Abstract
External quality assessment (EQA) for the Xpert MTB/RIF assay is part of the quality system required for clinical and laboratory practice. Five newly developed EQA panels that use different matrices, including a lyophilized sample (Vircell, Granada, Spain), a dried tube specimen (CDC), liquid (Maine Molecular Quality Control, Inc. [MMQCI], Scarborough, ME), artificial sputum (Global Laboratory Initiative [GLI]), and a dried culture spot (National Health Laboratory Services [NHLS]), were evaluated at 11 GeneXpert testing sites in South Africa. The panels comprised Mycobacterium tuberculosis complex (MTBC)-negative, MTBC-positive (including rifampin [RIF] susceptible and RIF resistant), and nontuberculosis mycobacterial material that was inactivated and safe for transportation. Twelve qualitative and quantitative variables were scored as acceptable (1) or unacceptable (0); the overall panel performance score for the Vircell, CDC, GLI, and NHLS panels was 9 of 12, while the MMQCI panel scored 6 of 12 (owing to the need for cold chain maintenance). All panels showed good compatibility with Xpert MTB/RIF testing, and none showed PCR inhibition. The use of a liquid or dry matrix did not appear to be a distinguishing criterion, as both matrices had reduced scores on insufficient volumes, a need for extra consumables, and the ability to transfer to the Xpert MTB/RIF cartridge. EQA is an important component of the quality system required for diagnostic testing programs, but it must be complemented by routine monitoring of performance indicators and instrument verification. This study aims to introduce EQA concepts for Xpert MTB/RIF testing and evaluates five potential EQA panels.
INTRODUCTION
The endorsement of the Xpert MTB/RIF (Xpert) assay by the World Health Organization (WHO) in December 2010 (see http://www.who.int/tb/laboratory/mtbrifrollout/en/index.html) led to an unprecedented commitment by tuberculosis (TB) programs and donors worldwide to implement this technology in efforts to improve the diagnosis of TB and initiate a continuum of care to reduce the burden of the disease. South Africa leads the implementation with more than 2,400,000 sputum specimens tested using the Xpert MTB/RIF assay (as of November 2013) on 284 instruments in 207 smear microscopy testing laboratories countrywide, with a further 6 Gx 80 instrument placements in the pipeline. The South African experience to date has highlighted several areas for program strengthening realized during the national phased implementation, including the need for improved technical and clinical training, the need for laboratory information systems for data management, and the need for quality system components such as an external quality assessment (EQA) program, to name a few (1, 2). The quality system needs to encompass the preanalytical, analytical, and postanalytical processes to ensure ongoing test quality, and an EQA program is one of the internationally recognized tools for this purpose (3, 4). In this context for Xpert MTB/RIF, a panel must require intact Mycobacterium tuberculosis that can be processed outside a biosafety level 3 (BSL3) (or even BSL2) laboratory infrastructure.
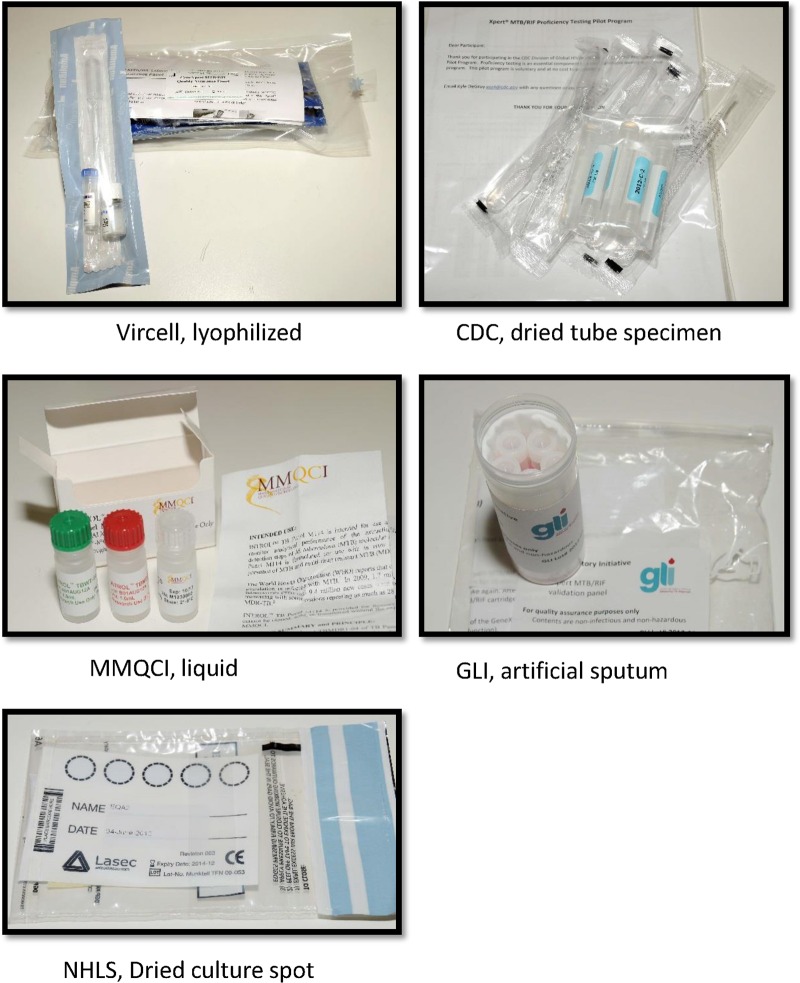
Currently, no EQA program exists for the Xpert MTB/RIF test, which represents a paradigm shift in molecular testing for TB. A methodology using dried culture spots (DCS) was developed in South Africa for verifying GeneXpert (5) instruments on installation to ensure that the instrumentation is fit for purpose and has been used as an EQA for a clinical trial with the AIDS Clinical Trial Group (ACTG) in sites in Africa, Brazil, Peru, and the United States. In this study, we investigated the performance of 5 EQA panels being developed for the Xpert MTB/RIF test. The five EQA panels consisting of various matrices developed specifically for the Xpert EQA were donated from both commercial and noncommercial manufacturers (Fig, 1). The DCS matrix was developed in South Africa for the National Health Laboratory Services (NHLS) National Priority Program (NPP) (5), the dried tube specimen (DTS) matrix