Abstract
Clinical echinocandin resistance among Candida glabrata strains is increasing, especially in the United States. Antifungal susceptibility testing is considered mandatory to guide therapeutic decisions. However, these methodologies are not routinely performed in the hospital setting due to their complexity and the time needed to obtain reliable results. Echinocandin failure in C. glabrata is linked exclusively to Fks1p and Fks2p amino acid substitutions, and detection of such substitutions would serve as a surrogate marker to identify resistant isolates. In this work, we report an inexpensive, simple, and quick classical PCR set able to objectively detect the most common mechanisms of echinocandin resistance in C. glabrata within 4 h. The usefulness of this assay was assessed using a blind collection of 50 C. glabrata strains, including 16 FKS1 and/or FKS2 mutants.
INTRODUCTION
Candida glabrata is a major agent of invasive candidiasis. It is considered the second-most-common Candida sp. isolated from blood samples in the United States and northern and eastern Europe and the third most common in the rest of the world (1–5). Its high frequency is, at least in part, associated with antifungal preexposure (6). Fluconazole resistance is common in C. glabrata, and echinocandins are recommended as first-line therapy. However, echinocandin resistance in C. glabrata is increasing (with rates ranging from 1% to 3% worldwide), making susceptibility testing mandatory to guide therapeutic decisions (1, 7–10). The Clinical and Laboratory Standards Institute (CLSI) and the European Committee on Antimicrobial Susceptibility Testing (EUCAST) established reference broth microdilution methods for Candida echinocandin susceptibility testing (11, 12). Moreover, the CLSI published revised interpretative guidelines in December 2012 that showed good performance in identifying echinocandin-resistant C. glabrata strains (7, 13). However, these methods have several limitations, including (i) a time-consuming and expensive methodology; (ii) the fact that standard echinocandin powders (indispensable for CLSI or EUCAST methods) are not commercially available; (iii) caspofungin MIC interlaboratory variability; (iv) overlapping susceptible and resistant populations; and (v) the need for 24 h of processing to obtain results (5, 11, 12, 14).
Clinical echinocandin resistance in C. glabrata is linked with substitutions in the hot spot regions of the Fks1p and Fks2p subunits of the β-d-1,3-glucan synthase complex (the target of echinocandins) (15–18). The detection of these FKS mutations has been considered the most accurate way to predict an echinocandin treatment failure (14, 18, 19). In an effort to improve the detection of echinocandin-resistant C. glabrata isolates, we developed a set of classical PCRs able to detect 10 of the most frequent mutations associated with clinical echinocandin resistance in less than 4 h. The sensitivity and specificity of the method were assessed using a blind collection of C. glabrata clinical isolates comprising echinocandin-resistant and -susceptible strains.
MATERIALS AND METHODS
Strains and blind study design.
Fifty C. glabrata strains were used throughout this work. All strains were isolated from patients with proven invasive fungal disease (20). Nineteen strains were obtained from the Public Health Research Institute (PHRI; Rutgers University, NJ), 20 from the Mycology laboratory of the Ramos Mejia Hospital (Buenos Aires, Argentina), and 11 from the Mycology and Molecular Diagnostics Laboratory (LMDM) (Santa Fe, Argentina). Sixteen strains showed FKS1 and/or FKS2 hot spot region mutations (Table 1). C. glabrata ATCC 90030 was used as the wild-type control strain to validate the PCRs. C. krusei ATCC 6258 and C. parapsilosis sensu stricto ATCC 22019 were used as susceptibility testing control strains (11, 13). The isolates were identified as C. glabrata by conventional phenotypic methods and by sequencing of the 5.8S RNA gene and adjacent internal transcribed spacer 1 (ITS1) and ITS2 regions (21, 22). The collection of strains was assembled at the PHRI center, and blind code numbers were assigned. Also, a set of C. glabrata strains with known FKS1 and/or FKS2 mutations were used to develop and test the proposed methodology before confirming its utility with the blind study.
TABLE 1.
