Abstract
The Xpert MTB/RIF (Xpert) assay is becoming a principal screening tool for diagnosing rifampin-resistant Mycobacterium tuberculosis complex (MTBC) infection. However, little is known about the performance of the Xpert assay in infections with both drug-sensitive and drug-resistant strains (mixed MTBC infections). We assessed the performance of the Xpert assay for detecting rifampin resistance using phenotypic drug sensitivity testing (DST) as the reference standard in 370 patients with microbiologically proven pulmonary tuberculosis. Mixed MTBC infections were identified genetically through 24-locus mycobacterial interspersed repetitive-unit–variable-number tandem-repeat (MIRU-VNTR) analysis. Logistic regression was used to identify the factors associated with poor (defined as treatment failure, default, and death from any cause) or good (defined as cure or successful treatment completion) clinical outcomes. The analytic sensitivity of the Xpert assay for detecting rifampin resistance was assessed in vitro by testing cultures containing different ratios of drug-sensitive and drug-resistant organisms. Rifampin resistance was detected by the Xpert assay in 52 (14.1%) and by phenotypic DST in 55 (14.9%) patients. Mixed MTBC infections were identified in 37 (10.0%) patients. The Xpert assay was 92.7% (95% confidence interval [CI], 82.4% to 97.9%) sensitive for detecting rifampin resistance and 99.7% (95% CI, 98.3% to 99.9%) specific. When restricted to patients with mixed MTBC infections, Xpert sensitivity was 80.0% (95% CI, 56.3 to 94.3%). False-negative Xpert results (adjusted odds ratio [aOR], 6.6; 95% CI,1.2 to 48.2) and mixed MTBC infections (aOR, 6.5; 95% CI, 2.1 to 20.5) were strongly associated with poor clinical outcome. The Xpert assay failed to detect rifampin resistance in vitro when <90% of the organisms in the sample were rifampin resistant. Our study indicates that the Xpert assay has an increased false-negative rate for detecting rifampin resistance with mixed MTBC infections. In hyperendemic settings where mixed infections are common, the Xpert results might need further confirmation.
INTRODUCTION
Tuberculosis (TB) is one of the leading causes of morbidity and mortality worldwide. Global efforts to control TB have been seriously challenged by the emergence of drug-resistant TB, including multidrug-resistant TB (MDR-TB) (1). MDR-TB, defined as TB caused by mycobacteria that are resistant to at least isoniazid and rifampin, is associated with worse clinical outcomes, and its treatment is expensive, lengthy, and complex. However, several studies have shown that high cure rates are achievable if appropriate treatment is initiated early (2, 3). Diagnostic delays with MDR-TB are associated with worse clinical outcomes and increased transmission (4).
Traditionally, a diagnosis of MDR-TB infection requires mycobacterial culture and phenotypic drug susceptibility testing (DST) (1). This approach requires relatively advanced laboratory capacity, is labor-intensive, and takes 1 to 3 months before the results are available. In 2011, the World Health Organization recommended the use of rapid molecular genotyping methods over conventional phenotypic methods for DST at the initial diagnosis (4). Rapid genotypic tests, which can diagnose resistance to rifampin alone or to rifampin and isoniazid within 2 h of testing, have demonstrated good overall concordance with phenotypic DSTs for MDR-TB (5). The Xpert MTB/RIF assay (Xpert) (Cepheid, Sunnyvale, CA, USA) is a rapid, automated, and cartridge-based genotypic test that can simultaneously detect Mycobacterium tuberculosis complex (MTBC) and rifampin resistance (6). Because of its ease of use and rapid results, the Xpert assay has been widely implemented, particularly in resource-limited settings in which TB is highly endemic.
Until recently, each TB episode was assumed to be caused by a single clonal MTBC strain. However, molecular-based studies have demonstrated that TB may be caused by multiple strains in the same patient (7–12). Far from being uncommon, mixed MTBC infections have been reported in up to 50% of TB cases from certain settings in which TB is endemic (7–12). Because drug-susceptible TB is still the most prevalent type of TB circulating in most communities, MDR-TB patients may have concurrent infections with drug-susceptible MTBC strains (13). Thus, establishing the performance of Xpert among patients with mixed infections is crucial. However, we are aware of only one study that investigated the performance of the Xpert assay in clinical sputum samples containing both sensitive and resistant strains (14). Here, we integrate clinical, epidemiologic, microbiological, and molecular approaches to illustrate the potential problems arising from the use of the Xpert assay for the routine detection of rifampin-resistant MTBC strains in settings in which TB is hyperendemic.
MATERIALS AND METHODS
Setting.
This study was conducted in Botswana, a sub-Saharan African country with a human immunodeficiency virus (HIV) prevalence of 18%, an annual TB incidence rate of 506/100,000 population, and increasing rates of MDR-TB. MDR-TB prevalence has increased by >12-fold from 1996 to 2008 among new TB patients, from 0.2% in 1996 to 2.5% in 2008 (15, 16). The Botswana National TB Program is in the process of scaling up the implementation of Xpert testing nationally. The Xpert assay results, including rifampin resistance test results, have been recommended for guiding initial therapy until phenotypic DST results are available (15).
Study patients.
This retrospective cohort study included adult patients (≥18 years old) who, between 1 January 2011 and 30 March 2012, were diagnosed with pulmonary TB (PTB) by positive Xpert assay results for M. tuberculosis and positive baseline sputum cultures (Fig. 1).
FIG 1.
