Abstract
Resistance to extended-spectrum β-lactam antibiotics has led to a greater reliance upon carbapenems, but the expression of carbapenemases threatens to limit the utility of these drugs. Current methods to detect carbapenemase activity are suboptimal, requiring prolonged incubations during which ineffective therapy may be prescribed. We previously described a sensitive and specific assay for the detection of carbapenemase activity using ertapenem and liquid chromatography-tandem mass spectrometry (LC-MS/MS). In this study, we assessed 402 Gram-negative rods, including both Enterobacteriaceae and non-Enterobacteriaceae expressing IMP, VIM, KPC, NDM, and/or OXA carbapenemases, by using imipenem, meropenem, and ertapenem with LC-MS/MS assays. LC-MS/MS methods for the detection of intact and hydrolyzed carbapenems from an enrichment broth were developed. No ion suppression was observed, and the limits of detection for all three drugs were below 0.04 μg/ml. The sensitivity and specificity of meropenem and ertapenem for carbapenemase activity among non-Enterobacteriaceae were low, but imipenem demonstrated a sensitivity and specificity of 96% and 95%, respectively, among all Gram-negative rods (GNR) tested, including both Enterobacteriaceae and non-Enterobacteriaceae. LC-MS/MS allows for the analysis of more complex matrices, and this LC-MS/MS assay could easily be adapted for use with primary specimens requiring growth enrichment.
INTRODUCTION
Resistance to β-lactam antibiotics among Gram-negative rods (GNRs) poses an increasingly severe clinical problem (1–3). Among β-lactams, carbapenems are resistant to hydrolysis by most β-lactamases, but carbapenemases capable of degrading all β-lactam antibiotics have been found in GNR throughout the world (4, 5). There are many different enzyme families that exhibit β-lactamase activity, and among related family members, enzymes can display β-lactamase, extended-spectrum β-lactamase (ESBL), or carbapenemase activity (6). The diversity of genes involved and potential phenotypic discordance between highly similar proteins have limited the development of rapid tests capable of identifying ESBL- or carbapenemase-producing organisms to better direct therapeutic interventions.
Given the constantly evolving genetic diversity of β-lactamases, phenotypic methods that detect carbapenem hydrolysis remain the ideal approach to identify resistance. Unlike molecular methods, phenotypic assays for β-lactam hydrolysis do not require knowledge of the genetic determinants of resistance and are not affected by mutations in primer targets. However, conventional phenotypic methods require up to 18 h or more.
β-Lactamase activity reliably leads to the hydrolysis of the β-lactam ring through the addition of H2O. We and others hypothesized that this mass change would be readily detectable by mass spectrometry (MS). Indeed, when examined by either matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) MS or liquid chromatography-coupled tandem MS (LC-MS/MS), hydrolyzed carbapenem antibiotics can be detected following incubation with carbapenemase-producing bacteria (7–11).
Several groups have described the use of MALDI-TOF MS for the phenotypic identification of carbapenemase activity (8–16). They have reported promising results, but MALDI-TOF MS is classically thought not to be well suited for small-molecule detection, and LC-MS/MS remains the gold standard method for small-molecule detection and quantitation. LC-MS/MS methods have excellent analytical sensitivity, and chromatographic retention time, precursor ion mass, and product ion mass combine to confer excellent analytical specificity. Additionally, nearly all papers have examined the use of ertapenem, meropenem, or imipenem individually, and the ideal substrate for carbapenemase detection remains unclear. We previously described an LC-MS/MS method for the detection of hydrolyzed ertapenem, and we have expanded those studies to include a larger, more diverse set of bacterial isolates and the use of meropenem and imipenem (7).
MATERIALS AND METHODS
Bacterial isolates.
Clinical isolates selected for carbapenem and/or extended-spectrum cephalosporin resistance were obtained from health care facilities through material transfer. These were supplemented with commercially available isolates obtained from International Health Management Associates Inc. (IHMA; Chicago, IL) or ATCC (Manassas, VA). The isolate set was assembled with the goal of maximizing enzyme and species diversity, and the number and type of isolates included were determined by their availability.
Phenotypic characterization of bacterial isolates.
Antimicrobial susceptibility testing (AST) was performed by disk diffusion assay as described previously (17). Disks were used as supplied (Thermo Scientific, Lenexa KS), or meropenem disks were supplemented with 3-aminophenylboronic acid (400 μg) or EDTA (100 mM) for KPC or metallo-β-lactamase screening, respectively (18). CLSI document M100-S24 interpretive criteria were used to determine susceptibility, and the modified Hodge test was performed as described previously (17).
Genotypic characterization of bacterial isolates.