Comparison of results from classical PCR set, DNA sequencing, and in vitro susceptibility determinations of the C. glabrata strains included in this study
| Straina | Classical PCR set resultb |
DNA sequencing result |
MIC (μg/ml)e |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1-F625 | 1-S629 | 1-D632 | 2-F659 | 2-S663 | Fks1pc | Fks2pd | ANF | CSF | MCF | |
| WT (n = 34) | + | + | + | + | + | WT | WT | 0.08 (S) | 0.09 (S) | 0.02 (S) |
| 42997 | − | + | + | + | + | F625S | WT | 2.00 | 2.00 | 0.50 |
| 5847 | + | − | + | + | + | S629P | WT | 4.00 | >8.00 | 2.00 |
| 3169 | + | + | − | + | + | D632E | WT | 2.00 | 2.00 | 2.00 |
| LMDM 37 | + | + | − | + | + | D632E | WT | 2.00 | 4.00 | 4.00 |
| 21900 | + | + | − | + | + | D632G | WT | 1.00 | 4.00 | 0.06 (S) |
| 42971 | + | + | − | + | + | D632Y | WT | 4.00 | 4.00 | 1.00 |
| 31498 | + | + | + | + | + | WT | F659del | 2.00 | 8.00 | 4.00 |
| 6183 | + | + | + | − | + | WT | F659S | 4.00 | >8.00 | 4.00 |
| M234 | + | + | + | − | + | WT | F659V | 1.00 | 4.00 | 1.00 |
| 20.551.099 | + | + | + | − | + | WT | F659Y | 1.00 | 2.00 | 0.12 (I) |
| 3.830 | + | + | + | + | − | WT | S663P | 2.00 | >8.00 | 1.00 |
| 37178 | + | + | + | + | − | W645STOP | S663P | 4.00 | >8.00 | 8.00 |
| M2798 | + | + | + | + | − | WT | S663P | 8.00 | >8.00 | 8.00 |
| 20.593.033 | + | + | + | + | − | W649STOP | S663P | 4.00 | >8.00 | 4.00 |
| LMDM 34 | + | + | + | + | − | WT | S663P | 2.00 | >8.00 | 2.00 |
| M2791 | + | + | + | + | − | WT | S663F | 4.00 | 4.00 | 4.00 |
Includes 34 wild-type C. glabrata strains and 16 FKS1 and/or FKS2 mutants.
Positive or negative signs indicate the presence or the absence of the corresponding PCR band in a electrophoresis gel.
WT, wild type at hot spots. Fks1p hot spot 1 includes amino acids between 625 and 633 (625-FLILSLRDP-633).
WT, wild type at hot spots. Fks2p hot spot 1 includes amino acids between 659 and 667 (659-FLILSLRDP-667).
Data represent geometric mean values. MICs were obtained on three separate days. ANF, anidulafungin. CSF, caspofungin. MCF, micafungin. (S) or (I) indicates that the strain is considered echinocandin susceptible or echinocandin intermediate, respectively (or is otherwise considered resistant), following the interpretative guidelines published in CLSI document M27-S4 (13).
Antifungals and susceptibility testing.
Caspofungin (CSF; Merck & Co. Inc., Rahway, NJ), anidulafungin (ANF; Pfizer, New York, NY), and micafungin (MCF; Astellas Pharma USA Inc., Deerfield, IL) were obtained as standard powder from their respective manufacturers. Echinocandin susceptibility testing was performed in triplicate in accordance with CLSI document M27-A3 and following the interpretive guidelines published in the M27-S4 document (11, 13).
DNA isolation, PCR conditions, and primer and PCR set design.
C. glabrata genomic DNAs were extracted with phenol-chloroform method (23) or with a Q-Biogene FastDNA kit (Q-Biogene). C. glabrata FKS1 and FKS2 genes with GenBank accession numbers XM_446406 and XM_448401, respectively, were used for primer design. Two groups of primers were used throughout this work. The primers in the first group (PCR control primers), consisting of primer pair 1-1670F and 1-2225R and primer pair 2-1619F and 2-2177R designed to specifically hybridize FKS1 and FKS2, respectively, were used as an amplification control for each of the five multiplex PCRs (Table 2). The second group of primers, named the mutation detection primers, included five oligonucleotides that were designed to detect the 10 most common mutations related with echinocandin resistance. These primers align the FKS1 and FKS2 hot spot 1 regions and were named 1-F625, 1-S629, 1-D632, 2-F659, and 2-S663. These primers were used in pairs with primers 1-1670F (1-S629 and 1-D632), 1-2225R (1-F625), 2-1619F (2-S663), and 2-2177R (2-F659). PCR primers were designed by using the oligonucleotide design tool of the IDT SciTools (Integrated DNA Technologies, Coralville, IA) and were purchased from Integrated DNA Technologies (IDT-Biodynamics, Buenos Aires, Argentina).
TABLE 2.