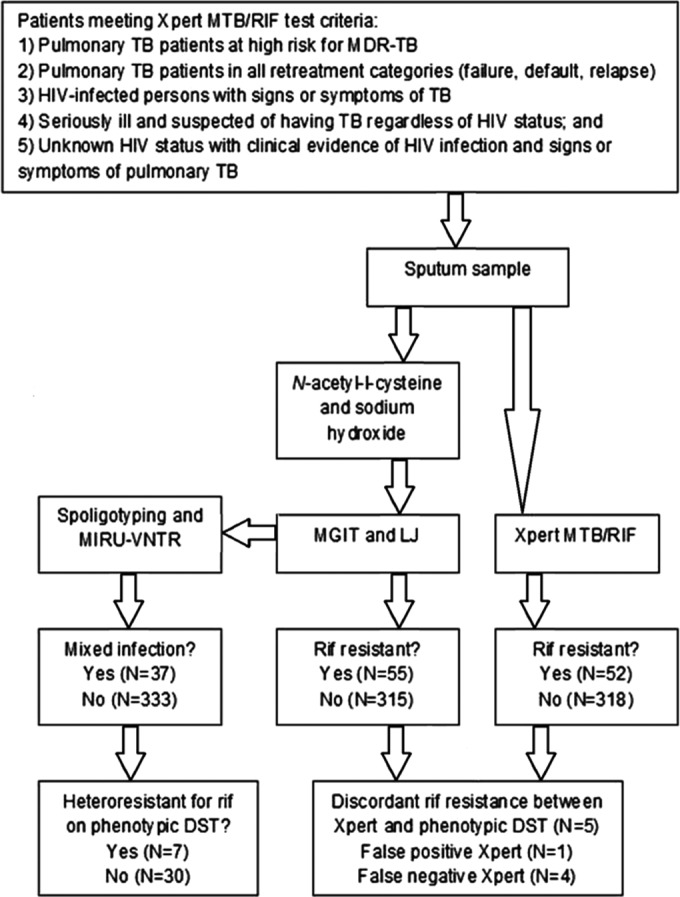
Diagnostic workflow for retrospective cohort of adult patients (≥18 years old) diagnosed with pulmonary tuberculosis based on Xpert MTB/RIF assay results and culture-positive baseline sputum cultures between 1 January 2012 and 30 March 2013. LJ, Lowenstein-Jen; RIF, rifampin.
Sputum sample collection and testing.
In accordance with the Botswana national guidelines for the laboratory diagnosis of PTB, two individual self-collected samples were collected on the first clinical encounter, and one overnight sample was provided by the patient on the following day. Sputum samples from patients at high risk for MDR-TB infections were routinely cultured on Mycobacteria Growth Indicator Tube (MGIT) medium (Fig. 1). All positive cultures underwent first-line DST (see below).
According to the national recommendations existing at that time, one of the individual spot samples from the first encounter was used for Xpert testing only, and the second individual spot sample was submitted to the National TB Reference Laboratory for both acid-fast bacilli (AFB) smear and culture. The indications for Xpert testing included: (i) individuals known or suspected to have PTB who were considered at high risk for MDR-TB, (ii) persons who had been treated with anti-TB drugs and in whom PTB had again been diagnosed, i.e., all retreatment categories (failure, default, and relapse), (iii) all persons who were living with HIV infection and had signs or symptoms of TB, (iv) patients who were seriously ill and suspected of having TB regardless of HIV status, and (v) patients with unknown HIV status presenting with clinical evidence of HIV infection and signs or symptoms of PTB. During the study period, all PTB patients who tested MTBC positive on the Xpert assay were also tested with phenotypic DST (15). All overnight samples were submitted to the National Reference TB Laboratory for AFB and culture. All samples were collected before treatment initiation. All Xpert testing and cultures were performed on individually collected samples. No sputum sample was split for simultaneous Xpert testing and culture.
Data collection.
The data were extracted from paper medical records and electronic databases at the TB clinics, the Botswana National Tuberculosis Program, and the Botswana National Tuberculosis Reference Laboratory. The data collected included the patient demographics, semiquantitative bacillary load by microscopy AFB, history of TB treatment, treatment category, clinical outcomes, HIV serostatus, and CD4+ cell count, along with the use of antiretroviral therapy (ART) in HIV-positive cases.
Procedures for Xpert testing.
Xpert testing was performed on sputum samples, using version 4 cartridges, according to the manufacturer's recommendations. First, the Xpert assay sample reagent (containing NaOH and isopropanol) was added in a 2:1 ratio to the tubes to kill the mycobacteria and liquefy the sample. The mixture was vigorously shaken and allowed to sit for 10 min before being shaken again and allowed to sit for another 5 min. Finally, 2 ml was pipetted into the Xpert assay cartridge and inserted into the GeneXpert instrument for PCR testing. The measurement and analysis were conducted automatically and reported by the GeneXpert Dx software (version 4.0).
Cultures and phenotypic DST.
The MTBC culture and first-line phenotypic DST were performed at the National Tuberculosis Reference Laboratory, Botswana. DST for second-line drugs was performed at the Regional TB Referral Laboratory in South Africa on all isolates that were found to be resistant to isoniazid and rifampin. All sputum samples were processed using the N-acetyl-l-cysteine and sodium hydroxide (NaOH) method, with a final concentration of 1% NaOH, and then cultured on MGIT 960 medium. Cultures failing to produce a positive result within 6 weeks were defined as negative for MTBC infection. We checked for contamination by identifying rapidly growing organisms and those with morphologies that were inconsistent with MTBC. MTBC strains grown on MGIT medium were tested for drug susceptibility using previously described methods (40, 41). Both laboratories used the proportion method on Lowenstein-Jensen (LJ) solid medium and the internationally recommended concentrations of antibiotics to determine drug susceptibilities for first- and second-line drugs. A breakpoint of 1.0 μg/ml was used to determine resistance to rifampin on MGIT 960.
DNA extraction and genotyping.
We extracted chromosomal DNA by boiling a suspension of mycobacteria in Tris-HCl and EDTA buffer, as previously described. We performed spoligotyping using previously described methods and assigned spoligotype families based on published definitions (17–19). We performed standardized 24-locus mycobacterial interspersed repetitive-unit–variable-number tandem-repeat (MIRU-VNTR) typing (20) and classified the patterns based on reference patterns in the MIRU-VNTRplus database (21, 22).
Definitions.
Definitive mixed MTBC infections were defined by the presence of strains with different 24-locus MIRU-VNTR patterns at ≥2 loci in the same sputum sample. Probable mixed MTBC infections were defined by the presence of distinct MIRU-VNTR patterns at a single double locus. A phenotypic discordant DST result for rifampin resistance was defined by the detection of at least one susceptible isolate and one resistant isolate by phenotypic DST within the same patient. Genotypic/phenotypic discordant DST results were defined as MTBC infection with rifampin resistance results that were different between the Xpert assay and phenotypic DST.
