 | N. otitidiscaviarum | GQ376191.1 | N. otitidiscaviarum | 2.138 | N. otitidiscaviarum | 2.044 |
| 66 | N. otitidiscaviarum | GQ376191.1 | N. otitidiscaviarum | 2.132 | N. otitidiscaviarum | 1.778 |
| 67 | N. otitidiscaviarum | GQ376191.1 | N. otitidiscaviarum | 2.114 | N. otitidiscaviarum (NRI) | 1.397 |
| 68 | N. abscessus | GU471235.1 | Lactobacillus fuchuensis (NRI) | 1.323 | Nocardia sp. (NRI) | 1.294 |
| 69 | N. abscessus | GU471235.1 | Clostridium tetani (NRI) | 1.277 | Lactobacillus amylovorus (NRI) | 1.274 |
| 70 | N. rhamnosiphila | EF418604.1 | Agromyces rhizospherae (NRI) | 1.325 | Lactobacillus fructivorans (NRI) | 1.266 |
| 71 | N. asteroides | AF430025.1 | N. asteroides | 1.777 | NRI | 1.580 |
| 72 | N. elegans | GQ376166.1 | N. nova | 2.071 | Lactobacillus paracasei (NRI) | 1.499 |
| 73 | N. carnea | GU433886.1 | Staphylococcus equorum (NRI) | 1.419 | Lactobacillus amylotrophicus (NRI) | 1.41 |
| 74 | N. transvalensis | AB084446.1 | L. paracasei (NRI) | 1.494 | Aeromonas salmonicida (NRI) | 1.416 |
| Kocuria species (15) | ||||||
| 75 | K. kristinae | EU379300.1 | K. kristinae | 1.992 | K. kristinae | 1.911 |
| 76 | K. kristinae | EU379300.1 | K. kristinae | 2.115 | K. kristinae | 2.085 |
| 77 | K. kristinae | EU379300.1 | K. kristinae | 1.973 | K. kristinae | 1.931 |
| 78 | K. kristinae | EU379300.1 | K. kristinae | 2.003 | K. kristinae | 1.944 |
| 79 | K. kristinae | EU379300.1 | K. kristinae | 2.005 | K. kristinae | 1.808 |
| 80 | K. kristinae | EU379300.1 | K. kristinae | 1.91 | K. kristinae | 1.823 |
| 81 | K. kristinae | EU379300.1 | K. kristinae | 1.984 | K. kristinae | 1.93 |
| 82 | K. kristinae | EU379300.1 | K. kristinae | 1.756 | K. kristinae | 1.747 |
| 83 | K. kristinae | EU379300.1 | K. kristinae | 2.113 | K. kristinae | 1.93 |
| 84 | K. kristinae | EU379300.1 | K. kristinae | 1.861 | NRI | 1.507 |
| 85 | K. kristinae | EU379300.1 | K. kristinae | 1.95 | K. kristinae | 1.81 |
| 86 | K. kristinae | EU379300.1 | K. kristinae | 1.717 | NRI | 1.667 |
| 87 | K. kristinae | EU379300.1 | K. kristinae | 1.831 | NRI | 1.639 |
| 88 | K. marina | KF306369.1 | K. marina | 1.955 | NRI | 1.507 |
| 89 | K. marina | KF306369.1 | K. marina | 1.824 | NRI | 1.477 |
| Rhodococcus species (10) | ||||||
| 90 | R. equi | JQ965789.1 | R. equi | 2.134 | R. equi | 2.010 |
| 91 | R. equi | JQ965789.1 | R. equi | 2.240 | R. equi | 2.202 |