Oligonucleotides primers used in this study
| Oligonucleotidea | Target gene | Purpose(s)b | 5′→3′ sequencec |
|---|---|---|---|
| 1-1670F | FKS1 | FKS1 HS1 AfS and AC | GTTGCTGCGGTCATGTTCTT |
| 1-2225R | FKS1 | FKS1 HS1 AfS and AC | GCGTTCCAGACTTGGGAAAT |
| 2-1619F | FKS2 | FKS2 HS1 AfS and AC | GAATGGTGGTTCGTTCCAAG |
| 2-2177R | FKS2 | FKS2 HS1 sequencing and AC | TGTTGCTTCTCAGACTTTCACC |
| 1-F625F | FKS1 | Mutation detection | CGCTGAATCATACTACTT |
| 1-S629R | FKS1 | Mutation detection | GATTGGATCTCTTGAGA |
| 1-D632R | FKS1 | Mutation detection | GACAAAATTCTGATTGGA |
| 2-F659F | FKS2 | Mutation detection | CTCTGAATCGTACTTCTT |
| 2-S663R | FKS2 | Mutation detection | GATAGGGTCTCTTAGAGA |
| 1-1776F | FKS1 | FKS1 HS1 sequencing | ACGTCGCTTCTCAAACCTTC |
| 1-2008R | FKS1 | FKS1 HS1 sequencing | CGGTAGCAATCATCAAACCC |
| 2-1881F | FKS2 | FKS1 HS1 sequencing | CGACGTTCAGCTTCAGAGTTT |
| 2-2513R | FKS2 | FKS2 HS1 AfS | CCAACAGAGAAGACAGTGTTGA |
| ITS1d | rDNAe | Molecular identification | TCCGTAGGTGAACCTGCGG |
| ITS4d | rDNA | Molecular identification | TCCTCCGCTTATTGATATGC |
F, sense; R, antisense.
AfS, amplification for subsequent sequencing; AC, amplification control; HS1, hot spot 1.
Nucleotides in bold show where a mutation could be present.
From reference 24.
rDNA, ribosomal DNA.
Amplifications were carried out in a 25-μl volume of a mixture containing 5 mM (NH4)2SO4, 5 mM KCl, 10 mM Tris-Cl (pH 8.8), 1 mM MgSO4, 5 ng of bovine serum albumin, 0.1% Triton X-100, 125 μM each dATP, dGTP, dCTP, and dTTP (Genbiotech, Buenos Aires, Argentina), a 0.5 μM concentration of each of the three primers, 1.25 U of Pegasus DNA polymerase (PBL, Buenos Aires, Argentina), and 10 to 25 ng of C. glabrata genomic DNA. Amplification was performed for one initial step of 2 min at 94°C followed by 25 cycles of 30 s at 94°C, 30 s at 56°C, and 1 min at 72°C and then a final cycle of 10 min at 72°C in an Applied Biosystems thermocycler (Tecnolab-AB, Buenos Aires, Argentina). The PCR products were analyzed by electrophoresis.
DNA sequencing.
The C. glabrata FKS1 hot spot 1 region (nucleotide [nt] 1776 to nt 2008), FKS2 hot spot 1 region (nt 1881 to nt 2177), and 5.8S RNA gene and adjacent internal transcribed spacer 1 (ITS1) and ITS2 regions were amplified and sequenced in both directions using the primers described in Table 2. For sequencing of the FKS1 and FKS2 hot spot 1 regions, primer pair 1-1670F and 1-2225R and primer pair 1-1619F and 1-2513R were used for PCR amplification, respectively. The purified fragments were then subjected to sequencing using primers 1-1776F and 1-2008R for FKS1 and 2-1881F and 2-2177R for FKS2 (Table 2). In Argentina, DNA sequencing was performed using a BigDye Terminator cycle sequencing ready-reaction system (Applied Biosystems, Buenos Aires, Argentina) according to the manufacturer's instructions. Sequence analysis was performed on an ABI Prism 310 DNA sequencer (Applied Biosystems) using the facilities available at Cromatida S.A. (Buenos Aires, Argentina). In the PHRI Center, DNA sequencing was performed with a CEQ dye terminator cycle sequencing QuickStart kit (Beckman Coulter, Fullerton, CA) according to the manufacturer's recommendations. Sequencing analyses were done with CEQ 8000 genetic analysis system software (Beckman Coulter) and with the BioEdit sequence alignment editor (Ibis Therapeutics, Carlsbad, CA).
RESULTS
Primer and PCR design for the detection of the molecular echinocandin resistance mechanism in C. glabrata.