was developed by the Centers for Disease Control and Prevention (CDC) (6), the artificial sputum matrix was developed by the Global Laboratory Initiative (GLI)/technical expert advisory group to the WHO and the Stop TB Partnership, the lyophilized sample matrix was developed by Vircell (Granada, Spain) in conjunction with the Foundation for Innovative New Diagnostics (FIND), and the liquid matrix specimens were developed by Maine Molecular Quality Controls, Inc. (MMQCI) (Scarborough, ME) (see http://www.mmqci.com/qc-m110.php). The performance evaluations of all five panels were conducted in South Africa at 11 Xpert testing sites. One reference laboratory tested each of the panels, and the results served as the reference standard against which the quantitative results from the 11 Xpert testing sites were compared.
FIG 1.
Representation of the five EQA panels designed for the Xpert MTB/RIF assay.
MATERIALS AND METHODS
Panel preparation and distribution.
Examples of the five panels are illustrated in Fig. 1. The NHLS DCS panel consisted of a characterized Mycobacterium tuberculosis complex (MTBC) rifampin (RIF)-susceptible isolate (H37Rv) grown in bulk single-cell cultures which had been inactivated using an Xpert MTB/RIF sample reagent (SR) buffer (a kit component), followed by quantification by flow cytometry and spotted with blue dye onto Whatman 903 filter cards (Lasec or LabMate, South Africa) (5). For the present study, the same technique was used to create an EQA panel consisting of 4 samples, 1 MTBC RIF susceptible, 1 MTBC RIF resistant, 1 nontuberculosis mycobacterium (NTM), and 1 negative (water), each spotted onto a separate Whatman filter paper card. The CDC panels were created by dilutions from known cultures grown in mycobacterial growth indicator tube (MGIT) (Becton Dickinson, Sparks, MD) media followed by chemical inactivation with Xpert MTB/RIF SR, glass bead disruption, and drying in a class II biosafety cabinet (6). This CDC panel consisted of 4 samples, 1 MTBC RIF susceptible, 2 MTBC RIF resistant, and 1 negative. The GLI panels were composed of artificial sputum containing heat-killed bacilli and consisted of 4 samples, 1 MTBC RIF susceptible, 2 MTBC RIF resistant, and 1 negative. A lyophilized panel from Vircell was developed in conjunction with the Foundation for Innovative New Technologies (FIND) and consisted of 4 samples, 1 MTBC RIF susceptible, 1 MTBC RIF resistant, 1 NTM, and 1 negative. The MMQCI liquid panel was composed of transfected MTBC DNA in Escherichia coli chemically fixed and killed. The MMQCI panel consisted of 3 samples, 1 MTBC RIF susceptible, 1 MTBC RIF resistant, and 1 negative.
All panels were centrally received and repackaged according to WHO safety protocols (see http://www.who.int/ihr/infectious_substances/en/) using gas-impermeable and sealed transport bags and boxes and couriered together to a convenience sample of 11 randomly selected NHLS Xpert laboratories. Testing sites represented a variety of testing volumes and levels of the TB laboratory network. All panels were transported at room temperature except the MMQCI panel which required cold chain maintenance. The Xpert sites varied in geography and service level and were visited prior to the commencement of the study to establish the feasibility of use and willingness of participants. The reference laboratory was the Research Diagnostic Laboratory in the Department of Molecular Medicine and Hematology, University of the Witwatersrand, Medical School in Johannesburg. To blind study participants to the panel compositions, barcodes were generated, encoding the panel matrix as well as the contents of each sample. Each panel box also contained panel-specific testing instructions produced by the developer. Extra consumables were required for the NHLS panel, including a universal container/Nunc tube/sputum jar into which the perforated spot is placed, and a pipette tip. An extra pipette was also required for the NHLS, GLI, and MMQCI panels to dispense the SR buffer into the containers. These were not supplied by the EQA program as they are standard laboratory consumables.