All clinical isolates were genotyped as previously described (7). Briefly, genomic DNA from each isolate was purified using PrepMan ultra reagent (Life Technologies, Grand Island, NY) according to the manufacturer's protocol. Five microliters of genomic DNA and primers specific for the amplification of KPC, NDM-1, IMP, VIM, or OXA was added to 25 μl of GeneAmp fast PCR master mix (Life Technologies, Grand Island, NY), and PCR was performed as described previously on a Mastercycler pro S system (7). The PCR mixtures were mixed with TrackIt cyan/orange loading buffer mix (Life Technologies, Grand Island, NY) and separated by gel electrophoresis on a 2% agarose gel containing the SYBR safe DNA gel stain (Life Technologies, Grand Island, NY). Gene-specific amplicons were visualized using the Safe Imager 2.0 (Life Technologies, Grand Island, NY).
LC-MS/MS methods for the detection of intact and hydrolyzed carbapenems.
The AB Sciex Triple Quad 5500 (AB Sciex, Framingham, MA) triple-quadrupole MS/MS system was operated in multiple reaction monitoring (MRM) mode. Optimal source settings for each drug-metabolite pair were determined and are listed in Table S1 in the supplemental material. The quadrupole was set for detection of a single drug-metabolite pair at a time, and MRM transitions are listed in Table S1 in the supplemental material. HPLC separation of intact and hydrolyzed ertapenem, meropenem, and imipenem was performed using Kinetex pentafluorophenyl (Phenomenex, Torrance, CA), Phalanx (Higgins Analytical, Mountain View, CA), and Targa (Higgins Analytical, Mountain View, CA) columns as listed in Table S1. All reagents were ultra-high-performance liquid chromatography (UHPLC)-grade and obtained from Sigma-Aldrich (St. Louis, MO). The total areas for all monitored peaks were integrated using the Analyst software package, version 1.6 (AB Sciex, Framingham, MA).
Detection of carbapenemase activity using mass spectrometry.
A 2 McFarland (McF) bacterial suspension in 2.0 ml of MZB (M/Z Diagnostics, New Haven, CT) was prepared from isolated bacterial colonies of freshly subcultured overnight cultures on Columbia blood agar (Thermo Scientific, Lenexa, KS). Either ertapenem (Merck, Whitehouse Station, NJ), meropenem (Sigma-Aldrich, St. Louis, MO), or imipenem (Sigma-Aldrich, St. Louis, MO) was added to a final concentration of 4 μg/ml. Cultures were incubated at 37°C for 1 h in a shaking incubator. Two-hundred-microliter aliquots of the mixture were added to 400 μl of methanol (Sigma-Aldrich, St. Louis, MO) in microcentrifuge tubes. Bacteria and precipitated protein were separated by centrifugation at 16,000 × g for 2 min. The supernatant was carefully removed, and 100-μl aliquots were mixed with 500 μl of deionized water for chromatographic-mass spectrometric analysis.
Five-microliter amounts of the solutions prepared as described above were injected onto appropriate LC columns using a Shimadzu Prominence UFLCXR LC system (Shimadzu, Columbia, MD), and analytes were detected with an AB Sciex Triple Quad 5500 MS/MS (AB Sciex, Framingham, MA) detector using the parameters described above. Total areas for the appropriate intact and hydrolyzed drug peak(s) were integrated using the Analyst software package (AB Sciex, Framingham, MA). The ratio of the total area of each hydrolyzed carbapenem metabolite and its corresponding parent carbapenem drug was used as an indication of carbapenemase activity. For all assays of carbapenemase activity, the non-carbapenemase-producing K. pneumoniae strain ATCC 1706 and the KPC-2-producing K. pneumoniae strain ATCC 1705 were included as negative and positive controls, respectively.
ROC analysis.
Receiver-operator curves (ROC) were calculated using GraphPad Prism (GraphPad Software, La Jolla, CA). Cutoffs were selected based on the lowest ratio cutoff that gave a specificity of at least 95%.
RESULTS
Development of LC methods for meropenem and imipenem.
We previously reported an LC-MS/MS assay for the detection of ertapenem hydrolysis from Enterobacteriaceae, but the performance of this assay was suboptimal for non-glucose-fermenting (NF) GNR expressing other carbapenemases (e.g., IMP, VIM, and OXA) (data not shown). We wished to develop an LC-MS/MS assay for carbapenemase detection that would be suitable for all clinically relevant GNR. We first determined the optimal tuning parameters for the detection of intact and hydrolyzed meropenem and imipenem on the AB Triple Quad 5500 MS/MS system. We manually tuned the source and MS/MS parameters by the infusion of a 0.1 μg/ml solution of meropenem or imipenem in 100% methanol with 0.1% formic acid using an integrated syringe pump. At the time of development, there were no published MRM transitions for the detection of intact or hydrolyzed forms of meropenem or imipenem. The most abundant precursor/product ion pairs (optimal MRM transition) were m/z 384.1 → m/z 68 for meropenem and m/z 300.1 → m/z 142.1 for imipenem (data not shown).