| 92 | R. equi | JQ965789.1 | R. equi | 2.087 | R. equi | 2.060 |
| 93 | R. equi | JQ965789.1 | R. equi | 2.188 | R. equi | 2.073 |
| 94 | R. equi | JQ965789.1 | R. equi | 2.255 | R. equi | 2.201 |
| 95 | R. equi | JQ965789.1 | R. equi | 2.258 | R. equi | 2.204 |
| 96 | R. equi | JQ965789.1 | R. equi | 2.278 | R. equi | 2.175 |
| 97 | R. equi | JQ965789.1 | R. equi | 2.192 | R. equi | 2.141 |
| 98 | R. kroppenstedtii | KC346296.1 | R. kroppenstedtii | 1.912 | NRI | 1.529 |
| 99 | R. kroppenstedtii | KC346296.1 | R. kroppenstedtii (NRI) | 1.595 | NRI | 1.519 |
| Gordonia species (7) | ||||||
| 100 | G. bronchialis | HQ316182.1 | G. bronchialis (NRI) | 1.662 | Morganella morganii (NRI) | 1.543 |
| 101 | G. bronchialis | HQ316182.1 | Morganella morganii (NRI) | 1.568 | Nocardia sp. (NRI) | 1.302 |
| 102 | G. bronchialis | HQ316182.1 | G. bronchialis (NRI) | 1.588 | Morganella morganii | 1.482 |
| 103 | G. amicalis | HQ842811.1 | G. rubripertincta (NRI) | 1.629 | G. rubripertincta (NRI) | 1.593 |
| 104 | G. amicalis | HQ842811.1 | G. rubripertincta (NRI) | 1.465 | G. terrae (NRI) | 1.404 |
| 105 | G. amicalis | HQ842811.1 | G. rubripertincta (NRI) | 1.436 | Staphylococcus chromogenes (NRI) | 1.406 |
| 106 | G. sputi | FJ536318.1 | G. sputi | 1.984 | G. sputi (NRI) | 1.617 |
| Tsukamurella species (2) | ||||||
| 107 | T. tyrosinosolvens | JX154557.1 | Tsukamurella paurometabola (NRI) | 1.598 | Tsukamurella sp. (NRI) | 1.511 |
| 108 | T. tyrosinosolvens | JX154557.1 | Tsukamurella sp. | 1.786 | T. inchonensis (NRI) | 1.622 |
The Bruker Biotyper database (DB 5627) does not include N. beijingensis, N. puris, N. rhamnosiphila, T. tyrosinosolvens, or G. amicalis.
NRI, not a reliable identification.
TABLE 2.
Serotypes, cluster analysis, and score values of MALDI-TOF MS spectra of 39 clinical isolates of L. monocytogenes
| Serotype | No. of isolates | Clustera | No. of isolates with indicated score values by MALDI-TOF Bruker Biotyper |
||
|---|---|---|---|---|---|
| <1.7 | 1.7–1.999 | ≥2.0 | |||
| 1/2a | 10 | A | 0 | 2 (1.889–1.902) | 8 (2.058–2.321) |
| 1/2b | 17 | B | 0 | 0 | 17 (2.045–2.375) |
| 4b | 12 | B | 0 | 2 (1.82–1.98) | 10 (2.069–2.437) |
See Fig. 2 for definition of clusters A and B based on cluster analysis by spectra generated by MALDI-TOF Bruker Biotyper.
Identification by the Bruker Biotyper.
