Abstract
Among 217 Aeromonas isolates identified by sequencing analysis of their rpoB genes, the accuracy rates of identification of A. dhakensis, A. hydrophila, A. veronii, and A. caviae were 96.7%, 90.0%, 96.7%, and 100.0%, respectively, by the cluster analysis of spectra generated by matrix-assisted laser desorption ionization–time of flight mass spectrometry.
TEXT
Most human infections caused by Aeromonas spp. have been associated with three species: Aeromonas hydrophila, A. veronii, and A. caviae (1–4). Recently, increasing evidence has demonstrated that A. dhakensis is an important species, which might cause severe soft tissue and bloodstream infections (5, 6). Although clinical infections caused by A. dhakensis have been reported in Taiwan (7), this species have been recovered from aquatic environments and clinical samples globally (8, 9). However, the prevalence of clinical infections caused by A. dhakensis is underestimated due to the possibility of this species being misidentified as A. hydrophila by the phenotype-based identification system (9, 10).
Previous publications suggested that A. hydrophila subsp. dhakensis and A. aquariorum represented the same taxon (11, 12). Although accurate identification of Aeromonas species could be achieved using the nucleotide sequences of housekeeping genes (7, 10), these molecular methods are labor-intensive and time-consuming. Recently, several studies have shown that the matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) can rapidly and accurately identify different Aeromonas species (13–15). However, among aeromonads that are studied using this method, species identification of A. dhakensis rarely has been discussed. In the present study, we assess the performance of two commercially available phenotypic identification systems, the Vitek 2 GN card and Phoenix system NMIC/ID-72 cards (Becton, Dickinson Microbiology Systems), and a MALDI-TOF MS system, the MALDI Biotyper system (microflex LT; Bruker Daltonik GmbH, Bremen, Germany), to identify the clinical Aeromonas species in Taiwan.
We analyzed a total of 217 nonduplicated clinical isolates of Aeromonas obtained from clinical specimens of the patients at the study hospital between 1998 and 2012, as well as 9 reference strains. All the isolates were stored at −70°C until use. Species identification of these clinical isolates was based on the sequence analysis of the partial rpoB gene (16). For the MALDI Biotyper system, the samples were prepared as previously described (15). The rpoB identification and MALDI-TOF procedures are described in the Materials and Methods in the supplemental material.
A. aquariorum, A. hydrophila subsp. dhakensis, A. hydrophila subsp. ranae, A. sanarellii, and A. taiwanensis are not included in the current database for Vitek 2, the Phoenix system, and the MALDI Biotyper. Reference strains of A. aquariorum and A. hydrophila subsp. dhakensis were classified as A. hydrophila in the Vitek 2 and Phoenix systems (Table 1). A. aquariorum strain MDC47T was misidentified as A. caviae (score value, 2.058) and A. hydrophila subsp. dhakensis strain LMG 19562 was identified as A. hydrophila (score value, 2.096). The A. hydrophila subsp. ranae BCRC 17768 strain was identified as A. hydrophila (Table 1).
TABLE 1.
Identification results of nine reference strains of Aeromonas species by two automatic identification systems and MALDI Biotyper system
| Reference Aeromonas strain | Results from MALDI Biotyper system |
Results from Phoenix system |
Results from Vitek 2 system |
|||
|---|---|---|---|---|---|---|
| Aeromonas sp. | Score | Aeromonas sp. | Identity (%) | Aeromonas sp. | Degree of discriminationa | |
| A. aquariorum MDC47T | A. caviae | 2.085 | A. hydrophila | 99 | A. hydrophila/caviae | ED |
| A. hydrophila subsp. dhakensis LMG 19562 | A. hydrophila | 2.096 | A. hydrophila | 99 | A. hydrophila/caviae | ED |
| A. hydrophila ATCC 7966T | A. hydrophila | 2.203 | A. sobria | 96 | A. hydrophila/caviae | ED |
| A. hydrophila BCRC 16704 | A. hydrophila | 2.228 | A. hydrophila | 99 | A. hydrophila/caviae | ED |
| A. hydrophila BCRC 13881 | A. hydrophila | 2.370 | A. veronii | 97 | A. hydrophila/caviae | ED |
| BCRC 17768 (A. hydrophila subsp. ranae Huys et al., 2003) | A. hydrophila | 2.358 | A. sobria | 96 | A. hydrophila/caviae | LD |
| A. veronii biovar sobria ATCC 9071T | A. veronii | 2.165 | A. sobria | 99 | A. hydrophila/caviae | LD |
| A. caviae ATCC 13136T | A. caviae | 2.290 | A. sobria | 94 | A. hydrophila/caviae | ED |
| A. bestiarum ATCC 13444 | A. bestiarum | 2.160 | A. veronii | 99 | A. hydrophila/caviae | ED |
ED, excellent discrimination; LD, low discrimination.