We used the World Health Organization (WHO) definitions for treatment outcomes based on routinely collected programmatic data (42). Cured was defined as a patient with a positive culture at the beginning of treatment but a negative culture in the last month of treatment and on at least one previous occasion. Treatment completion was defined as “a patient who completed treatment without evidence of failure but with no record to show that the sputum smear or culture results in the last month of treatment and on at least one previous occasion were negative, either because the tests were not done or because results are unavailable” (42). Treatment failure was defined as a patient with a positive culture ≥5 months after beginning treatment. Default from treatment was defined as a patient whose treatment was interrupted for ≥2 consecutive months. Dead was defined as death from any cause that occurred during the course of treatment. For analysis, we divided those outcomes into poor and good. Poor clinical outcomes were defined as treatment failure, default, or death from any cause. Good clinical outcomes were defined as bacteriologic cure or successful treatment completion based on routinely collected data.
Statistical analysis.
Patients were categorized based on the Xpert MTB/RIF assay results as rifampin resistant or rifampin susceptible. The patients were characterized using simple descriptive statistics. We calculated the sensitivity, specificity, positive predictive value, and negative predictive value of the Xpert assay for detecting rifampin resistance using phenotypic DST as the reference standard. We also stratified these estimates by the presence or absence of mixed MTBC infections. The characteristics and outcomes of patients with Xpert assay results showing rifampin resistance or rifampin susceptibility were compared using the Wilcoxon rank-sum test, the Student t test, and Fisher's exact test, as appropriate. Logistic regression analysis was used to determine the effect of false-negative Xpert assay results (for rifampin resistance) on poor clinical outcome.
In vitro mixed MTBC infection analysis.
Four different MTBC strains with high levels of rifampin resistance (by phenotypic DST) and an M. tuberculosis H37RV pansensitive control strain were used to calculate the in vitro Xpert assay performance for detecting rifampin resistance in mixed MTBC infections. Dilutions containing each strain were obtained through standard methods using 1 McFarland standard solution and sterile water. The cultures were quantitated by comparison to McFarland turbidity standards diluted in sterile water. The quantitated strains were serially diluted in sterile distilled water (dH2O), vortexing after each dilution, to final concentrations of 102, 103, 104, 106, and 108 CFU/ml. When making dilutions, the solutions were mixed by vortexing. Solutions containing mixtures of different concentrations of pansensitive and MDR strains were prepared and tested using the Xpert assay.
Ethics.
This study was approved by the Human Research Development Committee (HRDC) at the Botswana Ministry of Health, the Princess Marina Hospital Ethics Committee, and the University of Pennsylvania Institutional Review Board (IRB).
RESULTS
Determination of Xpert assay performance for the detection of rifampin resistance in sputum samples from culture-positive pulmonary TB patients from a setting in which TB and HIV are hyperendemic.
During the study period, 370 patients fulfilled the inclusion criteria for the study. All patients had at least one baseline Xpert assay result that was positive for MTBC and positive cultures, and 279 (75.4%) patients were infected with HIV. Fifty-two (14.1%) of the 370 patients tested positive for rifampin resistance on the Xpert assay (Table 1). Mixed MTBC infections were identified in 37 (10%) patients, all of whom had definitive mixed MTBC infections based on distinct patterns at two or more loci on the MIRU-VNTR analyses. Out of the 37 patients with mixed MTBC infections, 7 (18.9%) had phenotypic DST discordant results (at least one rifampin-susceptible and one rifampin-resistant isolate among the infecting strains). The other characteristics of the subjects are shown in Table 1.
TABLE 1.
Clinical characteristics of study patients
| Patient characteristic | No. (%) of patients (n = 370)a | Rifampin sensitive by Xpert (n = 318) | Rifampin resistant by Xpert (n = 52) | P value |
|---|---|---|---|---|
| Age (median [IQR]) (yr)a | 37 (31–44) | 38 (32–44) | 36 (30–43) | 0.51 |
| Age category (no. [%]) | 0.49 | |||
| 21–29 yr | 78 (21.1) | 63 (19.8) | 15 (28.9) | |
| 30–39 yr | 182 (49.2) | 159 (50.0) | 23 (44.2) | |
| 40–49 yr | 63 (17.0) | 56 (17.6) | 7 (13.5) | |
| >50 yr | 47 (12.7) | 40 (12.6) | 7 (13.5) | |
| Sex (no. [%]) | 0.22 | |||
| Male | 221 (59.7) | 186 (58.6) | 35 (67.3) | |
| Female | 149 (40.3) | 132 (41.4) | 17 (32.7) | |
| History of prior TB treatment (no. [%]) | <0.001 | |||
| Never treated for TB | 142 (38.4) | 131 (41.2) | 11 (21.2) | |
| Received only first-line drugs in the past | 204 (55.1) | 183 (57.6) | 21 (40.4) | |
| Received second-line drugs in the past | 24 (6.5) | 4 (1.3) | 20 (38.5) | |
| Treatment category (current episode) (no. [%])b | <0.001 | |||
| Initial treatment with first-line drug | 131 (35.4) | 131 (41.2) | 0 | |
| Retreatment regimen with first-line drug | 183 (49.5) | 183 (57.6) | 0 | |
| Second-line treatment regimen | 56 (15.1) | 4 (1.3) | 52 (100.0) | |
| HIV infected (no. [%]) | 279 (75.4) | 238 (74.8) | 41 (78.9) | 0.53 |
| CD4 cell count (median [IQR]) (cells/ml) | 209 (111–331) | 210 (120–342) | 198 (85–291) | 0.18 |
| CD4 cell count category (no. [%]) | 0.69 | |||
| <100 cells/ml | 54 (19.4) | 44 (18.1) | 10 (26.2) | |
| 100–199 cells/ml | 74 (26.5) | 64 (27.0) | 10 (23.8) | |
| 200–350 cells/ml | 88 (31.5) | 74 (31.2) | 14 (33.3) | |
| >350 cells/ml | 63 (22.6) | 56 (23.6) | 7 (16.7) | |
| Mixed TB infections (no. [%]) | 37 (10.0) | 20 (6.3) | 17 (32.7) | <0.001 |
| Clinical outcomes (no. [%]) | <0.001 | |||
| Cured | 251 (67.8) | 220 (69.2) | 31 (59.6) | |
| Completed treatment | 6 (1.6) | 0 | 6 (11.5) | |
| Death | 20 (5.4) | 19 (6.0) | 1 (1.9) | |
| Failed therapy | 31 (8.4) | 26 (8.2) | 5 (9.6) | |
| Defaulted | 16 (4.3) | 15 (4.7) | 1 (1.9) | |
| Still on treatment | 46 (12.4) | 38 (12.0) | 8 (15.4) |
IQR, interquartile range.