The MALDI-TOF MS spectra of six selected species of GPRs are shown in Fig. 1A and 2A. Of the 22 species of GPRs evaluated in this study, N. beijingensis, N. puris, Nocardia rhamnosiphila, Tsukamurella tyrosinosolvens, and G. amicalis are not included in the Bruker Biotyper database (DB 5627) (Table 1). Of the 20 isolates of N. brasiliensis, only one isolate was identified as N. brasiliensis and had an identification score value of 1.872. The other 19 isolates were reported as Nocardia species. However, 18 of the second-matched organisms were correctly identified (score values, 1.680 to 1.938). Of the 18 isolates of N. cyriacigeorgica, 15 were identified as N. cyriacigeorgica (one isolate with a score value of 2.177 and 14 with score values ranging from 1.601 to 1.924). Three N. cyriacigeorgica isolates were misidentified as Pseudomonas and Staphylococcus species. The rates of consistency between species using molecular identification and best-matched species by the Bruker Biotyper among other Nocardia isolates were 100% (6/6) for N. nova (score values, all >2.0) and N. otitidiscaviarum (3/3; score values, all >2.0), 87.5% (7/8) for N. farcinica (one isolate with a score value of 2.036 and 6 with score values ranging from 1.632 to 1.944), and 50% (2/4) for N. asiatica (score values, 1.843 and 1.859). Of the 13 isolates of Kocuria kristinae, all were correctly identified, including four with score values of 2.003 to 2.115 and nine with score values of 1.831 to 1.992. Two K. marina isolates were identified to the species level but had score values of 1.824 and 1.955. For the 10 Rhodococcus isolates, eight R. equi isolates were all correctly identified to the species level (score values, >2.0), and the remaining two R. kroppenstedtii isolates were also identified correctly but with low score values. The Bruker Biotyper was unable to correctly identify the other Nocardia and Tsukamurella species. Although two G. bronchialis isolates were correctly identified by the Bruker Biotyper, the score values were low (<1.7). All G. amicalis isolates were identified as Gordonia rubripertincta (score values, <1.7).
FIG 1.
Clustering analysis of spectra generated by the Bruker Biotyper MALDI-TOF MS system for six common Nocardia species with poor identification performance or absence in the Bruker Biotyper database. (A) Six clusters of Nocardia species spectra, i.e., cluster 1, N. cyriacigeorgica; cluster 2, N. brasiliensis; cluster 3, N. farcinica; cluster 4, N. puris; cluster 5, N. asiatica; and cluster 6, N. beijingensis. (B) The six peaks (indicated by numbers and arrows) used to define the six clusters which were generated by ClinProTools with the genetic algorithm are m/z 2,774.477 (cluster 1), m/z 5,389.792 (cluster 2), m/z 6,505.720 (cluster 3), m/z 5,428.795 (cluster 4), m/z 6,525.326 (cluster 5), and m/z 16,085.216 (cluster 6). The absolute intensities of the ions are shown on the y axis, and the masses (m/z) of the ions are shown on the x axis. The m/z values represent the mass-to-charge ratio.
FIG 2.
Clustering analysis of spectra generated by the Bruker Biotyper MALDI-TOF system for three different serotypes of 39 L. monocytogenes isolates. (A) Two clusters of L. monocytogenes spectra, i.e., cluster A (serotype 1/2a) and cluster B (serotypes 1/2b and 4b). Peaks are labeled with arrows. (B) The five peaks (indicated by numbers and arrows) used to define the two clusters that were generated by ClinProTools with the genetic algorithm are m/z 5,594.85, m/z 6,184.39, and m/z 11,187.31 for cluster 1, and m/z 5,601.21 and m/z 11,199.33 for cluster 2. The absolute intensities of the ions are shown on the y axis and the masses (m/z) of the ions are shown on the x axis. The m/z values represent the mass-to-charge ratio.
All isolates of L. monocytogenes were correctly identified by the Bruker Biotyper, and 89.7% had high identification score values (≥2.0) (Table 2).
Clustering analysis of the spectra of six Nocardia species and L. monocytogenes by the Bruker Biotyper.