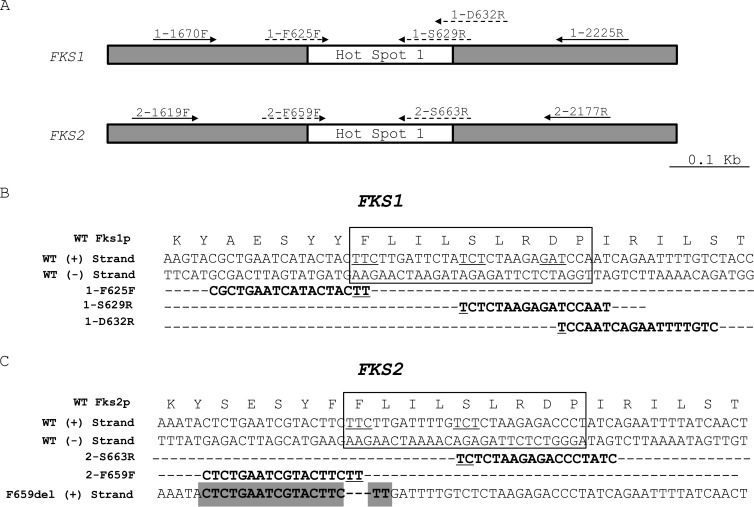
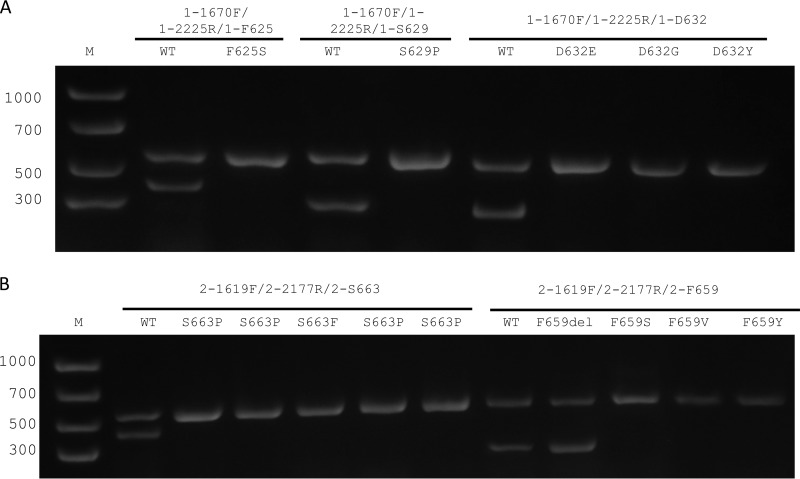
The C. glabrata FKS1 and FKS2 genes have high (>73%) homology, with portions with very low homology (lower than 50%) and others with the highest homology (>85% for the hot spot 1 regions of both genes) (Fig. 1). For this reason, we designed two groups of primers named PCR control primers and mutation detection primers (both groups are described above). The primers of the first group were designed to align the regions of lowest homology between the genes (hot spot 1 external region) with dual objectives: (i) to give the FKS1 or FKS2 gene specificity when used in combination with the mutation detection primers and (ii) to use them as internal controls for validation of the quality of DNA samples and the absence of PCR inhibitors, since the presence of a mutation is represented by a negative result in a PCR. On the other hand, primers 1-F625, 1-S629, 1-D632, 2-F659, and 2-S663 were designed specifically for priming FKS1 or FKS2 wild-type sequences, considering that a 3′ mismatch does not prime in a PCR under the appropriate conditions of stringent annealing temperatures (Fig. 1). Furthermore, other reaction variables such as annealing temperatures and MgSO4 and primer concentrations were taken into consideration for PCR design to allow the use of one PCR program irrespective of the primer set used. Under the PCR and reagent concentration conditions described above, all five PCRs could be run at the same time and with the same program in the thermocycler, showing excellent discrimination for both wild-type and mutant alleles. Therefore, for detection of the substitution at Fks1p residues F625, S629, and D632, the multiplex PCRs were performed using three primers per tube, including primers 1-1670F, 1-2225R, and 1-F625, primers 1-1670F, 1-2225R, and 1-S629, and primers 1-1670F, 1-2225R, and 1-D632, respectively. These PCRs gave one 555-bp band in all the tubes and 369-bp, 263-bp, and 252-bp bands when the isolate was wild type at residues F625, S629, and D632 at Fks1p, respectively. On the other hand, when a mutation is present in the codon that encodes any of the three amino acid residues listed above, a unique 555-bp band was observed after the electrophoresis (control PCR) (Fig. 2). The detection of amino acid substitutions at residues F659 and S663 of Fks2p was performed using a similar approach but with primers 2-1619F, 2-2177R, and 2-F659 and primers 2-1619F, 2-2177R, and 2-S663, respectively. In these cases, for a wild-type isolate, two bands (558 bp and 219 or 400 bp, respectively) were expected. For an echinocandin mutant with a substitution at F659 or S663 residues, a single 558-bp band was obtained (Fig. 2).
FIG 1.