Technologists were provided bench aids and received training on how to upload the results to TBGx Monitor (www.tbgxmonitor.com). TBGx Monitor is a web-based application developed by the Department of Molecular Medicine and Hematology of the NHLS and the University of the Witwatersrand team to manage their DCS verification program of all GeneXpert systems implemented in the field (5). An additional tab was included to allow the sites to upload the comma separated value (CSV) file format generated by the GeneXpert software after each EQA sample was analyzed on the GeneXpert instruments. The sites were also provided with a testing schedule, describing the order in which the tests were to be performed, to minimize any confounding variables. A qualitative questionnaire accompanied the five panels sent to each site and contained questions relating to the condition of the material, label clarity, ease of operation, sufficient material, and ease of protocol use. A general comments section allowed for narrative capture. The processing of the panels took place over 2 weeks, with three panels being performed in the first week and the remaining two panels in the second. This was done in order to minimize the interruption of daily workflow at these routine testing sites. The GeneXpert modules used for testing each sample were randomly assigned by the GeneXpert software and therefore not determined by the operators.
Panel evaluation and scoring.
A measure of overall panel performance was determined using a scoring system across the qualitative and quantitative variables. A score of 1 was awarded for acceptable performance and a score of 0 was awarded where issues were documented. Performance was calculated based on 10 qualitative and 2 quantitative evaluation criteria and summarized as the most frequent score reported across 11 testing sites. The qualitative criteria (also termed discrete variables) were (i) correct results reported for each panel compared to the results for the sample performed at the reference center, (ii) instrument errors related to sample volume (too little volume/too viscous/fluid transfer failed) testing, such as errors 5006 and 5007, (iii) the need for cold chain maintenance during transportation of a panel and therefore the need for additional packaging, (iv) the need for extra consumables to perform a panel test, (v) questionnaire observations related to the condition of the goods received, (vi) the clarity of the standard operating procedures (SOPs), (vii) ease of opening samples, (viii) the ease of handling samples, (ix) the ease of rehydration of sample, and (x) the ease of transferring samples to the Xpert testing cartridges. For example, (i small packaging and therefore a lower courier cost received a score of 1, while large packaging and a higher courier cost received a 0, (ii) the need for cold chain maintenance was scored as 0, while panels with no cold chain requirement received a score of 1, and (iii) in the case of the quantitative variables, a cycle threshold (CT) standard deviation (SD) similar to that of the majority group was awarded a score of 1, and an outlier (higher than the majority group) was awarded 0 for any probe (A to E); an invalid result was scored 0. These criteria may need to be refined with time and experience. In addition to the qualitative reported result (M. tuberculosis detected or M. tuberculosis not detected) generated by the Xpert MTB/RIF assay, a semiquantitative range (very low, low, medium, or high) based on the CT value was also reported (7). The CT is the threshold at which the fluorescence from the hybridization probes increases as the target region is amplified. This is further explained by the assay's molecular characteristics; Xpert is an automated molecular technology that amplifies the rpoB gene of Mycobacterium tuberculosis and detects this target using molecular beacons (7, 8). Resistance to rifampin (RIF) is determined by delayed or drop-out hybridization of the molecular beacon probes (5 probes span the rpoB target, and 1 probe hybridizes to the internal control). Delayed hybridization occurs when there is a >4 CT difference (for assay version G4) between any probes and dropout is reported when the CT is zero (9). The mean CT value and the SD of the CT for each probe (A to E) of each panel were calculated. The amount of variability in the CT values of each probe (A to E) across the panels was used as a quantitative evaluation criterion as a potential measure of bacterial consistency in each EQA matrix. For example, a qualitative Xpert result of RIF resistance might be generated by a delayed or a dropout probe(s). The final outcome result of RIF resistance is the same, but the SD for values ranging from 0 (dropout) to 40 (the highest CT value that could indicate delayed hybridization) would be high.
A sample from each panel was also processed at the reference laboratory and referred to as the reference result. Bar charts were used to visualize the SD of each probe set across all panels. PCR inhibition of the internal control (sample processing control [SPC]) was also reported if any sample yielded an invalid result. Quantitative criteria for scoring included (i) probe CT SD and (ii) PCR inhibition of any sample. Several other observations that would be considered important in managing a national EQA program but were not used to compare panel performance in this study were also noted during the study. These were number and proportion of returned results (categorized by results uploaded to TBGx Monitor, results returned by email, results received as paper returns, and results not returned) and issues such as unreadable samples (barcode scan errors) or incorrect labels.
Statistical analysis.
Summary descriptives and performance indicators were tabulated for each EQA panel. The means and standard deviations (SD) were calculated for the CT quantitative variable for each probe (A to E, and for the internal control [SPC]). The consensus (across all panels) SD of the CT was determined through visualization on a bar chart and used to separate the panel CT values into the consensus and outlier groups. MS Excel was used for all calculations. The performance scores are provided in binary format.
RESULTS
Panel compositions and general observations.
MTBC-positive RIF-resistant, MTBC-positive RIF-susceptible, and negative samples were included in all panels. The NHLS and Vircell panels were the only panels to also include an NTM sample. Two of the five panels (MMQCI and GLI) used a liquid matrix, and the remaining three (Vircell, CDC, and NHLS) used dried matrix formats. MMQCI was the only panel with 3 samples (only 3 were purchased for this study), while all the other panels consisted of 4 samples (Table 1). Results were returned for 96% (201/209) of the samples. All (100%) results were returned for the CDC, MMQCI, and NHLS panels, but results for only 91% of the GLI panels and 82% of the Vircell panels were returned. The “no returns” came from two sites which could not read the two-dimensional (2D) barcode labels affixed to the samples. All participants were encouraged to make use of the TBGx Monitor website for submission of their CSV files; however, only 33% of the MMQCI CSV files and 40 to 43% of the CSV files from all other panels were submitted online, suggesting some difficulty in use of TBGx Monitor. This was similarly noted in the narrative comments on the questionnaire. The Vircell panel had 4 unreadable 2D barcode scans. Two sites would have yielded nonconformances in an EQA program; one site did not return the questionnaire, and one site mixed all the labels across the panels and would have generated incorrect results. The latter samples were correctly identified through their CSV files and therefore were included in the data analysis.