For the detection of hydrolyzed meropenem and imipenem, a 0.1 μg/ml solution of chemically synthesized hydrolyzed metabolites (+18 Da form), m/z 402 and m/z 318.1, respectively (provided by Babu Purkayastha, personal communication), were also used for tuning MS parameters. During MS/MS analysis, the primary product ions detected for hydrolyzed meropenem and hydrolyzed imipenem were 358 Da and 103 Da, respectively. Thus, the final MRM transitions used for the detection of hydrolyzed meropenem were m/z 402 → m/z 358 (data not shown), and those for hydrolyzed imipenem were m/z 318.1 → m/z 103 (data not shown). The chromatographic parameters for the separation of intact and hydrolyzed meropenem and imipenem are shown in Table S1 in the supplemental material, and representative chromatograms are shown in Fig. 1.
FIG 1.
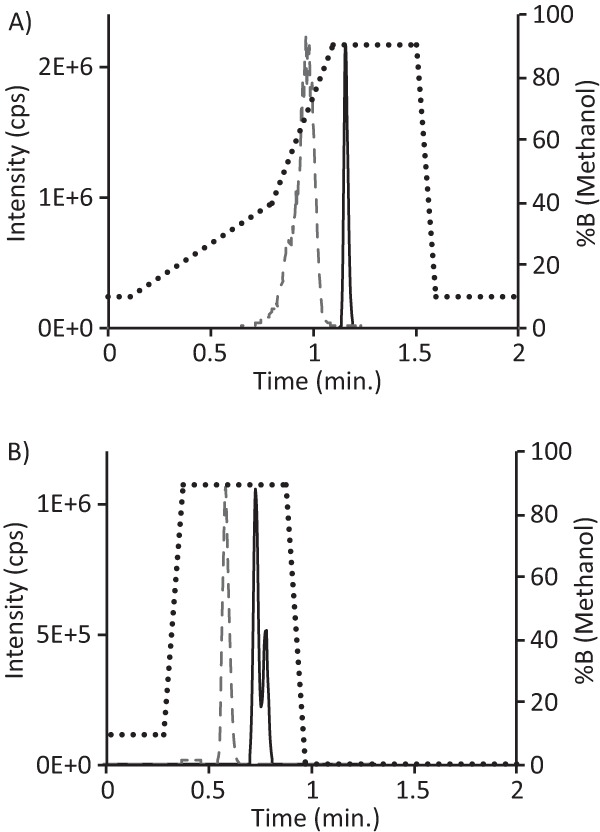
Results of LC-MS/MS method for detection of intact and hydrolyzed meropenem or imipenem. (A) MRM transitions were first-quadrupole (Q1) m/z = 384.1 Da for meropenem, Q1 m/z = 402 Da for hydrolyzed meropenem, and m/z = 68 Da and m/z = 358 Da for the respective predominant products (fragments) were monitored during a 2-min LC program (dotted trace) using deionized H2O plus 0.1% formic acid as solvent A and methanol plus 0.1% formic acid as solvent B. Meropenem (black solid trace) and hydrolyzed meropenem (gray dashed trace) were detected from a test mixture of intensity-matched samples of meropenem and hydrolyzed meropenem. (B) MRM transitions were Q1 m/z = 300.1 Da for imipenem, Q1 m/z = 318.1 Da for hydrolyzed imipenem, and m/z = 142.1 Da and m/z = 103 Da for the respective predominant products (fragments) were monitored during a 2-min LC program (dotted trace) using deionized H2O plus 0.1% formic acid as solvent A and methanol plus 0.1% formic acid as solvent B. Imipenem (black solid trace) and hydrolyzed imipenem (gray dashed trace) were detected from a test mixture of intensity-matched samples of imipenem and hydrolyzed imipenem.
To exclude the possibility of negative matrix effects on the ionization of analytes, we performed ion suppression studies using a blank matrix injected from the LC, while a mixture of intensity-matched intact and hydrolyzed meropenem or imipenem was introduced by direct infusion from a syringe pump. No significant ion suppression was observed for the meropenem/hydrolyzed meropenem or the imipenem/hydrolyzed imipenem in their retention time windows (see Fig. S1 in the supplemental material).
Optimization and retrospective validation of the assay.
To develop a robust assay for the detection of carbapenemase activity from GNR colonies, we optimized the assay parameters, including but not limited to broth composition (e.g., tryptic soy broth, Mueller-Hinton broth, or 0.45% NaCl), additives (e.g., lysozyme, detergents, or cations), drug concentration (2, 4, 10, or 20 μg/ml), incubation temperature (35, 37, or 42°C), incubation time (1, 2, 4, or 8 h), and bacterial inoculum (0.5, 2, 4, or 8 McF) (data not shown). The effects of varying these parameters were investigated using a set of 10 isolates known to be positive or negative for carbapenemase expression. Among carbapenemase-positive isolates, the diversity of hydrolysis activity was sought (data not shown). The final protocol uses a 2 McF suspension from fresh bacterial colonies prepared in MZB (based on Mueller-Hinton broth and developed in-house) and supplemented with a carbapenem at a physiologic concentration (e.g., 4 μg/ml ertapenem, meropenem, or imipenem). After 1 h of incubation with shaking at 37°C, a 200-μl aliquot is diluted 1:3 with methanol and centrifuged to remove precipitated protein, and the supernatant loaded onto the LC-MS/MS system (Fig. 2).