A clustering analysis was performed for the spectra generated by the Bruker Biotyper results for 63 isolates of six common Nocardia species with poor identification performance or that were absent in the Bruker database. Six clusters of Nocardia species spectra, i.e., cluster 1 (N. cyriacigeorgica), cluster 2 (N. brasiliensis), cluster 3 (N. farcinica), cluster 4 (N. puris), cluster 5 (N. asiatica), and cluster 6 (N. beijingensis), were identified based on the six peaks generated by ClinProTools with the genetic algorithm, i.e., m/z 2,774.477 (cluster 1), m/z 5,389.792 (cluster 2), m/z 6,505.720 (cluster 3), m/z 5,428.795 (cluster 4), m/z 6,525.326 (cluster 5), and m/z 16,085.216 (cluster 6) (Fig. 1A and B). A model was used that included the six specific peaks that were analyzed by the 63 spectra of six clusters. The recognition capacity for the 63 spectra was 100% correct in each cluster. Two clusters of L. monocytogenes spectra, i.e., cluster 1 (serotype 1/2a) and cluster 2 (serotypes 1/2b and 4b), were also found according to the five peaks, i.e., m/z 5,594.85, m/z 6,184.39, and m/z 11,187.31 for cluster 1, and m/z 5,601.21 and m/z 11,199.33 for cluster 2 (Fig. 2A and B).
DISCUSSION
Our analysis of the performance of the Bruker Biotyper MALDI-TOF system for identifying aerobically growing Gram-positive rod-shaped bacilli that had been confirmed by molecular identification methods disclosed several important findings. First, 89.7% of L. monocytogenes, 14.9% of Nocardia species, 80% of Rhodococcus species, and 26.7% of Kocuria species were correctly identified to the species level (score values, ≥2.0) by the Bruker Biotyper system. However, none of the Gordonia and Tsukamurella isolates were correctly identified. Second, although N. brasiliensis, N. cyriacigeorgica, and N. farcinica, the most commonly encountered Nocardia species in clinical settings, were listed in the updated Bruker Biotyper database (DB 5627), the system was not able to accurately identify them to the species level (score values, ≥2.0; identification rate, 10.9% [5/46]). Of the 47 isolates of the three species, only 35 (74.5%) were identified as the same species, and all had identification score values of ≥1.7 (both best-matched and second-matched organisms). Finally, six peaks generated by the cluster analysis of MALDI-TOF MS spectra were found to be markers that could differentiate among the six commonly encountered Nocardia species. Moreover, five peaks were also demonstrated to be characteristic of two clusters of L. monocytogenes based on the two serotype groups (serotype 1/2a in cluster A and serotypes 1/2b and 4b in cluster B).
The analysis of the 468-bp region of the secA1 gene has been demonstrated to be sufficient for the identification of all pathogenic species of Nocardia and may provide a more discriminative and precise method than 16S rRNA gene sequencing for identifying Nocardia isolates (28). In the present study, all but three of the Nocardia species identified by 16S rRNA gene sequencing were compatible with those identified by secA1 gene sequencing. Further molecular methods are needed to solve the discrepant results by 16S rRNA and secA1 gene sequencing for these three Nocardia species (28).
The Bruker Biotyper database used in this study (DS 5627) contains 32 different Nocardia species, 10 different Kocuria species, 27 different Rhodococcus species, seven different Gordonia species, two different Tsukamurella species, and six different Listeria species. Several species evaluated in the present study, including three Nocardia species (N. beijingensis, N. puris, and N. rhamnosiphila), T. tyrosinosolvens, and G. amicalis species were not listed in the database. Although the analyses of all isolates with discrepant identification results between the molecular and Bruker Biotyper methods were repeated twice, repeated analysis did not lead to better identification scores than analysis of single deposits. Our findings are consistent with those reported by Verroken et al. (27).
Farfour et al. (1) compared the accuracy of the Andromas MALDI-TOF MS system with that of a direct colony method to identify a set of 659 GPR isolates representing 16 bacterial genera and 72 species. Their bacterial collection included 32 isolates of L. monocytogenes (n = 32), 24 isolates of Listeria species (n = 24), and 46 isolates of Nocardia and other GPR bacterial species. In total, 98.5% of the non-Listeria GPR isolates were identified to the species level, and 1.2% were identified to the genus level. However, no isolates of Kocuria, Gordonia, or Tsukamurella species were evaluated in their study (1).