TABLE 1.
Description of the panels and summary of qualitative data
| Characteristic | Vircell panel | CDC panel | MMQCI panel | GLI panel | DCS panel |
|---|---|---|---|---|---|
| Matrix | Lyophilized | Dried tube specimen | Liquid (E. coli) | Artificial sputum | Dried culture spot |
| Sample(s) included in panel | |||||
| Negative | 1 | 1 | 1 | 1 | 1 |
| M. tuberculosis negative, NTM | 1 | 0 | 0 | 0 | 1 |
| M. tuberculosis positive, RIF sensitive | 1 | 1 | 1 | 1 | 1 |
| M. tuberculosis positive, RIF resistant | 1 | 2 | 1 | 2 | 1 |
| No. of testing sitesa | 12 | 12 | 12 | 12 | 12 |
| No. of samples sent to sites | 44 | 44 | 33 | 44 | 44 |
| Return of results | |||||
| Total returned results (no. [%]) | 36 (81.8) | 44 (100) | 33 (100) | 40 (90.9) | 44 (100) |
| CSV files uploaded onto TBGx Monitor (no. [%]) | 18 (40.9) | 19 (43) | 11 (33) | 18 (40.9) | 18 (40.9) |
| CSV returned by e-mail that could be uploaded (no. [%]) | 16 (36.4) | 16 (36.4) | 15 (45) | 16 (36.4) | 20 (45) |
| Paper returns (no. [%]) | 2 (4.5) | 9 (20) | 7 (21) | 6 (13.6) | 6 (13.6) |
| No returns (no. [%]) | 8 (18) | 0 | 0 | 4 (9) | 0 |
| Qualitative results | |||||
| No. of true negatives/total no. | 17/18 | 11/11 | 10/10 | 10/10 | 10/10 |
| No. of false negatives/total no. | 1/18b | 0/11 | 0/10 | 0/10 | 0/10 |
| True M. tuberculosis, RIF sensitive (no. detected/total no.) | 8/8 | 11/11 | 10/10 | 10/10 | 11/11 |
| False M. tuberculosis, RIF sensitive (no. detected/total no.) | 0/8 | 0/11 | 0/10 | 0/10 | 0/11 |
| True M. tuberculosis, RIF resistant (no. detected/total no.) | 9/9 | 22/22 | 10/11 | 18/18 | 11/11 |
| False M. tuberculosis, RIF resistant (no. detected/total no.) | 0/9 | 0/22 | 1/11c | 0/18 | 0/11 |
| True negative NTM (no. detected/total no. | NAd | NA | NA | NA | 11/11 |
| False negative, NTM (no. detected/total no.) | 0/11 | ||||
| Errors | 1e | 0 | 2f | 2g | 1h |
Eleven smear microscopy sites now performing Xpert MTB/RIF, and 1 reference site.
M. tuberculosis positive, RIF sensitive.
M. tuberculosis not detected.
NA, not applicable.
Error 2008 (instrument).
Error 5006 (insufficient volume).
Errors 2008 (instrument) and 5006 (insufficient volume).
Error 5007 (insufficient volume).
Qualitative panel performance.
The expected results were observed for the CDC, GLI, and NHLS panels. For the Vircell panels, one negative sample yielded a false-positive result and for the MMQCI panels, one positive result (with RIF resistance) was missed. For the CDC panels, no GeneXpert run errors were observed. The Vircell and GLI panels each had one sample where the instrument generated a 2008 error, which relates to syringe pressure and therefore is not related to the tested material. The MMQCI panels showed two insufficient volume errors (5007 and 5006) despite being a liquid matrix, and the GLI and NHLS panels each had 1 insufficient volume error (Table 1).
Of the qualitative questionnaire responses, the GLI panel had no issues, the NHLS panel had 1 issue (“not easy to open”), and the CDC panel had 2 issues (“not easy to open” and “not easy to handle”). The Vircell and MMQCI panels had three issues each, namely “not easy to transfer,” “material insufficient to test,” and “protocol not easy to follow.” The most expensive panel to transport was the MMQCI panel as this required cold chain maintenance. This was also the heaviest and largest package, due to the addition of cold packs. The courier costs depended on weight and distance and on whether road and/or air transportation was needed. The laboratory to which delivery was most expensive was the most remote, requiring both air and road transportation.
Quantitative performance of panels.
Table 2 presents the average and standard deviation (SD) of each probe's CT and SPC CT values across all panels. The sample size across panels per probe analyzed ranged from 7 to 16. The SD values for each probe across all panels are represented in two bar charts in Fig. 2 for M. tuberculosis-detected, RIF-resistant samples and RIF-susceptible samples. The Vircell followed by the MMQCI panels had the lowest SD on all probes compared to the other panels. The panel with the most variability (SD greater than the majority consensus SD of all panels [SD >2.3] for the RIF-susceptible samples was the CDC panel [probes A to E, SD >2.3] followed by the GLI panel [probes A and B SD >2.3]). The SD on the probes of the RIF-resistant sample was highest across 4 probes for the GLI panel (probes A to D, SD >2.3) and 1 probe for CDC panel (probe C, SD >2.3). Although the CDC, GLI, and NHLS panels showed more variability on their SPC, no invalid results (SPC inhibition) were reported for any sample across all panels, indicating that the panel materials did not cause any PCR inhibition.