FIG 2.
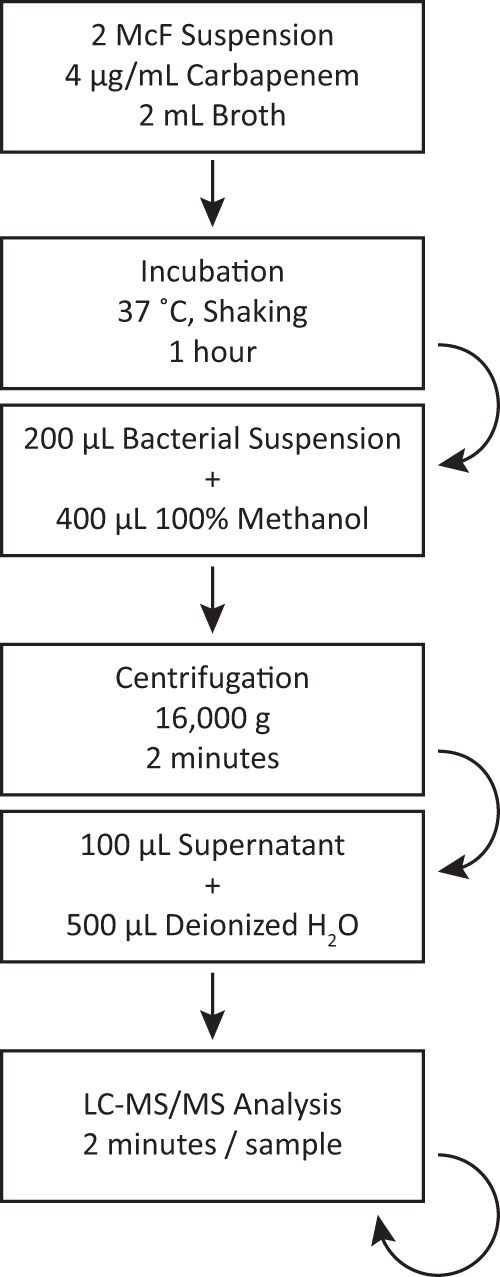
Final assay workflow. A 2 McF bacterial suspension in broth with 4 μg/ml carbapenem is incubated for 1 h with shaking at 37°C. A 200-μl aliquot is taken and added to 400 μl methanol. This is centrifuged for 2 min at 16,000 × g, and 100 μl of supernatant is combined with 500 μl of H2O for LC-MS/MS analysis.
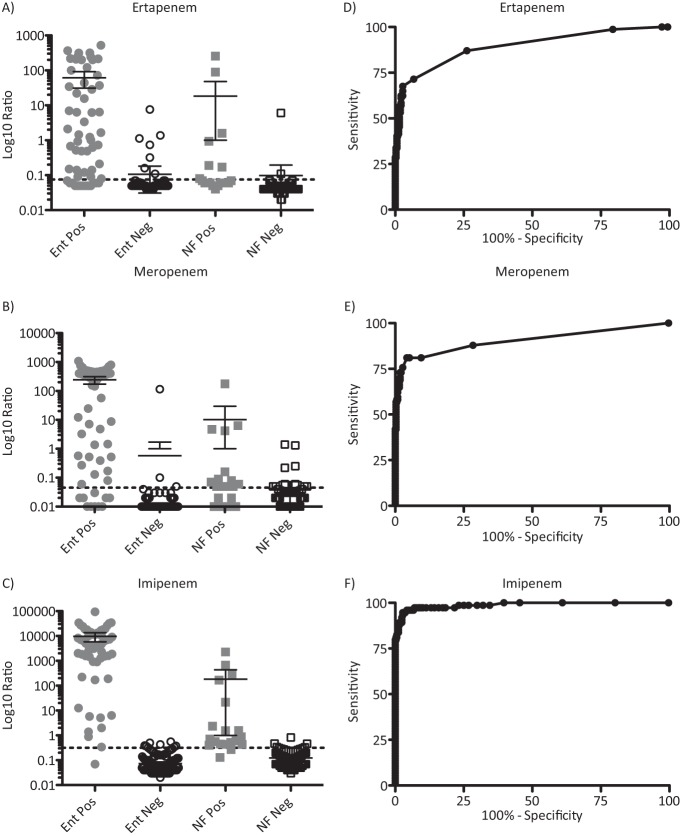
Using the incubation and LC-MS/MS protocols developed as described above, we compared the efficiency of carbapenem hydrolysis of ertapenem, meropenem, and imipenem by a set of 402 bacterial isolates (Table 1) that had been characterized for carbapenemase and ESBL genes by PCR, conventional AST by disk diffusion, phenotypic carbapenemase expression by the modified Hodge test (MHT), and AmpC detection by a double-disc synergy test, presented to the operator in a blinded fashion (19). Visual inspection of all the chromatograms revealed that the sensitivity values of the assay using ertapenem and meropenem were significantly lower than its sensitivity values using imipenem. For example, many carbapenemase-expressing isolates, including IMP-18-positive P. aeruginosa, VIM-27-positive K. pneumoniae, and OXA-24-positive A. baumannii, failed to efficiently hydrolyze either ertapenem or meropenem (Fig. 3A and B), while imipenem hydrolysis was readily observed (Fig. 3C). The hydrolysis ratio, defined as the calculated ratio of hydrolyzed drug peak area and intact drug peak area for each isolate, was compared with the predicted outcome; an isolate was classified as “true positive” if either (i) a known carbapenemase gene was detected by PCR and sequence verified or (ii) carbapenemase activity was detected in the MHT (7). The hydrolysis ratios for all isolates ranged from 0.00 to >100 for meropenem and from 0.02 to >100 for ertapenem and imipenem (Fig. 4A to C). Based on receiver-operator characteristics, optimal thresholds for positivity were established (Fig. 4D to F), and the cutoff, sensitivity, and specificity values for all isolates, Enterobacteriaceae, and NF GNR are listed in Table 2. Using the cutoffs determined as described above, imipenem demonstrated the greatest sensitivity for the detection of carbapenemase activity for the entire population of GNR, as well as for both Enterobacteriaceae and NF GNR when analyzed separately.