Of the 46 isolates of 12 different Nocardia species evaluated by Farfour et al. (1), 42 (91.3%) were identified to the species level by the Bruker Biotyper system, including six of seven isolates of N. brasiliensis, 10 of 11 isolates of N. farcinica, and all isolates of N. nova (n = 5), N. abscessus (n = 4), N. beijingensis (n = 3), N. asteroides (n = 2), and N. otitidiscaviarum (n = 2). In addition, Verroken et al. (27) reported that of 43 blind-coded Nocardia spectra after pretreatment by 30 min of boiling and ethanol-formic acid extraction of the organisms and alignment with the Bruker Biotyper database, 19 isolates (44%) were correctly identified, of which 10 (23%) were identified to the species level (log scores, ≥2) and 9 (21%) were identified to the genus level (log scores, ≥1.7 to 2) (27). The remaining 24 isolates (56%) yielded log scores of <1.7 and were thereby considered not identifiable (27). Of the 10 Nocardia species that were identified to the species level, nine were N. nova (n = 15), and one N. brasiliensis isolate was identified as Nocardia species, with a log score of 2.084 (27). The Bruker Biotyper failed to accurately identify all N. brasiliensis, N. cyriacigeorgica, and N. farcinica isolates. Of the remaining six N. nova isolates, one was identified as Nocardia aobensis (log score, 1.720) and five were identified as N. nova, with score values ranging from 1.87 to 1.988 (27). Their findings were partly in accordance with our results.
Correct identification to the genus level for L. monocytogenes isolates was reported by Farfour et al. (1) to be 100% (32/32) using the Bruker Daltonics system and by Rychert et al. (2) to be 76% (34/55) using the Vitek MS version 2.0 system (27). Farfour et al. (1) further demonstrated that with the exception of Listeria grayi isolates that were identified to the species level, all other Listeria isolates were identified to the genus level because of highly similar spectra. Barbuddhe et al. (33) demonstrated that the Bruker Daltonics MALDI-TOF system was an accurate system for the rapid identification of six Listeria species isolated from humans and various environmental sources. Of 86 L. monocytogenes isolates belonging to 15 different serotypes in their study, three lineages (I, II, and III) were identified and separated by two different peak pairs (5,590/11,179 Da and 5,597/11,193 Da) generated by MALDI-TOF. Furthermore, the MALDI-TOF lineages were in complete agreement with the pulsed-field gel electrophoresis lineage (33). They also found that the lineage of serotype 1/2a L. monocytogenes isolates (lineage II) differed from the lineage of serotypes 1/2b and 4b (lineage I). Their findings partially support our results, mainly because only isolates from human infection (bacteremia and/or meningitis) were evaluated in the present study.
Few studies have evaluated the accuracy of MALDI-TOF MS for the identification of Kocuria, Gordonia, Rhodococcus, and Tsukamurella species (1, 2). The consistent result of one bacteremic isolate of K. kristinae identified by MALDI-TOF MS and 16S rRNA gene sequencing analysis has been reported (10).
There are several limitations in this study. First, the number of isolates of several rare species tested in this study was small. For example, there are currently 86 recognized Nocardia species, >50% of which have been described during the last 10 years, and most have been implicated in various clinical infections in humans (19). Second, although several peaks were found to be characteristic of the six species of Nocardia and two clusters of serotypes of L. monocytogenes, further study is needed to establish in-house databases and to validate the accuracy of the performance of the Bruker Biotyper MALDI-TOF system.
In conclusion, our data suggest that the Bruker Biotyper MALDI-TOF system is ineffective for identifying Nocardia and other unusual GPRs (Gordonia and Tsukamurella species) because of the current database limitations. Taxonomic changes and characterization of novel species of GPRs are common. Therefore, it is necessary to continuously update the MALDI-TOF MS databases. Further expansion of the database with a larger number of recently described isolates of Nocardia species and other unusual GPRs is warranted.