TABLE 2.
Qualitative results describing the mean values and SD per probe compared to those of the reference
| Organism detected and probes used | Qualitative result for: |
Reference mean for: |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Vircell panel | CDC panel | MMQCI panel | GLI panel | DCS panel | Vircell panel | CDC panel | MMQCI panel | GLI panela | DCS panel | |
| M. tuberculosis, RIF sensitive | ||||||||||
| No. | 7 | 10 | 8 | 8 | 10 | 1 | 1 | 1 | 1 | 1 |
| Probe A (mean [SD]) | 19.6 (1.7) | 22.0 (2.9) | 24.1 (2.1) | 15.7 (2.3) | 19.4 (2.1) | 20.4 | 22.2 | 20.9 | 12.2 | 21.9 |
| Probe B (mean [SD]) | 21.4 (1.6) | 23.6 (3.0) | 25.5 (2.1) | 17.6 (2.5) | 21.0 (2.2) | 21.8 | 23.5 | 22.5 | 13.5 | 22.9 |
| Probe C (mean [SD]) | 20.0 (1.8) | 22.3 (3.0) | 24.5 (2.1) | 16.0 (2.2) | 19.8 (2.2) | 20.7 | 22.6 | 21.2 | 12.4 | 22.0 |
| Probe D (mean [SD]) | 21.2 (2.0) | 23.5 (2.8) | 25.5 (1.9) | 17.4 (2.3) | 20.8 (2.2) | 21.6 | 23.5 | 22.3 | 13.3 | 23.0 |
| Probe E (mean [SD]) | 20.9 (1.6) | 23.3 (2.8) | 25.2 (2.3) | 17.0 (2.2) | 20.8 (1.9) | 22.3 | 23.9 | 22.6 | 13.9 | 23.7 |
| SPC (mean [SD]) | 24.8 (2.1) | 26.8 (3.2) | 25.7 (2.1) | 27.8 (2.1) | 27.6 (3.1) | 26.7 | 24.3 | 26.6 | 28.7 | 30.5 |
| M. tuberculosis, RIF resistant | ||||||||||
| No. | 8 | 8 | 8 | 16a | 10 | 1 | 1 | 1 | 2 | 1 |
| Probe A (mean [SD]) | 19.3 (2.4) | 20.9 (1.8) | 0.0 (0.0) | 16.4 (4.9) | 20.9 (2.6) | 18.4 | 33.9 | 0.0b | 12.0 | 17.4 |
| 11.7 | ||||||||||
| Probe B (mean [SD]) | 20.9 (2.5) | 21.9 (1.8) | 0.0 (0.0) | 18.1 (4.9) | 22.7 (2.6) | 19.7 | 33.7 | 0.0b | 12.2 | 19.0 |
| 13.2 | ||||||||||
| Probe C (mean [SD]) | 19.9 (2.5) | 19.9 (21.3) | 22.7 (2.3) | 16.9 (5.0) | 21.2 (2.7) | 18.9 | 0.0b | 20.4 | 12.5 | 17.7 |
| 12.3 | ||||||||||
| Probe D (mean [SD]) | 21.1 (2.5) | 22.5 (1.5) | 0.0 (0.0) | 18.1 (4.7) | 0.0 (0.0) | 20.2 | 33.8 | 0.0b | 13.4 | 0.0b |
| 13.1 | ||||||||||
| Probe E (mean [SD]) | 0.0 (0.0) | 22.3 (1.9) | 23.2 (2.2) | 0.0 (0.0) | 22.1 (2.6) | 0.0b | 35.6 | 21.6 | 0.0b | 18.9 |
| 0.0b | ||||||||||
| SPC (mean [SD]) | 26.4 (3.5) | 26.0 (2.3) | 25.0 (2.0) | 29.4 (3.3) | 27.4 (3.4) | 23.6 | 31.7 | 28.3 | 26.9 | 27.0 |
| 28.4 | ||||||||||
| M. tuberculosis, RIF resistant | ||||||||||
| No. | NAc | 9 | NA | NA | NA | 1 | ||||
| Probe A (mean [SD]) | 26.4 (2.6) | 27.6 | ||||||||
| Probe B (mean [SD]) | 27.8 (2.4) | 27.9 | ||||||||
| Probe C (mean [SD]) | 26.8 (2.7) | 27.0 | ||||||||
| Probe D (mean [SD]) | 0.0 (0.0) | 0.0b | ||||||||
| Probe E (mean [SD]) | 27.6 (2.8) | 28.5 | ||||||||
| SPC (mean [SD]) | 27.3 (1.9) | 25.6 | ||||||||
| M. tuberculosis not detected, NTM | ||||||||||
| No. | All values negative | NA | NA | NA | 9 | 1 | ||||
| Probe A (mean [SD]) | 0.0 (0.0) | 0.0 | ||||||||
| Probe B (mean [SD]) | 0.0 (0.0) | 0.0 | ||||||||
| Probe C (mean [SD]) | 24.4 (18.4) | 0.0 | ||||||||
| Probe D (mean [SD]) | 0.0 (0.0) | 0.0 | ||||||||
| Probe E (mean [SD]) | 0.0 (0.0) | 0.0 | ||||||||
| SPC (mean [SD]) | 27.1 (1.7) | 28.5 | ||||||||
Two samples in the GLI panel were the same and were analyzed together, since they had the same Rif-resistant profile.