TABLE 1.
Characterization of bacterial isolates
| Species | No. of isolates with gene expressing indicated enzymea |
No. of carbapenem-sensitive isolates | Total no. of isolates | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbapenemase of type: |
β-Lactamase of type: |
||||||||||||
| KPC | IMP | VIM | NDM | OXA | Other | TEM | SHV | CTX-M | Multb | AmpCc | |||
| Klebsiella pneumoniae | 14 | 1 | 4 | 2 | 3 | 5 | 18 | 15 | 11 | 73 | |||
| Escherichia coli | 5 | 1 | 3 | 1 | 1 | 6 | 9 | 23 | 28 | 77 | |||
| Enterobacter species | 2 | 3 | 2 | 1 | 5 | 3 | 1 | 1 | 7 | 3 | 25 | 53 | |
| Citrobacter freundii | 2 | 1 | 1 | 1 | 2 | 7 | |||||||
| Serratia marcescens | 2 | 1 | 4 | 1 | 1 | 16 | 17 | 42 | |||||
| Pseudomonas aeruginosa | 4 | 2 | 4 | 2 | 1 | 75 | 88 | ||||||
| Acinetobacter baumannii | 11 | 1 | 2 | 1 | 9 | 24 | |||||||
| Other | 1 | 2 | 1 | 2 | 6 | 26 | 38 | ||||||
| Total | 26 | 6 | 10 | 10 | 17 | 8 | 26 | 23 | 11 | 69 | 3 | 193 | 402 |
Isolates were subjected to phenotypic and genotypic screening for β-lactamase genes. In total, 77 isolates expressed a carbapenemase and 132 isolates expressed a β-lactamase.
Mult, multiple different enzymes detected.
AmpC screening was performed phenotypically.
FIG 3.
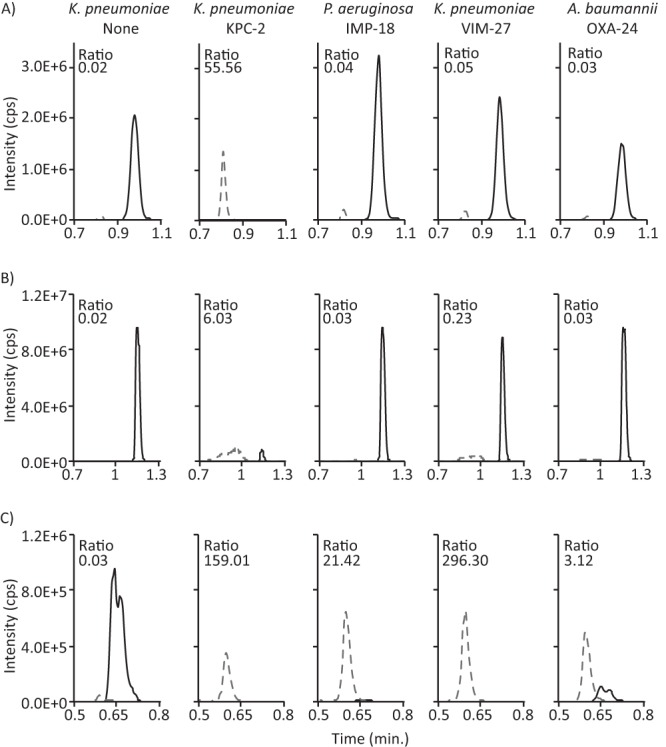
Drug hydrolysis efficiencies of carbapenemase-expressing bacteria. Chromatograms for parent drug (black solid trace) and hydrolyzed metabolite (gray dashed trace), obtained by performing the carbapenem hydrolysis protocol for 1 h using the indicated bacteria, are shown for ertapenem (A), meropenem (B), and imipenem (C). The non-carbapenemase-expressing (K. pneumoniae None) and KPC-2-expressing (K. pneumoniae KPC-2) K. pneumoniae strains were ATCC 1706 and ATCC 1705, respectively. The other isolates were newly described clinical isolates in this study. For clarity, only the relevant time window is shown. The calculated ratio of each hydrolyzed metabolite and parent drug is shown.