Footnotes
Published ahead of print 23 April 2014
REFERENCES
- 1. Farfour E, Leto J, Barritault M, Barberis C, Meyer J, Dauphin B, Le Guern AS, Leflèche A, Badell E, Guiso N, Leclercq A, Le Monnier A, Lecuit M, Rodriguez-Nava V, Bergeron E, Raymond J, Vimont S, Bille E, Carbonnelle E, Guet-Revillet H, Lécuyer H, Beretti JL, Vay C, Berche P, Ferroni A, Nassif X, Join-Lambert O. 2012. Evaluation of the Andromas matrix-assisted laser desorption ionization–time of flight mass spectrometry system for identification of aerobically growing Gram-positive bacilli. J. Clin. Microbiol. 50:2702–2707. 10.1128/JCM.00368-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rychert J, Burnham CA, Bythrow M, Garner OB, Ginocchio CC, Jennemann R, Lewinski MA, Manji R, Mochon AB, Procop GW, Richter SS, Sercia L, Westblade LF, Ferraro MJ, Branda JA. 2013. Multicenter evaluation of the Vitek MS matrix-assisted laser desorption ionization-time of flight mass spectrometry system for identification of Gram-positive aerobic bacteria. J. Clin. Microbiol. 51:2225–2231. 10.1128/JCM.00682-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Minero MV, Marín M, Cercenado E, Rabadán PM, Bouza E, Muñoz P. 2009. Nocardiosis at the turn of the century. Medicine (Baltimore, MD) 88:250–261. 10.1097/MD.0b013e3181afa1c8 [DOI] [PubMed] [Google Scholar]
- 4. Wilson JW. 2012. Nocardiosis: updates and clinical overview. Mayo Clin. Proc. 87:403–407. 10.1016/j.mayocp.2011.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu WL, Lai CC, Ko WC, Chen YH, Tang HJ, Huang YL, Huang YT, Hsueh PR. 2011. 1998. Clinical and microbiological characteristics of infections caused by various Nocardia species in Taiwan: a multicenter study from 1998 to 2010. Eur. J. Clin. Microbiol. Infect. Dis. 30:1341–1347. 10.1007/s10096-011-1227-9 [DOI] [PubMed] [Google Scholar]
- 6. Tan CK, Lai CC, Lin SH, Liao CH, Chou CH, Hsu HL, Huang YT, Hsueh PR. 2010. Clinical and microbiological characteristics of nocardiosis including those caused by emerging Nocardia species in Taiwan, 1998–2008. Clin. Microbiol. Infect. 16:966–972. 10.1111/j.1469-0691.2009.02950.x [DOI] [PubMed] [Google Scholar]
- 7. Lai CC, Tsai HY, Ruan SY, Liao CH, Hsueh PR. 2013. Fatal pneumonia and empyema thoracis caused by imipenem-resistant Nocardia abscessus in a cancer patient. J. Microbiol. Immunol. Infect., in press. 10.1016/j.jmii.2013.01.005 [DOI] [PubMed] [Google Scholar]
- 8. Lai CC, Lee LN, Teng LJ, Wu MS, Tsai JC, Hsueh PR. 2005. Disseminated Nocardia farcinica infection in a uraemia patient with idiopathic thrombocytopenia purpura receiving steroid therapy. J. Med. Microbiol. 54:1107–1110. 10.1099/jmm.0.46084-0 [DOI] [PubMed] [Google Scholar]
- 9. Lai CC, Wang JY, Lin SH, Tan CK, Wang CY, Liao CH, Chou CH, Huang YT, Lin HI, Hsueh PR. 2011. Catheter-related bacteraemia and infective endocarditis caused by Kocuria species. Clin. Microbiol. Infect. 17:190–192. 10.1111/j.1469-0691.2010.03211.x [DOI] [PubMed] [Google Scholar]
- 10. Citro R, Prota C, Greco L, Mirra M, Masullo A, Silverio A, Bossone E, Piscione F. 2013. Kocuria kristinae endocarditis related to diabetic foot infection. J. Med. Microbiol. 62:932–934. 10.1099/jmm.0.054536-0 [DOI] [PubMed] [Google Scholar]