(A) Representation of 1,000-nucleotide (nt) fragments of C. glabrata FKS genes, which include the hot spot 1 regions (white boxes). Filled arrows: oligonucleotide primers included in the PCR control group used as the reaction control. Dashed arrows: primers designed to detect C. glabrata FKS1 and FKS2 mutations (mutation detection group). (B) Alignment of primers 1-F625F, 1-S629R, and 1-D632R with the wild-type (WT) FKS1 gene. (C) Alignment of primers 2-S663R and 2-F659F with the wild-type FKS2 gene and primer 2-F659F with the FKS2 gene with the deletion of three nucleotides (from T1995 to C1997) (gray shading). Underlined nucleotides show the codons where the mutations are under cover by mutation detection primers. Boxes include the Fks1p and Fks2p hot spot 1 regions.
FIG 2.
Electrophoresis of the PCR set resolved in a 1.5% agarose gel. The three primers used in each of the tubes are named above the images. Lane M, molecular size marker. (A) PCRs designed to detect FKS1 mutant. Lanes 2, 4, and 6, C. glabrata ATCC 90030 (wild-type strain, echinocandin susceptible). Lane 3, C. glabrata strain 42997 (Fks1p-F625S). Lane 5, strain 5847 (Fks1p-S629P). Lane 6, strain LMDM37 (Fks1p-D632E). Lane 7, C. glabrata 21900 (Fks1p-D632G). Lane 8, isolate 42971 (Fks1p-D632Y). (B) Lanes 2 and 8, C. glabrata ATCC 90030 (echinocandin-susceptible wild-type strain). Lane 3, C. glabrata 3.830 (Fks2p-S663P). Lane 4, strain 37178 (Fks2p-S663P). Lane 5, strain M2791 (Fks2p-S663F). Lane 6, isolate 20.593.033 (Fks2p-S663P). Lane 7, strain LMDM 34 (Fks2p-S663P). Lane 9, strain 31498 (Fks2p-F659del). Lane 10, strain 6183 (Fks2p-F659S). Lane 11, strain M234 (Fks2p-F659V). Lane 12, isolate 20.551.099 (Fks2p-F659Y).
Validation of the multiplex PCR sets.
The utility of the PCR sets was evaluated by using a blind collection of 50 C. glabrata strains, including 16 echinocandin-resistant clinical isolates with different amino acid substitutions in both Fksp proteins (Table 1). Of the 50 isolates tested, 35 were considered wild-type strains by the proposed methodology since the 5 PCR tubes presented two bands in the electrophoresis. The rest were identified as FKS1 or FKS2 mutants with an amino acid substitution at residues Fks1p-F625 (n = 1), Fks1p-S629 (n = 1), Fks1p-D632 (n = 4), Fks2p-F659 (n = 4), and Fks2p-S663 (n = 5). A total of 49 of the 50 strains (98%) were correctly identified as echinocandin susceptible or resistant compared with the echinocandin susceptibility testing results. Also, we found 98% concordance between our proposed methodology and sequencing (Table 1). There was one false result, comprising a FKS2 mutant, in which Fks2p showed a deletion at the 659 residue (F659del). This deletion was not uncovered by the 2-F659 primer because three nucleotides were deleted and the nucleotide sequence where the primer was aligned was maintained (Fig. 2).
DISCUSSION
Prompt diagnosis and the correct treatment selection for invasive Candida infections significantly reduce mortality (25). Echinocandin drugs are now considered the best therapeutic option for C. glabrata infections since these yeasts are less susceptible to fluconazole and amphotericin B than other Candida spp. (10). Recent reports showed that the number of echinocandin-resistant isolates is increasing, making essential an accurate assessment of echinocandin susceptibility (7, 9). Whole-cell susceptibility testing using a reference protocol takes at least 48 h (11, 12). However, outside the United States, most of the susceptibility testing is being outsourced to reference laboratories due to the complexities of these methodologies, increasing the time needed to obtain reliable susceptibility data. To reduce this delay, we developed a simple set of multiplex PCRs able to objectively classify a C. glabrata strain as echinocandin susceptible or resistant in less than 4 h. The strict linkage between FKS1 and FKS2 hot spot region mutations and clinical echinocandin resistance provided the rationale for selecting the detection of these mutations as a surrogate marker for resistance. Twenty-three different amino acid substitutions in Fks1p and Fks2p hot spot regions were previously described (7, 8, 10, 15–17, 24, 26–31). However, 87.7% of the described clinically echinocandin-resistant strains showed substitutions at the Fks1p-F625 (2.46%), Fks1p-S629 (15.57%), Fks1p-D632 (5.74%), Fks2p-F659 (17.21%), and Fks2p-S663 (46.72%) residues (percentages were obtained over a total of 122 strains, 63 included in the cited reports plus 59 C. glabrata nonpublished echinocandin-resistant strains held in the Perlin's Echinocandin Resistance Reference Laboratory collection) (7, 8, 10, 15–17, 24, 26–31). Moreover, the strains harboring the most prevalent substitutions showed the highest echinocandin MIC values (7, 8, 10, 15–17, 24, 26–31). These data led us to decide to include the described five PCR assays to be able to detect the most common hot spot amino acid substitutions linked with echinocandin resistance in C. glabrata. In the blind study, we demonstrated that our set of PCRs was able to uncover mutants harboring Fks1p-F625S, Fks1p-S629P, Fks1p-D632G, Fks1p-D632E, Fks1p-D632Y, Fks2p-F659S, Fks2p-F659V, Fks2p-F659L, Fks2p-S663P, and Fks2p-S663F amino acid substitutions. Moreover, the designed primers would also potentially uncover less-common mutations as Fks1p-F625I (8) and Fks2p-F659Y (10, 24), since the primer's 3′ ends would not hybridize these mutated sequences.