FIG 4.
Comparative analysis of the ertapenem, meropenem, and imipenem hydrolysis assay results. (A to C) Hydrolysis ratios obtained from the ertapenem, meropenem, and imipenem assays for all isolates, segregated into carbapenemase-positive (Pos) or -negative (Neg) Enterobacteriaceae (Ent) or nonfermenting (NF) isolates and plotted on a log10 scale. Dashed lines represent the optimal threshold values obtained from ROC analysis (ertapenem, 0.0750; meropenem, 0.0450; and imipenem, 0.3150). Bars and whiskers represent means and 95% confidence intervals. (D to F) Receiver-operator curves for the 1-h carbapenem hydrolysis assay performed using 402 bacterial isolates (325 sensitive and 77 multidrug resistant) with 4 μg/ml ertapenem, meropenem, or imipenem.
TABLE 2.
Sensitivity and specificity results for the LC-MS/MS carbapenem hydrolysis assay
| Carbapenem | Threshold | Group of isolatesa | % Sensitivity (95% CIb) | % Specificity (95% CI) |
|---|---|---|---|---|
| Ertapenem | >0.0750 | All | 67.5 (55.9–77.7) | 97.2 (94.8–98.7) |
| Ent | 74.1 (61.0–84.8) | 96.6 (93.2–98.6) | ||
| NF | 36.8 (16.3–61.6) | 98.4 (94.2–99.8) | ||
| Meropenem | >0.0450 | All | 81.1 (70.3–89.3) | 95.7 (92.9–97.7) |
| Ent | 84.2 (72.1–92.5) | 98.5 (95.8–99.7) | ||
| NF | 68.4 (43.5–87.4) | 91.8 (85.4–96.0) | ||
| Imipenem | >0.3150 | All | 96.0 (88.6–99.2) | 95.1 (92.2–97.2) |
| Ent | 98.3 (90.6–100.0) | 95.6 (91.8–98.0) | ||
| NF | 89.5 (66.9–98.7) | 95.9 (90.7–98.7) |
Ent, Enterobacteriaceae; NF, non-glucose-fermenting Gram-negative rods.
CI, confidence interval.
Notably, of a total of six isolates (five Enterobacteriaceae isolates, including four Enterobacter sp. and one Escherichia coli isolate, and one Burkholderia cepacia isolate) that were positive by MHT and negative by PCR for the five major families of carbapenemase genes, all six were positive in the imipenem assay while only one and four were positive in the meropenem and ertapenem assays, respectively. These isolates presumably express one of the less commonly encountered carbapenemase genes.
DISCUSSION
A number of studies have described the use of MS instruments to detect carbapenemase activity. Most of these studies have used MALDI-TOF MS instruments, but they have varied by instrument type, carbapenem used, incubation conditions, and interpretive criteria, among other factors (8–16). Additionally, three studies have reported the use of LC-coupled MS analysis of carbapenemase activity either with a single-stage instrument or as tandem MS with a second-stage quadrupole or orbitrap (7, 11, 20). All of these factors can interact to affect assay performance, and the current study has several unique features compared to the previously published work.
Burckhardt and Zimmermann and Hrabák et al. first reported the use of MALDI-TOF for carbapenemase detection from isolated bacterial colonies, using ertapenem and meropenem, respectively (8, 9). Both reported excellent sensitivity and specificity for carbapenemase detection, but neither included Acinetobacter spp. and OXA carbapenemases. Kempf et al. used imipenem to study 149 bacterial isolates, including 63 OXA-producing A. baumannii isolates, and they reported 100% sensitivity and specificity at 4 h (12). Additional studies using ertapenem and meropenem have also reported good sensitivity and specificity for the detection of OXA activity (11, 13–15, 21).
We found that both ertapenem and meropenem lacked sensitivity for the detection of OXA, IMP, and VIM activity, especially among non-Enterobacteriaceae. This difference is likely due to differences in incubation protocols, including broth, inoculum, and time, rather than differences between LC-MS/MS and MALDI-TOF. Indeed, the analytic sensitivity was 40 ng/ml for all of the methods developed for intact ertapenem and intact and hydrolyzed meropenem. We found that incubation for longer time periods (up to 4 and 8 h) and the use of higher inocula increased the sensitivity of detection with these drugs (data not shown), but imipenem demonstrated superior performance under all conditions tested, including following a 1-h incubation.