- 11. Hsueh PR, Hung CC, Teng LJ, Yu MC, Chen YC, Wang HK, Luh KT. 1998. Report of invasive Rhodococcus equi infections in Taiwan, with an emphasis on the emergence of multidrug-resistant strains. Clin. Infect. Dis. 27:370–375. 10.1086/514667 [DOI] [PubMed] [Google Scholar]
- 12. Lai CC, Wang CY, Liu CY, Tan CK, Lin SH, Liao CH, Chou CH, Huang YT, Lin HI, Hsueh PR. 2010. Infections caused by Gordonia species at a medical centre in Taiwan, 1997 to 2008. Clin. Microbiol. Infect. 16:1448–1453. 10.1111/j.1469-0691.2010.03085.x [DOI] [PubMed] [Google Scholar]
- 13. Liu CY, Lai CC, Lee MR, Lee YC, Huang YT, Liao CH, Hsueh PR. 2011. Clinical characteristics of infections caused by Tsukamurella spp. and antimicrobial susceptibilities of the isolates. Int. J. Antimicrob. Agents. 38:534–537. 10.1016/j.ijantimicag.2011.07.018 [DOI] [PubMed] [Google Scholar]
- 14. Sheng WH, Huang YT, Chang SC, Hsueh PR. 2009. Brain abscess caused by Tsukamurella tyrosinosolvens in an immunocompetent patient. J. Clin. Microbiol. 47:1602–1604. 10.1128/JCM.01932-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang YT, Liao CH, Yang CJ, Teng LJ, Wang JT, Hsueh PR. 2011. Listeriosis, Taiwan, 1996–2008. Emerg. Infect. Dis. 17:1731–1733. 10.3201/eid1709.110093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hsieh WS, Tsai LY, Jeng SF, Hsu CH, Lin HC, Hsueh PR, Chen CY, Chou HC, Tsao PN, Yang PH. 2009. Neonatal listeriosis in Taiwan, 1990–2007. Int. J. Infect. Dis. 13:193–195. 10.1016/j.ijid.2008.06.006 [DOI] [PubMed] [Google Scholar]
- 17. Gaulin C, Gravel G, Bekal S, Currie A, Ramsay D, Roy S. 2014. Challenges in listeriosis cluster and outbreak investigations, province of Quebec, 1997–2011. Foodborne Pathog. Dis. 11:1–7. 10.1089/fpd.2013.1574 [DOI] [PubMed] [Google Scholar]
- 18. McCollum JT, Cronquist AB, Silk BJ, Jackson KA, O'Connor KA, Cosgrove S, Gossack JP, Parachini SS, Jain NS, Ettestad P, Ibraheem M, Cantu V, Joshi M, DuVernoy T, Fogg NW, Jr, Gorny JR, Mogen KM, Spires C, Teitell P, Joseph LA, Tarr CL, Imanishi M, Neil KP, Tauxe RV, Mahon BE. 2013. Multistate outbreaks of listeriosis associated with cantaloupe. N. Engl. J. Med. 369:944–953. 10.1056/NEJMoa1215837 [DOI] [PubMed] [Google Scholar]
- 19. Schlaberg R, Fisher MA, Hanson KE. 2014. Susceptibility profiles of Nocardia isolates based on current taxonomy. Antimicrob. Agents Chemother. 58:795–800. 10.1128/AAC.01531-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lai CC, Liu WL, Ko WC, Chen YH, Tang HJ, Huang YT, Hsueh PR. 2011. Antimicrobial-resistant Nocardia isolates, Taiwan, 1998–2009. Clin. Infect. Dis. 52:833–835. 10.1093/cid/ciq255 [DOI] [PubMed] [Google Scholar]
- 21. Lai CC, Liu WL, Ko WC, Chen YH, Tan HR, Huang YT, Hsueh PR. 2011. Multicenter study in Taiwan of the in vitro activities of nemonoxacin, tigecycline, doripenem, and other antimicrobial agents against clinical isolates of various Nocardia species. Antimicrob. Agents Chemother. 55:2084–2091. 10.1128/AAC.01808-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lai CC, Tan CK, Lin SH, Liao CH, Chou CH, Hsu HL, Huang YT, Hsueh PR. 2009. Comparative in vitro activities of nemonoxacin, doripenem, tigecycline and 16 other antimicrobials against Nocardia brasiliensis, Nocardia asteroides and unusual Nocardia species. J. Antimicrob. Chemother. 64:73–78. 10.1093/jac/dkp144 [DOI] [PubMed] [Google Scholar]