Recently, Pham et al. described a high-throughput microsphere-based assay using the Luminex MagPix technology suitable to identify C. glabrata FKS mutants (19). This method would be potentially used as a tool to evaluate a collection of strains in a reference laboratory. The advantage of the methodology that we are presenting is that it is based on the cheaper and commonly available classical PCR methodology, making it suitable to be used in a hospital setting for analyzing a few strains at a time. Moreover, this new method is able to uncover FKS mutations more quickly than other available molecular tools such as classical sequencing methods with no need for special equipment.
The main limitation of the proposed set of PCRs is its inability to detect the deletion of three nt at the codon which encoded the F659 at the Fks2p. This false result would be considered a very major error compared with whole-cell susceptibility testing since our proposed methodology would thus classify a resistant strain as susceptible. However, this molecular mechanism of echinocandin resistance has been described in only few strains worldwide and it is the least common substitution at this residue (8, 16, 19). Other limitations of this methodology are its inability to detect newly described mutations or other potential non-FKS-linked mechanisms associated with echinocandin resistance and the possibility of changes in epidemiology making the detection of the described mutations useless. However, these potential drawbacks are shared with any molecular method designed for the detection of mechanisms of resistance (32–34). Nevertheless, this methodology is suitable to be modified by adding or eliminating PCRs in order to adapt it to detect emerging mechanisms of resistance.
In conclusion, we present an inexpensive, simple, and quick molecular methodology able to objectively detect the most common mechanisms of echinocandin resistance in C. glabrata.
ACKNOWLEDGMENTS
This study was supported by Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) grant PIP2011/331 to G.G.-E. and R.G.V. and by a Universidad Nacional del Litoral (UNL) grant (CAI+D) to G.G.-E. and S.G. C.D. has a predoctoral fellowship from CONICET. F.L. had a fellowship from UNL. The Perlin laboratory was funded by grant AI069397 to D.S.P. and Pfizer.
Footnotes
Published ahead of print 14 May 2014
REFERENCES
- 1.Axner-Elings M, Botero-Kleiven S, Jensen RH, Arendrup MC. 2011. Echinocandin susceptibility testing of Candida isolates collected during a 1-year period in Sweden. J. Clin. Microbiol. 49:2516–2521. 10.1128/JCM.00201-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guinea J. 10 February 2014. Global trends in the distribution of Candida species causing candidemia. Clin. Microbiol. Infect. 10.1111/1469-0691.12539 [DOI] [PubMed] [Google Scholar]
- 3.Horn DL, Neofytos D, Anaissie EJ, Fishman JA, Steinbach WJ, Olyaei AJ, Marr KA, Pfaller MA, Chang CH, Webster KM. 2009. Epidemiology and outcomes of candidemia in 2019 patients: data from the prospective antifungal therapy alliance registry. Clin. Infect. Dis. 48:1695–1703. 10.1086/599039 [DOI] [PubMed] [Google Scholar]
- 4.Pfaller M, Neofytos D, Diekema D, Azie N, Meier-Kriesche HU, Quan SP, Horn D. 2012. Epidemiology and outcomes of candidemia in 3648 patients: data from the Prospective Antifungal Therapy (PATH Alliance(R)) registry, 2004–2008. Diagn. Microbiol. Infect. Dis. 74:323–331. 10.1016/j.diagmicrobio.2012.10.003 [DOI] [PubMed] [Google Scholar]
- 5.Rodriguez-Tudela JL, Gomez-Lopez A, Arendrup MC, Garcia-Effron G, Perlin DS, Lass-Florl C, Cuenca-Estrella M. 2010. Comparison of caspofungin MICs by means of EUCAST method EDef 7.1 using two different concentrations of glucose. Antimicrob. Agents Chemother. 54:3056–3057. 10.1128/AAC.00597-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lortholary O, Desnos-Ollivier M, Sitbon K, Fontanet A, Bretagne S, Dromer F. 2011. Recent exposure to caspofungin or fluconazole influences the epidemiology of candidemia: a prospective multicenter study involving 2,441 patients. Antimicrob. Agents Chemother. 55:532–538. 