As defined by the World Health Organization (WHO 2013).
Among the 52 patients who tested positive for rifampin resistance by the Xpert assay, 11 (21.2%) were new to treatment, 21 (40.4%) were retreatment cases with a history of prior use of first-line antituberculosis treatment only, and 20 (38.3%) cases had a prior MDR-TB history. In accordance with national guidelines, 131 (35.4%) were initiated on category I treatment and 183 (48.6%) were started on category II treatment. All patients who tested positive for rifampin resistance (52 [13.8%]) by the Xpert assay were initiated on MDR-TB treatment.
When the Xpert assay results for rifampin resistance were compared against phenotypic DST results, 5 results were discordant (Table 2). One sample showed resistance to rifampin in the Xpert results but not on phenotypic DST (false-positive Xpert assay result for rifampin resistance). Four samples had genotypic/phenotypic discordant DST results showing resistance to rifampin but negative Xpert assay results for such resistance (false-negative Xpert assay result for rifampin resistance). All five discordant samples had evidence of genotypic mixed MTBC infection, but only the four leading to false-negative Xpert assay results had phenotypic discordant DST patterns (one strain being rifampin susceptible and the other rifampin resistant).
TABLE 2.
Detection of rifampin resistance by the Xpert MTB/RIF assay and phenotypic drug susceptibility testing by mixed infection status and treatment outcomea
| Outcome and infection type by MIRU-VNTRa result | No. of samples with phenotypic rifampin susceptibility and indicated Xpert rifampin resistance result/total no. (%) |
No. of samples with phenotypic rifampin resistance and indicated Xpert rifampin resistance result/total no. (%) |
||
|---|---|---|---|---|
| Susceptible | Resistant | Susceptible | Resistant | |
| All outcomes | ||||
| All patients | 314/315 (99.7) | 1/315 (0.3) | 4/55 (7.2) | 51/55 (92.7) |
| Mixed infection | 16/17 (94.1) | 1/17 (5.9) | 4/20 (20.0) | 16/20 (80.0) |
| No evidence of mixed infection | 298/298 (100) | 0/298 (0) | 0/35 (0) | 35/35 (100) |
| Good (cure or successful treatment completion)b | ||||
| All patients | 220/220 (100) | 0/220 (0) | 0/37 (0) | 37/37 (100) |
| Mixed infection | 5/5 (100) | 0/5 (0) | 0/9 (0) | 9/9 (100) |
| No evidence of mixed infection | 215/215 (100) | 0/215 (0) | 0/28 (0) | 28/28 (100) |
| Poor (treatment failure, default, or death)b | ||||
| All patients | 56/57 (98.2) | 1/57 (1.8) | 4/10 (40.0) | 6/10 (60.0) |
| Mixed infection | 10/11 (90.9) | 1/11 (9.1) | 4/8 (50.0) | 4/8 (50.0) |
| No evidence of mixed infection | 46/46 (100) | 0/46 (0) | 0/2 (0) | 2/2 (100) |
Mycobacterial interspersed repetitive-unit–variable-number tandem-repeat (MIRU-VNTR) analysis.
Forty-six patients were still under treatment at the time of the analysis and were therefore not included in treatment outcome section (i.e., good or poor treatment outcome).
Using phenotypic DST as the reference standard, the overall sensitivity of the Xpert assay for detecting rifampin resistance in our population was 92.7% (n = 51/55; 95% confidence interval [CI], 82.4% to 97.9%), and its overall specificity was 99.7% (n = 314/315; 95% CI, 98.3% to 99.9%). In our study population, with a prevalence of 14.9% (n = 55/370; 95% CI, 11.4% to 18.9%) for rifampin resistance, the positive and negative predictive values were 98.1% (n = 51/52; 95% CI, 89.7% to 99.8%) and 98.7% (n = 314/318; 95% CI, 96.8% to 99.7%), respectively. However, when the analysis was restricted to patients with mixed MTBC infections, the Xpert assay showed a sensitivity of 80.0% (n = 16/20; 95% CI, 56.3% to 94.3%) for rifampin resistance (Table 2).
Sixty-seven of 370 (18.1%) patients had poor clinical outcomes during the follow-up: 20 (5.4%) died during treatment and 31 (8.4%) failed treatment. Forty-six (12.4%) patients were still on therapy at the time of this analysis and were excluded from analyses pertaining to treatment outcome. The multivariate analysis showed that a genotypic/phenotypic discordant DST result (adjusted odds ratio [aOR], 6.6; 95% CI, 1.2 to 48.2) and mixed MTBC infections (adjusted odds ratio [aOR], 6.5; 95% CI, 2.1 to 20.5) were significantly associated with poor clinical outcomes (Table 3). In HIV-infected subjects, a CD4 T-cell count of <100 cells/ml was also associated with poor outcome (aOR, 9.4; 95% CI, 2.7 to 32.6).
TABLE 3.
Predictors of poor clinical outcome
| Patient characteristic | Odds ratio | 95% CIa | P value |
|---|---|---|---|
| Male sex | 1.2 | 0.6–2.5 | 0.69 |
| Age category (yr) | |||
| 21–29 | Reference | ||
| 30–39 | 1.5 | 0.6–3.7 | 0.41 |
| 40–49 | 0.6 | 0.2–2.4 | 0.50 |
| >50 | 1.9 | 0.5–6.8 | 0.34 |
| History of prior TB treatment | |||
| Never treated for TB | Reference | ||
| Received only first-line drugs in the past | 1.9 | 0.2–19.3 | 0.60 |
| Received second-line drugs in the past | 1.1 | 0.1–13.5 | 0.99 |
| Treatment category (current episode) | |||
| Initial treatment with first-line drug | Reference | ||
| Retreatment regimen with first-line drug | 0.2 | 0.1–2.4 | 0.20 |
| Second-line treatment regimen | 0.2 | 0.1–1.6 | 0.12 |
| Semiquantitative acid-fast bacilli on baseline microscopy | |||
| Negative | Reference | ||
| Scanty or 1+ | 1.4 | 0.6–2.3 | 0.52 |
| 2+ or 3+ | 1.7 | 0.8–3.1 | 0.67 |
| CD4 cell count (cells/ml) | |||
| No HIV | Reference | ||
| >350 | 3.8 | 1.1–13.7 | 0.03 |
| 200–350 | 1.6 | 0.4–5.9 | 0.52 |
| 100–199 | 2.1 | 0.6–7.69 | 0.27 |
| <100 | 9.4 | 2.7–32.6 | <0.001 |
| Presence of mixed TB infections | 6.5 | 2.1–20.5 | 0.001 |
| False-negative result for rifampin resistance | 6.6 | 1.2–48.2 | 0.03 |
CI, confidence interval.