- 23. Welsh O, Vera-Cabrera L, Salinas-Carmona MC. 2013. Current treatment for Nocardia infections. Expert Opin. Pharmacother. 14:2387–2398. 10.1517/14656566.2013.842553 [DOI] [PubMed] [Google Scholar]
- 24. Dubois D, Grare M, Prere MF, Segonds C, Marty N, Oswald E. 2012. Performances of the Vitek MS matrix-assisted laser desorption ionization–time of flight mass spectrometry system for rapid identification of bacteria in routine clinical microbiology. J. Clin. Microbiol. 50:2568–2576. 10.1128/JCM.00343-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Seng P, Abat C, Rolain JM, Colson P, Lagier JC, Gouriet F, Fournier PE, Drancourt M, La Scola B, Raoult D. 2013. Identification of rare pathogenic bacteria in a clinical microbiology laboratory: impact of matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 51:2182–2194. 10.1128/JCM.00492-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Murray PR. 2010. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry: usefulness for taxonomy and epidemiology. Clin. Microbiol. Infect. 16:1626–1630. 10.1111/j.1469-0691.2010.03364.x [DOI] [PubMed] [Google Scholar]
- 27. Verroken A, Janssens M, Berhin C, Bogaerts P, Huang TD, Wauters G, Glupczynski Y. 2010. Evaluation of matrix-assisted laser desorption ionization–time of flight mass spectrometry for identification of Nocardia species. J. Clin. Microbiol. 48:4015–4021. 10.1128/JCM.01234-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Conville PS, Zelazny AM, Witebsky FG. 2006. Analysis of secA1 gene sequences for identification of Nocardia species. J. Clin. Microbiol. 44:2760–2766. 10.1128/JCM.00155-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Janda JM, Abbott SL. 2007. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J. Clin. Microbiol. 45:2761–2764. 10.1128/JCM.01228-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jadhav S, Bhave M, Palombo EA. 2012. Methods used for the detection and subtyping of Listeria monocytogenes. J. Microbiol. Methods 88:327–341. 10.1016/j.mimet.2012.01.002 [DOI] [PubMed] [Google Scholar]
- 31. Ketterlinus R, Hsieh SY, Teng SH, Lee H, Pusch W. 2005. Fishing for biomarkers: analyzing mass spectrometry data with the new ClinProTools software. Biotechniques 38:S37–S40 [DOI] [PubMed] [Google Scholar]
- 32. Teng SH, Chen CM, Lee MR, Lee TF, Chien KY, Teng LJ, Hsueh PR. 2013. Matrix-assisted laser desorption ionization–time of flight mass spectrometry can accurately differentiate between Mycobacterium masilliense (M. abscessus subspecies bolletti) and M. abscessus (sensu stricto). J. Clin. Microbiol. 51:3113–3116. 10.1128/JCM.01239-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Barbuddhe SB, Maier T, Schwarz G, Kostrzewa M, Hof H, Domann E, Chakraborty T, Hain T. 2008. Rapid identification and typing of Listeria species by matrix-assisted laser desorption ionization–time of flight mass spectrometry. Appl. Environ. Microbiol. 74:5402–5407. 10.1128/AEM.02689-07 [DOI] [PMC free article] [PubMed] [Google Scholar]