The characteristic spectra generated by the MALDI Biotyper for the six Aeromonas species are shown in Fig. S1 in the supplemental material. The accurate identification rates for species listed in the database (BD 5627) by the MALDI Biotyper system were 93.4% (57/61) for A. veronii, 97.1% (34/35) for A. hydrophila, and 83.9% (56/61) for A. caviae. All A. dhakensis isolates exhibited positive Voges-Proskauer (VP) but negative l-arabinose reactions. In the case of the Phoenix system, the accurate identification rate was 96.7% for 61 A. sobria isolates, 77.1% for 35 isolates of A. hydrophila, and 70.5% for 61 isolates of A. caviae (Table 2). In contrast, 83.4% (181/217) of the Aeromonas isolates were misidentified at the species level by the Vitek 2 system.
TABLE 2.
Performances of routine phenotypic identification method and MALDI Biotyper system for 217 clinical isolates of Aeromonas species
| Aeromonas species identified by gene sequencing (no. of isolates) | Results from MALDI Biotyper system |
Results from Phoenix system |
Results from Vitek 2 system |
|||
|---|---|---|---|---|---|---|
| Species (no. of isolates) | Correct IDa (%) to species level (score, ≥2.0) | Species (no. of isolates) | Correct ID (%) | Species (no. of isolates) | Correct ID (%) | |
| A. dhakensis (58) | A. hydrophila (52) | 0 | A. hydrophila (48) | 0.0 | A. hydrophila/A. caviae (58) | 0 |
| A. caviae (5) | A. veronii (4) | |||||
| A. jandaei (1) | A. sobria (4) | |||||
| Unidentified organism (2) | ||||||
| A. hydrophila subsp. hydrophila (35) | A. hydrophila (34) | 97.1 | A. hydrophila (27) | 77.1 | A. hydrophila/A. caviae (35) | 100 |
| A. caviae (1) | A. veronii (8) | |||||
| A. veronii (61) | A. veronii (57) | 93.4 | A. sobria (59) | 0 | A. hydrophila/A. caviae (21) | 1.6 |
| A. ichthiosmia (4) | Unidentified organisms (2) | A. sobria (34) | ||||
| A. veronii (1) | ||||||
| A. sobria/A. veronii (5) | ||||||
| A. caviae (61) | A. caviae (56) | 91.8 | A. caviae (43) | 70.5 | A. hydrophila/A. caviae (61) | 0 |
| A. hydrophila (4) | A. hydrophila (9) | |||||
| A. jandaei (1) | A. veronii (5) | |||||
| A. sobria (2) | ||||||
| Unidentified organism (2) | ||||||
| A. sanarellii (1) | A. caviae (1) | 0 | A. veronii | 0 | A. hydrophila/A. caviae | 0 |
| A. taiwanensis (1) | A. caviae (1) | 0 | A. veronii | 0 | A. hydrophila/A. caviae | 0 |
ID, identification.
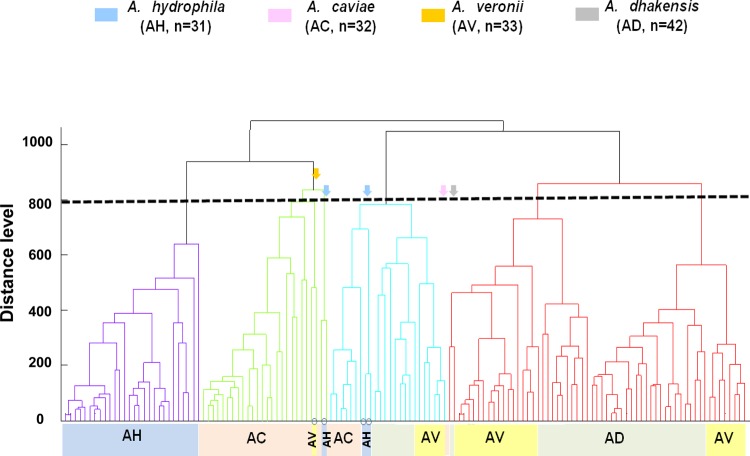
The dendrogram obtained from the MALDI Biotyper data of 123 genetically well characterized isolates is shown in Fig. 1, which shows five cluster groups with a default critical distance level of 850. These cluster groups are in accordance with those established by species identification by rpoB sequencing, with some variations. A. dhakensis was closer to A. veronii than other species in the MALDI-TOF dendrogram, with dividing branches linked at a distance level of 700.