Determination of in vitro performance of Xpert assay on samples with concurrent populations of rifampin-susceptible and rifampin-resistant M. tuberculosis.
The Xpert assay failed to identify subpopulations of rifampin-resistant strains of M. tuberculosis DNA mixed with a wild-type RNA polymerase beta (rpoB) sequence when they accounted for <90% of the mixture. The Xpert assay detected rifampin resistance only half the time when the concentrations of the rifampin-resistant subpopulations were between 90 and 99% of the mixture. The Xpert assay consistently identified the presence of rifampin-resistant subpopulations when their concentrations were >99%. Concentrations of rifampin-resistant isolates ≥108 CFU/ml consistently showed Xpert assay results for rifampin resistance regardless of the concentrations of the wild-type isolates.
DISCUSSION
The Xpert is a genotypic test that is increasingly used to screen for rifampin resistance (23). While the Xpert assay has the ability to simultaneously test for a larger number of rpoB mutations, it is not able to detect all mutations that cause rifampin resistance (6, 14). Furthermore, as shown in this study, the presence of wild-type sequences, which are detected by the probes, along with resistant sequences may make this approach intrinsically more susceptible to false-negative results in settings of mixed target MTBC populations.
Inconsistent results between the Xpert assay and phenotypic DST have been recognized (24–29). We identified only one case with positive Xpert assay and negative phenotypic DST results for rifampin resistance. While this might represent a true false-positive result, the lack of the rpoB gene sequence of the two strains identified in the sample precludes definitive conclusions. Prior studies reported that some patients with positive Xpert but negative phenotypic DST results for rifampin resistance (generally considered Xpert false positives) in fact have mutations associated with a rifampin-resistant phenotype (in spite of repeated and consistent negative results for rifampin resistance on phenotypic DST) (24–27), suggesting that some supposedly false-positive Xpert test results may be accurate and actually reflect insufficient phenotypic DST sensitivity. Low-level but probably clinically relevant rifampin resistance is sometimes missed by standard growth-based methods, particularly the automated broth-based systems (29). However, the clinical course and response to treatment of this subpopulation of patients remains largely unknown, highlighting some of the intrinsic difficulties of interpreting differences between genotypic and phenotypic resistance testing in clinical and public health practice (30, 31).
The causes of false-negative Xpert assay results for rifampin resistance have received less attention in the literature (32). The sensitivity of the Xpert assay for detecting rifampin resistance has been reported to be between 60% to almost 100%, depending on the characteristics of the population being tested and the bacterial loads in their samples (6, 24, 28, 33). However, the effect of mixed MTBC infections on the performance of the Xpert assay has not been fully studied. Although prior authors have shown decreased sensitivity with the Xpert assay for detecting rifampin resistance among samples with mixed MTBC strains (6, 14, 34, 35), to our knowledge, this is the first study to systematically look into this issue under programmatic conditions and with linkage to clinical outcomes.
Several authors have also shown a reduced sensitivity of the Xpert assay for detecting rifampin resistance among HIV-infected patients, occurring at increased frequency with advanced immunosuppression (24, 25, 28, 36). Our results also showed such an association in the univariate analyses. However, our multivariate analysis showed that mixed MTBC infections confounded the association between HIV infection and false-negative Xpert test results for rifampin resistance, although our ability to distinguish these factors is limited by statistical power (data not shown). In this context, it seems possible that the previously reported association between HIV infection and false-negative Xpert test results for rifampin resistance might reflect an increased prevalence of mixed MTBC infections among patients with advanced immunosuppression (11, 12).
Prior studies have shown that the Xpert assay is capable of detecting the presence of rifampin resistance mutations down to a concentration of 40% mutant DNA. The identification of the mutant strains becomes easier as more mutant DNA is present in the mixture, but mutant strains were not identifiable in DNA mixtures with 10, 20, and 30% mutant DNA (35). Our mixing studies are consistent with prior studies demonstrating that the proportion of mutant DNA required for detecting rifampin resistance was dependent on the type of mutation (6, 24, 35, 37). Thus, it is expected that test performance for detecting rifampin-resistant strains in mixed infections may vary depending on the specific mutations present in the community. We were not able to determine the specific rpoB mutations present in our subjects. Future studies are needed to determine whether the type of rpoB mutation impacts the performance of the Xpert assay in the context of mixed infections.
Most programs using Xpert testing as part of the diagnostic workup for patients with suspicion of TB use these results to guide the selection of TB treatment. If the Xpert assay detects rifampin resistance in patients considered to be at risk for MDR-TB, an appropriate MDR-TB regimen is started while additional sputum specimens are obtained for culture and phenotypic DST. However, phenotypic DST results become available several weeks after treatment initiation. Consistent with prior reports, we found that the failure to treat subpopulations of rifampin-resistant strains is strongly associated with poor clinical outcomes. In our study, all patients who started on a suboptimal antituberculosis treatment regimen due to failure to detect rifampin resistance at the time of treatment initiation had poor clinical outcomes. Thus, the clinical and public health importance of mixed MTBC infections are significant.
Our findings should be interpreted in the context of the study limitations. Given that the Botswana guidelines have specific criteria for the use of the Xpert assay in the workup of patients, our sample was highly selected and may not represent the general population of TB patients. HIV-infected patients, MDR-TB patients, and patients at higher risk for poor clinical outcomes are overrepresented in this study. The lack of data regarding the prevalence of mixed MTBC infections in Botswana precludes us from any comparison regarding their prevalence in our sample versus that in the general TB population. It also seems likely that mixed MTBC infections were overrepresented in our sample. A significant limitation of our mixing studies is the lack of genome sequencing to determine the rpoB mutation being tested. In addition, our assessments of phenotypic and genotypic resistance were performed on separate (individual) samples as opposed to split single samples, making it possible that the strain diversity and composition of the cultured samples (which eventually had phenotypic DST) were different than those tested by the Xpert assay. Further, our ability to detect mixed MTBC infections may have been compromised by not having tested unprocessed sputum samples for the presence of resistant strains (directly in the sputum sample), given that cultures can alter the strain composition present in the initial sample (8, 38, 39).