10.1128/AAC.01128-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alexander BD, Johnson MD, Pfeiffer CD, Jimenez-Ortigosa C, Catania J, Booker R, Castanheira M, Messer SA, Perlin DS, Pfaller MA. 2013. Increasing echinocandin resistance in Candida glabrata: clinical failure correlates with presence of FKS mutations and elevated minimum inhibitory concentrations. Clin. Infect. Dis. 56:1724–1732. 10.1093/cid/cit136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dannaoui E, Desnos-Ollivier M, Garcia-Hermoso D, Grenouillet F, Cassaing S, Baixench MT, Bretagne S, Dromer F, Lortholary O. 2012. Candida spp. with acquired echinocandin resistance, France, 2004–2010. Emerg. Infect. Dis. 18:86–90. 10.3201/eid1801.110556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu W, Tan J, Sun J, Xu Z, Li M, Yang Q, Shao H, Zhang L, Liu W, Wan Z, Cui W, Zang B, Jiang D, Fang Q, Qin B, Qin T, Li W, Guo F, Liu D, Guan X, Yu K, Qiu H, Li R. 2014. Invasive candidiasis in intensive care units in China: in vitro antifungal susceptibility in the China-SCAN study. J. Antimicrob. Chemother. 69:162–167. 10.1093/jac/dkt330 [DOI] [PubMed] [Google Scholar]
- 10.Pfaller MA, Castanheira M, Lockhart SR, Ahlquist AM, Messer SA, Jones RN. 2012. Frequency of decreased susceptibility and resistance to echinocandins among fluconazole-resistant bloodstream isolates of Candida glabrata. J. Clin. Microbiol. 50:1199–1203. 10.1128/JCM.06112-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clinical and Laboratory Standards Institute. 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts, 3rd ed. Document M27-A3 CLSI, Wayne, PA [Google Scholar]
- 12.European Committee on Antimicrobial Susceptibility Testing (EUCAST). 2012. Method for the determination of broth dilution minimum Inhibitory concentrations of antifungal agents for yeasts. EUCAST definitive document EDef 7.2 revision. EUCAST, Växjö, Sweden: [DOI] [PubMed] [Google Scholar]
- 13.Clinical and Laboratory Standards Institute. 2012. Reference method for broth dilution antifungal susceptibility testing of yeast—4th informational supplement—CLSI document M27-S4. CLSI, Wayne, PA [Google Scholar]
- 14.Arendrup MC, Garcia-Effron G, Lass-Florl C, Lopez AG, Rodriguez-Tudela JL, Cuenca-Estrella M, Perlin DS. 2010. Echinocandin susceptibility testing of Candida species: comparison of EUCAST EDef 7.1, CLSI M27-A3, Etest, disk diffusion, and agar dilution methods with RPMI and isosensitest media. Antimicrob. Agents Chemother. 54:426–439. 10.1128/AAC.01256-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cleary JD, Garcia-Effron G, Chapman SW, Perlin DS. 2008. Reduced Candida glabrata susceptibility secondary to an FKS1 mutation developed during candidemia treatment. Antimicrob. Agents Chemother. 52:2263–2265. 10.1128/AAC.01568-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Effron G, Lee S, Park S, Cleary JD, Perlin DS. 2009. Effect of Candida glabrata FKS1 and FKS2 mutations on echinocandin sensitivity and kinetics of 1,3-beta-D-glucan synthase: implication for the existing susceptibility breakpoint. Antimicrob. Agents Chemother. 53:3690–3699. 10.1128/AAC.00443-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katiyar S, Pfaller M, Edlind T. 2006. Candida albicans and Candida glabrata clinical isolates exhibiting reduced echinocandin susceptibility. Antimicrob. Agents Chemother. 50:2892–2894. 10.1128/AAC.00349-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perlin DS. 2007. Resistance to echinocandin-class antifungal drugs. Drug Resist. Updat. 10:121–130. 10.1016/j.drup.2007.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pham CD, Bolden CB, Kuykendall RJ, Lockhart SR. 2014. Development of a Luminex-based multiplex assay for detection of mutations conferring resistance to echinocandins in Candida glabrata. J. Clin. Microbiol. 52:790–795. 10.1128/JCM.03378-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ascioglu S, Rex JH, de Pauw B, Bennett JE, Bille J, Crokaert F, Denning DW, Donnelly JP, Edwards JE, Erjavec Z, Fiere D, Lortholary O, Maertens J, Meis JF, Patterson TF, Ritter J, Selleslag D, Shah PM, Stevens DA, Walsh TJ. 2002. Defining opportunistic invasive fungal infections in immunocompromised patients with cancer and hematopoietic stem cell transplants: an international consensus. Clin. Infect. Dis. 34:7–14. 10.1086/323335 [DOI] [PubMed] [Google Scholar]