FIG 1.
Principal component analysis (PCA) dendrogram generated by MALDI Biotyper mass spectra for 123 isolates of four Aeromonas species, including 118 clinical isolates and five reference strains. The four colors indicate the four Aeromonas species identified by gene sequencing analysis. The arrows indicate five isolates located in the incorrect clusters of species identified by gene sequencing analysis.
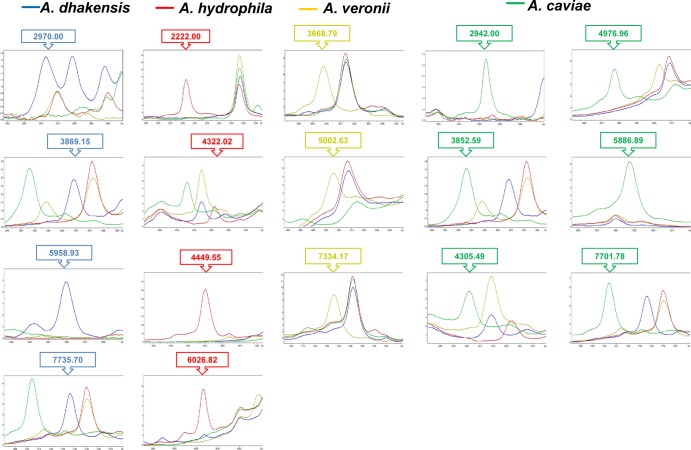
The above-mentioned 123 isolates used for dendrogram establishment were analyzed for specific signals by clustering analysis (Fig. 2). Another 100 isolates from four Aeromonas species were evaluated for external validation. The accuracy identification rates by MALDI-TOF for A. dhakensis, A. hydrophila, A. veronii, and A. caviae were 96.7%, 90.0%, 96.7%, and 100.0%, respectively (Table 3).
FIG 2.
Clustering analysis of MALDI Biotyper system results for four Aeromonas species. The signals generated by ClinProTools with the genetic algorithm were specific for identifying varied Aeromonas species: 2,970.00, 3,869.15, 5,958.93, and 7,735.70 m/z in A. dhakensis; 2,222.00, 4,322.02, 4,449.55, and 6,026.82 m/z in A. hydrophila isolates; 3668.79, 5,002.63, and 7,334.17 m/z in A. veronii; and 2,942, 3,852.59, 4,305.49, 4,976.96, 5,886.89, and 7,701.78 m/z in A. caviae.
TABLE 3.
Results of external validation of 100 isolates of four Aeromonas species by MALDI Biotyper system
| Aeromonas species | No. of isolates | No. (%) of isolates with indicated Aeromonas species by external validation |
|||
|---|---|---|---|---|---|
| A. dhakensis | A. hydrophila | A. veronii | A. caviae | ||
| A. dhakensis | 30 | 29 (96.7)a | 1 (3.3) | ||
| A. hydrophila subsp. hydrophila | 10 | 9 (90) | 1 (10) | ||
| A. veronii | 30 | 1 (3.3) | 29 (96.7) | ||
| A. caviae | 30 | 0 (0) | 30 (100) | ||
Bold type indicates rates of correct identification by external validation results.
Since there is no information regarding A. dhakensis in the database, some A. dhakensis isolates determined by gene sequencing are identified as A. hydrophila or A. caviae using the current MALDI-TOF database. The discrimination power for differentiating A. dhakensis from other species in the cluster analysis could be increased if the database were updated by the novel spectra of A. dhakensis isolates generated from this study.
Nearly all (97.0%) of Aeromonas isolates were correctly identified to the species level using the new model we generated. The discrepancy rate (3/100 [3%]) was comparable with the rate of 8.6% (12/139) reported by Lamy et al. (17). The advantages of identifying Aeromonas species by MALDI-TOF are enhanced compared with those of using commercial phenotypic systems. In the present study, the concordance rate of Vitek 2 GN and Phoenix NMIC/ID-72 at the species level to MALDI-TOF was 16.6% (36/217) and 59.4% (129/217), respectively. In contrast, in the study by Lamy et al. (17), in which A. dhakensis isolates were not included, the concordance rate of Vitek 2 GN and Phoenix NMIC/ID-72 was 82.7% and 73.5%, respectively (17).