By integrating epidemiologic, microbiological, and molecular strain typing data with in vitro experiments, our study demonstrates that the ability of the Xpert assay to detect rifampin resistance may be compromised in settings of mixed MTBC infection with strains possessing heterogeneous phenotypic DST patterns. In addition, we confirmed the major clinical and public health importance of such false-negative Xpert results for rifampin resistance, given its strong association with poor treatment outcomes. The increasing recognition of mixed MTBC infections in regions of high TB endemicity, which includes areas of high MDR-TB prevalence, represents a significant challenge for the use of the Xpert assay in these settings.
In conclusion, the Xpert assay represents a major advance in TB diagnostics and has shown good performance for diagnosing TB and detecting rifampin resistance in most settings. However, clinicians must be aware of the limitations of the assay when interpreting the Xpert test results. Given that mixed MTBC infections may be responsible for false-positive and -negative results, particularly in HIV-infected patients, caution is advised when using the Xpert test results to decide on the initial TB treatment regimen in settings in which TB and HIV are hyperendemic. In particular, clinicians should have a high index of suspicion for MDR-TB in patients failing first-line therapy even when the Xpert test results show rifampin susceptibility.
ACKNOWLEDGMENTS
We thank Rosemarie Kappes, Peter Mulcahy and Radisowa for their constant guidance, support, and input on the manuscript. We also thank the staff at the MDR-TB clinics, the resources and support provided by the Penn Center for AIDS Research, the Botswana Ministry for their constant support and, finally, all our patients that made this study possible.
We declare no competing interests or financial disclosures.
This work was supported in part by NIH grants R01AI097045, T32MH080634, and P30AI45008 (Penn Center for AIDS Research). The funders had no role in the study design, data collection or analysis, decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print 30 April 2014
REFERENCES
- 1. Zumla A, Abubakar I, Raviglione M, Hoelscher M, Ditiu L, McHugh TD, Squire SB, Cox H, Ford N, McNerney R, Marais B, Grobusch M, Lawn SD, Migliori GB, Mwaba P, O'Grady J, Pletschette M, Ramsay A, Chakaya J, Schito M, Swaminathan S, Memish Z, Maeurer M, Atun R. 2012. Drug-resistant tuberculosis–current dilemmas, unanswered questions, challenges, and priority needs. J. Infect. Dis. 205(Suppl 2):S228–S240. 10.1093/infdis/jir858 [DOI] [PubMed] [Google Scholar]
- 2. Daley CL, Caminero JA. 2013. Management of multidrug resistant tuberculosis. Semin. Respir. Crit. Care Med. 34:44–59. 10.1055/s-0032-1333546 [DOI] [PubMed] [Google Scholar]
- 3. Caminero JA. 2005. Management of multidrug-resistant tuberculosis and patients in retreatment. Eur. Respir. J. 25:928–936. 10.1183/09031936.05.00103004 [DOI] [PubMed] [Google Scholar]
- 4. Falzon D, Jaramillo E, Schünemann HJ, Arentz M, Bauer M, Bayona J, Blanc L, Caminero JA, Daley CL, Duncombe C, Fitzpatrick C, Gebhard A, Getahun H, Henkens M, Holtz TH, Keravec J, Keshavjee S, Khan AJ, Kulier R, Leimane V, Lienhardt C, Lu C, Mariandyshev A, Migliori GB, Mirzayev F, Mitnick CD, Nunn P, Nwagboniwe G, Oxlade O, Palmero D, Pavlinac P, Quelapio MI, Raviglione MC, Rich ML, Royce S, Rüsch-Gerdes S, Salakaia A, Sarin R, Sculier D, Varaine F, Vitoria M, Walson JL, Wares F, Weyer K, White RA, Zignol M. 2011. WHO guidelines for the programmatic management of drug-resistant tuberculosis: 2011 update. Eur. Respir. J. 38:516–528. 10.1183/09031936.00073611 [DOI] [PubMed] [Google Scholar]
- 5. Bwanga F, Hoffner S, Haile M, Joloba ML. 2009. Direct susceptibility testing for multi drug resistant tuberculosis: a meta-analysis. BMC Infect. Dis. 9:67. 10.1186/1471-2334-9-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Helb D, Jones M, Story E, Boehme C, Wallace E, Ho K, Kop J, Owens MR, Rodgers R, Banada P, Safi H, Blakemore R, Lan NT, Jones-López EC, Levi M, Burday M, Ayakaka I, Mugerwa RD, McMillan B, Winn-Deen E, Christel L, Dailey P, Perkins MD, Persing DH, Alland D. 2010. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J. Clin. Microbiol. 48:229–237. 10.1128/JCM.01463-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Glynn JR, Yates MD, Crampin AC, Ngwira BM, Mwaungulu FD, Black GF, Chaguluka SD, Mwafulirwa DT, Floyd S, Murphy C, Drobniewski FA, Fine PE. 2004. DNA fingerprint changes in tuberculosis: reinfection, evolution, or laboratory error? J. Infect. Dis. 190:1158–1166. 10.1086/423144 [DOI] [PubMed] [Google Scholar]
- 8. van Rie A, Victor TC, Richardson M, Johnson R, van der Spuy GD, Murray EJ, Beyers N, Gey van Pittius NC, van Helden PD, Warren RM. 2005. Reinfection and mixed infection cause changing Mycobacterium tuberculosis drug-resistance patterns. Am. J. Respir. Crit. Care Med. 172:636–642. 10.1164/rccm.200503-449OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. van Rie A, Warren R, Richardson M, Victor TC, Gie RP, Enarson DA, Beyers N, van Helden PD. 1999. Exogenous reinfection as a cause of recurrent tuberculosis after curative treatment. N. Engl. J. Med. 341:1174–1179. 10.1056/NEJM199910143411602 [DOI] [PubMed] [Google Scholar]