Probes that identified the result as MTBC detected, RIF resistant.
NA, not applicable.
FIG 2.
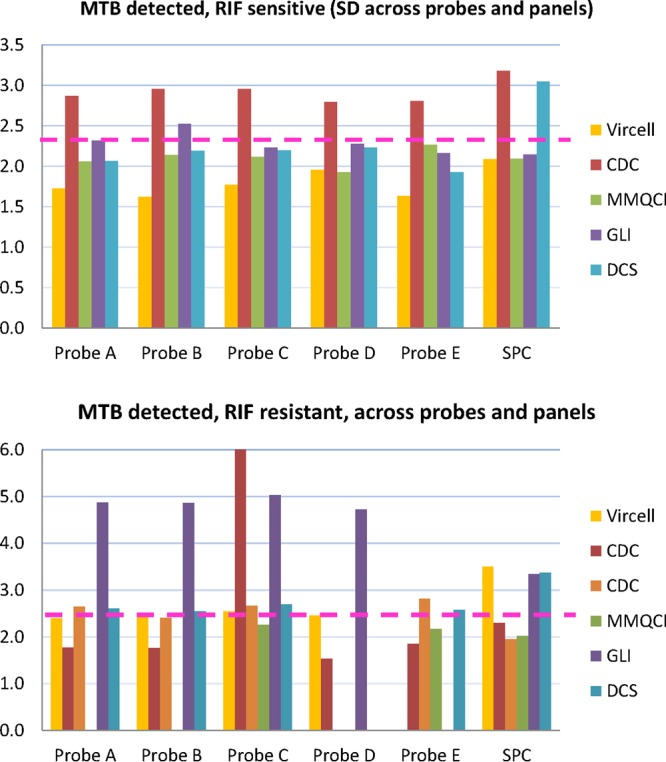
Bar charts of SD for each probe and internal control (SPC) across all panels for two groups of samples. (Top) M. tuberculosis detected, RIF susceptible; (bottom) M. tuberculosis detected, RIF resistant. The dotted line separates the SD values into two groups (>2.3 and <2.3).
Overall panel score.
The overall performance of the five panels using the scoring system for the main variables examined is summarized in Table 3. Overall, the Vircell, CDC, GLI, and NHLS panels all generated the same score of 9 out of a possible score of 12. The MMQCI panel was the only panel with a lower score of 6. Further details on the panels are as follows. The Vircell panel yielded an incorrect result, the standard operating procedure (SOP) was unclear, and material was not easy to handle. The CDC panel was not easy to open or handle, and there was greater variability on amplification and probe hybridization. The GLI panel had an insufficient volume, required extra consumables, and had greater variability on amplification and probe binding. The NHLS panel had an insufficient volume, required extra consumables, and was not easy to open. The MMQCI panel yielded an incorrect result, had an insufficient volume, and required cold chain maintenance and extra consumables, the SOP was unclear, and the material was not easy to transfer to the Xpert cartridge.
TABLE 3.
Overall performance scores of the five EQA pilot panels
| Variable | Performance score for: |
||||
|---|---|---|---|---|---|
| Vircell panel | CDC panel | MMQCI panel | GLI panel | DCS panel | |
| Qualitative analysis (n = 11) | |||||
| Correct result | 0 | 1 | 0 | 1 | 1 |
| Instrument error related to volume | 1 | 1 | 0 | 0 | 0 |
| Cold chain maintenance and special packaging required | 1 | 1 | 0 | 1 | 1 |
| Extra consumables required | 1 | 1 | 0 | 0 | 0 |
| Questionnaire (n = 10) | |||||
| Received in good condition | 1 | 1 | 1 | 1 | 1 |
| SOP clarity | 0 | 1 | 0 | 1 | 1 |
| Easy to open | 1 | 0 | 1 | 1 | 0 |
| Easy to handle | 0 | 0 | 1 | 1 | 1 |
| Easy to rehydrate and dissolves fully | 1 | 1 | 1a | 1a | 1 |
| Easy to transfer to Xpert cartridge | 1 | 1 | 0 | 1 | 1 |
| Quantitative analysis | |||||
| Increased probe Ct SD above pool | 1 | 0 | 1 | 0 | 1 |
| PCR inhibition | 1 | 1 | 1 | 1 | 1 |
| Subtotal | 9 | 9 | 6 | 9 | 9 |
Already in liquid format.
DISCUSSION
The implementation of a new assay in settings with users who are often not experienced with laboratory testing creates many challenges in the quality system (10), even though the test in question is designed for users with minimal expertise. The quality system, as described by the Clinical and Laboratory Standards Institute (CLSI), is intended to ensure the quality of the overall testing process (from specimen collection to result reporting for patient management), to detect and reduce errors, to improve consistency within and between laboratories, to contain costs, and to ensure customer satisfaction. This system encompasses 12 elements such as organization, personnel, equipment, quality control, assessment, facility, and safety (11, 12).