The correct identification of A. dhakensis among Aeromonas isolates is clinically important for optimizing antimicrobial therapy, because a significant proportion of A. dhakensis isolates in Taiwan were found to carry resistance genes (18), such as AmpC-like β-lactamase, which is responsible for cephalosporin resistance (19), and cphA metallo-β-lactamase, which is responsible for carbapenem resistance (20). The MIC data determined by the Phoenix NMIC/ID-72 system are reported to have a good correlation with the results of the CLSI reference broth microdilution method in Aeromonas isolates (21, 22). We found the resistance rates of ertapenem and gentamicin among A. dhakensis isolates to be 12.1% and 6.9%, respectively (see Table S1 in the supplemental material), highlighting its potential for antimicrobial resistance.
The dendrogram in Fig. 1 shows the heterogeneity of the protein “fingerprint” in A. dhakensis and A. veronii isolates. The MALDI-TOF MS method has the potential to group the isolates below the species level. However, the MALDI-TOF dendrogram does not have genetically discriminative information the way other molecular typing methods, such as multilocus sequence typing or pulsed-field gel electrophoresis, do. Further studies comparing the results between MALDI-TOF and other methods at the subspecies level are warranted.
In summary, MALDI-TOF MS might have the potential to be incorporated into the routine microbiology laboratory workflow for rapid identification of aeromonads.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the grants from National Science Council, Taiwan (NSC 99-2628-B-006-014-MY3 and NSC 102-2314-B-006-055), and National Cheng Kung University Hospital (NCKUH-10307014).
Footnotes
Published ahead of print 23 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01025-14.
REFERENCES
- 1.Chao CM, Lai CC, Tang HJ, Ko WC, Hsueh PR. 2013. Biliary tract infections caused by Aeromonas species. Eur. J. Clin. Microbiol. Infect. Dis. 32:245–251. 10.1007/s10096-012-1736-1 [DOI] [PubMed] [Google Scholar]
- 2.Chuang HC, Ho YH, Lay CJ, Wang LS, Tsai YS, Tsai CC. 2011. Different clinical characteristics among Aeromonas hydrophila, Aeromonas veronii biovar Sobria and Aeromonas caviae monomicrobial bacteremia. J. Korean Med. Sci. 26:1415–1420. 10.3346/jkms.2011.26.11.1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chao CM, Lai CC, Gau SJ, Hsueh PR. 2013. Skin and soft tissue infection caused by Aeromonas species in cancer patients. J. Microbiol. Immunol. Infect. 46:144–146. 10.1016/j.jmii.2013.02.006 [DOI] [PubMed] [Google Scholar]
- 4.Chao CM, Lai CC, Tang HJ, Ko WC, Hsueh PR. 2012. Skin and soft-tissue infections caused by Aeromonas species. Eur. J. Clin. Microbiol. Infect. Dis. 32:543–547. 10.1007/s10096-012-1771-y [DOI] [PubMed] [Google Scholar]
- 5.Ko WC, Chuang YC. 1995. Aeromonas bacteremia: review of 59 episodes. Clin. Infect. Dis. 20:1298–1304. 10.1093/clinids/20.5.1298 [DOI] [PubMed] [Google Scholar]
- 6.Wu CJ, Wu JJ, Yan JJ, Lee HC, Lee NY, Chang CM, Shih HI, Wu HM, Wang LR, Ko WC. 2007. Clinical significance and distribution of putative virulence markers of 116 consecutive clinical Aeromonas isolates in southern Taiwan. J. Infect. 54:151–158. 10.1016/j.jinf.2006.04.002 [DOI] [PubMed] [Google Scholar]
- 7.Chen PL, Wu CJ, Chen CS, Tsai PJ, Tang HJ, Ko WC. 2013. A comparative study of clinical Aeromonas dhakensis and Aeromonas hydrophila isolates in southern Taiwan: A. dhakensis is more predominant and virulent. Clin. Microbiol. Infect., in press. 10.1111/1469-0691.12456 (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 8.Esteve C, Alcaide E, Blasco MD. 2012. Aeromonas hydrophila subsp. dhakensis isolated from feces, water and fish in Mediterranean Spain. Microbes Environ. 27:367–373. 10.1264/jsme2.ME12009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aravena-Román M, Harnett GB, Riley TV, Inglis TJ, Chang BJ. 2011. Aeromonas aquariorum is widely distributed in clinical and environmental specimens and can be misidentified as Aeromonas hydrophila. J. Clin. Microbiol. 49:3006–3008. 10.1128/JCM.00472-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morinaga Y, Yanagihara K, Eugenin FL, Beaz-Hidalgo R, Kohno S, Figueras Salvat MJ. 2013. Identification error of Aeromonas aquariorum: A causative agent of septicemia. Diagn. Microbiol. Infect. Dis. 76:106–109. 10.1016/j.diagmicrobio.2013.01.019 [DOI] [PubMed] [Google Scholar]