- 10. Huyen MN, Kremer K, Lan NT, Cobelens FG, Buu TN, Dung NH, Caws M, Tiemersma EW, van Soolingen D. 2012. Mixed tuberculosis infections in rural South Vietnam. J. Clin. Microbiol. 50:1586–1592. 10.1128/JCM.00434-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cohen T, van Helden PD, Wilson D, Colijn C, McLaughlin MM, Abubakar I, Warren RM. 2012. Mixed-strain Mycobacterium tuberculosis infections and the implications for tuberculosis treatment and control. Clin. Microbiol. Rev. 25:708–719. 10.1128/CMR.00021-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cohen T, Wilson D, Wallengren K, Samuel EY, Murray M. 2011. Mixed-strain Mycobacterium tuberculosis infections among patients dying in a hospital in KwaZulu-Natal, South Africa. J. Clin. Microbiol. 49:385–388. 10.1128/JCM.01378-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zignol M, van Gemert W, Falzon D, Sismanidis C, Glaziou P, Floyd K, Raviglione M. 2012. Surveillance of anti-tuberculosis drug resistance in the world: an updated analysis, 2007–2010. Bull. World Health Organ. 90:111D–119D. 10.2471/BLT.11.092585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Blakemore R, Story E, Helb D, Kop J, Banada P, Owens MR, Chakravorty S, Jones M, Alland D. 2010. Evaluation of the analytical performance of the Xpert MTB/RIF assay. J. Clin. Microbiol. 48:2495–2501. 10.1128/JCM.00128-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Botswana Ministry of Health. 2009. Botswana National Tuberculosis Programme Report. Botswana Ministry of Health, Gaborone, Botswana [Google Scholar]
- 16.Botswana Ministry of Health. 2008. Ministry of Health of Botswana. BIAS 3 Report. Botswana Ministry of Health, Gaborone, Botswana [Google Scholar]
- 17. van der Zanden AG, Kremer K, Schouls LM, Caimi K, Cataldi A, Hulleman A, Nagelkerke NJ, van Soolingen D. 2002. Improvement of differentiation and interpretability of spoligotyping for Mycobacterium tuberculosis complex isolates by introduction of new spacer oligonucleotides. J. Clin. Microbiol. 40:4628–4639. 10.1128/JCM.40.12.4628-4639.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brudey K, Driscoll JR, Rigouts L, Prodinger WM, Gori A, Al-Hajoj SA, Allix C, Aristimuño L, Arora J, Baumanis V, Binder L, Cafrune P, Cataldi A, Cheong S, Diel R, Ellermeier C, Evans JT, Fauville-Dufaux M, Ferdinand S, Garcia de Viedma D, Garzelli C, Gazzola L, Gomes HM, Guttierez MC, Hawkey PM, van Helden PD, Kadival GV, Kreiswirth BN, Kremer K, Kubin M, Kulkarni SP, Liens B, Lillebaek T, Ho ML, Martin C, Martin C, Mokrousov I, Narvskaïa O, Ngeow YF, Naumann L, Niemann S, Parwati I, Rahim Z, Rasolofo-Razanamparany V, Rasolonavalona T, Rossetti ML, Rüsch-Gerdes S, Sajduda A, Samper S, et al. 2006. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol. 6:23. 10.1186/1471-2180-6-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Filliol I, Driscoll JR, Van Soolingen D, Kreiswirth BN, Kremer K, Valétudie G, Anh DD, Barlow R, Banerjee D, Bifani PJ, Brudey K, Cataldi A, Cooksey RC, Cousins DV, Dale JW, Dellagostin OA, Drobniewski F, Engelmann G, Ferdinand S, Gascoyne-Binzi D, Gordon M, Gutierrez MC, Haas WH, Heersma H, Källenius G, Kassa-Kelembho E, Koivula T, Ly HM, Makristathis A, Mammina C, Martin G, Moström P, Mokrousov I, Narbonne V, Narvskaya O, Nastasi A, Niobe-Eyangoh SN, Pape JW, Rasolofo-Razanamparany V, Ridell M, Rossetti ML, Stauffer F, Suffys PN, Takiff H, Texier-Maugein J, Vincent V, De Waard JH, Sola C, Rastogi N. 2002. Global distribution of Mycobacterium tuberculosis spoligotypes. Emerg. Infect. Dis. 8:1347–1349. 10.3201/eid0811.020125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Supply P, Lesjean S, Savine E, Kremer K, van Soolingen D, Locht C. 2001. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J. Clin. Microbiol. 39:3563–3571. 10.1128/JCM.39.10.3563-3571.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rüsch-Gerdes S, Willery E, Savine E, de Haas P, van Deutekom H, Roring S, Bifani P, Kurepina N, Kreiswirth B, Sola C, Rastogi N, Vatin V, Gutierrez MC, Fauville M, Niemann S, Skuce R, Kremer K, Locht C, van Soolingen D. 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 44:4498–4510. 10.1128/JCM.01392-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Allix-Béguec C, Harmsen D, Weniger T, Supply P, Niemann S. 2008. Evaluation and strategy for use of MIRU-VNTRplus, a multifunctional database for online analysis of genotyping data and phylogenetic identification of Mycobacterium tuberculosis complex isolates. J. Clin. Microbiol. 46:2692–2699. 10.1128/JCM.00540-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vassall A, van Kampen S, Sohn H, Michael JS, John KR, den Boon S, Davis JL, Whitelaw A, Nicol MP, Gler MT, Khaliqov A, Zamudio C, Perkins MD, Boehme CC, Cobelens F. 2011. Rapid diagnosis of tuberculosis with the Xpert MTB/RIF assay in high burden countries: a cost-effectiveness analysis. PLoS Med. 8:e1001120. 10.1371/journal.pmed.1001120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Boehme CC, Nicol MP, Nabeta P, Michael JS, Gotuzzo E, Tahirli R, Gler MT, Blakemore R, Worodria W, Gray C, Huang L, Caceres T, Mehdiyev R, Raymond L, Whitelaw A, Sagadevan K, Alexander H, Albert H, Cobelens F, Cox H, Alland D, Perkins MD. 2011. Feasibility, diagnostic accuracy, and effectiveness of decentralised use of the Xpert MTB/RIF test for diagnosis of tuberculosis and multidrug resistance: a multicentre implementation study. Lancet 377:1495–1505. 10.1016/S0140-6736(11)60438-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marlowe EM, Novak-Weekley SM, Cumpio J, Sharp SE, Momeny MA, Babst A, Carlson JS, Kawamura M, Pandori M. 2011. Evaluation of the Cepheid Xpert MTB/RIF assay for direct detection of Mycobacterium tuberculosis complex in respiratory specimens. J. Clin. Microbiol. 49:1621–1623. 10.1128/JCM.02214-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Theron G, Peter J, van Zyl-Smit R, Mishra H, Streicher E, Murray S, Dawson R, Whitelaw A, Hoelscher M, Sharma S, Pai M, Warren R, Dheda K. 2011. Evaluation of the Xpert MTB/RIF assay for the diagnosis of pulmonary tuberculosis in a high HIV prevalence setting. Am. J. Respir. Crit. Care. 184:132–140. 10.1164/rccm.201101-0056OC [DOI] [PubMed] [Google Scholar]