Many quality system components have often been lacking in resource-poor settings in general (13, 14), and the quality assessment programs for the Xpert MTB/RIF assay have been no exception. In particular, there are three components that appear critical for ensuring the quality and accuracy of Xpert MTB/RIF testing, (i) verification of each GeneXpert module (on installation or repair) to ensure that the instrument is fit for purpose before testing and reporting on clinical specimens, (ii) external quality assessment to ensure that the entire testing process (preanalytical, analytical, and postanalytical) is managed for quality results, and (iii) continuous performance monitoring (error rate, potential for contamination, usage, etc.). Verification should be performed on each module, and although published evidence on the use of EQA panels for instrument verification is currently available for only the NHLS panel (5), the other panels evaluated in the current study may also prove suitable for verification.
This feasibility study on five EQA panels addressed the gap in EQA for the Xpert MTB/RIF assay and showed little overall differences in scores between the Vircell, CDC, GLI, or NHLS panels, indicating that all of these panels are likely to be suitable for use in an EQA program. The MMQCI liquid panel received a lower score as it was the only panel that required cold chain maintenance, and the material was not easy to transfer to the Xpert cartridge. This cold chain requirement is likely to be a challenge in many low-resource settings. All panels were received in good condition and therefore were good for shipping across distances, and all were easy to rehydrate and dissolve fully in the SR buffer, showing good compatibility with the Xpert testing process. In addition, none of the panel materials caused any PCR inhibition. The matrix requirement (liquid or dry) for the EQA material did not appear to be a distinguishing criterion as both liquid and dried formats received reduced scores due to insufficient volumes, the need for extra consumables, and ease of transfer to the Xpert cartridge. The increased variation (increased SD) in the CT values of the CDC and GLI panels (due to some Xpert MTB/RIF tests reporting delayed probe hybridization and some reporting dropout probes with no hybridization) may be attributed to the use of MGIT bulk stock, which may have increased clumping of bacterial cells compared to that of the bulk stock manufactured in the single-cell format used in the NHLS product or other factors inherent to their specific preparation. Minimal variations may, however, be more acceptable in an EQA program monitoring RIF resistance rates using differences in probe dropout or probe delayed hybridization as well as differences in CT values across the reported semiquantitative categories, since studies have already described the potential use of the Xpert assay for patient monitoring using CT values (9, 15).
In keeping with the international recommendations for an EQA that a preferred matrix bear resemblance to the clinical sample and be relatively cost-effective to produce, easy, stable, and safe to transport, and accurate and precise (11, 12, 16), a liquid specimen format for the Xpert MTB/RIF EQA may prove more suitable. Xpert programs would also need guidance on EQA testing schedules and perhaps could draw from other existing molecular EQA schemes; the NHLS National Priority Program in South Africa, for example, subscribes to international schemes that provide three to four panels per year for molecular testing such as HIV load.
Factors such as SOP clarity, label bar code scanning, and the use of the web-based program for result entry highlight the need for any EQA program to be accompanied by training and on-going improvements. The web-based program TBGx Monitor was under development during this study, and some South African Xpert users raised concerns about the difficulty of its use. At the same time, TBGx Monitor was also being used in a pilot study conducted in collaboration with ACTG experienced clinical trial sites. The ACTG trial included 6 sites in the United States, South Africa, Brazil, and Peru. The NHLS panels were tested, and despite English not being the first language in some sites, none of the sites reported difficulty in uploading their results. Efficient use of both the GeneXpert instrument and the TBGx Monitor software requires basic computer skills. Thus, additional computer training may be required for technicians with limited previous experience with computers.
Another component in the quality system identified as important to Xpert testing is continuous performance monitoring, since EQA only provides a snapshot. Connectivity standards (17) have made possible several commercial products for hospital, clinic, and laboratory information systems as well as instrument information systems, and these should be applied to all testing platforms. In South Africa, for example, 17 national HIV load molecular testing laboratories comprising two platforms (the CAP/CTM assay from Roche Molecular Systems, Branchburg, NJ, and m2000sp/m2000rt systems from Abbott Molecular Inc., Des Plaines, IL) are centrally monitored in real time for instrument down time, utility, error rate, contamination, etc., which results in rapid response to problems. A similar concept should be applied to the GeneXpert as part of managing the entire quality system. Such a remote monitoring system is under development by the GeneXpert manufacturer, Cepheid (Sunnyvale, CA, USA).
ACKNOWLEDGMENTS
The opinions expressed herein are those of the authors and do not necessarily represent the official position of the U.S. Centers for Disease Control and Prevention.
This project was funded in part by the President's Emergency Plan for AIDS Relief (PEPFAR) through the U.S. Centers for Disease Control and Prevention, under the terms of grant U2GPS0001328-05. The Grand Challenges Canada (grant 0007-02-01-01-01 [to Wendy Stevens]) also provided support.
We thank the South African National Health Laboratory Service GeneXpert participating sites.
This work was performed in the Department of Molecular Medicine and Haematology, School of Pathology, Faculty of Health Sciences, University of the Witwatersrand, Johannesburg, South Africa.
Lesley Scott discloses that she is the inventor of the dried culture spot product (18).