The effective use of MALDI-TOF MS for the detection of intact and hydrolyzed β-lactam antibiotics is somewhat surprising. LC-MS/MS is considered the gold standard method for small-molecule detection, and its performance is well suited to the detection of small amounts of compounds present in highly complex matrices, such as serum or bacterial growth broths. To achieve the level of performance reported, carbapenemase assays using MALDI-TOF MS require the use of high bacterial inocula (exceeding 8 McF in most cases), high drug concentrations (10- to 100-fold higher than used here), and simple incubation broths (0.45% saline and/or 20 mM Tris-HCl). Despite these limitations, the performance of MALDI-TOF for carbapenemase detection from isolated colonies seems quite effective.
No studies of MALDI-TOF have investigated the analytic performance of the instruments for the detection of small molecules, and most have relied solely upon the disappearance of intact carbapenem to classify an isolate as positive. Additionally, the effects of ion suppression on carbapenem detection have not been reported for MALDI-TOF, and this could be particularly important when test interpretation relies upon the lack of detection of a compound. In several studies, the matrix peak has been used as an internal standard, but the chemical differences between MALDI matrix and carbapenems are substantial, and differential ionization effects may not be effectively determined without an appropriate internal standard.
Our method has a lower limit of quantitation of 0.02 μg/ml for both intact and hydrolyzed imipenem, and this allows for strict quantitative criteria to be developed for classification as carbapenemase positive or negative. Indeed, ours is the first study to perform ROC analysis to objectively determine optimal, quantitative cutoffs for isolate classification. Kempf et al. used a requirement of either imipenem disappearance or an intact/hydrolyzed ratio of <0.5 to classify isolates, while Wang et al. developed several models to classify isolates (12, 14). Finally, in a study using LC-MS/MS, 18O-isotopically labeled internal standard was generated that allowed a quantitative cutoff to be generated based solely upon the amount of hydrolyzed meropenem detected (20). Regardless of the methodology used, strict quantitative criteria and the use of isotopically labeled internal standards will likely be required for wider implementation of MS-based detection of β-lactamase activity.
MALDI-TOF has been used to detect carbapenemase activity from positive blood cultures, but an enrichment step may be required for the timely detection of β-lactamase activity from other primary specimens, including surveillance swabs, urine cultures, or wound specimens (10). Depending on local trends, carbapenemase expression may contribute little to observed resistance to carbapenems, and traditional phenotypic antimicrobial susceptibility methods would be required to capture all resistance. In this sense, carbapenemase activity assays would simply be an adjunct to traditional testing, reducing the time to detection by 18 h or more.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grant R43AI102541 (M.V.K., M.E.H., and D.R.P.) and a grant from Connecticut Innovations.
We thank Babu Purkayastha of AB Sciex for purified hydrolyzed carbapenem antibiotics and Alidad Mireskandari of M/Z Diagnostics for administrative support.
At the time of the performance of this work, M.V.K. and A.N.Z. were employees of M/Z Diagnostics. T.S.M., M.E.H., and D.R.P. are founders and equity holders of M/Z diagnostics.
Footnotes
Published ahead of print 30 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00547-14.
REFERENCES
- 1.Giske CG, Monnet DL, Cars O, Carmeli Y. 2008. Clinical and economic impact of common multidrug-resistant gram-negative bacilli. Antimicrob. Agents Chemother. 52:813–821. 10.1128/AAC.01169-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marchaim D, Gottesman T, Schwartz O, Korem M, Maor Y, Rahav G, Karplus R, Lazarovitch T, Braun E, Sprecher H, Lachish T, Wiener-Well Y, Alon D, Chowers M, Ciobotaro P, Bardenstein R, Paz A, Potasman I, Giladi M, Schechner V, Schwaber MJ, Klarfeld-Lidji S, Carmeli Y. 2010. National multicenter study of predictors and outcomes of bacteremia upon hospital admission caused by Enterobacteriaceae producing extended-spectrum beta-lactamases. Antimicrob. Agents Chemother. 54:5099–5104. 10.1128/AAC.00565-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mauldin PD, Salgado CD, Hansen IS, Durup DT, Bosso JA. 2010. Attributable hospital cost and length of stay associated with health care-associated infections caused by antibiotic-resistant gram-negative bacteria. Antimicrob. Agents Chemother. 54:109–115. 10.1128/AAC.01041-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ho J, Tambyah PA, Paterson DL. 2010. Multiresistant Gram-negative infections: a global perspective. Curr. Opin. Infect. Dis. 23:546–553. 10.1097/QCO.0b013e32833f0d3e [DOI] [PubMed] [Google Scholar]