- 21.Kurzman CP, Fell JW, Boekhout T. 2011. The yeast—a taxonomic study. Elsevier, London, United Kingdom [Google Scholar]
- 22.White TJ, Bruns TD, Lee SB, Taylor JW. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 315–322 Innis MA, Gelfand DH, Sninsky JJ, White TJ. (ed), PCR protocols: a guide to methods and applications. Academic Press, Inc., San Diego, CA [Google Scholar]
- 23.Sambrook J, Fritsch EF, Maniatis T. 1998. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 24.Zimbeck AJ, Iqbal N, Ahlquist AM, Farley MM, Harrison LH, Chiller T, Lockhart SR. 2010. FKS mutations and elevated echinocandin MIC values among Candida glabrata isolates from U.S. population-based surveillance. Antimicrob. Agents Chemother. 54:5042–5047. 10.1128/AAC.00836-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morrell M, Fraser VJ, Kollef MH. 2005. Delaying the empiric treatment of candida bloodstream infection until positive blood culture results are obtained: a potential risk factor for hospital mortality. Antimicrob. Agents Chemother. 49:3640–3645. 10.1128/AAC.49.9.3640-3645.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bizerra FC, Jimenez-Ortigosa C, Souza AC, Breda Queiroz-Telles F, Perlin DS, Colombo AL. 2014. Breakthrough candidemia due to multidrug-resistant Candida glabrata during prophylaxis with a low dose of micafungin. Antimicrob. Agents Chemother. 58:2438–2440. 10.1128/AAC.02189-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castanheira M, Woosley LN, Diekema DJ, Messer SA, Jones RN, Pfaller MA. 2010. Low prevalence of fks1 hot spot 1 mutations in a worldwide collection of Candida strains. Antimicrob. Agents Chemother. 54:2655–2659. 10.1128/AAC.01711-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia-Effron G, Chua DJ, Tomada JR, Dipersio J, Perlin DS, Ghannoum M, Bonilla H. 2010. Novel FKS mutations associated with echinocandin resistance in Candida species. Antimicrob. Agents Chemother. 54:2225–2227. 10.1128/AAC.00998-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katiyar SK, Alastruey-Izquierdo A, Healey KR, Johnson ME, Perlin DS, Edlind TD. 2012. Fks1 and Fks2 are functionally redundant but differentially regulated in Candida glabrata: implications for echinocandin resistance. Antimicrob. Agents Chemother. 56:6304–6309. 10.1128/AAC.00813-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Messer SA, Jones RN, Moet GJ, Kirby JT, Castanheira M. 2010. Potency of anidulafungin compared to nine other antifungal agents tested against Candida spp., Cryptococcus spp., and Aspergillus spp.: results from the global SENTRY Antimicrobial Surveillance Program (2008). J. Clin. Microbiol. 48:2984–2987. 10.1128/JCM.00328-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson GR, III, Wiederhold NP, Vallor AC, Villareal NC, Lewis JS, Patterson TF. 2008. Development of caspofungin resistance following prolonged therapy for invasive candidiasis secondary to Candida glabrata infection. Antimicrob. Agents Chemother. 52:3783–3785. 10.1128/AAC.00473-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balashov SV, Park S, Perlin DS. 2006. Assessing resistance to the echinocandin antifungal drug caspofungin in Candida albicans by profiling mutations in FKS1. Antimicrob. Agents Chemother. 50:2058–2063. 10.1128/AAC.01653-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia-Effron G, Dilger A, Alcazar-Fuoli L, Park A, Mellado E, Perlin DS. 2008. Rapid detection of triazole antifungal resistance in Aspergillus fumigatus. J. Clin. Microbiol. 46:1200–1206. 10.1128/JCM.02330-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perlin DS, Balashov S, Park S. 2008. Multiplex detection of mutations. Methods Mol. Biol. 429:23–31. 10.1007/978-1-60327-040-3_2 [DOI] [PubMed] [Google Scholar]
- 35.Quindos G. 2014. Epidemiology of candidaemia and invasive candidiasis. A changing face. Rev. Iberoam. Micol. 31:42–48. 10.1016/j.riam.2013.10.001 [DOI] [PubMed] [Google Scholar]