- 11.Beaz-Hidalgo R, Martinez-Murcia A, Figueras MJ. 2013. Reclassification of Aeromonas hydrophila subsp. dhakensis Huys et al. 2002 and Aeromonas aquariorum Martínez-Murcia et al. 2008 as Aeromonas dhakensis sp. nov. comb nov. and emendation of the species Aeromonas hydrophila. Syst. Appl. Microbiol. 36:171–176. 10.1016/j.syapm.2012.12.007 [DOI] [PubMed] [Google Scholar]
- 12.Martinez-Murcia AJ, Monera A, Saavedra MJ, Oncina R, Lopez-Alvarez M, Lara E, Figueras MJ. 2011. Multilocus phylogenetic analysis of the genus Aeromonas. Syst. Appl. Microbiol. 34:189–199. 10.1016/j.syapm.2010.11.014 [DOI] [PubMed] [Google Scholar]
- 13.Bizzini A, Greub G. 2010. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry, a revolution in clinical microbial identification. Clin. Microbiol. Infect. 16:1614–1619. 10.1111/j.1469-0691.2010.03311.x [DOI] [PubMed] [Google Scholar]
- 14.Donohue MJ, Smallwood AW, Pfaller S, Rodgers M, Shoemaker JA. 2006. The development of a matrix-assisted laser desorption/ionization mass spectrometry-based method for the protein fingerprinting and identification of Aeromonas species using whole cells. J. Microbiol. Methods 65:380–389. 10.1016/j.mimet.2005.08.005 [DOI] [PubMed] [Google Scholar]
- 15.Lamy B, Kodjo A, Laurent F, ColBVH Study Group 2011. Identification of Aeromonas isolates by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Diagn. Microbiol. Infect. Dis. 71:1–5. 10.1016/j.diagmicrobio.2011.04.014 [DOI] [PubMed] [Google Scholar]
- 16.Küpfer M, Kuhnert P, Korczak BM, Peduzzi R, Demarta A. 2006. Genetic relationships of Aeromonas strains inferred from 16S rRNA, gyrB and rpoB gene sequences. Int. J. Syst. Evol. Microbiol. 56:2743–2751. 10.1099/ijs.0.63650-0 [DOI] [PubMed] [Google Scholar]
- 17.Lamy B, Laurent F, Verdier I, Decousser JW, Lecaillon E, Marchandin H, Roger F, Tigaud S, de Montclos H, colBVH Study Group. Kodjo A. 2010. Accuracy of 6 commercial systems for identifying clinical Aeromonas isolates. Diagn. Microbiol. Infect. Dis. 67:9–14. 10.1016/j.diagmicrobio.2009.12.012 [DOI] [PubMed] [Google Scholar]
- 18.Chen PL, Ko WC, Wu CJ. 2012. Complexity of β-lactamases among clinical Aeromonas isolates and its clinical implications. J. Microbiol. Immunol. Infect. 45:398–403. 10.1016/j.jmii.2012.08.008 [DOI] [PubMed] [Google Scholar]
- 19.Wu CJ, Wang HC, Chen PL, Chang MC, Sunny Sun H, Chou PH, Ko WC. 2013. AQU-1, a chromosomal class C β-lactamase, among clinical Aeromonas dhakensis isolates: distribution and clinical significance. Int. J. Antimicrob. Agents 42:456–461. 10.1016/j.ijantimicag.2013.08.002 [DOI] [PubMed] [Google Scholar]
- 20.Wu CJ, Chen PL, Wu JJ, Yan JJ, Lee CC, Lee HC, Lee NY, Chang CM, Lin YT, Chiu YC, Ko WC. 2012. Distribution and phenotypic and genotypic detection of a metallo-β-lactamase, CphA, among bacteraemic Aeromonas isolates. J. Med. Microbiol. 61:712–719. 10.1099/jmm.0.038323-0 [DOI] [PubMed] [Google Scholar]
- 21.Clinical and Laboratory Standards Institute (CLSI). 2010. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria; approved guideline, 2nd ed. CLSI M45–A2. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 22.Liu ZK, Ling TK, Cheng AF. 2005. Evaluation of the BD Phoenix automated microbiology system for identification and antimicrobial susceptibility testing of common clinical isolates. Med. Princ. Pract. 14:250–254. 10.1159/000085744 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.