- 5.Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 17:1791–1798. 10.3201/eid1710.110655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bush K, Jacoby GA. 2010. Updated functional classification of β-lactamases. Antimicrob. Agents Chemother. 54:969–976. 10.1128/AAC.01009-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peaper DR, Kulkarni MV, Tichy AN, Jarvis M, Murray TS, Hodsdon ME. 2013. Rapid detection of carbapenemase activity through monitoring ertapenem hydrolysis in Enterobacteriaceae with LC-MS/MS. Bioanalysis 5:147–157. 10.4155/bio.12.310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burckhardt I, Zimmermann S. 2011. Using matrix-assisted laser desorption ionization-time of flight mass spectrometry to detect carbapenem resistance within 1 to 2.5 hours. J. Clin. Microbiol. 49:3321–3324. 10.1128/JCM.00287-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hrabák J, Walková R, Studentová V, Chudácková E, Bergerová T. 2011. Carbapenemase activity detection by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 49:3222–3227. 10.1128/JCM.00984-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sparbier K, Schubert S, Weller U, Boogen C, Kostrzewa M. 2012. Matrix-assisted laser desorption ionization-time of flight mass spectrometry-based functional assay for rapid detection of resistance against β-lactam antibiotics. J. Clin. Microbiol. 50:927–937. 10.1128/JCM.05737-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carvalhaes CG, Cayô R, Assis DM, Martins ER, Juliano L, Juliano MA, Gales AC. 2013. Detection of SPM-1-producing Pseudomonas aeruginosa and class D β-lactamase-producing Acinetobacter baumannii isolates by use of liquid chromatography-mass spectrometry and matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 51:287–290. 10.1128/JCM.02365-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kempf M, Bakour S, Flaudrops C, Berrazeg M, Brunel J-M, Drissi M, Mesli E, Touati A, Rolain J-M. 2012. Rapid detection of carbapenem resistance in Acinetobacter baumannii using matrix-assisted laser desorption ionization-time of flight mass spectrometry. PLoS One 7:e31676. 10.1371/journal.pone.0031676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alvarez-Buylla A, Picazo JJ, Culebras E. 2013. Optimized method for Acinetobacter spp carbapenemase detection and identification by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 51:1589–1592. 10.1128/JCM.00181-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L, Han C, Sui W, Wang M, Lu X. 2013. MALDI-TOF MS applied to indirect carbapenemase detection: a validated procedure to clearly distinguish between carbapenemase-positive and carbapenemase-negative bacterial strains. Anal. Bioanal. Chem. 405:5259–5266. 10.1007/s00216-013-6913-2 [DOI] [PubMed] [Google Scholar]
- 15.Lee W, Chung H-S, Lee Y, Yong D, Jeong SH, Lee K, Chong Y. 2013. Comparison of matrix-assisted laser desorption ionization-time-of-flight mass spectrometry assay with conventional methods for detection of IMP-6, VIM-2, NDM-1, SIM-1, KPC-1, OXA-23, and OXA-51 carbapenemase-producing Acinetobacter spp., Pseudomonas aeruginosa, and Klebsiella pneumoniae. Diagn. Microbiol. Infect. Dis. 77:227–230. 10.1016/j.diagmicrobio.2013.07.005 [DOI] [PubMed] [Google Scholar]
- 16.Hoyos-Mallecot Y, Cabrera-Alvargonzalez JJ, Miranda-Casas C, Rojo-Martín MD, Liebana-Martos C, Navarro-Marí JM. 2014. MALDI-TOF MS, a useful instrument for differentiating metallo-β-lactamases in Enterobacteriaceae and Pseudomonas spp. Lett. Appl. Microbiol. 58:325–329. 10.1111/lam.12203 [DOI] [PubMed] [Google Scholar]
- 17.Clinical and Laboratory Standards Institute. 2014. Performance standards for antimicrobial susceptibility testing; 24th informational supplement. CLSI document M100-S24. CLSI, Wayne PA [Google Scholar]
- 18.Tsakris A, Poulou A, Pournaras S, Voulgari E, Vrioni G, Themeli-Digalaki K, Petropoulou D, Sofianou D. 2010. A simple phenotypic method for the differentiation of metallo-beta-lactamases and class A KPC carbapenemases in Enterobacteriaceae clinical isolates. J. Antimicrob. Chemother. 65:1664–1671. 10.1093/jac/dkq210 [DOI] [PubMed] [Google Scholar]
- 19.Black JA, Moland ES, Thomson KS. 2005. AmpC disk test for detection of plasmid-mediated AmpC beta-lactamases in Enterobacteriaceae lacking chromosomal AmpC beta-lactamases. J. Clin. Microbiol. 43:3110–3113. 10.1128/JCM.43.7.3110-3113.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang M, Shen Y, Turko IV, Nelson DC, Li S. 2013. Determining carbapenemase activity with 18O labeling and targeted mass spectrometry. Anal. Chem. 85:11014–11019. 10.1021/ac402627k [DOI] [PubMed] [Google Scholar]
- 21.Hrabák J, Studentová V, Walková R, Zemlicková H, Jakubu V, Chudácková E, Gniadkowski M, Pfeifer Y, Perry JD, Wilkinson K, Bergerová T. 2012. Detection of NDM-1, VIM-1, KPC, OXA-48, and OXA-162 carbapenemases by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 50:2441–2443. 10.1128/JCM.01002-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.