Footnotes
Published ahead of print 30 April 2014
REFERENCES
- 1.Stevens W. 2012. Analysis of needed interface between molecular diagnostic tests and conventional mycobacteriology: country experience in South Africa. In 43rd Union World Conference on Lung Health, 13 to 17 November 2012, Kuala Lumpur, Malaysia [Google Scholar]
- 2.Boehme C. 2012. Xpert MTB/RIF: technology update. In Workshop on TB Diagnostics and Laboratory Services: Actions for Care Delivery and Sustainability. Global Laboratory Initiative, WHO, Annecy, France [Google Scholar]
- 3.Parsons LM, Somoskovi A, Gutierrez C, Lee E, Paramasivan CN, Abimiku A, Spector S, Roscigno G, Nkengasong J. 2011. Laboratory diagnosis of tuberculosis in resource-poor countries: challenges and opportunities. Clin. Microbiol. Rev. 24:314–350. 10.1128/CMR.00059-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palamountain KM, Baker J, Cowan EP, Essajee S, Mazzola LT, Metzler M, Schito M, Stevens WS, Young GJ, Domingo GJ. 2012. Perspectives on introduction and implementation of new point-of-care diagnostic tests. J. Infect. Dis. 205(Suppl. 2):S181–S190. 10.1093/infdis/jis203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scott LE, Gous N, Cunningham BE, Kana BD, Perovic O, Erasmus L, Coetzee GJ, Koornhof H, Stevens W. 2011. Dried culture spots for Xpert MTB/RIF external quality assessment: results of a phase 1 pilot study in South Africa. J. Clin. Microbiol. 49:4356–4360. 10.1128/JCM.05167-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeGruy K, Zilma R, Alexander H. 2012. Dried tube specimens (DTS) for Xpert MTB/RIF proficiency panels. In First African Society for Laboratory Medicine (ASLM) Conference, Cape Town, South Africa [Google Scholar]
- 7.Helb D, Jones M, Story E, Boehme C, Wallace E, Ho K, Kop J, Owens MR, Rodgers R, Banada P, Safi H, Blakemore R, Lan NT, Jones-Lopez EC, Levi M, Burday M, Ayakaka I, Mugerwa RD, McMillan B, Winn-Deen E, Christel L, Dailey P, Perkins MD, Persing DH, Alland D. 2010. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J. Clin. Microbiol. 48:229–237. 10.1128/JCM.01463-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boehme CC, Nabeta P, Hillemann D, Nicol MP, Shenai S, Krapp F, Allen J, Tahirli R, Blakemore R, Rustomjee R, Milovic A, Jones M, O'Brien SM, Persing DH, Ruesch-Gerdes S, Gotuzzo E, Rodrigues C, Alland D, Perkins MD. 2010. Rapid molecular detection of tuberculosis and rifampin resistance. N. Engl. J. Med. 363:1005–1015. 10.1056/NEJMoa0907847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blakemore R, Nabeta P, Davidow AL, Vadwai V, Tahirli R, Munsamy V, Nicol M, Jones M, Persing DH, Hillemann D, Ruesch-Gerdes S, Leisegang F, Zamudio C, Rodrigues C, Boehme CC, Perkins MD, Alland D. 2011. A multisite assessment of the quantitative capabilities of the Xpert MTB/RIF assay. Am. J. Respir. Crit. Care Med. 184:1076–1084. 10.1164/rccm.201103-0536OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parekh BS, Kalou MB, Alemnji G, Ou CY, Gershy-Damet GM, Nkengasong JN. 2010. Scaling up HIV rapid testing in developing countries: comprehensive approach for implementing quality assurance. Am. J. Clin. Pathol. 134:573–584. 10.1309/AJCPTDIMFR00IKYX [DOI] [PubMed] [Google Scholar]
- 11.Clinical and Laboratory Standards Institute. 2013. Quality management system: development and management of laboratory documents; approved guidelines—6th ed. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 12.Noble MA. 2013. An overview of the essential elements of a PT/EQA program. U.S. Centers for Disease Control and Prevention, Atlanta, GA: http://wwwn.cdc.gov/mlp/pdf/GAP/Noble.ppt [Google Scholar]
- 13.Nkengasong JN, Nsubuga P, Nwanyanwu O, Gershy-Damet GM, Roscigno G, Bulterys M, Schoub B, DeCock KM, Birx D. 2010. Laboratory systems and services are critical in global health: time to end the neglect? Am. J. Clin. Pathol. 134:368–373. 10.1309/AJCPMPSINQ9BRMU6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petti CA, Polage CR, Quinn TC, Ronald AR, Sande MA. 2006. Laboratory medicine in Africa: a barrier to effective health care. Clin. Infect. Dis. 42:377–382. 10.1086/499363 [DOI] [PubMed] [Google Scholar]
- 15.Theron G, Pinto L, Peter J, Mishra HK, Mishra HK, van Zyl-Smit R, Sharma SK, Dheda K. 2012. The use of an automated quantitative polymerase chain reaction (Xpert MTB/RIF) to predict the sputum smear status of tuberculosis patients. Clin. Infect. Dis. 54:384–388. 10.1093/cid/cir824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noble MA. 2007. Does external evaluation of laboratories improve patient safety? Clin. Chem. Lab Med. 45:753–755. 10.1515/CCLM.2007.166 [DOI] [PubMed] [Google Scholar]
- 17.Clinical and Laboratory Standards Institute. 2008. Point of care connectivity; approved standard—2nd ed. CLSI document POCT1-A2 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 18.Scott L. 8,709,712 US patent. 2014 Apr;