- 27. Van Rie A, Mellet K, John MA, Scott L, Page-Shipp L, Dansey H, Victor T, Warren R. 2012. False-positive rifampicin resistance on Xpert MTB/RIF: case report and clinical implications. Int. J. Tuberc. Lung Dis. 16:206–208. 10.5588/ijtld.11.0395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lawn SD, Brooks SV, Kranzer K, Nicol MP, Whitelaw A, Vogt M, Bekker LG, Wood R. 2011. Screening for HIV-associated tuberculosis and rifampicin resistance before antiretroviral therapy using the Xpert MTB/RIF assay: a prospective study. PLoS Med. 8:e1001067. 10.1371/journal.pmed.1001067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Van Deun A, Barrera L, Bastian I, Fattorini L, Hoffmann H, Kam KM, Rigouts L, Rüsch-Gerdes S, Wright A. 2009. Mycobacterium tuberculosis strains with highly discordant rifampin susceptibility test results. J. Clin. Microbiol. 47:3501–3506. 10.1128/JCM.01209-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Scott LE, McCarthy K, Gous N, Nduna M, Van Rie A, Sanne I, Venter WF, Duse A, Stevens W. 2011. Comparison of Xpert MTB/RIF with other nucleic acid technologies for diagnosing pulmonary tuberculosis in a high HIV prevalence setting: a prospective study. PLoS Med. 8:e1001061. 10.1371/journal.pmed.1001061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Van Deun A, Aung KJ, Bola V, Lebeke R, Hossain MA, de Rijk WB, Rigouts L, Gumusboga A, Torrea G, de Jong BC. 2013. Rifampin drug resistance tests for tuberculosis: challenging the gold standard. J. Clin. Microbiol. 51:2633–2640. 10.1128/JCM.00553-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chakravorty S, Boehme C, Lee J. 2012. Tuberculosis diagnostics in the new millennium: role in TB identification and control. Tuberc. Res. Treat. 2012:768603. 10.1155/2012/768603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.World Health Organization. 2011. Rapid implementation of the Xpert MTB/RIF diagnostic test: technical and operational ‘how-to' practical considerations. World Health Organization, Geneva, Switzerland: 2011 http://whqlibdoc.who.int/publications/2011/9789241501569_eng.pdf [Google Scholar]
- 34. Chakravorty S, Aladegbami B, Thoms K, Lee JS, Lee EG, Rajan V, Cho EJ, Kim H, Kwak H, Kurepina N, Cho SN, Kreiswirth B, Via LE, Barry CE, III, Alland D. 2011. Rapid detection of fluoroquinolone-resistant and heteroresistant Mycobacterium tuberculosis by use of sloppy molecular beacons and dual melting-temperature codes in a real-time PCR assay. J. Clin. Microbiol. 49:932–940. 10.1128/JCM.02271-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chakravorty S, Kothari H, Aladegbami B, Cho EJ, Lee JS, Roh SS, Kim H, Kwak H, Lee EG, Hwang SH, Banada PP, Safi H, Via LE, Cho SN, Barry CE, III, Alland D. 2012. Rapid, high-throughput detection of rifampin resistance and heteroresistance in Mycobacterium tuberculosis by use of sloppy molecular beacon melting temperature coding. J. Clin. Microbiol. 50:2194–2202. 10.1128/JCM.00143-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Boehme C. 2010. Feasibility and impact of using Xpert MTB/RIF: results from demonstration studies. FIND and Partners Symposium, Berlin, Germany, 12 November 2010 http://r4d.dfid.gov.uk/PDF/Outputs/FIND/CatharinaBoehme_FeasibilityAndImpact.pdf [Google Scholar]
- 37. Boehme CC, Nabeta P, Hillemann D, Nicol MP, Shenai S, Krapp F, Allen J, Tahirli R, Blakemore R, Rustomjee R, Milovic A, Jones M, O'Brien SM, Persing DH, Ruesch-Gerdes S, Gotuzzo E, Rodrigues C, Alland D, Perkins MD. 2010. Rapid molecular detection of tuberculosis and rifampin resistance. N. Engl. J. Med. 363:1005–1015. 10.1056/NEJMoa0907847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hingley-Wilson SM, Casey R, Connell D, Bremang S, Evans JT, Hawkey PM, Smith GE, Jepson A, Philip S, Kon OM, Lalvani A. 2013. Undetected multidrug-resistant tuberculosis amplified by first-line therapy in mixed infection. Emerg. Infect. Dis. 19:1138–1141. 10.3201/eid1907.130313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Martin A, Herranz M, Ruiz Serrano MJ, Bouza E, García de Viedma D. 2010. The clonal composition of Mycobacterium tuberculosis in clinical specimens could be modified by culture. Tuberculosis (Edinb.) 90:201–207. 10.1016/j.tube.2010.03.012 [DOI] [PubMed] [Google Scholar]
- 40. Bemer P, Palicova F, Rüsch-Gerdes S, Drugeon HB, Pfyffer GE. 2002. Multicenter evaluation of fully automated BACTEC Mycobacteria Growth Indicator Tube 960 system for susceptibility testing of Mycobacterium tuberculosis. J. Clin. Microbiol. 40:150–154. 10.1128/JCM.40.1.150-154.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rüsch-Gerdes S, Domehl C, Nardi G, Gismondo MR, Welscher HM, Pfyffer GE. 1999. Multicenter evaluation of the Mycobacteria Growth Indicator Tube for testing susceptibility of Mycobacterium tuberculosis to first-line drugs. J. Clin. Microbiol. 37:45–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.World Health Organization. 2013. Revised definitions and reporting framework for tuberculosis. World Health Organization, Geneva, Switzerland: http://www.euro.who.int/en/health-topics/communicable-diseases/tuberculosis/news/news/2013/04/revised-definitions-and-reporting-framework-for-tuberculosis [Google Scholar]


